Abstract
The recent discovery that GRP78/BiP, a typical endoplasmic reticulum (ER) lumenal chaperone, can be expressed on the cell surface, interacting with an increasing repertoire of surface proteins and acting as receptor in signaling pathways, represents a paradigm shift in its biological function. However, the mechanism of GRP78 trafficking from the ER to the cell surface is not well understood. Using a combination of cellular, biochemical, and mutational approaches, we tested multiple hypotheses. Here we report that ER stress actively promotes GRP78 localization on the cell surface, whereas ectopic expression of GRP78 is also able to cause cell surface relocation in the absence of ER stress. Moreover, deletion of the C-terminal ER retention motif in GRP78 alters its cell surface presentation in a dose-dependent manner; however, mutation of the putative O-linked glycosylation site Thr648 of human GRP78 is without effect. We also identified the exposure of multiple domains of GRP78 on the cell surface and determined that binding of extracellular GRP78 to the cell surface is unlikely. A new topology model for cell surface GRP78 is presented.
Keywords: Cell Surface, Endoplasmic Reticulum (ER), Mutant, Protein Cross-linking, Protein Domains, GRP78/BiP
Introduction
Endoplasmic reticulum (ER)2 chaperones are essential for the normal function of the ER (1). One of the best characterized ER chaperones is the 78-kDa glucose-regulated protein (GRP78), which is also referred to as BiP or HSPA5. GRP78 is involved in many cellular processes, including translocating newly synthesized polypeptides across the ER membrane, facilitating the folding and assembly of proteins, targeting misfolded proteins for ER-associated protein degradation, regulating calcium homeostasis, and serving as an ER stress sensor (2, 3). GRP78 is a master regulator for ER stress due to its role as a major ER chaperone with antiapoptotic properties as well as its ability to control the activation of the unfolded protein response signaling. In the tumor microenvironment, tumor cells undergo ER stress due to hypoxia and nutrient deprivation. ER stress induction of GRP78 in cancer cells favors cell survival (4, 5) and contributes significantly to tumor progression and drug resistance in both proliferating and dormant cancer cells, as well as tumor-associated endothelial cells (6–11).
Traditionally, GRP78 is regarded as an ER lumen-localized chaperone protein due to the retrieval capacity through the KDEL retention motif present on its C terminus (12). However, it has been reported that GRP78 can be detected in the nucleus and mitochondria (13, 14). Recently, an isoform of GRP78 generated by alternative splicing is localized to the cytosol (15). Additionally, emerging evidence suggests that a subfraction of the GRP78 cellular pool can localize to the surface in specific cell types, in particular cancer cells (16–20). Global profiling of cell surface proteome of tumor cells revealed a relative abundance of heat shock chaperones and glucose-regulated proteins, including GRP78 (21). The preferential expression of GRP78 on the surface of tumor cells, but not in normal organs, enables specific tumor targeting by circulating ligands as well as other cytotoxic agents for cancer therapy without harmful effect on normal tissues (17, 18, 22). In another example, surface GRP78 mediates the antiangiogenic and proapoptotic activity of Kringle 5 through high affinity binding interaction of Kringle 5 with GRP78 exposed on the surface of stimulated endothelial cells and on hypoxic and cytotoxic stressed tumor cells (23, 24).
Although the physiological function of cell surface GRP78 is still emerging, evidence is accumulating that GRP78 can form cell surface complexes with specific proteins that in turn play an important role in signal transduction (19, 20). It has been reported that GRP78 is an interactive partner of the low density lipoprotein receptor-related protein, and knockdown of GRP78 by small interfering RNA attenuates activated α2-macroglobulin-induced signal transduction, impacting survival and metastasis of prostate cancer cells (25–27). Cripto, a multifunctional cell surface protein that is key to vertebrate embryogenesis and human tumor progression, was bound to cell surface GRP78, and blockade of this interaction prevented oncogenic Cripto signaling (28, 29). Additionally, GRP78 associates with GPI-anchored T-cadherin on the surface of vascular endothelial cells, promoting their survival (30). GRP78 interacts with the major histocompatibility complex class I molecules and is implicated as a co-receptor for viral entry (31, 32). Surface GRP78 is also required for the activation of an extrinsic apoptotic pathway mediated by extracellular Par-4 and TRAIL (33).
