Abstract
Obesity represents a state of chronic, low grade inflammation and is associated with infiltration of increased numbers of adipose tissue macrophages (ATMs). Diet-induced obesity leads to an increase in non-inflammatory M1-like ATMs displaying the CD11c surface marker. We assessed the function of CD11c-positive ATMs when insulin resistant high fat diet (HFD) mice become insulin-sensitive after switching from HFD to normal chow (NC). HFD mice rapidly become insulin-sensitive in all major insulin-target tissues, including muscle, liver, and adipose tissue, after the diet switch. In adipose tissue the CD11c-positive macrophages remain constant in number despite the presence of insulin sensitivity, but these macrophages now assume a new phenotype in which they no longer exhibit increased inflammatory pathway markers. Adipose tissue markers of apoptosis and necrosis were elevated on HFD and remain high after the HFD → NC diet switch. Furthermore, ATM accumulation preceded detectable adipocyte necrosis at the early phase of HFD. Together, these results indicate that 1) CD11c-positive M1-like ATMs can exhibit phenotypic plasticity and that the polarization of these cells between inflammatory and non-inflammatory states is well correlated to the presence of absence of insulin resistance, and 2) adipocyte necrosis and apoptosis can be dissociated from ATM accumulation.
Keywords: Adipose Tissue, Diabetes, Inflammation, Insulin Resistance, Macrophage, Obesity
Introduction
It is now clear that obesity gives rise to a state of chronic, low grade inflammation that contributes to insulin resistance and type-2 diabetes (1). In both humans and rodents, macrophages (M) accumulate in adipose tissue (AT) with increasing body weight (2–4), and recent evidence implicates ATMs2 as major contributors to tissue inflammation and insulin resistance in obesity (5, 6). For example, mouse models have demonstrated that ATMs can be both necessary and sufficient (1, 7–9) for the development of insulin resistance in obesity. Specifically, disabling the inflammatory pathway within macrophages by creating myeloid cell-specific knockouts of IκB kinase β or JNK1 protected mice from diet-induced insulin resistance (5, 6). Macrophages have broad functions in the maintenance of tissue homeostasis through the clearance of senescent cells and remodeling and repair of tissues after inflammation (10). It has been shown that infiltrating ATMs play an important role in obesity-associated AT remodeling, based on the observations that ATMs in obese mice and humans localize around dead adipocytes, which are more prevalent in obesity (11). At these sites of adipocyte death, ATMs aggregate to form “crown-like structures” that envelope and ingest the large dead or dying adipocytes. In obesity, these ATMs secrete increased amounts of proinflammatory cytokines, such as TNFα and IL-6, which are implicated in the development of insulin resistance (12–15).
Macrophages show significant functional heterogeneity, as local environmental factors can shape their properties and activation state (16, 17). Different stimuli activate macrophages to express distinct patterns of chemokines, surface markers, and enzymes that ultimately generate the diversity of macrophage function seen in inflammatory and non-inflammatory settings. This is reflected in macrophage cell types with tissue specific functions, such as osteoclasts, Kupffer cells, and alveolar macrophages. Macrophage heterogeneity can also be observed within a single tissue type. In general, ATMs have been operationally defined across M1-like to M2 polarization states. These states have largely been defined in vitro, but tissue macrophages most likely operate along a continuum between these states in vivo. M1-like or “classically activated” macrophages are induced by proinflammatory mediators such as lipopolysaccharide and IFNγ. These cells secrete high levels of proinflammatory cytokines (TNFα, IL-6, IL-12) and generate reactive oxygen species through the actions of inducible nitric-oxide synthase (Nos2). M2 or “alternatively activated” macrophages can be generated in vitro by exposure to IL-4 and IL-13 (18). M2 macrophages secrete low levels of proinflammatory cytokines and high levels of anti-inflammatory cytokines. M1-like cells but not M2 cells express the CD11c surface marker and are the ones that produce the high levels of pro-inflammatory cytokines that are linked to the development of obesity-associated insulin resistance (19). Recent evidence shows that the majority of the ATMs that accumulate in obese adipose tissue are proinflammatory and express the cell surface markers F4/80, CD11b, CD11c, and are M1-like. In contrast, ATMs, which display F4/80 and CD11b but are negative for CD11c, do not exhibit inflammatory pathway activation and are M2-like. Lumeng et al. (19) showed that ATMs undergo a phenotypic switch from the M2 polarization state to a more M1-like, CD11c+ polarization state upon high fat feeding, and Patsouris et al. have shown that selective depletion of CD11c+ ATMs reverses insulin resistance even in the presence of a high fat diet (HFD) and obesity (20). Furthermore, over the past few years, several studies have reported increased TH1 lymphocyte and decreased content of Tregs in obese adipose tissue, and these adipose tissue lymphocytes may provide local tissue signals promoting ATM chemotaxis and activation (21, 22).
Although much is known about the accumulation of ATMs during the development of obesity and insulin resistance, relatively little is known about ATM fate and function during the resolution of obesity and insulin resistance. Therefore, in this study we explored whether the number of CD11c-positive ATMs decreases or whether these cells acquire a new phenotype when obese, insulin-resistant HFD mice become insulin-sensitive after their diet is changed from HFD back to normal chow (NC).
EXPERIMENTAL PROCEDURES
Animals and Animal Care
Male C57BL/6J mice were purchased from The Jackson Laboratory at 12 weeks of age and were rendered insulin resistant by feeding an HFD (60% kcal from fat; D12492, Research Diets) for 20 weeks. Then the diet was switched to NC (12% kcal from fat; Purina 5001, LabDiet) for another 3 weeks. Control mice were fed a NC all the time. Animals were maintained on a 12-h/12-h light/dark cycle with free access to food and water. All animal procedures were in accordance with University of California San Diego research guidelines for the care and use of laboratory animals.
Metabolism Insulin Tolerance Tests (ITTs), Glucose Tolerance Tests (GTTs), Hyperinsulinemic Euglycemic Clamp
Glucose and insulin tolerance tests were performed on 4-h-fasted mice. For GTT, animals were injected intraperitoneally with dextrose (1 g/kg, Hospira, Inc), whereas for ITT 0.5 units/kg insulin (Novolin R, Novo-Nordisk) was injected intraperitoneal glucose excursion after injection was monitored over time. Blood samples were drawn at 0, 15, 30, 60, and 120 min after dextrose injection or 0, 15, 30, 60, and 90 min after insulin injection. And glucose was measured using a One-Touch glucose-monitoring system (Lifescan). Mouse clamp was performed as previously described (23).
Stromal Vascular Cell (SVC) Isolation and FACS Analysis
Epididymal fat pads were weighed, rinsed 3 times in phosphate-buffered saline (PBS), and then minced in FACS buffer (PBS + 1% low endotoxin bovine serum albumin). Tissue suspensions were centrifuged at 500 × g for 5 min and then collagenase-treated (1 mg/ml, Sigma) for 30 min at 37 °C with shaking. Cell suspensions were filtered through a 100-μm filter and centrifuged at 500 × g for 5 min. SVC pellets were then incubated with RBC lysis buffer (eBioscience) for 5 min before centrifugation (300 × g for 5 min) and resuspended in FACS buffer. SVC were incubated with Fc Block (BD Biosciences) for 20 min at 4 °C before staining with fluorescently labeled primary antibodies or control IgGs for 30 min at 4 °C. F4/80-APC FACS antibody was purchased from AbD Serotec (Raleigh, NC); FITC-CD11b and PE-CD11c FACS antibodies were from BD Biosciences. Cells were gently washed twice and resuspended in FACS buffer with propidium iodide (Sigma). For FITC-Galectin-3 staining (Cedarlane Laboratories Ltd.), SVCs were fixed with 4% paraformaldehyde in phosphate-buffered saline for 5 min at 4 °C and wash twice, and resuspended SVC were treated with 100 μl of 0.1% saponin in Hanks' balanced salt solution for 5 min at 4 °C, washed once, then incubated with the FITC-Galectin-3 antibody for 30 min at 4 °C. SVCs were analyzed using a FACSAria flow cytometer (BD Biosciences). Unstained, single stains and Fluorescence Minus One controls were used for setting compensation and gates. The events are first gated based on Forward-area versus Side scatter-area as well as Side scatter-height versus Side scatter-width and Forward scatter-height versus Forward scatter-width for a total of three dual-parameter plots to gate out aggregates and debris. We used single color controls to calculate compensation using the FACSDiva software. A plot of Forward scatter versus propidium iodide was used as the fourth gate to identify individual, live cells. To measure markers with the maximum sensitivity, each fluorochrome is plotted versus propidium iodide, and polygons are drawn, angled with the aid of the Fluorescence Minus One controls. This excludes dead and autofluorescent cells but includes dim positives. By using polygon gates in combination with logical gates, inclusion of false positive cells in the gates is reduced.
