Abstract
In a mature and infectious retroviral particle, the capsid protein (CA) forms a shell surrounding the genomic RNA and the replicative machinery of the virus. The irregular nature of this capsid shell precludes direct atomic resolution structural analysis. CA hexamers and pentamers are the fundamental building blocks of the capsid, however the pentameric state, in particular, remains poorly characterized. We have developed an efficient in vitro protocol for studying the assembly of Rous sarcoma virus (RSV) CA that involves mild acidification and produces structures modeling the authentic viral capsid. These structures include regular spherical particles with T = 1 icosahedral symmetry, built from CA pentamers alone. These particles were subject to cryoelectron microscopy (cryo-EM) and image processing, and a pseudo-atomic model of the icosahedron was created by docking atomic structures of the constituent CA domains into the cryo-EM-derived three-dimensional density map. The N-terminal domain (NTD) of CA forms pentameric turrets, which decorate the surface of the icosahedron, while the C-terminal domain (CTD) of CA is positioned underneath, linking the pentamers. Biophysical analysis of the icosahedral particle preparation reveals that CA monomers and icosahedra are the only detectable species and that these exist in reversible equilibrium at pH 5. These same acidic conditions are known to promote formation of a RSV CA CTD dimer, present within the icosahedral particle, which facilitates capsid assembly. The results are consistent with a model in which RSV CA assembly is a nucleation-limited process driven by very weak protein-protein interactions.
Keywords: Biophysics, Electron Microscopy (EM), Protein Self-assembly, Retrovirus, Virus Assembly, Capsid Protein, Rous Sarcoma Virus
Introduction
Retroviruses undergo a complex, multistaged assembly process directed by a precursor polyprotein Gag (1, 2). Several thousand copies of Gag assemble into a spherical immature virion; their N-termini associated with inner viral membrane and their C termini contacting the RNA genome in the particle interior. Sometime after particle budding, in a process termed maturation, Gag is cleaved by the viral protease, releasing the matrix, capsid (CA),4 and nucleocapsid proteins, the major structural proteins of the mature virus. CA reorganizes to form the external shell of the viral core, inside which the genomic RNA and the replicative enzymes of the virus are packaged. During the early stages of viral infection, the core is delivered into the cytoplasm of the host cell where uncoating of the core and reverse transcription take place (3). Regulated dissociation of the capsid shell surrounding the core appears to be critical for progression of the infection cycle (4–6) and interference with this process forms part of the innate immune response in many species (7–10). Because of its central role in the viral replication cycle, the organization of the retroviral core, and the pathways through which CA assembles during particle maturation have been widely investigated.
Application of cryoelectron microscopy (cryo-EM) techniques has enabled visualization of authentic retroviral cores; however the resolution achieved to date does not enable a molecular interpretation. Cores within mature virus particles are polymorphic (11–17). They have a conical appearance in human immunodeficiency virus (HIV-1), and a more polyhedral shape in Rous sarcoma virus (RSV). The polymorphism of viral cores precludes averaging, and limits the resolution of structural analysis. However, under appropriate conditions, CA can be assembled in vitro into more regular structures that mimic the authentic core (18–26). High-resolution cryo-EM studies of the in vitro assembly products, in combination with x-ray diffraction and solution NMR spectroscopy studies of CA and its constituent domains have helped clarify the basic aspects of capsid architecture.
CA is composed of two independently folded, predominantly α-helical domains, connected by a short and flexible linker. Despite morphological differences, the mature capsid has a common molecular organization across all retroviral genera. It is built in large part from an array of hexameric CA (24, 27–29), where the hexameric rings of the N-terminal domain (NTD) are linked to neighboring rings via dimerization of the C-terminal domain (CTD) (18, 30, 31). The NTD thereby forms the capsid exterior, while the CTD is positioned underneath, facing the capsid interior. Heterotypic NTD-CTD interactions are also critical for capsid formation and stability (27, 29, 32, 33). For HIV-1, structural descriptions of the mature hexameric capsid array have advanced to atomic resolution (29).
Given that the capsid cannot be closed with hexamers alone, it has been conjectured that capsid pentamers must exist to enable closure of the shell (24, 34). Very recently, this notion received direct experimental support, through a cryo-EM study that visualized RSV CA pentamers within icosahedral assemblies, as well as the interactions between CA pentamers and hexamers (19). Pentameric and hexameric CA rings are constructed through slight variation of essentially similar interfaces. Diversity in the positioning of the pentameric rings within the capsid, and in the intrinsic curvature of the hexagonally packed capsid surface, is now hypothesized to give rise to the observed differences in core morphology between the retroviral genera (19, 29, 34).
Less is known about the intermolecular forces which help form and stabilize the capsid, and the pathways through which the capsid assembles. HIV-1 CA exists in a reversible monomer-dimer equilibrium at neutral pH, with dimerization occurring via the CTD. In the presence of salt or crowding agent it can be induced to further assemble in vitro into tubular and conical structures, which resemble the viral core (20, 21, 23, 24). RSV CA, in contrast, remains monomeric at neutral pH. It can however be induced to assemble in vitro either by mild acidification (18, 22), or by the addition of anions such as phosphate (25, 26). In the case of mild acidification, the assembly products are a mixture of planar sheets composed purely of CA hexamers; “T = 1” icosahedra composed purely of CA pentamers; and Core-Like Particles (CLPs), which are inferred to contain both hexamers and pentamers (supplemental Figs. S1 and S2).
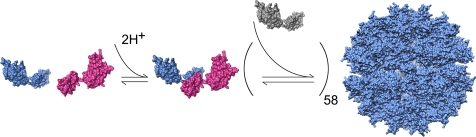
The promotion of capsid assembly by acidification is understood in structural terms. Protonation of an aspartic acid residue within the CTD facilitates dimerization of CA (18), which in turn drives higher-order capsid assembly (Fig. 1). Addition of sodium phosphate to RSV CA at neutral pH results in the formation of similar assembly products although the underlying mechanism is not completely defined. However, both proton-driven and phosphate-driven assembly of RSV CA seem to involve a nucleation step (18, 25). This also appears to be the case for in vitro assembly of HIV-1 CA (35).
FIGURE 1.
