Abstract
Fibroblast growth factors (FGFs) are key regulators of cell proliferation, tumor-induced angiogenesis, and migration. FGFs are essential for early embryonic development, organ formation, and angiogenesis. FGF1 also plays an important role in inflammation, wound healing, and restenosis. The biological effects of FGF1 are mediated through the activation of the four transmembrane phosphotyrosine kinase fibroblast growth factor receptors in the presence of heparin sulfate proteoglycans and, therefore, require the release of the protein into the extracellular space. FGF1 is exported through a non-classical release pathway involving the formation of a specific multiprotein complex. The protein constituents of this complex include FGF1, S100A13, and the p40 form of synaptotagmin 1 (Syt1). Because FGF1 plays an important role in tumor formation, it is clear that preventing the formation of the multiprotein complex would be an effective strategy to inhibit a wide range of cancers. To understand the molecular events in the FGF1 release pathway, we studied the FGF1-S100A13 tetrameric and FGF1-S100A13-C2A hexameric complex structures, which are both complexes possibly formed during the non-classical pathway of FGF1 release.
Keywords: Protein, Protein/Protein-protein Interactions, Protein/Export, Protein/Intracellular Trafficking, Protein/Structure, Signal Transduction
Introduction
Acidic fibroblast growth factor (FGF1)2 belongs to a large family of heparin-binding growth factors. Apart from their mitogenic activity, FGFs are the key activators of tumor-induced angiogenesis (1, 2). The majority of the members of the FGF family protein are exported via the ER/Golgi-dependent secretory pathway. Soluble secretory proteins characteristically include N-terminal signal peptides that direct protein localization to the ER translocation apparatus (3). Following vesicular transport from the ER via the Golgi apparatus to the cell surface, luminal proteins are released into the extracellular space by fusion of the Golgi-derived secretory vesicles to the plasma membrane. This pathway of protein expression from eukaryotic cells is known as the classical ER/Golgi-dependent secretory pathway. However, proteins such as fibroblast growth factor (FGF1 and FGF2), sphingosine kinase 1, interleukin 1α and 1β, gelectin-1, and the extravesicular p40 fragment of Syt1 all lack the classical signal sequence and can be exported from the cell in the absence of a functional ER/Golgi system (4–6). Most of these proteins are associated with key cellular processes, such as angiogenesis, inflammation, tumor growth, cell proliferation, or differentiation. In this context, the possible release pathways used by proteins that lack signal peptides have been the subject of recent investigation. FGF1 has been shown to be secreted by an alternative pathway activated by different forms of stress, such as heat shock (7, 8), serum starvation (9), or hypoxia (10). Although it was first assumed that angiogenic growth factors might be released from mechanically injured tissues to promote wound healing, a process that requires angiogenesis, various lines of evidence demonstrated that FGF1 is exported from cultured cells in the absence of appropriate amounts of cell death (7, 8).
The secreted FGF1 isolated from cell culture supernatants represents a latent homodimer that can also be formed upon chemical oxidation of FGF1 in vitro (11). Upon heat shock, two intracellular proteins have been shown to associate with FGF1 in the cytoplasm. These are the cleavage products of the transmembrane protein synaptotagmin, consisting of its cytoplasmic domain (p40Syt1) and the Ca2+ binding protein S100A13 (7–8, 12–15). Apparently, they are exported together with FGF1. Direct roles for Syt1 and S100A13 in FGF1 export have been demonstrated by both antisense RNA-mediated knockdown of Syt1 expression and the expression of a dominant negative S100A13 mutant that attenuates the export of FGF1 (12, 15).
S100A13 is a newly discovered member of the S100 protein family. It is characterized by its specificity for different forms of cancer (16–17). S100A13 has been reported to co-express with fibroblast growth factor 1 (FGF1) in brain tumors (16), demonstrating a perivascular distribution. S100A13 is a member of the family of Ca2+-binding proteins that are characterized by the absence of a classical signal peptide sequence and the presence of two EF-hand domains. S100A13 is a novel member of the S100 gene family that encodes a highly charged C-terminal domain that could be involved in specific protein interactions. S100 proteins are characterized by two distinct EF-hand motifs displaying different Ca2+ affinities (18). Maciag and co-workers (8, 13) demonstrated that S100A13 is involved in the regulation of the release of FGF1 in response to stress, independent of the conventional ER-Golgi pathway. When S100A13 is expressed in FGF1-free cells, it exhibits a spontaneous non-classical release both at 37 and 42 °C, but, when it is expressed in cells with FGF1, it is released only by heat shock (12). An additional indication of the participation of S100A13 in the export of proinflammatory cytokines is its specificity in binding to anti-inflammatory drugs, such as amlexanox and chromolyn (19). S100A13 appears to be the central player in the formation of this complex.
Synaptotagmin 1 is a synaptic vesicle protein that is essential for the fast component of neurotransmitter release (20). Synaptotagmin exists as a large gene family in mammals and is characterized by a common structure; an N-terminal transmembrane sequence is joined to a variable length linker, followed by two tandemly arranged, distinct C2 domains, C2A and C2B. Based on the results of mutation analysis, it has been suggested that the C2A domain (127 amino acids) of Syt1 provides the interface for the binding of FGF1 and S100A13 (16). The C2A domain is the membrane-proximal region, and it mediates the interaction of Syt1 with phosphatidylserine (21). This interaction is proposed to contribute to the lipid rearrangements that are essential for membrane fusion (22). However, under conditions of cell stress, Syt1, similar to S100A13 and FGF1, localizes at the inner leaflet of the plasma membrane, where the formation of the release complex apparently takes place (23). Using an in vitro model of secretion, it has been demonstrated that FGF1 is exported as a non-covalent complex containing S100A13 and p40 Syt1 (13, 15). The release of FGF1 in response to stress is dependent on Syt1 expression. The expression of either a deletion mutant (lacking 95 amino acids from the extravesicular portion of Syt1) or the presence of an antisense Syt1 gene is able to repress FGF1 release in NIH3T3 cells (8, 15). In addition, FGF1 purified from ovine brain as a molecular weight aggregate exists as a component of the non-covalent heparin-binding complex with Syt1 and S100A13. FGF1 is released under temperature stress as a multiprotein complex consisting of FGF1, S100A13, and Syt1.
These results suggest that FGF1-S100A13-C2A complex formation is the first step in the FGF1 export pathway, followed by direct translocation of this protein complex across the plasma membrane. In this paper, we describe the interfacial regions of the solution structures of the FGF1-S100A13 and FGF1-S100A13-C2A complexes. Our results demonstrate that S100A13 acts as a template for the formation of the multiprotein complex.
EXPERIMENTAL PROCEDURES
Materials
Ingredients for Luria Broth were obtained from AMRESCO (USA). Aprotinin, pepstatin, leupeptin, phenylmethylsulfonyl fluoride, Triton X-100, and β-mercaptoethanol were obtained from Sigma. Heparin- and glutathione-Sepharoses were obtained from Amersham Biosciences. 15NH4Cl, 13C-labeled glucose, and D2O were purchased from Cambridge Isotope Laboratories. All other chemicals used were of high quality analytical grade. Unless specified, all solutions were made in 25 mm sodium phosphate containing 100 mm NaCl and 2 mm CaCl2.
Expression and Purification of the FGF1, C2A, and S100A13 Domains
Human FGF1 cDNA encoding the recombinant protein were subcloned in to pET(20+) expression vector. FGF1 was overexpressed and purified using methods reported by Arunkumar et al. (24). Human S100A13 and C2A cDNA encoding the recombinant protein were subcloned into the pGEX expression vector. C2A and S100A13 were expressed in Escherichia coli (BL21DE3). The unlabeled protein was expressed in Luria Broth (LB) medium. The soluble portion of the cell lysate was loaded onto a GST-Sepharose column. Nonspecifically bound proteins were removed by washing the column with phosphate-buffered saline buffer. The bound GST-C2A/GST-S100A13 protein was eluted with 10 mm glutathione and 50 mm Tris-HCl buffer (pH 8.0). The GST-fused S100A13 or C2A proteins were exchanged with phosphate-buffered saline buffer, and then the solution was treated with 50 μg of thrombin for 10–12 h. The GST portion of the protein was cleaved during thrombin digestion, and then the whole solution was reloaded onto the glutathione S-transferase column to obtain pure C2A or S100A13. The S100A13 or C2A domain that was obtained was further purified by gel filtration on a Superdex-75 (Amersham Biosciences) column using fast protein liquid chromatography and 10 mm sodium phosphate (pH 7.0) containing 100 mm NaCl as the eluent. The purity of the protein was checked by SDS-PAGE, and the molecular weight was confirmed by electrospray mass analysis.