Protein and Lipid Analyses
Plasma insulin levels were measured by enzyme-linked immunosorbent assay (ALPCO). Plasma FFA levels were measured enzymatically using a commercially available kit (NEFA C; Wako Chemicals USA). Glycerol was determined using the free glycerol reagent (Sigma). Adiponectin was measured using a kit (Bridge International). Leptin, resistin, MCP-1, and PAI-1 serum levels were measured using a multiplex enzyme-linked immunosorbent assay (Millipore/Linco research). IL-12p70, IFNγ, IL-6, IL-10, keratinolyte-derived chemokine, and TNF-α levels in epididymal white adipose tissue (epi-WAT) were measured using a multiplex enzyme-linked immunosorbent assay (Meso Scale Discovery).
Immunohistochemistry and Adipocyte Cell Sizing
Mice were euthanized by CO2 narcosis/cervical dislocation. Epi-WAT, liver and muscle from quadriceps were dissected, fixed, and embedded in paraffin and sectioned. Sections from at least 4 mice per group were stained with hematoxylin and eosin or incubated with perilipin antibody (Abcam) at a 1:50 dilution overnight at 4 °C. Subsequently, a biotinylated anti-rat secondary antibody (Pharmingen) was used at 1:100 dilution followed by 1:500 horseradish peroxidase-streptavidin (The Jackson Laboratory) and development in substrate chromogen. Slides were counterstained with Mayer's solution and mounted with Vectashield mounting media. Brightfield pictures were taken of 3 representative fields per slide using a fluorescent microscope (10× objective). Using the apoptosis detection kit (Chemicon), TUNEL staining was performed. Using the Image J software, adipocyte (n = 100 adipocytes) diameter was measured, and an average diameter was recorded for each animal. Values were recorded in arbitrary units.
Cathepsin D Activity
Cathepsin D activity was determined by the SensoLyte 520 cathepsin D assay kit (Anaspec). Frozen adipose tissue was grinded in liquid nitrogen, and then dithiothreitol-containing assay buffer was added. Supernatant from centrifuging at 12,000 rpm for 10 min at 4 °C was used for the assay according to the kit instructions. The data were normalized to 10 μg of protein (relative fluorescence units/10 μg).
mRNA Isolation and qPCR
Total RNA was extracted from skeletal muscle, liver, and adipose tissue using the RNA purification kit (RNeasy Plus, Qiagen). First-strand cDNA was synthesized using SuperScript III and random hexamers (Invitrogen). Samples were run in 25-μl reactions using an iTaq SYBR Green supermix (Bio-Rad) on an MJ Research Chromo4 Real Time PCR system (Bio-Rad). Samples were incubated at 95 °C for 3 min followed by 40 cycles at 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s. Genes were expressed as mRNA level normalized to a standard housekeeping gene (glyceraldehyde-3-phosphate dehydrogenase or 36B4) using the ΔΔCT method. The specificity of the PCR amplification was verified by melting curve analysis of the final products using Opticon 3 software (Bio-Rad). Primer sequences were: glyceraldehyde-3-phosphate dehydrogenase (forward, 5′-AATGTGTCCGTCGTGGATCT-3′; reverse, 5′-CATCGAAGGTGGAAGAGTGG-3′); 36B4 (forward, 5′-TGGCCAATAAGGTGCCAGCTGCTG-3′; reverse, 5′-CTTGTCTCCAGTCTTTATCAGCTGCAC-3′); F4/80 (forward, 5′-CTTTGGCTATGGGCTTCCAGTC-3′; reverse, 5′-GCAAGGAGGACAGAGTTTATCGTG-3′); CD11c (forward, 5′-ACACAGTGTGCTCCAGTATGA-3′; reverse, 5′-GCCCAGGGATATGTTCACAGC-3′); (forward, 5′-ATGAAGAACCTCCGGGAAAT3′; reverse, 5′-GCTTAGATCATGGCGTGGTT-3′); TNFα (forward, 5′-CCAGACCCTCACACTCAGATC-3′; reverse, 5′-CACTTGGTGGTTTGCTACGAC-3′); IL-1β (forward, 5′-AAATACCTGTGGCCTTGGGC-3′; reverse, 5′-CTTGGGATCCACACTCTCCAG-3′); IL-4 (forward, 5′-ATGGAGCTGCAGAGACTCTT-3′; reverse, 5′-AAAGCATGGTGGCTCAGTAC-3′); IL-6 (forward, 5′-CCAGAGATACAAAGAAATGATGG-3′; reverse, 5′-ACTCCAGAAGACCAGAGGAAAT-3′); IL-10 (forward, 5′-TGAATTCCCTGGGTGAGAAG-3′; reverse, 5′-TCACTCTTCACCTGCTCCACT-3′); INFγ (forward, 5′-TCAAGTGGCATAGATGTGGAAGAA-3′; reverse, 5′-TGGCTCTGCAGGATTTTCATG-3′); PGC1α (forward, 5′-AGACGGATTGCCCTCATTTGA-3′; reverse, 5′-TGTAGCTGAGCTGAGTGTTGG-3′); PDK4 (forward, 5′-CCGCTTAGTGAACACTCCTTC-3′; reverse, 5′-TCTACAAACTCTGACAGGGCTTT-3′); galectin-3 (forward, 5′-ATGAAGAACCTCCGGGAAAT-3′; reverse, 5′-GCTTAGATCATGGCGTGGTT-3′).
Muscle and Liver Lipid Analyses
The lipids from plasma and tissues are extracted in the presence of authentic internal standards by the method of Folch et al. (24). Using chloroform:methanol (2:1 v/v), individual lipid classes within each extract are separated by liquid chromatography (Agilent Technologies model 1100 series). Each lipid class is trans-esterified in 1% sulfuric acid in methanol in a sealed vial under a nitrogen atmosphere at 100 °C for 45 min. The resulting fatty acid methyl esters are extracted from the mixture with hexane containing 0.05% butylated hydroxytoluene and prepared for gas chromatography by sealing the hexane extracts under nitrogen. Fatty acid methyl esters are separated and quantified by capillary gas chromatography (Agilent Technologies model 6890) equipped with a 30-m DB-88MS capillary column (Agilent Technologies) and a flame-ionization detector.
Statistical Analyses
Data are presented as the means ± S.E. The significance of differences between groups was evaluated using analysis of variance. The p value <0.05 was considered significant.
RESULTS
Switching from HFD to NC Improves Glucose Tolerance and Insulin Sensitivity
As shown in Fig. 1, A and B, the diet switch caused a 15% decrease in body weight and 25% decrease in epi-WAT mass. We evaluated the effect of switching from (20 weeks, 60%) HFD to NC on glucose homeostasis and insulin sensitivity. ITTs and GTTs were performed 3 weeks after the dietary switch. As shown in Fig. 1, C and D, 20 weeks of HFD led to significant insulin and glucose intolerance in the C57BL/6J mice compared with the NC diet, and this effect was markedly attenuated in the HFD → NC group. Thus, basal glucose levels (Fig. 1E) and glucose tolerance were nearly normalized, and insulin tolerance was markedly improved. This was accompanied by an ∼80% decrease in basal insulin values (Fig. 1F), indicating improved insulin sensitivity.