A model for proton-driven assembly of the icosahedral particles. Proton uptake by an aspartic acid located on the surface of the CTD promotes dimerization of RSV CA monomers. The dimer acts as a nucleating species. Subsequent addition of CA monomers results, in some instances, in the formation of T = 1 icosahedra. UCSF Chimera was used to generate this figure and Figs. 2 and 3.
The T = 1 icosahedron generated by mild acidification, and composed purely of RSV CA pentamers, is the subject of this study. We report a moderate resolution three-dimensional reconstruction, derived from cryo-EM images of such icosahedral particles. By docking known atomic structures of the NTD and CTD into the density map, a pseudo-atomic model of the icosahedron is constructed. Comparison with a model previously generated for the phosphate-assembled T = 1 icosahedral particles, shows that regardless of the conditions used to initiate assembly, there is a common structural outcome. Our structural model is consistent with the finding that proton-driven dimerization of the CTD facilitates capsid assembly. Biophysical characterization of the icosahedral particle preparation shows that assembly proceeds to completion once initiated, as only monomer and icosahedral can be detected in solution. These species exist in reversible equilibrium at pH 5. Additional experiments highlight the general importance of nucleation in initiating RSV capsid assembly in vitro.
EXPERIMENTAL PROCEDURES
Expression and Purification of CA
Full-length RSV CA (Prague C strain, GenbankTM Accession Number V01197) was produced by heterologous expression in Escherichia coli and purified to homogeneity as previously described (18). Following purification the protein was dialyzed into a standard storage buffer (10 mm MOPS/KOH (pH 7.0), 50 mm NaCl, 0.5 mm sodium azide, 0.25 mm TCEP-HCl) and stored at 4 °C. Protein concentrations were estimated using UV absorption measurements at 280 nm (36).
In Vitro Assembly of CA
In vitro assembly of RSV CA was initiated by transferring the protein into a high-salt, mildly acidic buffer. Sitting drop vapor diffusion, dialysis, and jump dilution were used for CA assembly. Among these methods, jump-dilution was used exclusively for the production of icosahedra, as this method was most efficient and reproducible. Typically, the assembly buffer (0.2 m citric acid/KOH (pH 4.9), 1.4 m NaCl) was added to an equal volume of CA solution (400–600 μm in standard storage buffer), followed by 2–3 h incubation at 18 °C (a 1:1 mixture of storage and assembly buffer has a pH of 5.0). Icosahedral particles could be efficiently separated from sheets and CLPs using differential centrifugation. Spinning the assembled material at 25,000 × g (15 min, 18 °C) in a benchtop centrifuge pelleted the larger assembly products. The supernatant, which we term the icosahedral particle preparation, was used for cryo-EM or biophysical analysis.
For the CA nucleation experiment, the icosahedral particle preparation (in a 1:1 mixture of assembly and storage buffer) was first dialyzed into the storage buffer, and then mixed with unassembled CA, also in storage buffer. 2 μl of the icosahedral particle preparation (total [CA] = 34 μm) was mixed with 18 μl of the unassembled monomer (total [CA] = 547 μm). Control experiments were performed by using pure storage buffer in place of the icosahedral particle preparation. Following mixing, the solutions were incubated at 18 °C for 48 h before examination using transmission electron microscopy (TEM).
Electron Microscopy
For routine TEM analysis, 5 μl of sample was applied onto a 300 mesh EM-grid covered with plastic-supported carbon film that was rendered hydrophilic by glow discharge in air and then negatively stained using 1.5% uranyl acetate. For cryo-EM of the T = 1 icosahedra, 5 μl of the icosahedral particle preparation was applied to a holey EM grid (Quantifoil R2/2) that had been rendered hydrophilic by glow discharging in the presence of n-amylamine. Vitrified specimens were prepared by using Vitrobot Mark IV (FEI) at 4 °C and 90–100% relative humidity. Focal pairs with defocus values of ∼3.0 μm and 0.8 μm were imaged under minimal electron dose using a Philips Tecnai 12 electron microscope equipped with a Lab6 gun and operated at 120 kV. The images were recorded on SO-163 (Kodak) films at a nominal magnification of 42,000, and developed in D19 (Kodak) diluted 1:1 with deionized water for 10 min.
Image Processing
Electron micrographs displaying minimal drift and astigmatism were digitized using Nikon LS-9000 film scanner at a step size 10.5 μm, which corresponded to 2.5 Å on the specimen. Unless otherwise specified, image processing was carried out using the Bsoft package (37, 38). Images of icosahedral particles were manually selected from high-defocused micrographs in Bshow, and then the location of the corresponding particles in the close-to-focus images were searched and extracted using Bmgalign and Bpick, respectively. The Contrast Transfer Function (CTF) was estimated for individual micrographs using Bshow, and then the particle images were phase-flipped using Bctf. Origin and orientation of particles in the low-defocused images were iteratively searched and the three-dimensional reconstruction was generated using PFT2 and EM3DR2 (39–41). The resolution of the three-dimensional map was estimated using the 0.5 Fourier shell coefficient cutoff criterion (42).
Atomic structures of the NTD (PDB ID code: 1EM9) and the low pH CTD dimer (PDB ID code: 3G21) from RSV CA were manually docked into the density ascribed to a CA monomer in the three-dimensional reconstruction using UCSF Chimera (43). Initially, the NTD structure was placed into one of the five density volumes that protrude around the local 5-fold axis and then the CTD dimer was docked into the density linking neighboring pentameric rings at the local 2-fold axis. The docking was refined using Sculptor (44), and the symmetrized, complete pseudo-atomic model for the icosahedron was generated using UROX (45). The cross-correlation between the cryo-EM density map and the fitted x-ray structures were calculated in Sculptor. The image reconstruction and pseudo-atomic model have been deposited in the EM Data Bank (entry code: EMD-1710).
Size Exclusion Chromatography
Small zone size exclusion chromatography was performed using an Äkta purifier (GE Healthcare). Icosahedral particle preparations (500 μl; total [CA] 47 μm) were injected onto a Superose 6 10/300 column (GE Healthcare) equilibrated with 1:1 mixture of assembly buffer and storage buffer. The column was calibrated using protein standards (Sigma) in 20 mm HEPES/NaOH (pH 7.5), 150 mm NaCl. Injections of tobacco Mosaic virus and l-tyrosine were used to estimate the void volume (V0) and included volume (Vi) of the column. All chromatography runs were carried out at 5.5 °C, with a flow rate of 0.25 ml/min. Elution of proteins was monitored by following UV absorbance at 280 nm.