Preparation of Isotope-enriched FGF1, C2A, and S100A13
Uniform 15N labeling and 15N plus 13C labeling of FGF1, C2A, and S100A13 was achieved by culturing the cells in M9 minimal medium containing either 15NH4Cl for single (15N) labeling or 15NH4Cl and [13C]glucose for double (15N and 13C) labeling. To achieve maximal expression yields, the composition of the M9 medium was modified by the addition of a vitamin mixture. The expression host strain E. coli BL21 (DE3) pLysS is a vitamin B1-deficient host; therefore, the medium was supplemented with thiamine (vitamin B1).
Isothermal Titration Calorimetry
Protein-protein binding was characterized by measuring the heat changes during the titration of its partner protein into the protein solution using a Microcal VP titration calorimeter. S100A13, FGF1, C2A, and S100A13-FGF1 solutions were centrifuged and degassed under vacuum before use. Titrations were performed by injecting 8-μl aliquots of protein (30 times; 1 mm concentration) into 0.1 mm partner protein solution. The titrations were performed at 25 °C by dissolving the proteins in 25 mm sodium phosphate (pH 6.0) containing 100 mm NaCl and 2 mm calcium chloride. Results of the titration curves were corrected using buffer-protein and protein-buffer controls and analyzed using Origin software supplied by Microcal.
1H-15N HSQC Titration
NMR data were recorded at 25 °C on Bruker 800-, 600-, and 500-MHz spectrometers equipped with cryogenic probes. For the two-dimensional heteronuclear experiments, the concentration of the proteins used was ∼0.6 mm. All of the protein samples were prepared in 25 mm phosphate buffer (in 90% H2O, 10% D2O) containing 100 mm sodium chloride and 2 mm calcium chloride. The spectra were recorded at 25 °C at pH ∼6.0. The 15N-labeled proteins were titrated with unlabeled proteins at 1:1 molar ratios in the binary complexes and 1:1:1 molar ratios in the ternary complexes. A plot of the weighted average of the (15N and 1H) chemical shift perturbations of the residues of the protein was calculated using the equation, Δδ = ((δ1H)2 + 0.2 (δ15N)2)½. Amide proton exchange rates were monitored by acquiring a series of 1H-15N HSQC spectra of proteins in their free states and upon their interaction with other protein partners involved in the multiprotein release complex. The spectra were processed with Topspin and analyzed with Sparky (25).
Three-dimensional NMR Experiments
The FGF1 and S100A13 resonances in the FGF1-S100A13 tetrameric complex and the C2A resonances in the FGF1-S100A13-C2A heterohexameric complex were assigned using various multidimensional NMR experiments. Assignments for the backbone 1H, 13C, and 15N resonances in the complexes containing FGF1, S100A13, and C2A were obtained through three-dimensional HNCA and HNCOCA experiments (26). The side chain resonances were assigned using three-dimensional 15N-edited TOCSY-HSQC and HCCH-TOCSY data sets supplemented with other experiments, including CBCACONH (27) and HBHACONH (28). HNCO spectra were used to assign the carbonyl carbons (29). The aromatic resonances of FGF1, S100A13, and C2A were assigned using simultaneous 13C/15N-edited NOESY-HSQC spectra (30). Intermolecular distance restraints were derived from the three-dimensional, 13C(ω2)-edited, 12C(ω3)-filtered NOESY-HSQC spectrum (31) of the 1:1 15N/13C/1H FGF1, 14N/12C/1H S100A13, and 15N/13C/1H S100A13, 14N/12C/1H FGF was used for the FGF1-S100A13 complex, and a 1:1 15N/13C/1H C2A to 14N/12C/1H FGF-S100A13 complex was used for FGF1-S100A13-C2A complexes.
Structure Calculations
Structures of FGF1 and S100A13 in the FGF1-S100A13 tetrameric complex and C2A domain in the FGF1-S100A13-C2A complex were calculated iteratively using ARIA/CNS (version 2.2) using the PARALLHDG 5.3 force field in the PARALLHDG mode (32, 33). Preliminary structure calculations based on intramolecular nuclear Overhauser effect (NOE) data and TALOS (34) data established that the backbone folds of FGF1, S100A13, and C2A are not substantially altered by the formation of the protein complexes. Distance restraints, dihedral angle, and hydrogen-bonding constraints were used in the structure calculation. Interproton distance restraints for the structure calculations were derived from the 15N-separated NOESY-HSQC and 13C-separated NOESY-HSQC experiments. The quality of the calculated structures was assessed using PROCHECK (35).
Docking Studies
HADDOCK (36–40) was applied to dock FGF1 and S100A13 for the FGF1-S100A13 tetrameric complex and to dock C2A and the FGF1-S100A13 tetrameric complex for the FGF1-S100A13-C2A heterohexameric complex using the previously determined structures established using ARIA and intermolecular NOEs. Intermolecular distance restraints were derived from a three-dimensional, 13C and 15N (F1)-filtered, 13C (F2)-edited, 12C (F3)-filtered NOESY experiment. A scaling factor was determined by comparing the intensities of the resolved peaks with those of the corresponding peaks in the 13C-edited NOESY spectrum acquired for the FGF1-S100A13-C2A complex. The chemical shift perturbations observed upon complex formation were used to define ambiguous interaction restraints (AIRs) for residues at the interface. Active residues were defined as those having both chemical shift perturbations and a residue-accessible relative surface area larger than 50% for either side-chain or backbone atoms as calculated with NACCESS (41). Passive residues were defined as all other surface non-accessible residues (having a relative residue-accessible surface area smaller than 50% for their side-chain or backbone atoms). AIRs were defined between every active residue of the first protein and all active and passive residues of second protein and vice versa. A total of 5000 rigid body docking trials were carried out using the standard HADDOCK protocol, with the 100 lowest energy solutions used for subsequent semiflexible simulated annealing and water refinement. The 20 structures with the lowest energies were taken to represent the structure of the complex. The structures were analyzed with PROCHECK (35).
RESULTS
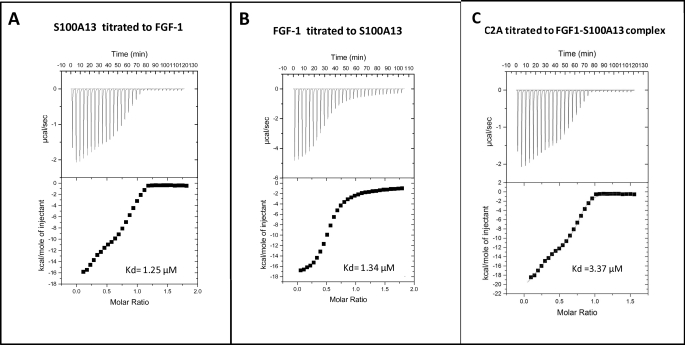
Isothermal titration calorimetry (ITC) is a very useful technique to study protein-protein interactions (42). ITC may be reliably used to measure the binding constants and energy changes that accompany the interactions of proteins with other proteins. Most importantly, ITC measurements provide information on the number of protein binding sites. We have measured the binding affinities of FGF1 to S100A13, S100A13 to FGF1, and C2A to the FGF1-S100A13 complex by ITC (Fig. 1). The binding constants are in the range 1.25–3.37 × 10−6 m.
FIGURE 1.
A, isothermogram representing the binding of S100A13 to FGF1 at 25 °C. The raw data of the titration of S100A13 with FGF1 are shown in the top, whereas the bottom shows the integrated data obtained after subtracting the heat of dilution. B, isothermogram representing the binding of FGF1 to S100A13 at 25 °C. The raw data of the titration of FGF1 with S100A13 are shown in the top, whereas the bottom shows the integrated data obtained after subtracting the heat of dilution. The titrations were performed in 25 mm phosphate-buffered saline (pH 6.0) containing 100 mm NaCl. C, isothermogram representing the binding of C2A to FGF1-S100A13 complex at 25 °C. The raw data of the titration of C2A with FGF1-S100A13 are shown in the top, whereas the bottom shows the integrated data obtained after subtracting the heat of dilution. The titrations were performed in 25 mm sodium phosphate (pH 6.0) containing 100 mm NaCl and 2 mm calcium chloride.