FIGURE 1.
Insulin resistance was improved in the HFD → NC-switched mice. C57BL/6J mice were fed a HFD for 23 or 20 weeks then a NC diet for 3 or 23 weeks. Body weight (A) and epi-WAT weight (B) were measured. ITTs (C) and GTTs (D) were performed by intraperitoneal injection of insulin (0.5 units/kg of body weight) or glucose (1 g/kg of body weight) after 4 h of fasting. Basal glucose (E) and insulin (F) were measured just before the insulin injection during ITTs. Hyperinsulinemic euglycemic clamp were performed after a 6-h fast. Glucose infusion rates (GIR, G) and glucose disposal rates (GDR, H) indicate whole body insulin sensitivity. Insulin-stimulated glucose disposal rate (IS-GDR; I) is indicative of insulin sensitivity in skeletal muscle, and liver insulin sensitivity is reflected by suppression of hepatic glucose production (HGP, J and K). Blood levels of FFA (L) and glycerol (M) reflect insulin sensitivity in adipose tissue. Data are expressed as the mean ± S.E., n = 10 per group in A–D, and n = 6 per group in E–K. *, p < 0.05; **, p < 0.01 (compared with the HFD group).
To more accurately quantify whole body alterations in insulin sensitivity, hyperinsulinemic euglycemic clamp studies were performed (Fig. 1, G–L). The amount of exogenous glucose required to maintain euglycemia (GIR) and the glucose disposal rate (GDR) during the clamp studies were substantially lower in the HFD group compared with both the HFD → NC and NC mice (Fig. 1, G and H). The insulin-stimulated glucose disposal rate (IS-GDR), which primarily reflects skeletal muscle insulin sensitivity, was markedly blunted in the HFD group, reflecting HFD-induced muscle insulin resistance. This defect was essentially normalized in the HFD → NC mice, demonstrating that the dietary change reversed the skeletal muscle insulin-resistant state (Fig. 1I). The ability of insulin to suppress hepatic glucose production (HGP), which reflects hepatic insulin sensitivity, was markedly impaired in the HFD/obese mice (Fig. 1J), as evidenced by decreased suppression of HGP by insulin during the clamp study compared with NC mice. After the dietary change (HFD → NC), HGP suppression was greater, indicating improved hepatic insulin sensitivity. The basal HGP values were also reduced (Fig. 1K) in the HFD-NC mice, consistent with the reduction in basal glucose values (Fig. 1E) and improved hepatic insulin sensitivity. Interestingly, the resolution of muscle insulin resistance (∼90%) was somewhat greater than the improvement in hepatic insulin resistance (∼50%). Insulin-induced suppression of plasma FFA and glycerol levels are indicators of adipose tissue insulin sensitivity, and as shown in Fig. 1, L and M, HFD-fed mice exhibited impaired FFA suppression (32 versus 63%) and a higher plasma glycerol level (0.13 ± 0.01 versus 0.05 ± 0.004 mg/ml) compared with NC fed mice. The HFD → NC diet switch resulted in normalization of glycerol and FFA suppression by insulin. Together, these data demonstrate that changing from HFD to NC leads to reduction of systemic insulin resistance, and these insulin-sensitizing effects occur in all three major insulin target tissues, with the improvement in adipose tissue and muscle, which is somewhat greater than in liver.
Macrophage Content of WAT after the HFD → NC Diet Switch
Large numbers of ATMs accumulate in obesity, and substantial evidence exists pointing to an etiologic role for these cells in the development of chronic tissue inflammation and insulin resistance/diabetes. Therefore, we quantitated the effect of these dietary manipulations on ATM subpopulations. SVC of epi-WAT depots from the three groups were isolated and stained with F4/80 and CD11b antibodies. A large increase in F4/80 and CD11b double-positive macrophages was observed in the HFD mice (1.25 ± 0.20 × 106/g of fat HFD versus 0.41 ± 0.07 × 106/g of fat NC; p < 0.01; Fig. 2, A and B, and supplemental Fig. 1), and this is consistent with previous studies (25). The increase in ATM content in the epididymal fat pad was comparable with accumulation of ATMs in other visceral depots such as mesenteric and retroperitoneal fat. Interestingly, as previously noted (26, 27), ATM content in subcutaneous adipose tissue did not increase nearly as much as in the visceral fat (supplemental Fig. 2). Importantly, the HFD → NC mice exhibited the same number of doubly positive macrophages as the HFD group (1.16 ± 0.14 × 106/g of fat, Fig. 2, A and B). These comparisons were fully consistent with the relative measurements of adipose tissue F4/80 mRNA expression in HFD, HFD → NC and NC mice (Fig. 2C), which show equally high values in HFD and HFD → NC mice compared with NC. These findings indicate that ATM content does not diminish 3 weeks after switching from HFD to NC.
FIGURE 2.
Macrophages maintained in the adipose tissue of HFD → NC-switched mice. SVCs were isolated from the epi-WAT samples then were stained with F4/80, CD11b, and CD11c and analyzed by FACS for F4/80+CD11b+ (A and B). Samples were gated for F4/80+CD11b+ and examined for coexpression of CD11c conjugated with phycoerythrin (PE) (A and E). The relative mRNA levels of the macrophage maker F4/80 (C) and M1-like marker CD11c (D) were analyzed by qPCR. Data are expressed as the mean ± S.E., n = 4 per group, **, p < 0.01 compared with the HFD group.
ATMs are heterogeneous, and we and others have found two subpopulations of F4/80+CD11b+ ATMs, one of which is positive and the other negative for CD11c. The CD11c+ (triply positive, F4/80+, CD11b+, CD11c+ cells) M1-like macrophages account for the majority of the increase in ATMs in obesity and overexpress proinflammatory cytokines compared with the CD11c− ATMs (19, 28). As shown in Fig. 2D, there was a 42-fold increase in CD11c mRNA in HFD compared with NC mice, and the HFD → NC mice exhibited the same level of CD11c as HFD mice. FACS analyses showed a much greater number of F4/80, CD11b, and CD11c triple-positive ATMs in the epi-WAT of HFD mice versus NC mice (0.486 ± 0.034 × 106/g of fat versus 0.038 ± 0.007 × 106/g of fat; p < 0.01, Fig. 2, A and E), and ∼ 40% of the F4/80+, CD11b+ cells also displayed CD11c on HFD compared with <10% on NC. No decrease in the number of CD11c-positive ATMs was seen in the HFD → NC group (0.483 ± 0.021 × 106/g of fat, Fig. 2, A and E) and, as on HFD, ∼40% of F4/80, CD11b+ cells displayed CD11c. These results show that total ATMs as well as CD11c+ M1-like cells remained constant in number after the 3-week dietary change despite resolution of the glucose intolerance and insulin resistance in the HFD → NC group. Interestingly, by 5 weeks of the HFD → NC diet switch, CD11c-positive ATM content decreased toward normal (supplemental Fig. 3).