Distribution coefficients (σ) for all proteins were calculated from elution volumes (Ve) as σ = (Ve − V0)/Vi (46). The Hydrodynamic radii of the protein standards were calculated from the published translational diffusion coefficients using the Stokes-Einstein relationship (supplemental Table S1) and a linear calibration plot was constructed (cube root of the distribution coefficient versus hydrodynamic radius) (47, 48). This plot was used to estimate the sizes of the species present in the icosahedral particle preparation.
Dynamic Light Scattering
All dynamic light scattering measurements were made at a wavelength of 831.1 nm using a Dynapro Titan (Wyatt Technology). Before data collection, solutions were filtered (0.2 μm) and spun at high speed (25,000 × g) in a bench top centrifuge to remove dust and other particulate matter. Intensity Autocorrelation Functions (ACFs) were measured for both unassembled CA (in storage buffer) and icosahedral particle preparations (in a 1:1 mixture of storage buffer and assembly buffer). ACFs were also recorded for the buffer solutions alone, to detect any correlated light scattering by the buffer components. All samples were maintained at 18 °C during the data collection.
For unassembled CA, measurements were made using protein solutions of varying concentration (40–670 μm). The normalized ACFs, G(τ), were fitted using Equation 1 appropriate for scattering of non-interacting protein molecules of fixed size (49, 50),
using non-linear least squares methods. This yielded estimates for the translational diffusion coefficient (D). Here τ is the decay time, A is a constant dependent on the experimental conditions, and q = (4πn)/λ)sin(θ/2) is the length of the scattering vector, which is a function of the solvent refractive index n, the wavelength of the incident light λ, and the scattering angle θ. At each protein concentration, several hundred ACFs were accumulated and analyzed. Linear extrapolation of the mean, apparent D to infinite protein dilution (51) was then used to estimate the true translational diffusion coefficient (D0) of unassembled CA. The hydrodynamic radius (RH) of unassembled CA was then calculated from D0 using the Stokes-Einstein relationship.
For the icosahedral particle preparations, measurements were made on solutions with a total [CA] = 47 μm. Normalized ACFs were fitted with a generalization of Equation 2, appropriate when there are several non-interacting species of fixed size present in solution (49, 50).
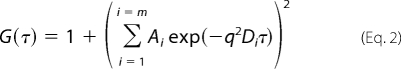 |
Associated with each scattering species i, are two model parameters Ai and Di. To account for weak correlated light scattering from the buffer, a 3 species model was fitted (2 CA species plus buffer), fixing the diffusion coefficient (D) for the buffer contribution, and allowing only the corresponding amplitude (A) to vary. It is the very disparate sizes of the scattering species, which enable straightforward application of Equation 2. The appropriate value for the buffer diffusion coefficient was determined from measurements made on the buffer alone. Several thousand individual ACFs were analyzed to derive the mean reported model parameters. This analysis ignores any chemical exchange between the scattering species (i.e. monomer and icosahedra), and also assumes that the species diffuse independently (i.e. that their diffusion is uncoupled, and all cross-diffusion coefficients are negligible (see Annunziata et al. (52) for discussion).
Quantitative interpretation of the DLS data required that the refractive index, kinematic viscosity, and density of all buffers be determined at 18 °C (supplemental Table S2). Buffer refractive index at 831.1 nm was measured using a critical angle digital refractometer (Schmidt and Haensch DSR-λ). Kinematic viscosity was measured using a Cannon-Ubbelohde viscometer (Cannon Instrument Corporation). Buffer density was measured using a Gay-Lussac specific gravity bottle (Wilmad-Labglass) and a standard mass balance (Sartorius CPA225D), making the required corrections for air buoyancy (53).
The expected hydrodynamic radius of monomeric CA was calculated based on the atomic model using the precise boundary element method of Aragon and Hahn (54) as implemented in the program BEST. It should be noted that the calculation neglects the contribution of the last 10 amino acid residues of CA, which are disordered and not included in the structural model.
RESULTS
RSV CA Forms T = 1 Icosahedra upon Acidification
We have previously shown that mild acidification causes RSV CA to assemble into complexes that model the authentic viral core (18). Together with CLPs, planar sheets, and small spherical particles result from the in vitro assembly process (supplemental Fig. S1). Class averages generated from the cryo-EM images of the spherical particles exhibit distinct 2-, 3-, and 5-fold rotational symmetry, consistent with these objects possessing icosahedral symmetry (supplemental Fig. S2).
A three-dimensional map of the icosahedron was reconstructed at 18.3 Å resolution by single particle image analysis. A total of 1,610 icosahedral particles were picked from the cryo-EM images out of which 1,310 particles contributed to the final reconstruction. It was clear from the reconstruction that the icosahedron has a “T = 1” architecture (55) i.e. is composed of 12 identical pentameric CA rings. The maximum diameter of the icosahedron is ∼23 nm, with an outer layer composed of protruding lobes of density; five at each vertex, and a continuous layer of inner density surrounding a ∼10 nm diameter cavity. The three-dimensional map was thresholded such that the density volume was 120% of the expected molecular mass of 60 CA monomers (25.5 kDa), a typical contour level used in moderate resolution cryo-EM analysis (55).
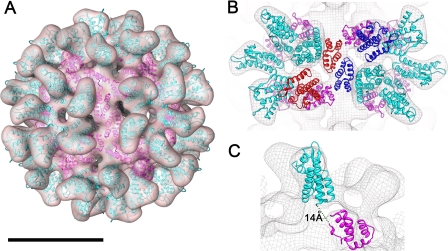
Although the resolution of the three-dimensional map was insufficient to visualize the boundaries between the CA monomers, a plausible pseudo-atomic model was derived through the docking of atomic models for the NTD and CTD of RSV CA (Fig. 2A). Initially, manual docking of the domains was guided by previously published pseudo-atomic models of the HIV-1 CA hexamer (PDB ID code: 3DIK) (27) and the RSV CA pentamer (19). The hand of the three-dimensional reconstruction was ascertained during the docking exercise. The atomic structure of the NTD (PDB ID code: 1EM9) fitted into the protruding densities in the outer layer of the three-dimensional reconstruction showed a good agreement. We examined the fitting of the x-ray crystal structures of CTD dimers determined at alkaline pH (PDB ID codes: 3G1G and 3G1I) and at acidic pH (PDB ID code: 3G21) in the three-dimensional map. Consistent with the conditions used for assembly, only the low-pH dimer showed a good fit into the density that links two pentamers (Fig. 2B). After refinement using Sculptor, the cross-correlation coefficient between the docked model and the density map was 91.1% at 18 Å resolution. In the final pseudo-atomic model, the N terminus of each CTD is separated from the C terminus of a neighboring NTD by a distance of ∼14 Å. Assuming some small structural rearrangement, this distance could be readily spanned by the stretch of 3 amino acids (Glu-Pro-Ala) defining the flexible interdomain linker (Fig. 2C).