Chemical shift perturbation is the most widely used NMR method to map protein interfaces (43, 44). Briefly, the 1H-15N HSQC spectrum of a labeled protein is monitored while the unlabeled interaction partner is titrated against it, and the perturbations of the chemical shifts are recorded. The cross-peaks in the 1H-15N HSQC spectrum, which are perturbed upon the addition of the protein, often represent the binding site(s) of the protein to the target protein. Therefore, monitoring 1H-15N chemical shift perturbations by observing the shifts in the 1H-15N HSQC spectrum provides residue level information about the protein-protein interface. The interaction causes changes in the chemical environment of the protein interfaces and therefore affects the chemical shifts of the nuclei in that area. The 15N chemical shift perturbation technique has been used in many cases to map protein-protein interactions (43–45). To understand the mechanism of the FGF1 non-classical pathway at the molecular level, we solved the FGF1-S100A13 tetrameric and FGF1-S100A13-C2A hexameric complex structures, which are possibly the key complexes formed in the non-classical pathway of FGF1.
In the FGF1-S100A13 binary complex, we mapped FGF1 binding sites on S100A13 and S100A13 binding sites on FGF1 based on the chemical shift perturbations observed in the 1H-15N HSQC spectrum during the titration. 1H-15N HSQC spectra of free FGF1 and free S100A13 are well dispersed, and the cross-peaks representing the backbone amide protons of the 127 residues of FGF1 and the 96 residues of S100A13 were unambiguously assigned. At an FGF1 (15N-labeled)/S100A13 (unlabeled) mixing ratio of 1:1, cross-peaks corresponding to 14 residues in FGF1 (Lys-98, Asn-99, Trp-100, Phe-101, Arg-112, Gly-113, Arg-115, Thr-116, His-117, Tyr-118, Gly-119, Gln-120, Lys-121, and Ala-122) are perturbed upon complexation, whereas the other residues retain chemical shifts identical to those of the free FGF1 (supplemental Fig. 1A). These 14 residues possibly constitute the S100A13 binding site(s) in FGF1. The plot of the weighted average of the 15N and 1H chemical shift perturbations of the residues in FGF1 upon complex formation with S100A13 also shows that a maximum of 14 residues are perturbed (supplemental Fig. 1B). These residues are distributed in two loop regions of FGF1 (Lys-98 to Phe-101 and Arg-112 to Ala-122) and are close to helix 1, loop 1, and helix 2 of S100A13. At an FGF1 (unlabeled)/S100A13 (15N-labeled) ratio of 1:1, cross-peaks corresponding to 13 residues, including Thr-15′, Thr-18′, Phe-21′, Thr-22′, Phe-23′, Arg-25′, Gln-26′, Glu-27′, Arg-29′, Lys-30′, Asp-31′, Asn-36′ and Glu-40′ (where a prime indicates S100A13 residues), are perturbed upon complexation, whereas the other residues remain at the same chemical shift as in free S100A13. These residues possibly constitute the FGF1 binding site(s) in S100A13. A plot of the weighted average of the 15N and 1H chemical shift perturbation of residues in S100A13 upon complex formation with FGF1 shows that the S100A13 binding region is distributed in helix 1, loop 1, and helix 2 (supplemental Fig. 1D).
Comparison of the hydrogen/deuterium exchange rates of the individual amide protons in their free and bound states confirmed the binding interfaces in the complex FGF1-S100A13. Amide protons in proteins dissolved in D2O can readily be exchanged with deuterons. However, amide protons that are involved in backbone hydrogen bonding are more resistant to exchange than those that are located in the unstructured portions of the protein molecule (46).
After mapping the interfaces with HSQC perturbation data and H-D exchange data, we focused on solving the solution structures of the FGF1-S100A13 tetrameric complex in order to understand the molecular interactions in more detail. Triple-resonance experiments were performed by mixing doubly labeled (15N and 13C) protein with the corresponding unlabeled partner(s) to form the appropriate complexes. We have assigned the FGF1 and S100A13 resonances in the FGF1-S100A13 tetrameric complex. An unambiguous way of mapping biomolecular interactions is by using the intermolecular NOE (47). The intensity of the NOE is proportional to the sixth root of the interproton distance (r−6 distance proportionality). We obtained intramolecular NOEs from isotope-edited NOE spectra using 15N-edited NOESY and 13C-edited NOESY experiments. We observed the intermolecular NOEs in the FGF1-S100A13 (supplemental Fig. 2) complexes by mixing a doubly labeled (15N, 13C) protein with its corresponding unlabeled protein partner(s).
Structure of the FGF1-S100A13 Tetrameric Complex
The resonance assignments in the FGF1-S100A13 tetrameric complex were obtained using standard strategies based on triple resonance experiments. In total, 95% of the backbone amide resonances in the 1H-15N HSQC spectrum and their α-carbons were sequentially assigned, based on the analysis of the HNCA and HN(CO)CA experiments. The backbone carbonyl carbons were assigned using the HNCO experiment. The α and side chain proton resonances were assigned based on three-dimensional 15N-edited TOCSY-HSQC, three-dimensional HCCH-TOCSY, and 15N-edited NOESY-HSQC experiments. The NH2 groups of Gln and Asn residues were connected to their side chain γ and β protons using 15N-edited NOESY-HSQC.
Structure of FGF1 in the FGF1-S100A13 Complex
A set of 1646 intramolecular NOEs were assigned from the analysis of the three-dimensional 15N-edited NOESY-HSQC and converted to 1468 relevant distance restraints. In addition, 38 hydrogen bonds, identified from deuterium exchange experiments, were also used. Thus, a total of 1506 distance restraints were used for the final structure calculations (Table 1). The final representative ensemble of structures shows few molecular and constraint violations greater than 5 Å. The average r.m.s. deviation value for the secondary structure region was 0.56 ± 0.02 Å for the backbone atoms and 0.85 ± 0.02 Å for all heavy atoms. PROCHECK analysis of the structure indicated good stereochemistry for the bond angles and bond lengths and showed that 99.4% of all of the non-glycine residues fall within the allowed region of the Ramachandran plot (Table 1).
TABLE 1.
Structural statistics for the final 20 simulated annealing structures of FGF1, S100A13, and the FGF-S100A13 tetrameric complex
| Parameters | Values |
|---|---|
| Structural statistics from ARIA/CNS restrained calculations (FGF1 in the FGF1-S100A13 tetrameric complex) | |
| Protein distance restraints | |
| Total | 1506 |
| Intraresidue | 285 |
| Sequential | 392 |
| Medium range 1 < |i − j| < 5 | 175 |
| Long range |i − j| ≥ 5 | 536 |
| Hydrogen bond restraints | 38 |
| Dihedral angle restraints | 110 |
| Structural statistics for 20 structures | |
| Average r.m.s. deviation | |
| Backbone r.m.s. deviation to mean (Å) | 0.85 ± 0.02 Å |
| Heavy atom r.m.s. deviation to mean (Å) | 1.32 ± 0.06 Å |
| Average r.m.s. deviation (structured region) | |
| Backbone r.m.s. deviation to mean (Å) | 0.56 ± 0.02 Å |
| Heavy atom r.m.s. deviation to mean (Å) | 1.10 ± 0.09 Å |
| Ramachandran analysis | |
| Residues in most favored regions | 75.1% |
| Residues in additional allowed regions | 20.5% |
| Residues in generously allowed regions | 3.8% |
| Residues in disallowed regions | 0.6% |
| Structural statistics from ARIA/CNS restrained calculations (S100A13 domain in the FGF1-S100A13 tetrameric complex) | |
| Protein distance restraints | |
| Total | 1675 |
| Intraresidue | 311 |
| Sequential | 433 |
| Medium range 1 < |i − j| < 5 | 169 |
| Long range |i − j| ≥ 5 | 576 |
| Hydrogen bond restraints | 40 |
| Dihedral angle restraints | 95 |
| Structural statistics for 20 structures | |
| Average r.m.s. deviation | |
| Backbone r.m.s. deviation to mean (Å) | 0.82 ± 0.02 Å |
| Heavy atoms r.m.s. deviation to mean (Å) | 1.31 ± 0.04 Å |
| Average r.m.s. deviation (structured region) | |
| Backbone r.m.s. deviation to mean (Å) | 0.42 ± 0.02 Å |
| Heavy atom r.m.s. deviation to mean (Å) | 0.94 ± 0.03 Å |
| Ramachandran analysis | |
| Residues in most favored regions | 73.0% |
| Residues in additional allowed regions | 24.5% |
| Residues in generously allowed regions | 2.0% |
| Residues in disallowed regions | 0.5% |
| Structural statistics from HADDOCK restrained calculations (FGF1-S100A13 tetrameric complex); structural statistics for 20 structures | |
| Average r.m.s. deviation at backbone to mean (Å) | 0.76 ± 0.08 Å |
| Average r.m.s. deviation at FGF1 and S100A13 interface | 0.52 ± 0.07 Å |
| Average r.m.s. deviation at FGF1 interface | 0.50 ± 0.02 Å |
| Average r.m.s. deviation at S100A13 interface | 0.56 ± 0.02 Å |
| Ramachandran analysis | |
| Residues in most favored regions | 73.4% |
| Residues in additional allowed regions | 22.7% |
| Residues in generously allowed regions | 3.3% |
| Residues in disallowed regions | 0.6% |
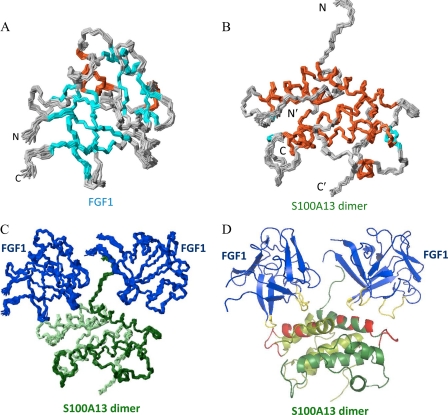
Fig. 2A shows the superposition of an ensemble of 20 structures of the FGF1 complex with S100A13. The FGF1 structure is β-sheet-rich, containing 12 β-strands and three α-helices. The FGF1 structure has a similar conformation to the unbound state. The S100A13 binding site on FGF1 is distributed over two regions, the first in the IX loop and the second in the XI loop region. The majority of the residues in the interfacial region are highly solvent-accessible. When the free FGF1 structure and the FGF1 complex structures are compared, the first and second binding regions show some conformational change, possibly attributable to the binding of S100A13. The IX loop of FGF1 is near the S100A13 loop 1, and loop XI is near helix 1 of S100A13.