Inflammatory Pathway Activation in WAT after the HFD → NC Diet Switch
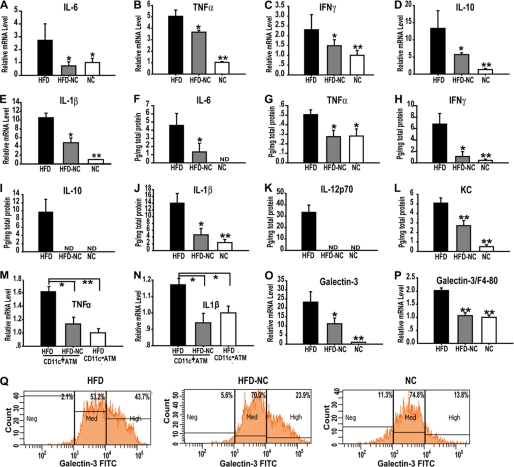
The CD11c+ ATM content increases markedly during HFD, peaking at ∼12 weeks (Fig. 2 and supplemental Fig. 4), and because these cells are important contributors to the increased expression of pro-inflammatory genes in obese AT (19, 28), we quantified the expression of several of these genes in epi-WAT. As shown in Fig. 3, A–E, HFD led to a marked increase in the mRNA levels of cytokines such as IL-6, TNFα, IFNγ, IL-10, and IL-1β compared with NC, and this increase was markedly blunted in the HFD → NC mice. We also measured tissue protein levels of these cytokines, which revealed high levels on HFD compared with NC, with near normalization of IL-6, TNFα, IFNγ, IL-10, IL-1β, and IL-12p70 along with a marked decrease in keratinocyte-derived chemokine levels in the HFD → NC mice (Fig. 3, F–L). These results are consistent with the qPCR data and demonstrate that the dietary change from HFD to NC resulted in decreased proinflammatory responses in AT despite the fact that the CD11c+, M1-like ATMs remain constant in number. To test if the CD11c+ macrophages in the HFD → NC mice contribute to the reduction of inflammation, CD11c+ macrophages and CD11c− macrophages were sorted by FACS from epi-WAT of 35 HFD and 34 HFD → NC mice. As shown in Fig. 3, M and N, on HFD the CD11c+ macrophages showed a marked increase in mRNA levels of TNFα and IL-1β compared with CD11c− cells, and after the dietary switch to NC, the increase in gene expression in the CD11c+ cells was markedly blunted, demonstrating that CD11c+ macrophages from the HFD → NC mice reverted to a less inflammatory phenotype.
FIGURE 3.
Cytokine levels in the epi-WAT. Shown are the relative mRNA levels of inflammatory cytokines IL-6 (A), TNFα (B), IFNγ (C), IL-10 (D), and IL-1β (E) in the epi-WAT of HFD, HFD → NC, and NC mice, as measured by qPCR. Cytokine protein levels were also measured in epi-WAT lysates from HFD, HFD → NC, and NC mice including IL-6 (F), TNFα (G), IFNγ (H), IL-10 (I), IL-1β (J), IL-12p70 (K), and keratinocyte-derived chemokine (KC) (L). CD11c+ macrophages (F4/80+CD11b+CD11C+) and CD11C-macrophages (F4/80+CD11b+CD11C−) were FACS-sorted from 35 HFD mice and 34 HFD-NC mice. mRNA levels of TNFα in the sorted cells (M) and IL-1β (N) were measured by qPCR. Galectin-3 levels were measured using qPCR (O and P). FACS analysis measured the intensity of galectin-3 in the CD11c+ macrophages in epi-WAT (Q). Data are expressed as the mean ± S.E., n = 9–10 per group. *, p < 0.05; **, p < 0.01 (compared with the HFD group). ND means not detectable.
Galectin-3 is a lectin expressed by activated macrophages that mediates macrophage chemotactic, phagocytic, and inflammatory responses. As shown in Fig. 3O, ATM from HFD mice expressed more galectin-3 compared with the NC mice, and the diet switch significantly decreased galectin-3 levels. To relate galectin-3 expression to ATM number, we normalized the results to F4/80 mRNA levels, and with this analysis, galectin-3 levels were high in the HFD mice and fell to the same levels in the NC and HFD → NC mice (Fig. 3P). FACS analysis also showed that the CD11c+ macrophages in HFD mice express a higher intensity of galectin-3 than NC mice, and this increase was blunted in the HFD → NC mice (Fig. 3Q).
Adipocyte Size and Adipokine Secretion
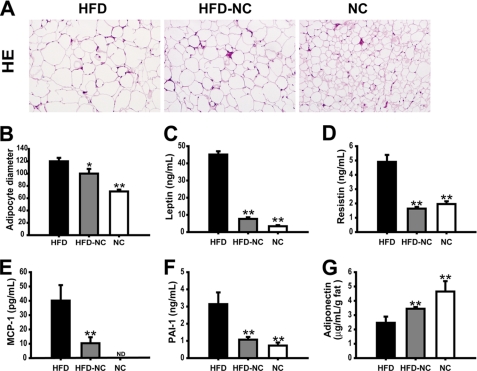
It is well known that HFD increases adipocyte size and adipose tissue mass and that enlarged adipocytes are more insulin-resistant. As shown in Fig. 4, A and B, adipocyte size in the epi-WAT of the HFD mice is much greater than in NC mice, and some adipocytes from the HFD mice can be 5 times bigger than in NC mice. This increase was partially reversed in the HFD → NC mice. Adipose tissue can secrete a variety of adipokines and cytokines that modulate insulin sensitivity and ATM recruitment. As shown in Fig. 4, C–F, adipocytes from HFD mice secrete more leptin, resistin, PAI-1, and MCP-1 than NC, and this increase was normalized in the HFD → NC group. Also, adipocytes from HFD mice secrete less adiponectin per gram of fat than NC, and this decrease was ameliorated in HFD → NC group (Fig. 4G).
FIGURE 4.
Morphology of epi-WAT and plasma adipokine levels. Paraffin-imbedded epi-WAT sections were stained with hematoxylin and eosin (A), and the relative adipocyte diameter was measured by using Image J software (B). Multiple enzyme-linked immunosorbent assays were used to measure the concentration of leptin (C), resistin (D), MCP-1 (E), PAI-1 (F), and adiponectin (G) in serum of HFD, HFD → NC, and NC mice. Data are expressed as the mean ± S.E., n = 9–10 per group in B–G and J. **, p < 0.01 compared with the HFD group. ND means not detectable.
Adipocyte Necrosis and Apoptosis
Strissel et al. (29) suggested that adipocyte death is an early, progressive, and depot-dependent event with up to 80% of fat cells undergoing necrosis in diet-induced obese mice. In our models we assessed apoptosis in epi-WAT by TUNEL staining. As shown in Fig. 5A, there was no detectable TUNEL staining in NC, whereas there was an equivalent severalfold increase in the number of cells positive for TUNEL staining in both HFD and HFD → NC. Thus, HFD led to an increase in adipocyte apoptosis that was not alleviated by switching to chow diet at 3 weeks.
FIGURE 5.
Adipose tissue death. TUNEL (A) staining was used to analyze apoptosis in epi-WAT of HFD, HFD → NC, and NC mice. Perilipin (B) staining and cathepsin D activity (C) were used to assess necrosis in epi-WAT of HFD, HFD → NC, and NC mice. F4/80 (D) mRNA level and cathepsin D activity (E) were used to assess macrophage infiltration and necrosis in epi-WAT of 1-week-old HFD mice and NC mice. Data are expressed as the mean ± S.E., n = 4 per group in A–C, n = 5 per group in D and E. *, p < 0.05; **, p < 0.01 (compared with the HFD group).
Loss of perilipin staining is a general marker for adipocyte death, and we assessed the degree of adipocyte necrosis by immunostaining adipose tissue sections for perilipin, which surrounds live adipocytes. Adipose tissue from NC mice was characterized by greater perilipin staining compared with HFD mice, and the HFD → NC diet switch did not restore perilipin content (Fig. 5B). We also measured cathepsin D activity in the epi-WAT as an additional measure of necrosis. As shown in Fig. 5C, adipose tissue from HFD mice displayed increased cathepsin D activity compared with NC mice, and the HFD → NC diet switch did not cause a reduction in cathepsin D activity. These results indicate that HFD → NC mice exhibit the same extent of necrosis in the epi-WAT as HFD mice. This indicates that the improvement in insulin resistance and reversal of proinflammatory effects are independent of the presence of adipocyte necrosis. To further explore this dissociation between necrosis and macrophage-mediated inflammation, we assessed these variables at an early stage of HFD. As seen in Fig. 5D, after 1 week of HFD, an increase in ATM content was clearly observed. However, despite the initial increase in ATMs, we found no evidence for increased cathepsin D activity (Fig. 5E), indicating that macrophage immigration begins before the onset of HFD-induced adipocyte necrosis.