FIGURE 2.
A pseudo-atomic model of the RSV CA T = 1 icosahedron generated at low pH, based on three-dimensional reconstruction from cryo-EM images. A, pseudo-atomic model of the RSV CA icosahedron produced by docking high-resolution structures of the constituent domains into the three-dimensional map calculated at ∼18 Å resolution. The domains are shown in a ribbon representation. B, detail of two CA pentamers, related by the 2-fold symmetry of the icosahedron. C, detail of a monomer. The three-dimensional density map enclosing the model is depicted as an isosurface in A and isomesh in B and C. The NTD and CTD are colored cyan and magenta, respectively. Monomers involved in linking two CA pentamers together via CTD dimerization are shown in blue and red in B. Scale bar is 10 nm.
Description of the Icosahedral Particle Structure
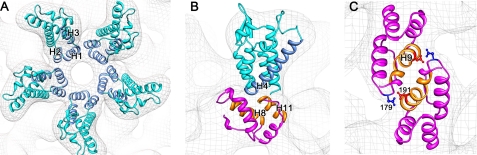
The three interdomain interfaces i.e. NTD-NTD, NTD-CTD, and CTD-CTD that have been identified in other retroviral CA assemblies are all involved in the formation of the icosahedron. We inspected, in particular, the dispositions of the α-helices at the interdomain interfaces. For clarity, the α-helices within a monomer are numbered sequentially, beginning at the N terminus.
NTD-NTD Interface
The pentameric CA turrets are formed through NTD-NTD interactions. Helices 1, 2, and 3 form a loosely packed 15 helix bundle disposed around the local 5-fold axis (Fig. 3A). As can be seen, in this bundle, helices 1 and 2 are proximal while helix 3 is distal from the 5-fold axis suggesting that helix 3 may play a less direct role in forming the pentameric CA assembly. In contrast, helix 3 is critical for the water-mediated NTD-NTD interactions found in the HIV-1 and murine leukemia virus CA hexamers (28, 29).
FIGURE 3.
Interdomain contacts within the RSV CA T = 1 icosahedron. A, NTD-NTD interface; B, NTD-CTD interface; and C, CTD-CTD dimerization interface. The density map is depicted as an isomesh. The NTD and CTD are colored in cyan and magenta, respectively. Helices involved in the interdomain contacts are colored in light blue and orange for the NTD and CTD, respectively. These are labeled with a capital letter H and the helix number. Two pairs of interacting aspartic acid residues that stabilize the CTD-CTD dimer are shown in ball-and-stick representation in C.
NTD-CTD Interface
NTD-CTD interdomain contacts help stabilize the RSV CA pentamer. The N terminus of helix 4, from the NTD, is proximal to helices 8 and 11 from the CTD. The arrangement of helices at the NTD-CTD interface, with the points of contact involving helix termini, is similar to that seen in HIV-1 CA hexamer (29). However in the HIV-1 CA hexamer, helix 3 contacts helix 8 through an extended hydrogen-bonding network. However, in the RSV CA pentamer, helices 3 and 8 are too far apart to harbor such interactions (Fig. 3B).
CTD-CTD Interface
The low-pH CTD dimer x-ray structure used in building the pseudo-atomic model is stabilized primarily through two hydrogen bonds that results from protonation of aspartate Asp-191 (18). These hydrogen bonds link helix 9 of each monomer with the loop connecting helices 8 and 9 (Fig. 3C).
Biophysical Analysis of the Icosahedral Particle Preparation
With a structural model of the icosahedral particles determined, we performed experiments to help understand how the particles assemble from CA monomers. There is suggestive evidence that the icosahedral particles can form in situ (15, 19), and the CA pentamers have a clear role in helping form the authentic core (24, 34). Hence we wanted to establish if the icosahedra were in reversible equilibrium with unassembled CA, and if any other species of intermediate size could be detected in the particle preparations. For this purpose, we employed two biophysical techniques: small-zone size exclusion chromatography (SEC) and dynamic light scattering (DLS).
Size Exclusion Chromatography
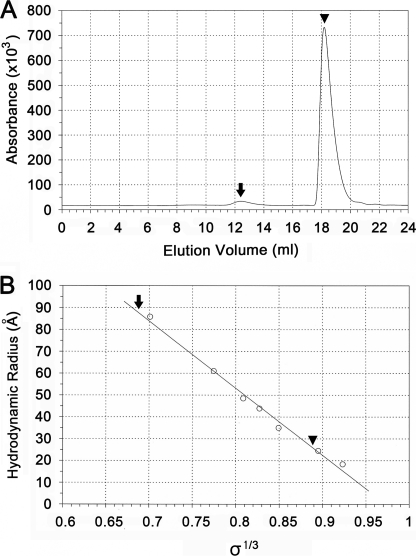
After separating the icosahedra from the other assembly products using high-speed centrifugation, the icosahedral particle preparation (total [CA] = 47 μm) was loaded onto a Superose 6 SEC column capable of resolving large protein complexes. The resulting chromatogram showed two well-resolved peaks (Fig. 4A). TEM of fractions taken from the 2 peaks, demonstrated the presence of icosahedra in the fast-eluting peak but not in the slow-eluting peak. Through comparison with standards of known size (Fig. 4B), the hydrodynamic radius (RH) of the smaller species was estimated to be 2.62 nm, consistent with the size of the CA monomer, while RH for the larger species was estimated to be 8.90 nm, approximately the size expected for the icosahedral particles (see “Discussion”). Hence only monomers and icosahedra, and no intermediate species, could be detected in solution using SEC. The experiments also established that (a) the kinetics of exchange between monomer and icosahedra must be relatively slow, because they could be isolated using a size exclusion column (56–58) and (b) that the majority of CA in the icosahedral particle preparations remains as a monomer.