FIGURE 2.
A, superposition of the backbone (N, Cα, and C′) atoms of the 20 final solution structures of FGF1 in the FGF1-S100A13 tetrameric complex. Orange, cyan, and gray, helix, β-strand, and loop regions, respectively. B, superposition of the backbone (N, Cα, and C′) atoms of the 20 final solution structures of the S100A13 dimer in the FGF1-S100A13 tetrameric complex. Orange, cyan, and gray, helix, β-strand, and loop regions respectively. C, superposition of the backbone (N, Cα, and C′) atoms of the 20 final solution structures of the FGF1-S100A13 tetrameric complex (two monomers are shown in light green and dark green). D, MOLMOL representation of the FGF1-S100A13 tetrameric complex structure; FGF1 is shown in blue, and two S100A13 monomers are shown in light green and dark green. The S100A13 binding region on FGF1 is shown in yellow, and the FGF1 binding region on S100A13 is shown in red.
Structure of S100A13 in the FGF1-S100A13 Complex
A set of 1649 intramolecular NOEs were assigned from the analysis of the three-dimensional 15N-edited NOESY-HSQC spectrum and converted to 1635 relevant distance restraints. Additionally, 40 hydrogen bonds that were identified from the deuterium exchange experiments were also used. Thus, a total of 1675 distance restraints were used for the final structure calculations (Table 1). The 20 structures of the S100A13 homodimer complex with FGF1 (Fig. 2B) that had the lowest energies were taken to represent the structure of the tetrameric complex. The average r.m.s. deviation value for the secondary structure region was 0.42 ± 0.02 Å for the backbone atoms and 0.82 ± 0.02 Å for all heavy atoms. A PROCHECK analysis of the structure indicated good stereochemistry for the bond angles and bond lengths and showed that 99.5% of all of the non-glycine residues fall within the allowed region of the Ramachandran plot. Fig. 2B shows the superposition of an ensemble of 20 structures of S100A13 in the tetrameric complex with FGF1. The S100A13 structure is a homodimer, with each monomer encompassing four α-helices and two β-strands, in agreement with the chemical shift indices. The FGF1 binding site on S100A13 is distributed over two regions, the first on helix 1 and the second region in loop 1. The majority of the residues in the interfacial region are highly solvent-accessible.
Structure of the FGF1-S100A13 Complex
Calculating the structures of protein-protein complexes using intermolecular data, chemical shift data, or both has recently been highly successful in generating structures when using the strong modeling program HADDOCK (36–40). HADDOCK was used to dock the previously determined structures of FGF1 and S100A13 (in FGF1-S100A13 complex) from ARIA/CNS. The structure of the binary complex of FGF1 and S100A13 was calculated using 30 intermolecular NOEs obtained by analyzing the filtered NOE data from the complex. Based on chemical shift perturbations of FGF1 and S100A13 upon complex formation, AIRs were defined for the residues at the interface. The active and passive residues were used to generate AIRs (Table 2). A total of 5000 rigid body docking trials were carried out using the standard HADDOCK protocol, with the 100 lowest energy structures used for subsequent semiflexible simulated annealing and water refinement. The 20 structures with the lowest energy were taken to represent the structure of the complex (Fig. 2C). The average r.m.s. deviation value for the backbone is 0.76 ± 0.08 Å. The average r.m.s. deviation at the interface between FGF1 and S100A13 is 0.52 ± 0.07. The average r.m.s. deviation at the FGF1 interface is 0.50 ± 0.02 Å. The average r.m.s. deviation at the S100A13 interface is 0.56 ± 0.08 Å. The complex structure was analyzed using PROCHECK (35).
TABLE 2.
List of the active and passive residues used to define the ambiguous interaction restraints for the docking of S100A13 with FGF1 and FGF1-S100A13 complex with C2A
The prime and double prime indicate residues of S100A13 and C2A, respectively.
| Complex | Residues |
|---|---|
| FGF1-S100A13 tetrameric complex | |
| FGF1 | |
| Active residues | Lys-98, Asn-99, Arg-112, Arg-115, His-117, Tyr-118, Gly-119, Gln-120, Lys-121 |
| Passive residues | Trp-100, Phe-101, Gly-113, Thr-116, Ala-122 |
| S100A13 | |
| Active residues | Thr-15′, Phe-21′, Thr-22′, Phe-23′, Arg-25′, Gln-26′, Lys-30′, Asn-36′ |
| Passive residues | Thr-18′, Glu-27′, Arg-29′, Asp-31′ |
| FGF1-S100A13 C2A hexameric complex | |
| FGF1-S100A13 complex | |
| Active residues | Thr-23, Thr-27, Arg-28, Arg-30, Ser-31, Asp-32, Gln-33, Lys-105, Asn-107, Lys-111, Pro-114, Arg-115, His-48′, Lys-51′, Val-53′, Gly-54′, Asp-57′, Glu-58′, Lys-91′, Lys-94′, Ile-95′, Arg-96′, Lys-97′ |
| Passive residues | Val-24, Asp-25, Gly-26, Asp-29, Lys-106, Gly-108, Ser-109, Cys-110, Arg-112, Leu-46′, Pro-47′, Leu-49′, Ser-55′, Leu-56′, Asp-92′, Leu-93′ |
| C2A | |
| Active residues | Leu-32″, Asp-33″, Met-34″, Gly-35″, Gly-36″, Leu-47″, Pro-48″, Asp-49″, Lys-50″, His-59″, Arg-60″, Lys-61″, Gly-82″, Phe-92″, Arg-94″, Phe-95″, Ser-96″, Lys-97″, His-98″, Asp-99″, Gly-103″, Lys-105″, Pro-107″, Ser-125″, Glu-127″ |
| Passive residues | Ala-31″, Thr-37″, Ser-38″, Thr-62″, Leu-63″, Lys-83″, Thr-84″, Asp-93″, Gly-102″, Glu-103″, Lys-105″, Pro-107″, Ser-125″ |
Fig. 2D shows the ribbon representation of the FGF1-S100A13 tetrameric complex. S100A13 is a homodimer. In this complex, each monomer binds one FGF1 molecule, and the complex appears as like two symmetric units. The yellow region on the FGF1 molecule shows the S100A13 binding region. The red region on S100A13 shows the FGF1 binding region.