Lipid and Inflammatory Changes in Liver and Muscle
HFD leads to full blown hepatosteatosis (Fig. 6A) as previously reported. Interestingly, 3 weeks after switching from HFD to NC, hepatic fat accumulation was largely resolved, and this was further demonstrated by biochemical measurements showing normalization of triacylglycerol, diacylglycerol, and cholesterol ester content in the HFD → NC mice (Fig. 6, B–D). Furthermore, we found that HFD led to an increase in hepatic expression of F4/80 and CD11c (Fig. 6, E and F), suggesting an increase in Kupffer cells or newly recruited cells. In contrast to what was observed in adipose tissue, switching from HFD to NC led to a substantial decrease in F4/80 and CD11c expression. We also found that the diet switch increased IL-4 expression, a TH2 cytokine, compared with HFD mice, suggesting that the remaining Kupffer cells or newly recruited macrophages acquire a less proinflammatory state (Fig. 6G).
FIGURE 6.
Liver data. A, hematoxylin and eosin (HE) staining of the paraffin-embedded liver sections of HFD, HFD → NC, and NC mice is shown. Liver samples were dehydrated and analyzed for accumulation of lipid intermediates including triacylglycerol (B), cholesterol ester (C) and diacylglycerol (D). Relative mRNA levels of F/80 (E), CD11c (F), and IL-4 (G) were analyzed by qPCR. Data are expressed as the mean ± S.E., n = 4 per group; *, p < 0.05; **, p < 0.01 (compared with the HFD group).
With respect to macrophages, skeletal muscle exhibited similar changes as seen in liver, in contrast to the relative persistence of ATMs in adipose tissue. As previously reported, HFD can cause an increase in macrophage content of intermuscular fat deposits (3, 30). In the current studies HFD led to an increase in F4/80 and CD11c expression in skeletal muscle, which was reduced in the HFD → NC mice (Fig. 7, A and B). Consistent with this decrease in macrophage content, inflammatory cytokine expression, such as IL-6 and TNFα, was also markedly decreased (Fig. 7, C and D). Interestingly, PGC1α expression increased in the HFD → NC mice (Fig. 7E), consistent with improved mitochondrial function and enhanced insulin sensitivity. It is known that PDK4 can impair glucose oxidation followed by increased fatty acid oxidation (31). As shown in Fig. 7F, HFD resulted in 3-fold higher PDK4 expression compared with the NC fed mice, and this increase was fully normalized by diet switching. We also found that HFD led to the expected increase in triacylglycerol, diacylglycerol, and cholesterol ester content in muscle, and these biochemical abnormalities were largely normalized back to NC levels in the HFD → NC mice (Fig. 7, G–I). All of these changes are fully consistent with the marked improvement in skeletal muscle insulin sensitivity observed in the euglycemic hyperinsulinemic clamp studies.
FIGURE 7.
Muscle data. Muscle (quadriceps) macrophage content was calculated by measuring the relative mRNA levels of F4/80 (A) and CD11c (B) by qPCR. Relative mRNA levels of inflammatory cytokines IL-6 (C) and TNFα (D) in quadriceps were determined by qPCR. Mitochondria function-related genes PDK4 (E) and PGC1α (F) were also analyzed by qPCR. Muscle samples were dehydrated and analyzed for accumulation of lipid intermediates including triacylglycerol (G), cholesterol ester (H), and diacylglycerol (I). Data are expressed as the mean ± S.E., n = 4 per group; *, p < 0.05; **, < 0.01 (compared with the HFD group).
Lipid Class Composition in Liver, Muscle, Epi-WAT, and Serum
Lipid accumulation was much greater in both liver and muscle on HFD compared with NC and was greatly reduced after the HFD→ NC switch (Figs. 6 and 8). We also measured fatty acid composition of the various tissues and plasma in the NC, HFD, and HFD → NC mice utilizing a high density, quantitative lipidomic analysis. The results show that the dietary manipulations led to major changes in lipid composition in all tissues (Fig. 8 and supplemental Figs. 5–7). For example, across the different lipid classes, there were generally less ω3 fatty acids and more ω6 fatty acids in HFD mice in all three tissues. Switching from HFD to NC led to an increase in ω3 and a decrease in ω6 fatty acids in most lipid classes in liver and muscle with little if any changes in adipose tissue (Fig. 8 and supplemental Figs. 5–7). As seen, analysis of the specific lipid classes shows that the tissue levels of ω3 and 6 fatty acids moieties contained in the diets (18:3n3 and 18:2n6) differed minimally, or not at all, between HFD and NC, whereas the more downstream ω3 and 6 FA showed large changes. This indicates that the different dietary conditions modified the activity of the metabolic pathways, which generate the ω3 and 6 FA products, including arachidonic acid, eicosapentaenoic acid, and docosahexaenoic acid. The tissue content of the ω3 and six FAs goes in opposite directions, and it is possible that this reflects alterations in subcellular localization of key biosynthetic enzymes or differential affinity of one or more of these enzymes for substrate. In liver, muscle, fat, and blood, stearic acid (18:0) levels were higher on HFD compared with NC, and the diet switch reversed this increase (Fig. 8D). In all three tissues as well as in blood, palmitoleate (16:1n7) levels were higher on NC compared with HFD, and this is consistent with a recent report that palmitoleate may be an insulin-sensitizing lipokine (32). Palmitoleate levels increased after 3 weeks on NC in liver and blood (Fig. 9, A and D) but did not change in the triacylglycerol fraction and only slightly in the phospholipid fraction in WAT and muscle (Fig. 9, B and C).
FIGURE 8.
Lipid class composition analysis for fatty acids in HFD, HFD-NC and NC mice. Eicosapentaenoic acid (EPA) (C20:5n3), docosahexaenoic acid (DHA) (C22:6n3), and arachidonic acid (AA) (C20:4n6) fatty acid composition in liver (A), muscle (B), and epi-WAT (C) of HFD, HFD → NC, and NC mice is shown. Also, shown are saturated fatty acid C18:0 levels in liver, muscle, epi-WAT, and serum (D). Data are expressed as the mean ± S.E., n = 4 per group; *, p < 0.05; **, p < 0.01 (compared with the HFD group).
FIGURE 9.
Palmitoleate analysis for fatty acids in HFD, HFD-NC and NC mice. C16:1n7 composition in liver (A), muscle (B), epi-WAT (C), and serum (D) of HFD, HFD → NC, and NC mice is shown. Data are expressed as the mean ± S.E., n = 7–10 per group; *, p < 0.05; **, p < 0.01 (compared with the HFD group).
DISCUSSION
It is now generally accepted that chronic tissue inflammation is an important cause of insulin resistance in obesity (1, 3, 4) and that the proinflammatory tissue macrophage, particularly in adipose tissue, can be an initiating cell type in this inflammatory response (1, 7–9). In addition, we and others have shown that a specific subpopulation of macrophages, characterized by cell surface positivity for the markers F4/80, CD11b, and CD11c, accounts for the majority of the increased macrophage influx into adipose tissue (19, 28). These cells assume a highly proinflammatory state, releasing large amounts of tissue cytokines that can cause insulin resistance through paracrine mechanisms. In addition, genetic deletion of CD11c-positive macrophages led to near reversal of HFD-induced inflammation with near complete normalization of glucose tolerance, hyperinsulinemia, and insulin sensitivity (20). In the current study we have examined the kinetics of accumulation of these proinflammatory CD11c+ macrophages in adipose tissue during the course of diet-induced obesity and have also assessed their disappearance once high fat feeding is discontinued and normal chow diet resumed. We found that CD11c+ macrophages rapidly influx into adipose tissue, reaching a peak by ∼12 weeks. After switching from high fat diet to normal chow, ATMs remain constant in number for 3 weeks and then decline toward the values observed in NC fed mice by 5 weeks.