FIGURE 4.
Size exclusion chromatography of the icosahedral particle preparation. A, peaks corresponding to the icosahedra and to the unassembled monomer are indicated by arrow and arrowhead, respectively. B, plot of cube root of distribution coefficient (σ1/3) as a function of hydrodynamic radius for the protein size standards. The distribution coefficients for the icosahedron (σ = 0.319) and CA monomer (σ = 0.700) are indicated by arrow and arrowhead. The resultant estimates for the hydrodynamic radius are 8.90 nm (icosahedron) and 2.62 nm (CA monomer).
Although kinetically slow, we demonstrated that the monomer and icosahedra exist in reversible equilibrium. Thus, when the CA monomers or icosahedra isolated by SEC were subject to concentration and reinjection onto a SEC column, chromatograms containing both species were again obtained (supplemental Fig. S3). This indicates that in the time required to concentrate and re-inject the samples (∼2 h), equilibrium was partially or completely re-established.
Dynamic Light Scattering
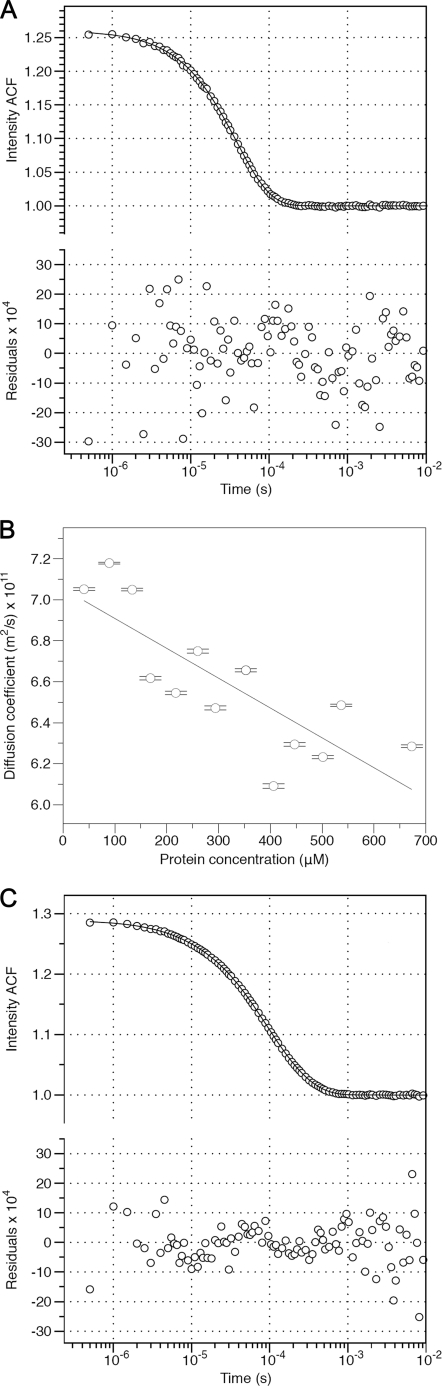
To obtain more precise estimates for the sizes of the species present in solution, and confirm the results obtained using SEC, we characterized both unassembled CA and the icosahedral particle preparation using DLS. The ACFs for the unassembled CA solutions could be fit with a simple model, allowing for a single scattering species of fixed size (Fig. 5A). Model fitting resulted in estimates for the translational diffusion coefficient of the protein. Linear extrapolation of the results to infinite protein dilution (Fig. 5B), produced an estimate of 7.05 × 10−11 m2/s (18 °C, standard storage buffer) for the true translation diffusion coefficient of unassembled CA, which corresponds to a hydrodynamic radius of 2.74 nm. As a point of comparison we calculated the expected RH for the CA monomer. The NTD and CTD of CA are known to be flexibly tethered (59), so we took the domain configuration found within the icosahedra as representative of a conformation accessible in solution. After modeling the three missing residues of the interdomain linker, the hydrodynamic properties of the monomer were calculated from the atomic model using a precise boundary element method (54). The calculation yielded RH = 2.76 nm, in excellent agreement with the experimentally determined value.
FIGURE 5.
Dynamic light scattering studies of the CA monomer and the icosahedral particle preparation. A, typical intensity auto-correlation function (ACF) accumulated from measurements for a CA monomer solution ([CA] = 670 μm) and the fit to a one component scattering model (Equation 1). The hollow circles are the experimentally determined ACF; the solid line represents the fitted model. The residual differences between model and experimental data are indicated in the bottom of the panel. B, plot of the mean, apparent, translational diffusion coefficient of monomeric CA, in standard storage buffer, as a function of protein concentration. Error bars correspond to ± S.E. The value at “zero” protein concentration reflects the true translational diffusion coefficient of monomeric CA. C, typical intensity auto-correlation function accumulated from an icosahedral particle preparation (total [CA] = 47 μm) and its fit to a three component scattering model (Equation 2). The buffer scattering contribution was constrained during fitting (see text). The hollow circles are the experimentally determined ACF; the solid line represents the fitted model. The residual differences between model and experimental data are indicated in the bottom of the panel.
ACFs resulting from the icosahedral particle preparations were fit to an extension of the simple model, allowing for the presence of multiple independent scattering species with fixed sizes (Equation 2). Based on the results of the SEC analysis, a 2-species model seemed appropriate. However the assembly buffer itself contributed weak correlated light scattering, hence a 3-species model was fitted, with experimentally derived constraints placed on the buffer scattering contribution (see “Experimental Procedures”). This resulted in an excellent fit to the data, with random residuals (Fig. 5C). We estimated that for the smaller protein species RH = 2.82 nm, whereas for the larger protein species RH = 11.66 nm. These are, within the likely error in the measurements, the expected values for the monomer and icosahedra respectively. It was not possible to make measurements over an extended concentration range and extrapolate the results to zero protein concentration since the icosahedral particles dominate the light scattering.
Role of Nucleation in Initiating in Vitro Assembly of RSV CA
Our DLS analysis confirms that RSV CA exists as a monomer in standard storage buffers at neutral pH, consistent with prior observation (18, 22, 59). Capsid assembly can be initiated by lowering the pH (18, 22) or introducing anions such as phosphate (19, 25, 26). In each case, perturbation of the solution conditions is posited to result in formation of a nucleating species, which can be extended into capsid-like structures through addition of monomeric CA.