Structure of the FGF-S100A13-C2A Heterohexameric Complex
We were interested in determining the interface regions in the FGF1-S100A13-C2A hexameric complex; therefore, we mapped the C2A-binding sites on the FGF1-S100A13 tetrameric complex. We mapped the FGF1-S100A13 binding sites on C2A on the basis of the 1H-15N HSQC chemical shift perturbations in the FGF1-S100A13 (15N-labeled)/C2A (unlabeled) and FGF1-S100A13 (unlabeled)/C2A (15N-labeled) complex spectra (supplementalFig. 3). These complexes were characterized before the advent of the TROSY technique. With TROSY, NMR lines remain narrow for large systems, and there is an approximate upper size limit of 100 kDa (48). Comparison of the hydrogen/deuterium exchange rates of the individual amide protons in their free and bound states confirmed the binding interfaces in the complex FGF1-S100A13-C2A.
After confirming the interface regions by HSQC and hydrogen/deuterium exchange data on C2A and C2A binding sites on the FGF1-S100A13 complex, we attempted to solve the solution structure of the FGF1-S100A13-C2A heterohexameric complex structure. To solve the FGF1-S100A13-C2A heterohexameric complex three-dimensional structure, we added doubly labeled C2A to the unlabeled FGF1-S100A13 tetrameric complex and solved the C2A structure in the complex using all of the required multidimensional NMR experiments. We have assigned the C2A resonances in the FGF1-S100A13-C2A hexameric complex. We observed intermolecular NOEs between C2A and (FGF1-S100A13) complex by mixing the double labeled C2A with an unlabeled FGF1-S100A13 tetrameric complex.
Assignments of C2A in the FGF1-S100A13-C2A Hexameric Complex
The resonances of the C2A domain of the synaptotagmin in the complex were assigned using standard strategies based on triple-resonance experiments. The backbone amide resonances in the 1H-15N HSQC spectrum and their α-carbons were sequentially assigned based on the analysis of HNCA and HN(CO)CA experiments. The backbone carbonyl resonance assignments were determined from HNCO experiments. The α and side chain carbon resonances were assigned using three-dimensional CBCA(CO)NH, three-dimensional HCCH-TOCSY, and CCC(CO)NH spectra. The α and side chain proton resonances were assigned using three-dimensional 15N-edited TOCSY-HSQC, three-dimensional HCCH-TOCSY, and 15N-edited NOESY-HSQC spectra. The NH2 groups of Gln and Asn residues were connected to their side chain γ and β protons using 15N-edited NOESY-HSQC.
Structure of the C2A Domain in the FGF1-S100A13-C2A Hexameric Complex
A set of 1589 intramolecular NOEs were assigned by analyzing the three-dimensional 15N-edited NOESY-HSQC spectrum, and they were converted to 1525 relevant distance restraints. In addition, 41 hydrogen bonds identified from deuterium exchange experiments were also used. Thus, 1566 distance restraints were used for the final structure calculations (Table 3). The final representative ensemble of structures shows few molecular and constraint violations greater than 5 Å. The average r.m.s. deviation value for the secondary structure region is 0.62 ± 0.02 Å for the backbone atoms and 0.92 ± 0.02 Å for all heavy atoms. PROCHECK analysis of the structure indicated good stereochemistry for the bond angles and bond lengths and showed that 99.6% of all non-glycine residues fall within the allowed region of the Ramachandran plot (Table 3).
TABLE 3.
Structural statistics for the final 20 simulated annealing structures of C2A and the FGF1-S100A13-C2A heterohexameric complex
| Parameters | Values |
|---|---|
| Structural statistics from ARIA/CNS restrained calculations (the C2A domain in the FGF1-S100A13-C2A heterohexameric complex) | |
| Protein distance restraints | |
| Total | 1566 |
| Intraresidue | 285 |
| Sequential | 392 |
| Medium range 1 < |i − j| < 5 | 175 |
| Long range |i − j| ≥ 5 | 536 |
| Hydrogen bond restraints | 41 |
| Dihedral angle restraints | 110 |
| Structural statistics for 20 structures | |
| Average r.m.s. deviation | |
| Backbone r.m.s. deviation to mean (Å) | 0.92 ± 0.02 Å |
| Heavy atoms r.m.s. deviation to mean (Å) | 1.13 ± 0.04 Å |
| Average r.m.s. deviation (structured region) | |
| Backbone r.m.s. deviation to mean (Å) | 0.62 ± 0.02 Å |
| Heavy atom r.m.s. deviation to mean (Å) | 1.15 ± 0.07 Å |
| Ramachandran analysis | |
| Residues in most favored regions | 73.0% |
| Residues in additional allowed regions | 24.5% |
| Residues in generously allowed regions | 2.1% |
| Residues in disallowed regions | 0.4% |
| Structural statistics from HADDOCK restrained calculations (the FGF1-S100A13-C2A heterohexameric complex); structural statistics for 20 structures | |
| Average r.m.s. deviation at backbone to mean (Å) | 0.28 ± 0.02 Å |
| Average r.m.s. deviation at FGF1 and C2A interface | 0.29 ± 0.02 Å |
| Average r.m.s. deviation at C2A and S100A13 interface | 0.31 ± 0.03 Å |
| Average r.m.s. deviation at FGF1 and S100A13 interface | 0.24 ± 0.03 Å |
| Average r.m.s. deviation at FGF1 interface | 0.20 ± 0.02 Å |
| Average r.m.s. deviation at S100A13 interface | 0.36 ± 0.02 Å |
| Average r.m.s. deviation at C2A interface | 0.23 ± 0.02 Å |
| Ramachandran analysis | |
| Residues in most favored regions | 73.4% |
| Residues in additional allowed regions | 22.7% |
| Residues in generously allowed regions | 3.3% |
| Residues in disallowed regions | 0.6% |
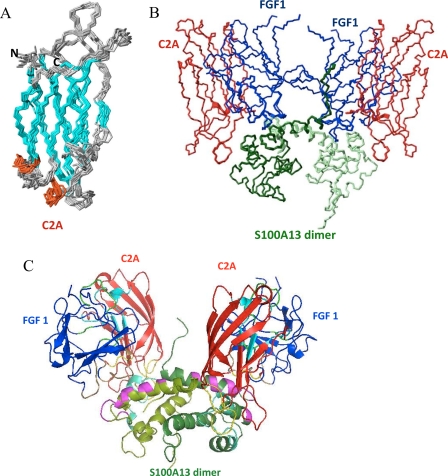
Fig. 3A shows the superposition of an ensemble of 20 structures of the C2A domain in the FGF1-S100A13-C2A complex. The structure of the C2A domain of synaptotagmin consists of a compact β-sandwich formed by two four-stranded β-sheets, with loops emerging at the bottom and top of the sandwich. The backbone r.m.s. deviation for the superposition of the C2A domain complex structure with free C2A is within the range 1.1–1.83 Å. The FGF1 binding site on C2A is distributed over four regions on FGF1. The first two regions are in loop 3, loop 5, and β-strand VI; the third region is from β-strand VII to the third α-helix; and the fourth region is at the C-terminal end. The S100A13 binding site on C2A is distributed over three regions in S100A13. The first region is on loop 1, and the second and third regions are on loop 3 and loop 6, respectively. The majority of the residues in the interfacial region are highly solvent-accessible.
FIGURE 3.
A, superposition of the backbone (N, Cα, and C′) atoms of the 20 final solution structures of C2A in the FGF1-S100A13-C2A heterohexameric complex. B, superposition of the backbone (N, Cα, and C′) atoms of the 20 final solution structures of the FGF1-S100A13-C2A heterohexameric complex. C, MOLMOL representation of the FGF1-S100A13-C2A heterohexameric complex; FGF1 is shown in blue, two S100A13 monomers are shown in light and dark green, and the C2A domain is shown in red. C2A and S100A13 binding regions on FGF1 are shown in green and brown. FGF1 and C2A binding regions on S100A13 are shown in magenta and cyan. FGF1 and S100A13 binding regions on C2A are shown in light blue and yellow.