To better understand the role of the CD11c+ macrophage in the etiology of insulin resistance, we examined the time course of reversion of glucose intolerance, insulin resistance, ATM content, and inflammation after discontinuing HFD. Understanding the temporal sequence of the events during reversion of insulin resistance is a powerful tool to gain insights into the underlying mechanisms, as events that occur after insulin sensitivity is restored cannot be the cause of the reversion. The major findings are that insulin resistance and glucose intolerance return to normal, or near normal, levels after 3 weeks of the diet switch, whereas total macrophage content in adipose tissue as well as CD11c+ macrophage content remains unchanged for 3 weeks but then declines toward normal by 5 weeks. Thus, at the time that glucose intolerance/insulin resistance normalized (3 weeks post-diet switch), CD11c+ and total ATM content remains unchanged. Remarkably, however, we found that despite the unchanged macrophage content, markers of inflammation (i.e. tissue cytokines, and inflammatory pathway gene expression) were largely normalized in the 3-week HFD → NC mice. Thus, despite the ongoing presence of the CD11c+ ATM subpopulation, these cells reorient their phenotype from a highly proinflammatory state to a non-inflammatory state, consistent with the role of these cells and tissue inflammation in the etiology of insulin resistance.
After 3 weeks of the diet switch from HFD to NC, the mice showed a 15% decrease in body weight and a 25% decrease in fat pad mass (Fig. 1, A and B). This suggests that the total number of CD11c+ ATMs per mouse could be decreased after the diet switch, whereas the content of these ATMs per gram of fat was unchanged, and this was accompanied by a large decrease in a variety of tissue inflammatory markers. On the other hand, the diet-switched mice were still obese compared with the NC group, but insulin sensitivity was largely normalized, at least in muscle and fat. This suggests potential effects of dietary nutrient content independent of adiposity.
There are a number of chemotactic factors that can stimulate macrophage migration (7, 33, 34). Adipocytes secrete MCP-1 as well as other chemotactic factors, leading to initial macrophage influx. Once within the adipose tissue, the resident ATMs also secrete chemotactic factors, causing further macrophage in migration, perpetuating the inflammatory state. In the context of chronic obesity-related inflammation, it is important to recall that acute inflammatory responses normally dissipate via the process of inflammation resolution. Because obesity-induced adipose tissue inflammation is a chronic, low grade tissue phenotype, which likely lasts as long as the obesity and high caloric state persist, it seems probable that some elements of the resolution process are attenuated in obesity. In this event, events which initiate inflammatory responses as well as those which maintain it become of etiologic interest. We found that after 3 weeks of the diet switch, total ATM content remained constant, although the CD11c+ macrophages exhibited an entirely different phenotype. This suggests that the CD11c+ ATMs present at the end of the HFD period responded to NC-derived dietary or tissue cues that caused them to change their polarization state to a less inflammatory cell type, whereas the cell surface markers we measured remain unchanged. Because ATM content eventually falls by 5 weeks, it is probable that influx of macrophages is eventually curtailed by the diet switch. Our data do not prove that the switch in ATM phenotype is not due to an influx of non-inflammatory macrophages bearing CD11c. However, our overall interpretation is that this scenario is less likely than a polarization switch of the ATMs, which were already present in the adipose tissue, from a proinflammatory to a less inflammatory state while still expressing CD11c. There are several reasons for this interpretation. First, because total ATM content had not changed after 3 weeks of the diet switch, this would mean that influx of this new cell type would have to be balanced completely by efflux of the original ATMs. This seems less likely, as it would require two processes to cancel each other out, and the influx would be in the context of an adipose tissue depot in which MCP1 expression is decreased. Second, to our knowledge, a non-inflammatory monocytic cell type expressing CD11c has not been identified (19). For these reasons, we favor the repolarization concept.
ATMs typically form crown-like structures surrounding large or dying adipocytes (11); it is logical to propose that signals derived from these dying adipocytes are responsible for recruiting macrophages into adipose tissue. However, no direct evidence for this exists, and our current results suggest that other factors may contribute to this process. With respect to apoptosis, although our data do not allow identification of which cell types within the adipose tissue undergo apoptosis, they do show that the overall level of apoptosis is higher in adipose tissue from HFD versus NC mic, and that this level of apoptosis remains unchanged after switching from HFD to NC for 3 weeks. Likewise, the number of necrotic cells, as measured by decreased perilipin staining (11) and increased cathepsin D activity, is increased on HFD compared with NC, but after 3 weeks of the diet switch, overall perilipin staining and cathepsin D activity remained the same. These events occurred at the same time that the CD11c+ ATMs underwent the phenotypic change to a relatively non-inflammatory profile, which was accompanied by normalization of glucose intolerance and insulin sensitivity. Interestingly, 1 week of HFD feeding can cause insulin resistance and increased ATM content in adipose tissue (30, 35). However, our data show no evidence of adipocyte necrosis at this early stage of ATM accumulation. These findings suggest that the tissue cues, which influence the inflammatory macrophage phenotype, can be dissociated from the processes of necrosis/apoptosis and may be dietary in origin or represent signals emanating from living adipocytes and/or other constituents of adipose tissue such as lymphocytes.
The above considerations raise the question as to what causes the CD11c+ macrophages to assume their proinflammatory phenotype in the first place. Lumeng et al. (19) have presented data indicating that these cells exhibit their proinflammatory phenotype at the time they arrive in the adipose tissue. Our data indicate that these cells retain a significant degree of plasticity and that local environmental factors can alter their phenotype, at least from the proinflammatory to the non-inflammatory state. Given this degree of plasticity, it seems likely that during the course of obesity and HFD, local environmental signals are involved in the maintenance of the proinflammatory state of CD11c+ macrophages even though they might exhibit this state after initial entry. It seems likely that the adipocyte, adipose tissue lymphocytes, or some other adipose cell type is the source of these local signals.
One of the potential tissue environmental signals is free fatty acids. Within adipose tissue, macrophages are surrounded by adipocytes constantly releasing free fatty acids, and previous studies have shown that saturated fatty acids exert potent proinflammatory effects in macrophages by signaling through the TLR4 pathway (30, 36–39). In these studies we showed that switching the diet from HFD to NC led to normalization of basal blood glycerol levels and normalization of insulin-mediated suppression of circulating FFAs. Based on this, the ATMs exist in a less proinflammatory environment in the 3-week HFD → NC mice, likely contributing to the change in the CD11c+ cell phenotype. In addition to FFAs, adipose tissue secretes a variety of chemokines and cytokines, and going from HFD → NC has a profound effect on expression of these factors (Fig. 3). Clearly one or more of these chemokines/cytokines could serve as diet-sensitive environmental signals.
Based on the results of the glucose clamp data, switching from HDF → NC for 3 weeks led to complete normalization of skeletal muscle and adipose tissue insulin sensitivity, whereas hepatic insulin sensitivity was only improved by ∼50%. This occurred at the same time that liver and muscle triglyceride and diacylglyceride concentrations returned to normal. Although the mechanism for this apparent delay in normalization of hepatic insulin sensitivity remains to be determined, this result is consistent with other studies showing dissociation of hepatic steatosis from insulin resistance (6, 40, 41).