The proton-driven assembly process leads to the production of fully assembled icosahedra in appreciable quantity. The icosahedral particles are metastable when transferred back into a neutral pH, low salt buffer (18). We hypothesized that dissociation of the icosahedra might create transient, low-abundance species that are capable of nucleating further capsid assembly without addition of phosphate or protons (i.e. under conditions, which are not normally permissive for capsid assembly).
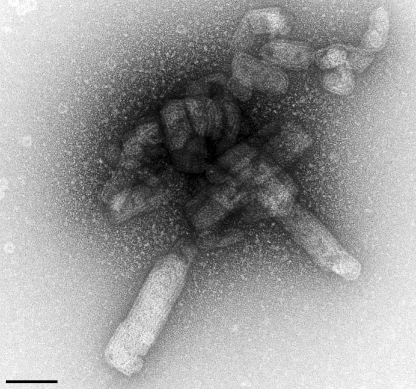
Hence we tested whether introducing trace amounts of the icosahedral particle preparation into a pool of unassembled CA could seed further CA assembly. For this purpose, the icosahedral particle preparation was dialyzed into a low molarity neutral pH buffer and used to spike a concentrated solution of CA monomer in the same buffer. After 48 h of incubation at 18 °C, the sample was examined by TEM. With a small number of icosahedra in the background, large tubular assembly products and core-like particles were observed (Fig. 6). Control experiments in which the CA monomer solution was spiked with buffer alone resulted in no assembly.
FIGURE 6.
Assembly of RSV CA nucleated by the icosahedral particle preparation. An electron micrograph showing tubular assembly products that were formed when a small amount of the icosahedral particle preparation was added to a solution of monomeric CA in low molarity buffer at neutral pH. The specimen was visualized by negative staining. Scale bar is 100 nm.
DISCUSSION
Upon acidification RSV CA assembles into various morphologically diverse complexes, which model the authentic viral core. Prominent among these complexes are spherical particles with T = 1 icosahedral symmetry. We have generated a structural model for the acid-induced icosahedron, and have biophysically characterized the icosahedral particle preparation. The basic building block of the icosahedron is a CA pentamer, which is stabilized by both NTD-NTD and NTD-CTD interdomain contacts. The pentamers are linked together by CTD homodimers (Figs. 2 and 3). Postassembly, once the larger complexes such as sheets and core-like particles are removed, only icosahedra and CA monomer remain. These species are present in reversible equilibrium at pH 5, with no intermediates detectable (Figs. 4 and 5). Addition of small amounts of the icosahedral particle preparation to a pool of CA monomers promotes further assembly into capsid-like structures, under conditions that are otherwise not permissive to any CA assembly (Fig. 6). This highlights the importance of nucleation in controlling assembly of RSV CA in vitro and possibly also in vivo.
The Initiation of RSV CA Assembly in Vitro
We have previously shown that CTD dimerization is promoted by acidification (18), with the underlying mechanism involving proton uptake by an aspartate on the surface of the domain. A causal relationship between CTD dimerization and higher-order CA assembly was confirmed by introducing a covalent bond across the dimer interface, which created a super assembling capsid mutant (18). These observations lead to the general model of in vitro capsid assembly, and in particular the formation of icosahedra, that is presented in Fig. 1. The good correspondence between the low pH CTD dimer structure, determined using x-ray crystallography (18), and the three-dimensional reconstruction of the acid-induced T = 1 icosahedron (Fig. 2) is fully supportive of such a model. The CTD dimer bridges adjacent pentamers in the icosahedron (Fig. 2B), helping explain why dimerization is so effective in promoting capsid assembly.
Protonation of the CTD is not, however, obligate for formation and maintenance of the T = 1 icosahedral particles. Multivalent anions, such as phosphate also promote assembly of RSV CA in vitro (25, 26), resulting in the formation of icosahedra at neutral pH (19). Once assembled, the acid-induced particles can be transferred into a neutral pH buffer, where they are metastable. The pseudo-atomic model for the acid-induced particles, determined in this study, appears essentially identical to the model for the phosphate-induced particles reported earlier by Cardone et al. (19). The same low pH CTD dimer is present in both models. It remains possible that protonation and deprotonation of the critical aspartate within the CTD is associated with some subtle change in dimer organization. This cannot be determined from current three-dimensional image reconstructions of the icosahedral particles, both because of limited resolution, and because of the averaging involved in the image reconstruction process.
In the case of the phosphate-assembled icosahedra, it has been proposed that the addition of anions facilitates CA assembly by reducing electrostatic repulsion between clusters of positively charged residues at the NTD-NTD and NTD-CTD interfaces, that are involved in the formation of both hexamers and pentamers (19). Our study suggests that these repulsive electrostatic forces are delicately balanced with weak attractive forces. If the balance is perturbed, either by diminishing the repulsive forces (as in the addition of phosphate), or by strengthening the attractive forces (as in acidification, see Fig. 1), then capsid assembly results in vitro. For some small spherical viruses, empty capsids can be assembled from coat proteins in the absence of any nucleic acid, a process fundamentally similar to that studied here. Assembly is often strongly influenced by pH and ionic strength (60–63), though in some cases this may simply reflect the presence of basic nucleic acid binding domains within these proteins.
Although it is apparent that phosphate-induced assembly of RSV CA at neutral pH also involves a nucleation step, the nature of the nucleating species has not yet been fully defined. There is some suggestive evidence that it may be a dimer (25) whose structure, given the elevated pH, is likely to be different from the CTD-mediated dimer that initiates assembly at low pH. Regardless, our study makes it clear that in vitro assembly of RSV CA can be triggered by different molecular forces and proceed by differing pathways, while still arriving at the same basic structural outcome.
We have also highlighted the importance of nucleation in this system with a simple experiment as follows. In a neutral pH, low-salt storage buffer, RSV CA is monomeric and does not assemble. However, under these same conditions, when trace amounts of an icosahedral particle preparation were introduced into the CA monomer solution, large, tubular structures and core-like particles resulted (Fig. 6). Our interpretation of this result is that transient dissociation of the icosahedral particles creates species capable of nucleating further capsid assembly without the addition of either phosphate or protons, the otherwise obligate triggers for efficient RSV CA association in vitro.