Structure of the FGF1-S100A13-C2A Complex
We used the ARIA-calculated C2A domain structure in the FGF1-S100A13-C2A complex and the FGF1-S100A13 tetrameric complex structure in conjunction with the HADDOCK program to calculate the structure of the FGF1-S100A13-C2A complex. The hexameric complex was calculated using 38 intermolecular NOEs between C2A and the FGF1-S100A13 complex, which were obtained from analysis of 13C-filtered NOE complex data. Based on chemical shift perturbations in the FGF1-S100A13 complex and in C2A upon formation of the hexameric complex, AIRs were defined for residues at the interface. The active and passive residues were used to generate AIR restraints (Table 2). These restraints were used as ambiguous restraints. A total of 5000 rigid body docking trials were carried out using the standard HADDOCK protocol, with the 100 lowest energy structures used for subsequent semiflexible simulated annealing and water refinements. The 20 structures with the least energy were taken to represent the structure of the complex (Fig. 3B). The average r.m.s. deviation value for the backbone was 0.28 ± 0.02 Å. The average r.m.s. deviation at the FGF1 and C2A interface was 0.29 ± 0.02 Å. The average r.m.s. deviation at the FGF1 and S100A13 interface was 0.24 ± 0.03 Å. The average r.m.s. deviation at the S100A13/C2A interface was 0.31 ± 0.03 Å. The complex structure was analyzed using PROCHECK.
Fig. 3C shows a ribbon representation of the FGF1-S100A13-C2A heterohexameric complex. It clearly shows that the S100A13 homodimer acts as template for the whole complex formation. The two FGF1 and two C2A molecules bind to the homodimer of S100A13, and the whole complex appears like two symmetric units (Fig. 3C). The green region depicted on the FGF1 molecule is C2A binding, and the brown region represents the S100A13 binding region. The magenta region on S100A13 represents the FGF1 binding region, and the cyan region represents the C2A binding region. The light blue region depicted on the C2A molecule is the FGF1 binding region, and the yellow region shows the S100A13 binding region.
DISCUSSION
ITC experiments provide direct information on the stoichiometry, binding affinity, and heat changes that occur during protein-protein binding reactions in solution. Here, we monitored the binding affinity of FGF1 to S100A13, S100A13 to FGF1, and C2A to the FGF1-S100A13 tetrameric complex. The isothermograms in Fig. 1, A and B, represent the binding between FGF1 and S100A13. The binding constant between FGF1 and S100A13 is moderately strong (1.2–1.34 μm; Fig. 1). The isothermogram represents the titration of C2A to the FGF1-S100A13 complex (Fig. 1C). The decrease in entropy (Δs = 38.5 cal mol−1 K−1) indicates that the primary binding between the C2A and FGF1-S100A13 is through charge-charge interactions.
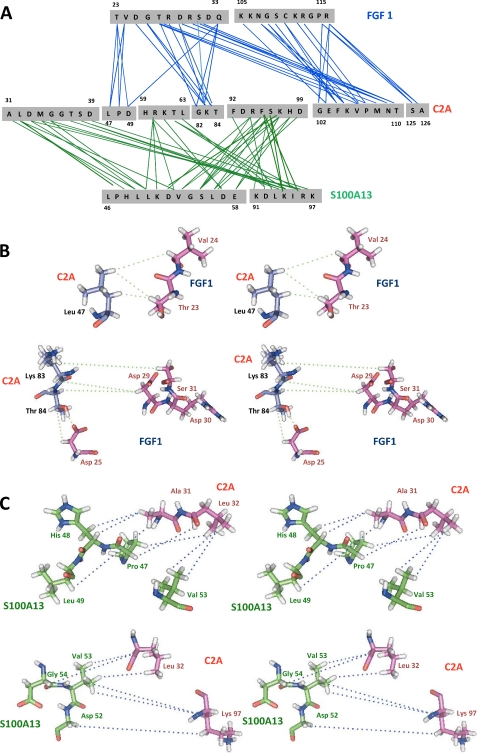
S100A13-FGF1 Interface
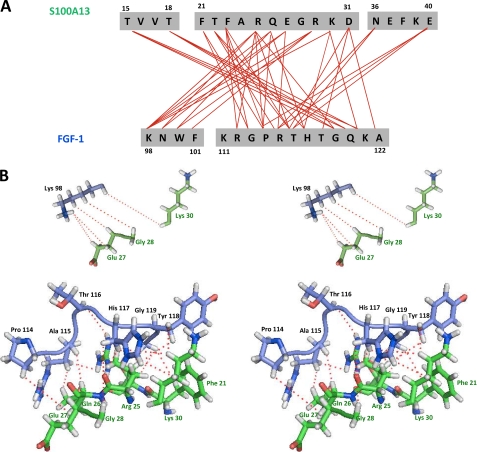
A detailed summary of the intermolecular contacts between S100A13 and FGF1 is shown in Fig. 4. There are 36 residues at the interface region, 20 from S100A13 and 16 from FGF1. The majority of the interactions between the two proteins are either hydrophobic or electrostatic. Key residues that are involved in two or more intermolecular contacts include Thr-15′, Phe-21′, Thr-22′, Phe-23′, Arg-25′, Gln-26′, Lys-30′, and Asn-36′ of S100A13 and Lys-98, Asn-99, Arg-112, Arg-115, His-117, Tyr-118, Gly-119, Gln-120, and Lys-121 of FGF1. Based on these results, the contact between the two proteins S100A13 and FGF1 is mainly the result of a combination of hydrophobic and polar interactions (Fig. 4B).
FIGURE 4.
A, summary of the intermolecular contacts between FGF1 and S100A13 in the FGF1-S100A13 complex. B, stereo view of intermolecular contacts (as indicated by dotted lines) between FGF1 and S100A13 in the FGF1-S100A13 heterotetrameric complex.
In addition, eight hydrogen bonds between the side chains and the backbone were observed; the side chain of Arg-25′(NE) hydrogen-bonded to the backbone nitrogen of His-117, as did Arg-25′(O) with His-117 HD1, Gln-26′(O) with Trp-100 HE1, Glu-40′ OE1 with Arg-115 HD1, and Gly-113 O with Arg-25′(HH2). The large interfacial region between FGF1 and S100A13 (∼3322 Å) contains intermolecular salt bridges, hydrogen bonds, and hydrophobic contacts, all of which combined provide the information for binding recognition between FGF1 and S100A13.
FGF1-S100A13-C2A Interface
Detailed summaries of the intermolecular contacts between FGF1-C2A and S100A13-C2A in the FGF1-S100A13-C2A hexameric complex are shown in Fig. 5A. There are 42 residues at the interface: 22 from FGF1 and 20 from S100A13 with C2A. The following residues are involved in two or more intermolecular contacts: Thr-22, Val-23, Thr-26, Arg-27, Ser-30, Gln-32, Lys-104, Asn-107, Ser-109, and Arg-115 of FGF1; Leu-46′, His-48′, Leu-49′, Asp-49″, Leu-50′, Val-53′, Gly-54′, Ser-55′, Leu-56′, Lys-83″, Thr-84″, Lys-91′, Lys-94′, Ile-95′, and Arg-96′ of S100A13; and Leu-32″, Asp-33″, Gly-35″, Ser-38″, Arg-60″, Lys-61″, Arg-94″, Phe-95″, Ser-96″, Lys-97″, Glu-103″, Lys-105″, Val-106″, Met-108″, Asn-109″, Ser-125″, and Ala-126″ of C2A. The interface region of FGF1 is distributed over two loops and three β-strands and is close to the C2A molecule. In C2A, the interfacial residues are mainly distributed in two loop regions, one β-sheet and the C-terminal end of the protein. In S100A13, the interfacial region is distributed over helix 1, loop 2, loop 3, and the C terminus.
FIGURE 5.
A, summary of the intermolecular contacts between C2A and FGF1-S100A13 in the FGF1-S100A13-C2A heterohexameric complex. B, stereo view of intermolecular contacts (as indicated by dotted lines) between C2A and FGF1 in the FGF1-S100A13-C2A heterohexameric complex. C, stereo view of intermolecular contacts (as indicated by dotted lines) between C2A and S100A13 in the FGF1-S100A13-C2A heterohexameric complex.