Polyunsaturated fatty acids can be classified into ω3 and ω6 categories, and we found that HFD led to increased ω6 and decreased ω3 fatty acid content in liver, muscle, and fat compared with NCs. However, the levels of 18:3n3 and 18:2n6 FAs, which are the actual ω3 and ω6 constituents in the diets, were very similar, whereas the levels of the downstream ω3 and ω6 products were altered by the different dietary conditions. This indicates that it is the biochemical pathways which metabolize dietary lipids that were altered by the HFD and NC conditions. It has been shown that both ω3 and ω6 fatty acids can be precursors for signaling molecules with opposing effects. For example, ω6 fatty acids such as arachidonic acid (C20:4n6), which was increased by HFD, can be converted to prostaglandins, leukotrienes, and other lipoxygenase or cyclooxygenase products. These products are important regulators of cellular function and can cause proinflammatory, atherogenic, and prothrombotic effects (42). On the other hand, ω3 fatty acids, such as docosahexaenoic acid (C22:6n3) and eicosapentaenoic acid (C20:5n3), which were decreased on HFD, can antagonize the pro-inflammatory effects of ω6 fatty acids and down-regulate inflammatory gene expression (43–47). Furthermore, treatment of obese mice and rats with ω3 FAs improves insulin resistance (48, 49). Our lipodomic data showed that ω3 and ω6 fatty acid composition was normalized in liver and muscle, but not adipose tissue, 3 weeks after the diet switch, and these changes may contribute to the improved insulin sensitivity. Our lipodomic data also showed that stearic acid (C18:0) was generally increased in most lipid classes and reverted to normal after the diet switch. It is interesting to note that C16:0 did not increase on HFD (supplemental Fig. 5). Which means that the ratio of C18:0 to C16:0 actually went up. This is consistent with Matsuzaka et al. (50), who showed that HFDs increase Elovl6 activity, which increases the ratio of C18:0 to C16:0. Because saturated FAs can activate macrophages and ω3 FAs are anti-inflammatory, it seems possible that these dietary/obesity-induced alterations in the mixtures of adipose tissue FAs could be one of the tissue cues responsible for determining ATM content and phenotype. In addition, Cao et al. (32) have recently reported that palmitoleate may function as a lipokine with insulin-sensitizing properties. Interestingly, we found that blood, muscle, and liver palmitoleate concentrations were depressed on HFD compared with NC mice and increased in the 3-week HFD → NC group. The relative lack of change in 16:1n7 composition in adipose tissue suggests that adipocyte de novo fatty acid synthesis is suppressed on HFD, as previously reported (32), and does not appreciably increase after switching to NC for 3 weeks. Because muscle FAs are largely derived from FAs released from adipocytes, this could also explain why 16:1n7 values are largely unchanged in muscle after the diet switch. This would also mean that the increase in palmitoleate levels in liver are the product of de novo fatty acid synthesis in hepatocytes. Apart from changes in fatty acid composition, we also observed normalization of intrahepatic and intramyocellular triacylglycerol and diacylglycerol levels after 3 weeks of the diet switch.
In summary, these results demonstrate the phenotypic plasticity of CD11c+ ATMs elicited by changing from HFD to NC. On HFDs, these cells are highly proinflammatory, leading to tissue inflammation, which contributes to the insulin resistant state. Three weeks after switching from HFD to NC, the insulin resistance was nearly normalized, but CD11c+ ATM content remained unchanged. However, these CD11c+ ATMs now assume a non-inflammatory phenotype, showing that these cells retain a degree of phenotypic plasticity and that it is the inflammatory state of these cells rather than simply their presence in adipose tissue that influences overall insulin sensitivity. In addition, adipose tissue markers of apoptosis and necrosis were elevated on HFDs and remained equally high in the 3-week HFD → NC mice despite normalization of insulin sensitivity. This suggests that ATMs can modify their cellular phenotype in response to nutritional or local tissue signals apart from the apoptosis/necrosis pathways.
Acknowledgments
We thank Carlos Castorena from University of California San Diego for assistance with animal maintenance and Jachelle M. Ofrecio for assay insulin measurements.
This work was supported, in whole or in part, by National Institutes of Health Grants DK033651, DK074868, DK063491, DK020541, and DK048321 and Eunice Kennedy Shriver NICHD/National Institutes of Health Cooperative Agreement U54 HD 012303 as part of the specialized Cooperative Centers Program in Reproduction and Infertility Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7.
- ATM
- adipose tissue macrophage
- ITT
- Insulin tolerance test
- GTT
- glucose tolerance test
- SVC
- stromal vascular cell
- FA
- fatty acid
- FFA
- free FA
- WAT
- white adipose tissue
- epi-WAT
- epididymal WAT
- HGP
- hepatic glucose production
- NC
- normal chow
- TNF
- tumor necrosis factor
- IL
- interleukin
- IFN
- interferon
- HFD
- high fat diet
- FACS
- fluorescence-activated cell sorter
- FITC
- fluorescein isothiocyanate
- TUNEL
- terminal dUTP nick-end labeling
- qPCR
- quantitative PCR.
REFERENCES
- 1.Shoelson S. E., Herrero L., Naaz A. (2007) Gastroenterology 132, 2169–2180 [DOI] [PubMed] [Google Scholar]
- 2.Cancello R., Henegar C., Viguerie N., Taleb S., Poitou C., Rouault C., Coupaye M., Pelloux V., Hugol D., Bouillot J. L., Bouloumié A., Barbatelli G., Cinti S., Svensson P. A., Barsh G. S., Zucker J. D., Basdevant A., Langin D., Clément K. (2005) Diabetes 54, 2277–2286 [DOI] [PubMed] [Google Scholar]
- 3.Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., Ferrante A. W., Jr. (2003) J. Clin. Invest. 112, 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H., Barnes G. T., Yang Q., Tan G., Yang D., Chou C. J., Sole J., Nichols A., Ross J. S., Tartaglia L. A., Chen H. (2003) J. Clin. Invest. 112, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arkan M. C., Hevener A. L., Greten F. R., Maeda S., Li Z. W., Long J. M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M. (2005) Nat. Med. 11, 191–198 [DOI] [PubMed] [Google Scholar]
- 6.Solinas G., Vilcu C., Neels J. G., Bandyopadhyay G. K., Luo J. L., Naugler W., Grivennikov S., Wynshaw-Boris A., Scadeng M., Olefsky J. M., Karin M. (2007) Cell Metab. 6, 386–397 [DOI] [PubMed] [Google Scholar]
- 7.Kanda H., Tateya S., Tamori Y., Kotani K., Hiasa K., Kitazawa R., Kitazawa S., Miyachi H., Maeda S., Egashira K., Kasuga M. (2006) J. Clin. Invest. 116, 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamei N., Tobe K., Suzuki R., Ohsugi M., Watanabe T., Kubota N., Ohtsuka-Kowatari N., Kumagai K., Sakamoto K., Kobayashi M., Yamauchi T., Ueki K., Oishi Y., Nishimura S., Manabe I., Hashimoto H., Ohnishi Y., Ogata H., Tokuyama K., Tsunoda M., Ide T., Murakami K., Nagai R., Kadowaki T. (2006) J. Biol. Chem. 281, 26602–26614 [DOI] [PubMed] [Google Scholar]