Biophysical Analysis of the Assembly Products and Its Implications for Capsid Formation
Biophysical analysis of the assembly products present at equilibrium has provided further insight into the assembly process. Overall, the results from SEC and DLS are self-consistent. They confirm that RSV CA exists as a monomer in solution prior to assembly. Post assembly, following removal of the larger assembly products, only monomer and fully assembled icosahedra are present at appreciable concentration. These species are in reversible equilibrium at pH 5, although it should be noted that exchange is slow, and appears to involve a nucleation step in the forward direction. The observation that intact icosahedra and free monomer are the dominant species present at equilibrium is consistent with a simple equilibrium model of capsid assembly (64), which predicts that assembly intermediates will be found in only trace concentrations. Overall the reversibility of assembly demonstrates that the CA-CA interactions which direct capsid formation must be weak, involving very small association energies when considered individually.
Based on the DLS data (Fig. 4C) we estimate the hydrodynamic radius of the icosahedral particles to be 11.66 nm, in good agreement with the measured 11.5 nm outer radius of the particles determined by 3D image reconstruction (Fig. 2). In contrast, SEC provides a significantly smaller estimate of the particle size (RH = 8.90 nm). We suggest two reasons for this discrepancy. Firstly, icosahedral particle formation is reversible. While the rates of inter-conversion between monomer and icosahedron are clearly slow, they will not be negligible relative to the time taken for passage of the sample through the column (∼37 min for the monomer). The theory of migration for such slowly interconverting systems is complicated (56–58); however transient dissociation of icosahedral particles will clearly result in longer average retention times, and hence a lowered estimate for RH. Secondly, the T = 1 CA icosahedron is a hollow object with an inner cavity radius of ∼10 nm and is easily compressed and flattened when preserved under heavy metal stain on continuous carbon film (data not shown). Thus, it is quite possible that intact icosahedral particles can undergo mechanical compression and/or distortion during transport down the SEC column, which would also lead to a lowered estimate for the hydrodynamic radius. DLS measurements, are made on solutions at equilibrium, and are likely to yield more reliable size estimates than SEC, where mass transport effects as well as the mechanical forces acting on the capsid, cannot be ignored.
CONCLUSION
This study demonstrates that in vitro assembly of RSV CA under mildly acidic conditions results in the formation of T = 1 icosahedral particles containing CA pentamers. These particles provide an excellent model for studying the pentamers, which are critical for closure of the authentic viral core. At the resolution achieved in this study, the structure of the low-pH icosahedral particles appears essentially identical to that of the particles assembled at neutral pH in the presence of phosphate (19). Hence, although the intermediates involved in the proton-driven and phosphate-driven assembly pathways may differ (18, 25), these pathways can converge to a common structural outcome. Our biophysical experiments highlight the importance of nucleation in influencing RSV capsid assembly, and demonstrate the reversibility of the underlying molecular interactions. Such reversibility is important, as controlled disassembly of the capsid shell, which surrounds the viral core is a critical early step in the viral replication cycle. Proton binding may play a role in regulating capsid assembly or stability in vivo, and acidification can be used as an in vitro tool, producing structures which model the authentic viral capsid, and therefore facilitating the study of retroviral capsid assembly.
Acknowledgments
We thank Graham Bailey for technical assistance with purification and characterization of protein. We thank Adrian Turner for maintenance of the electron microscope.
This work was supported, in whole or in part, by National Institutes of Health R21 Grant (to A. K. M.). This work was also supported by the New Zealand Health Research Council Sir Charles Hercus Health Research Fellowship (to R. L. K.).
The image reconstruction and pseudo-atomic model have been deposited in the EM Data Bank (entry code EMD-1710).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- CA
- capsid protein
- RSV
- Rous sarcoma virus
- cryo-EM
- cryoelectron microscopy
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- HIV-1
- human immunodeficiency virus type-1
- CLP
- core-like particle
- TEM
- transmission electron microscopy
- CTF
- contrast transfer function
- ACF
- intensity autocorrelation function
- SEC
- size exclusion chromatography
- DLS
- dynamic light scattering
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Ganser-Pornillos B. K., Yeager M., Sundquist W. I. (2008) Curr. Opin. Struct. Biol. 18, 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mateu M. G. (2009) FEBS J. 276, 6098–6109 [DOI] [PubMed] [Google Scholar]
- 3.Nisole S., Saïb A. (2004) Retrovirology 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auerbach M. R., Brown K. R., Singh I. R. (2007) J. Virol. 81, 12337–12347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dismuke D. J., Aiken C. (2006) J. Virol. 80, 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forshey B. M., von Schwedler U., Sundquist W. I., Aiken C. (2002) J. Virol. 76, 5667–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huthoff H., Towers G. J. (2008) Trends Microbiol 16, 612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman R. M., Johnson W. E. (2007) AIDS Rev. 9, 114–125 [PubMed] [Google Scholar]
- 9.Ozato K., Shin D. M., Chang T. H., Morse H. C., 3rd (2008) Nat. Rev. Immunol. 8, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towers G. J. (2007) Retrovirology 4, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamin J., Ganser-Pornillos B. K., Tivol W. F., Sundquist W. I., Jensen G. J. (2005) J. Mol. Biol. 346, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs J. A., Grünewald K., Glass B., Förster F., Kräusslich H. G., Fuller S. D. (2006) Structure 14, 15–20 [DOI] [PubMed] [Google Scholar]