The large interfacial region in the FGF1-S100A13-C2A heterohexameric complex (∼4250 Å; Fig. 5) contains the intermolecular salt bridges and hydrogen bonds that provide the information for binding recognition between FGF1 and C2A, together with a large number of hydrophobic contacts. The side chain of Thr-37″ (OG1 and HG1) is hydrogen-bonded to Arg-17 HH12 and Asp-25 OD1, Lys-105″ HZ3 to Gln-124 OE2, Lys-105″ HZ1 to Gly-108 O, Gln-124″ HE22 to Asn-107 O, Lys-128″ HZ1 to Gly-108 O, Gln-124″ OE1 to Lys-105 HZ1, Thr-110″ OG1 and HG1 to Arg-28 NH and O, Gln-124″ OE1 to Lys-105 HZ2, Glu-127″ OE1 to Arg-115 HE and HH21, Arg-112 HH21 to Glu-124″ OE1, and Arg-112 HH2 to Glu-127″ OE2. The side chain of Lys-97′ (HZ1 and HZ2) is hydrogen-bonded to the backbone carbonyls of Asp-33″, Thr-37″, and Asp-93″ and to the backbone nitrogen of Thr-37″. The backbone HN of Val-53′ and Gly-54′ is hydrogen-bonded to the backbone carbonyl and nitrogen of Ser-96′. The backbone HN of Ala-31″ and Gly-35″ are hydrogen-bonded to the backbone nitrogen and carbonyl of Pro-47′ and Lys-51′. The positively charged residues of S100A13 at the C-terminal end (Lys-91′, Lys-94′, Arg-96′, and Lys-97′) have intermolecular contacts with Leu-32″, Asp-33″, Gly-35″, Thr-37″, and Ser-38″. These amino acids are crucial for the formation of the FGF1-releasing multiprotein complex, because mutation/deletion of these positive residues results in a failure to secrete FGF1. Based on the above results, FGF-C2A and S100A13-C2A contact each other in the hexameric complex mainly through a combination of hydrophobic and charge-charge interactions (Fig. 5, B and C).
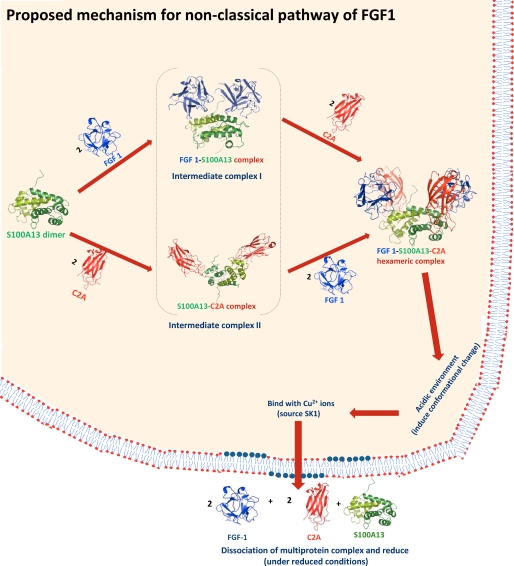
Mechanism for the Non-classical Secretory Pathway of FGF1
In this paper, we describe the solution structure of the FGF1-S100A13-C2A heterohexameric complex, which is the core component in the acidic fibroblast growth factor non-classical pathway. S100A13 plays a crucial role in the entire non-classical pathway. S100A13 is a homodimer and acts as a template for the formation of the Cu2+-induced FGF1 dimer. Two cysteines from the two FGF1 molecules (which are bound to S100A13) come closer in the presence of Cu2+ on the surface of S100A13, to allow the formation of a disulfide bond under acidic conditions. Copper is known to bind with S100A13 and C2A molecules and to induce the formation of the FGF1 homodimer (49, 50). Maciag and co-workers (14, 15) described how copper plays a crucial role in the non-classical pathway of acidic fibroblast growth factor release.
The formation of the multiprotein complex takes place in the vicinity of the cell membrane (4). The cell membrane is asymmetric, with acidic phospholipids, such as phosphatidylglycerol, phosphatidylserine, and phosphatidylinositol, in the inner leaflet of the cell membrane (51). Extracellular stimuli induce acidic phospholipid flipping to the cell surface (52), where phosphotidylserine can be detected using fluorescently tagged recombinant annexin V (53). This externalization mechanism is reversible. FGF1, S100A13, and Syt1 (C2A) specifically bind acidic phospholipids and have been demonstrated to destabilize the liposomes, which are composed of acidic phospholipids (23).
Based on the present results and evidence from the literature, we propose a mechanism for the non-classical secretory pathway of FGF1 (Fig. 6). First, the FGF1-S100A13 or S100A13-C2A (54) tetrameric complexes are formed. These complexes are intermediate state complexes for the FGF1-S100A13-C2A hexameric complex. These complexes then bind to C2A/FGF1 to form the hexameric complex (Fig. 1C and supplemental Fig. 4), which is the core component in the multiprotein complex. This hexameric complex formation is the key step in the non-classical pathway of FGF1. Later, this complex moves close to the acidic environment of the inner leaflet of the cell membrane. The structure of the FGF1-releasing complex under the acidic conditions changes as the negative membrane potential creates an acidic microenvironment that causes a partial denaturation of the proteins. Later, this complex interacts with Cu2+ ions (carried by SK1) (55) and moves close to the acidic environment of the inner leaflet of the cell membrane. FGF1 forms a disulfide-bridged homodimer in the presence of copper (14). The conformational change could occur in the hexameric complex under the acidic conditions after the formation of the disulfide bridge between two FGF1 in the presence of Cu2+ ions. These partially structured states of the complex that are generated at the membrane are highly competent to traverse across the membrane bilayers because the partial unfolding results in the exposure of normally hidden hydrophobic residues (56). The lipid binding abilities of the FGF1 and C2A domains of Syt1 are significantly higher in the partially structured state than in the native state (57). FGF1 is secreted as a homodimer in an inactive conformation (14), but under reducing conditions, such as those found outside cell membrane, this disulfide bond will break.
FIGURE 6.
The proposed mechanism of the acidic fibroblast growth factor in the non-classical pathway. First, the FGF1-S100A13 or S100A13-C2A tetrameric complexes are formed. These complexes are intermediate state complexes for the FGF1-S100A13-C2A hexameric complex. These complexes then bind to C2A/FGF1 to form the hexameric complex, which is the core component in the multiprotein complex. Later, this complex moves close to the acidic environment of the inner leaflet of the cell membrane. Here, this complex interacts with Cu2+ ions (from SK1) and moves close to the acidic environment of the inner leaflet of the cell membrane. These partially structured states of the complex that are generated at the membrane are highly competent to traverse across the membrane bilayers. Under reducing conditions, such as those found outside cell membrane, this complex will dissociate.
In this paper, we explicate the interfacial regions of the proteins in the hexameric complex, which is the core component in the acidic fibroblast growth factor non-classical secretory pathway. These findings may prove useful in attempts to understand the mechanism of the non-classical pathway of FGF1 secretion at the molecular level. The biological effects of FGF1 are mediated through the activation of four transmembrane phosphotyrosine kinase fibroblast growth factor receptors in the presence of heparin sulfate proteoglycans and therefore require the release of these proteins into the extracellular space. The information provided here may provide clues to how to stop the multiprotein complex formation that is essential for FGF1 transport, thereby assisting in rational drug design for FGF1-induced angiogenesis and cell proliferation.
Acknowledgment
We thank the high field Nuclear Magnetic Resonance Center in the National Core Facility of the National Research Program for Genomic Medicine, Academia Sinica (Taipei, Taiwan).
This work was supported by National Science Council of Taiwan Grant NSC 98-2113-M-007-018-MYZ.
The atomic coordinates and structure factors (codes 2ki4 and 2ki6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- FGF
- fibroblast growth factor
- ER
- endoplasmic reticulum
- NOE
- nuclear Overhauser effect
- NOESY
- NOE spectroscopy
- AIR
- ambiguous interaction restraint
- ITC
- isothermal titration calorimetry
- r.m.s.
- root mean square.