- 9.Hirasaka K., Kohno S., Goto J., Furochi H., Mawatari K., Harada N., Hosaka T., Nakaya Y., Ishidoh K., Obata T., Ebina Y., Gu H., Takeda S., Kishi K., Nikawa T. (2007) Diabetes 56, 2511–2522 [DOI] [PubMed] [Google Scholar]
- 10.Gordon S. (1998) Res. Immunol. 149, 685–688 [DOI] [PubMed] [Google Scholar]
- 11.Cinti S., Mitchell G., Barbatelli G., Murano I., Ceresi E., Faloia E., Wang S., Fortier M., Greenberg A. S., Obin M. S. (2005) J. Lipid Res. 46, 2347–2355 [DOI] [PubMed] [Google Scholar]
- 12.Hotamisligil G. S. (1999) Exp. Clin. Endocrinol. Diabetes 107, 119–125 [DOI] [PubMed] [Google Scholar]
- 13.Rotter V., Nagaev I., Smith U. (2003) J. Biol. Chem. 278, 45777–45784 [DOI] [PubMed] [Google Scholar]
- 14.Kim H. J., Higashimori T., Park S. Y., Choi H., Dong J., Kim Y. J., Noh H. L., Cho Y. R., Cline G., Kim Y. B., Kim J. K. (2004) Diabetes 53, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 15.Hotamisligil G. S., Spiegelman B. M. (1994) Diabetes 43, 1271–1278 [DOI] [PubMed] [Google Scholar]
- 16.Gordon S., Taylor P. R. (2005) Nat. Rev. Immunol. 5, 953–964 [DOI] [PubMed] [Google Scholar]
- 17.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. (2004) Trends Immunol. 25, 677–686 [DOI] [PubMed] [Google Scholar]
- 18.Gordon S. (2003) Nat. Rev. Immunol. 3, 23–35 [DOI] [PubMed] [Google Scholar]
- 19.Lumeng C. N., Bodzin J. L., Saltiel A. R. (2007) J. Clin. Invest. 117, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patsouris D., Li P. P., Thapar D., Chapman J., Olefsky J. M., Neels J. G. (2008) Cell Metab. 8, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura S., Manabe I., Nagasaki M., Eto K., Yamashita H., Ohsugi M., Otsu M., Hara K., Ueki K., Sugiura S., Yoshimura K., Kadowaki T., Nagai R. (2009) Nat. Med. 15, 914–920 [DOI] [PubMed] [Google Scholar]
- 22.Feuerer M., Herrero L., Cipolletta D., Naaz A., Wong J., Nayer A., Lee J., Goldfine A. B., Benoist C., Shoelson S., Mathis D. (2009) Nat. Med. 15, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hevener A. L., He W., Barak Y., Le J., Bandyopadhyay G., Olson P., Wilkes J., Evans R. M., Olefsky J. (2003) Nat. Med. 9, 1491–1497 [DOI] [PubMed] [Google Scholar]
- 24.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 25.Inouye K. E., Shi H., Howard J. K., Daly C. H., Lord G. M., Rollins B. J., Flier J. S. (2007) Diabetes 56, 2242–2250 [DOI] [PubMed] [Google Scholar]
- 26.Surmi B. K., Hasty A. H. (2008) Future Lipidol. 3, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harman-Boehm I., Blüher M., Redel H., Sion-Vardy N., Ovadia S., Avinoach E., Shai I., Klöting N., Stumvoll M., Bashan N., Rudich A. (2007) J. Clin. Endocrinol. Metab. 92, 2240–2247 [DOI] [PubMed] [Google Scholar]
- 28.Hevener A. L., Olefsky J. M., Reichart D., Nguyen M. T., Bandyopadyhay G., Leung H. Y., Watt M. J., Benner C., Febbraio M. A., Nguyen A. K., Folian B., Subramaniam S., Gonzalez F. J., Glass C. K., Ricote M. (2007) J. Clin. Invest. 117, 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strissel K. J., Stancheva Z., Miyoshi H., Perfield J. W., 2nd, DeFuria J., Jick Z., Greenberg A. S., Obin M. S. (2007) Diabetes 56, 2910–2918 [DOI] [PubMed] [Google Scholar]
- 30.Nguyen M. T., Favelyukis S., Nguyen A. K., Reichart D., Scott P. A., Jenn A., Liu-Bryan R., Glass C. K., Neels J. G., Olefsky J. M. (2007) J. Biol. Chem. 282, 35279–35292 [DOI] [PubMed] [Google Scholar]
- 31.Majer M., Popov K. M., Harris R. A., Bogardus C., Prochazka M. (1998) Mol. Genet. Metab. 65, 181–186 [DOI] [PubMed] [Google Scholar]
- 32.Cao H., Gerhold K., Mayers J. R., Wiest M. M., Watkins S. M., Hotamisligil G. S. (2008) Cell 134, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sell H., Eckel J. (2007) Curr. Opin. Lipidol. 18, 258–262 [DOI] [PubMed] [Google Scholar]
- 34.Nomiyama T., Perez-Tilve D., Ogawa D., Gizard F., Zhao Y., Heywood E. B., Jones K. L., Kawamori R., Cassis L. A., Tschöp M. H., Bruemmer D. (2007) J. Clin. Invest. 117, 2877–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheja L., Heese B., Zitzer H., Michael M. D., Siesky A. M., Pospisil H., Beisiegel U., Seedorf K. (2008) Exp. Diabetes Res. 2008, 230837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim F., Pham M., Luttrell I., Bannerman D. D., Tupper J., Thaler J., Hawn T. R., Raines E. W., Schwartz M. W. (2007) Circ. Res. 100, 1589–1596 [DOI] [PubMed] [Google Scholar]
- 37.Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., Flier J. S. (2006) J. Clin. Invest. 116, 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong S. W., Kwon M. J., Choi A. M., Kim H. P., Nakahira K., Hwang D. H. (2009) J. Biol. Chem. 284, 27384–27392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J. Y., Hwang D. H. (2006) Mol. Cells 21, 174–185 [PubMed] [Google Scholar]
- 40.Monetti M., Levin M. C., Watt M. J., Sajan M. P., Marmor S., Hubbard B. K., Stevens R. D., Bain J. R., Newgard C. B., Farese R. V., Sr., Hevener A. L., Farese R. V., Jr. (2007) Cell Metab. 6, 69–78 [DOI] [PubMed] [Google Scholar]
- 41.Lesniewski L. A., Hosch S. E., Neels J. G., de Luca C., Pashmforoush M., Lumeng C. N., Chiang S. H., Scadeng M., Saltiel A. R., Olefsky J. M. (2007) Nat. Med. 13, 455–462 [DOI] [PubMed] [Google Scholar]
- 42.Bagga D., Wang L., Farias-Eisner R., Glaspy J. A., Reddy S. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1751–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee T. H., Hoover R. L., Williams J. D., Sperling R. I., Ravalese J., 3rd, Spur B. W., Robinson D. R., Corey E. J., Lewis R. A., Austen K. F. (1985) N. Engl. J. Med. 312, 1217–1224 [DOI] [PubMed] [Google Scholar]
- 44.Endres S., Ghorbani R., Kelley V. E., Georgilis K., Lonnemann G., van der Meer J. W., Cannon J. G., Rogers T. S., Klempner M. S., Weber P. C. (1989) N. Engl. J. Med. 320, 265–271 [DOI] [PubMed] [Google Scholar]
- 45.Meydani S. N., Lichtenstein A. H., Cornwall S., Meydani M., Goldin B. R., Rasmussen H., Dinarello C. A., Schaefer E. J. (1993) J. Clin. Invest. 92, 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hudert C. A., Weylandt K. H., Lu Y., Wang J., Hong S., Dignass A., Serhan C. N., Kang J. X. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11276–11281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo C. J., Chiu K. C., Fu M., Lo R., Helton S. (1999) J. Surg. Res. 82, 216–221 [DOI] [PubMed] [Google Scholar]
- 48.González-Périz A., Horrillo R., Ferré N., Gronert K., Dong B., Morán-Salvador E., Titos E., Martínez-Clemente M., López-Parra M., Arroyo V., Clària J. (2009) FASEB J. 23, 1946–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kusunoki M., Tsutsumi K., Hara T., Ogawa H., Nakamura T., Miyata T., Sakakibara F., Fukuzawa Y., Suga T., Kato K., Hirooka Y., Nakaya Y. (2003) Metabolism 52, 30–34 [DOI] [PubMed] [Google Scholar]
- 50.Matsuzaka T., Shimano H., Yahagi N., Kato T., Atsumi A., Yamamoto T., Inoue N., Ishikawa M., Okada S., Ishigaki N., Iwasaki H., Iwasaki Y., Karasawa T., Kumadaki S., Matsui T., Sekiya M., Ohashi K., Hasty A. H., Nakagawa Y., Takahashi A., Suzuki H., Yatoh S., Sone H., Toyoshima H., Osuga J., Yamada N. (2007) Nat. Med. 13, 1193–1202 [DOI] [PubMed] [Google Scholar]