- 13.Briggs J. A., Watson B. E., Gowen B. E., Fuller S. D. (2004) J. Virol. 78, 2606–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggs J. A., Wilk T., Welker R., Kräusslich H. G., Fuller S. D. (2003) EMBO J. 22, 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butan C., Winkler D. C., Heymann J. B., Craven R. C., Steven A. C. (2008) J. Mol. Biol. 376, 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kingston R. L., Olson N. H., Vogt V. M. (2001) J. Struct Biol. 136, 67–80 [DOI] [PubMed] [Google Scholar]
- 17.Yeager M., Wilson-Kubalek E. M., Weiner S. G., Brown P. O., Rein A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7299–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey G. D., Hyun J. K., Mitra A. K., Kingston R. L. (2009) Structure 17, 737–748 [DOI] [PubMed] [Google Scholar]
- 19.Cardone G., Purdy J. G., Cheng N., Craven R. C., Steven A. C. (2009) Nature 457, 694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Alamo M., Rivas G., Mateu M. G. (2005) J. Virol. 79, 14271–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrlich L. S., Agresta B. E., Carter C. A. (1992) J. Virol. 66, 4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingston R. L., Fitzon-Ostendorp T., Eisenmesser E. Z., Schatz G. W., Vogt V. M., Post C. B., Rossmann M. G. (2000) Structure 8, 617–628 [DOI] [PubMed] [Google Scholar]
- 23.Lanman J., Sexton J., Sakalian M., Prevelige P. E., Jr. (2002) J. Virol. 76, 6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Hill C. P., Sundquist W. I., Finch J. T. (2000) Nature 407, 409–413 [DOI] [PubMed] [Google Scholar]
- 25.Purdy J. G., Flanagan J. M., Ropson I. J., Craven R. C. (2009) J. Mol. Biol. 389, 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdy J. G., Flanagan J. M., Ropson I. J., Rennoll-Bankert K. E., Craven R. C. (2008) J. Virol. 82, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganser-Pornillos B. K., Cheng A., Yeager M. (2007) Cell 131, 70–79 [DOI] [PubMed] [Google Scholar]
- 28.Mortuza G. B., Haire L. F., Stevens A., Smerdon S. J., Stoye J. P., Taylor I. A. (2004) Nature 431, 481–485 [DOI] [PubMed] [Google Scholar]
- 29.Pornillos O., Ganser-Pornillos B. K., Kelly B. N., Hua Y., Whitby F. G., Stout C. D., Sundquist W. I., Hill C. P., Yeager M. (2009) Cell 137, 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamble T. R., Yoo S., Vajdos F. F., von Schwedler U. K., Worthylake D. K., Wang H., McCutcheon J. P., Sundquist W. I., Hill C. P. (1997) Science 278, 849–853 [DOI] [PubMed] [Google Scholar]
- 31.Worthylake D. K., Wang H., Yoo S., Sundquist W. I., Hill C. P. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 85–92 [DOI] [PubMed] [Google Scholar]
- 32.Bowzard J. B., Wills J. W., Craven R. C. (2001) J. Virol. 75, 6850–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanman J., Lam T. T., Emmett M. R., Marshall A. G., Sakalian M., Prevelige P. E., Jr. (2004) Nat. Struct. Mol. Biol. 11, 676–677 [DOI] [PubMed] [Google Scholar]
- 34.Heymann J. B., Butan C., Winkler D. C., Craven R. C., Steven A. C. (2008) Comput. Math. Methods Med. 9, 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barklis E., Alfadhli A., McQuaw C., Yalamuri S., Still A., Barklis R. L., Kukull B., López C. S. (2009) J. Mol. Biol. 387, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heymann J. B. (2001) J. Struct Biol. 133, 156–169 [DOI] [PubMed] [Google Scholar]
- 38.Heymann J. B., Belnap D. M. (2007) J. Struct. Biol. 157, 3–18 [DOI] [PubMed] [Google Scholar]
- 39.Baker T. S., Cheng R. H. (1996) J. Struct. Biol. 116, 120–130 [DOI] [PubMed] [Google Scholar]
- 40.Crowther R. A., Amos L. A., Finch J. T., De Rosier D. J., Klug A. (1970) Nature 226, 421–425 [DOI] [PubMed] [Google Scholar]
- 41.Fuller S. D., Butcher S. J., Cheng R. H., Baker T. S. (1996) J. Struct. Biol. 116, 48–55 [DOI] [PubMed] [Google Scholar]
- 42.Van Heel M. (1987) Ultramicroscopy 21, 95–100 [DOI] [PubMed] [Google Scholar]
- 43.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 44.Wriggers W., Milligan R. A., McCammon J. A. (1999) J. Struct. Biol. 125, 185–195 [DOI] [PubMed] [Google Scholar]
- 45.Navaza J., Lepault J., Rey F. A., Alvarez-Rúa C., Borge J. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1820–1825 [DOI] [PubMed] [Google Scholar]
- 46.Ackers G. K. (1970) Adv. Protein Chem. 24, 343–446 [DOI] [PubMed] [Google Scholar]
- 47.Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 48.Winzor D. J. (2003) J. Biochem. Biophys. Methods 56, 15–52 [DOI] [PubMed] [Google Scholar]
- 49.Finsy R. (1994) Adv. Colloid Interface Sci. 52, 79–143 [Google Scholar]
- 50.Lomakin A., Teplow D. B., Benedek G. B. (2005) Methods Mol. Biol. 299, 153–174 [DOI] [PubMed] [Google Scholar]
- 51.Xu R. (2000) Particle Characterization: Light Scattering Methods, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 52.Annunziata O., Buzatu D., Albright J. G. (2005) Langmuir 21, 12085–12089 [DOI] [PubMed] [Google Scholar]
- 53.Battino R., Williamson A. G. (1984) J. Chem. Ed. 61, 51–52 [Google Scholar]
- 54.Aragon S., Hahn D. K. (2006) Biophys. J. 91, 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker T. S., Olson N. H., Fuller S. D. (1999) Microbiol. Mol. Biol. Rev. 63, 862–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cann J. R., Kegeles G. (1974) Biochemistry 13, 1868–1874 [DOI] [PubMed] [Google Scholar]
- 57.Stevens F. J. (1989) Biophys. J. 55, 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmerman J. K. (1974) Biochemistry 13, 384–389 [DOI] [PubMed] [Google Scholar]
- 59.Campos-Olivas R., Newman J. L., Summers M. F. (2000) J. Mol. Biol. 296, 633–649 [DOI] [PubMed] [Google Scholar]
- 60.Adolph K. W., Butler P. J. (1974) J. Mol. Biol. 88, 327–341 [DOI] [PubMed] [Google Scholar]
- 61.Salunke D. M., Caspar D. L., Garcea R. L. (1989) Biophys. J. 56, 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lepault J., Petitpas I., Erk I., Navaza J., Bigot D., Dona M., Vachette P., Cohen J., Rey F. A. (2001) EMBO J. 20, 1498–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ceres P., Zlotnick A. (2002) Biochemistry 41, 11525–11531 [DOI] [PubMed] [Google Scholar]
- 64.Zlotnick A. (1994) J. Mol. Biol. 241, 59–67 [DOI] [PubMed] [Google Scholar]