REFERENCES
- 1.Raman R., Venkataraman G., Ernst S., Sasisekharan V., Sasisekharan R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellegrini L. (2001) Curr. Opin. Struct. Biol. 11, 629–634 [DOI] [PubMed] [Google Scholar]
- 3.Blobel G. (2000) Chembiochem 1, 86–102 [DOI] [PubMed] [Google Scholar]
- 4.Prudovsky I., Mandinova A., Soldi R., Bagala C., Graziani I., Landriscina M., Tarantini F., Duarte M., Bellum S., Doherty H., Maciag T. (2003) J. Cell Sci. 116, 4871–4881 [DOI] [PubMed] [Google Scholar]
- 5.Nickel W. (2005) Traffic 6, 607–614 [DOI] [PubMed] [Google Scholar]
- 6.Prudovsky I., Tarantini F., Landriscina M., Neivandt D., Soldi R., Kirov A., Small D., Kathir K. M., Rajalingam D., Kumar T. K. (2008) J. Cell. Biochem. 103, 1327–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson A., Friedman S., Zhan X., Engleka K. A., Forough R., Maciag T. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10691–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson A., Tarantini F., Gamble S., Friedman S., Maciag T. (1995) J. Biol. Chem. 270, 33–36 [DOI] [PubMed] [Google Scholar]
- 9.Shi J., Friedman S., Maciag T. (1997) J. Biol. Chem. 272, 1142–1147 [DOI] [PubMed] [Google Scholar]
- 10.Mouta Carreira C., Landriscina M., Bellum S., Prudovsky I., Maciag T. (2001) Growth Factors 18, 277–285 [DOI] [PubMed] [Google Scholar]
- 11.Engleka K. A., Maciag T. (1992) J. Biol. Chem. 267, 11307–11315 [PubMed] [Google Scholar]
- 12.Landriscina M., Soldi R., Bagalá C., Micucci I., Bellum S., Tarantini F., Prudovsky I., Maciag T. (2001) J. Biol. Chem. 276, 22544–22552 [DOI] [PubMed] [Google Scholar]
- 13.Mouta Carreira C., LaVallee T. M., Tarantini F., Jackson A., Lathrop J. T., Hampton B., Burgess W. H., Maciag T. (1998) J. Biol. Chem. 273, 22224–22231 [DOI] [PubMed] [Google Scholar]
- 14.Landriscina M., Bagalá C., Mandinova A., Soldi R., Micucci I., Bellum S., Prudovsky I., Maciag T. (2001) J. Biol. Chem. 276, 25549–25557 [DOI] [PubMed] [Google Scholar]
- 15.LaVallee T. M., Tarantini F., Gamble S., Mouta Carreira C., Jackson A., Maciag T. (1998) J. Biol. Chem. 273, 22217–22223 [DOI] [PubMed] [Google Scholar]
- 16.Hayrabedyan S., Kyurkchiev S., Kehayov I. (2005) J. Rep. Immunol. 67, 87–101 [DOI] [PubMed] [Google Scholar]
- 17.Sparvero L. J., Asafu-Adjei D., Kang R., Tang D., Amin N., Im J., Rutledge R., Lin B., Amoscato A. A., Zeh H. J., Lotze M. T. (2009) J. Transl. Med. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya S., Chazin W. J. (2003) Structure 11, 738–740 [DOI] [PubMed] [Google Scholar]
- 19.Shishibori T., Oyama Y., Matsushita O., Yamashita K., Furuichi H., Okabe A., Maeta H., Hata Y., Kobayashi R. (1999) Biochem. J. 338, 583–589 [PMC free article] [PubMed] [Google Scholar]
- 20.Geppert M., Goda Y., Hammer R. E., Li C., Rosahl T. W., Stevens C. F., Südhof T. C. (1994) Cell 79, 717–727 [DOI] [PubMed] [Google Scholar]
- 21.Garcia J., Gerber S. H., Sugita S., Südhof T. C., Rizo J. (2004) Nat. Struct. Mol. Biol. 11, 45–53 [DOI] [PubMed] [Google Scholar]
- 22.Gerber S. H., Rizo J., Südhof T. C. (2002) Diabetes 51, S12–S18 [DOI] [PubMed] [Google Scholar]
- 23.Graziani I., Bagalá C., Duarte M., Soldi R., Kolev V., Tarantini F., Kumar T. K., Doyle A., Neivandt D., Yu C., Maciag T., Prudovsky I. (2006) Biochem. Biophys. Res. Commun. 349, 192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arunkumar A. I., Kumar T. K., Kathir K. M., Srisailam S., Wang H. M., Leena P. S., Chi Y. H., Chen H. C., Wu C. H., Wu R. T., Chang G. G., Chiu I. M., Yu C. (2002) Protein Sci. 11, 1050–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goddard T. D., Kneller D. G.SPARKY 3, University of California, San Francisco, CA [Google Scholar]
- 26.Grzesiek S., Bax A. (1993) J. Magn. Reson. B 102, 103–106 [Google Scholar]
- 27.Wittekind M., Mueller L. (1993) J. Magn. Reson. B 101, 201–205 [Google Scholar]
- 28.Grzesiek S., Bax A. (1993) J. Biomol. NMR 3, 185–204 [DOI] [PubMed] [Google Scholar]
- 29.Kay L. E., Xu G. Y., Yamazaki G. (1994) J. Magn. Reson. A 109, 129–133 [Google Scholar]
- 30.Pascal S. M., Muhandiram D. R., Yamazaki T., Forman-Kay J. D., Kay L. E. (1994) J. Magn. Reson. B 103, 197–201 [Google Scholar]
- 31.Breeze A. L. (2000) Prog. Nucl. Magn. Res. Spectr. 36, 323–372 [Google Scholar]
- 32.Linge J. P., O'Donoghue S. I., Nilges M. (2001) Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 33.Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., Nilges M. (2007) Bioinformatics 23, 381–382 [DOI] [PubMed] [Google Scholar]
- 34.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 35.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 36.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 37.Diaz A. R., Stephenson S., Green J. M., Levdikov V. M., Wilkinson A. J., Perego M. (2008) J. Biol. Chem. 283, 2962–2972 [DOI] [PubMed] [Google Scholar]
- 38.Williams C., Rezgui D., Prince S. N., Zaccheo O. J., Foulstone E. J., Forbes B. E., Norton R. S., Crosby J., Hassan A. B., Crump M. P. (2007) Structure 15, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 39.Ababou A., Gautel M., Pfuhl M. (2007) J. Biol. Chem. 282, 9204–9215 [DOI] [PubMed] [Google Scholar]
- 40.De Vries S. J., Van Dijk A. D. J., Krzeminski M., Van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 41.Hubbard S. J., Thornton J. M. (1993) NACCESS, University College London, London, UK [Google Scholar]
- 42.Pierce M. M., Raman C. S., Nall B. T. (1999) Methods 19, 213–221 [DOI] [PubMed] [Google Scholar]
- 43.Garrett D. S., Seok Y. J., Peterkofsky A., Clore G. M., Gronenborn A. M. (1997) Biochemistry 36, 4393–4398 [DOI] [PubMed] [Google Scholar]
- 44.Hall D. A., Vander Kooi C. W., Stasik C. N., Stevens S. Y., Zuiderweg E. R., Matthews R. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9521–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y. G., Song A. X., Gao Y. G., Shi Y. H., Lin X. J., Cao X. T., Lin D. H., Hu H. Y. (2006) Protein Sci. 15, 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wand A. J., Englander S. W. (1996) Curr. Opin. Biotechnol. 7, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piotto M., Saudek V., Sklenár V. (1992) J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 48.Riek R., Pervushin K., Wüthrich K. (2000) Trends Biochem. Sci. 25, 462–468 [DOI] [PubMed] [Google Scholar]
- 49.Sivaraja V., Kumar T. K., Rajalingam D., Graziani I., Prudovsky I., Yu C. (2006) Biophys. J. 91, 1832–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajalingam D., Kumar T. K., Yu C. (2005) Biochemistry 44, 14431–14442 [DOI] [PubMed] [Google Scholar]
- 51.Pomorski T., Hrafnsdottir S., Devaux O., Van Meer G. (2001) Semin. Cell. Dev. Biol. 12, 139–148 [DOI] [PubMed] [Google Scholar]
- 52.Bevers E. M., Comfurius P., Dekkers D. W., Zwaal R. F. (1999) Biochim. Biophys. Acta 1439, 317–330 [DOI] [PubMed] [Google Scholar]
- 53.Fischer K., Voelkl S., Berger J., Andreesen R., Pomorski T., Mackensen A. (2006) Blood 108, 4094–4101 [DOI] [PubMed] [Google Scholar]
- 54.Mohan S. K., Rani S. G., Kumar S. M., Yu C. (2009) Biochem. Biophys. Res. Commun. 380, 514–519 [DOI] [PubMed] [Google Scholar]
- 55.Soldi R., Mandinova A., Venkataraman K., Hla T., Vadas M., Pitson S., Duarte M., Graziani I., Kolev V., Kacer D., Kirov A., Maciag T., Prudovsky I. (2007) Exp. Cell Res. 313, 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ptitsyn O. B. (1995) Adv. Protein Chem. 47, 83–229 [DOI] [PubMed] [Google Scholar]
- 57.Rajalingam D., Graziani I., Prudovsky I., Yu C., Kumar T. K. (2007) Biochemistry 46, 9225–9238 [DOI] [PMC free article] [PubMed] [Google Scholar]