Recently, a new 82-kDa tumor-specific variant of GRP78 containing an O-linked carbohydrate moiety specific to malignant cells was discovered (34). This finding raises the possibility of targeting tumor cells via a specific cell surface variant of GRP78 (34). Further, phage display-derived human monoclonal antibodies isolated by binding to the surface of live primary breast cancer cells recognize a modified form of surface GRP78 at the C terminus (35). Despite these advances, the extent and mechanism of relocalization of GRP78/BiP from the ER to the cell surface at the biochemical level is not well understood. Previously, through limited trypsin digestion and biochemical extractions of microsomes, we provided evidence that a subpopulation of GRP78 exists constitutively as a transmembrane protein, consistent with hydrophobicity predictions (6). Because the ER membrane is a source for the plasma membrane, this could provide a plausible explanation for GRP78 cell surface localization. On the other hand, ER lumenal GRP78, by itself or through interacting with other proteins trafficking through the ER, could also cycle to the cell surface by escaping the ER retrieval mechanism. Here we report on the characterization of cell surface relocalization of GRP78 through a combination of cellular, biochemical, and mutational approaches and test various hypotheses that could help explain how GRP78 is expressed at the cell surface. The current studies provide quantitative analysis of GRP78 localization to the cell surface and reveal that this process involves multiple mechanisms. We demonstrate that ER stress can actively promote GRP78 surface expression, and overexpression of GRP78 can also lead to cell surface localization independent of ER stress. Further, based on analysis of domains of GRP78 that are exposed at the cell surface, we predict a topology for surface GRP78.
EXPERIMENTAL PROCEDURES
Cell Culture
Human cell lines 293T, HeLa, and MCF7 were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin antibiotics. All cells were maintained at 37 °C in a humidified atmosphere of 5% CO2, 95% air.
Reagents
Avidin-horseradish peroxidase conjugate was purchased from BD Biosciences, fluorescein anti-rabbit IgG was from Vector Laboratories (Burlingame, CA), 4′,6-diamidino-2-phenylindole was from Sigma, high capacity Neutravidin-agarose resin was from Thermoscientific (Rockford, IL), PE anti-mouse IgG1 was from BD Biosciences, normal mouse IgG (sc-3877) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and recombinant full-length hamster GRP78 protein (SPP-765) was from Stressgen (Ann Arbor, MI).
Plasmids
The construction of the expression plasmid for full-length human GRP78 (amino acids (aa) 1–654) has been described (10). Briefly, human GRP78 cDNA was prepared by reverse transcription of total HEK293T RNA, followed by a two-step PCR amplification and subcloning into the BamHI/XhoI sites of pcDNA3 (Invitrogen) to generate pcDNA3-hGRP78, which contains 5′-untranslated region, complete open reading frame of full-length GRP78, and partial 3′-untranslated region. For the epitope tagging of GRP78, the FLAG sequence was inserted after the ER signal peptide at residue 19, and six tandem copies of histidine were added after the KDEL motif. For generation of the KDEL deletion mutant and exchange of threonine (residue 648) to alanine, the protocols described in the QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) were followed. The generation of the plasmid for secreted GRP78 also utilized the pcDNA3.1 as vector, and two extra amino acids (GS) were inserted as linker prior to the His tag epitope.
Transfection Conditions
After the cells grew to 60–80% confluence, the expression vectors were transiently transfected into the indicated cell lines with BioT (Bioland Scientific, Cerritos, CA) or PolyFect (Qiagen, Valencia, CA) following the manufacturer's instructions, and the cells were harvested after 48 h.
Immunoblot Analysis
Cells were lysed in radioimmune precipitation buffer supplemented with competent protease inhibitor tablet (Roche Applied Science). The cell lysates were subjected to Western blot analysis as described (36). The proteins were resolved on 10% SDS-polyacrylamide gels, and then transferred to nitrocellulose membrane at 4 °C at 20 V overnight. Targeted protein was probed by the indicated antibody as follows: anti-FLAG mouse antibody (Sigma), 1:1,000; anti-His rabbit antibody (Santa Cruz Biotechnology, Inc.), 1:500; anti-GRP78 mouse antibody (BD Biosciences), 1:1,000; anti-PDI antibody (Stressgen, Ann Arbor, MI), 1:2,000; anti-KDEL mouse antibody (Stressgen), 1:1,000; anti-GRP94 rabbit antibody (Stressgen, SPA-851), 1:2,000; anti-CHOP mouse antibody (GADD153 B-3) (Santa Cruz Biotechnology, Inc.), 1:1,000; anti-β-actin mouse antibody (Sigma), 1:5,000; and the anti-sodium potassium ATPase mouse antibody (NKA α1, 464.6, gift of Dr. Alicia McDonough, University of Southern California), 1:1,000. Experiments were repeated 2–4 times. Protein levels were either visualized by autoradiography of Western blots or by the Bio-Rad Fluor-S Max MultiImager. The protein band intensities were quantitated using the Quantity One software package.
Cell Surface Protein Biotinylation
The cells were grown to 60–70% confluence. Prior to biotinylation, the used medium was discarded, and the cells were rinsed with chilled PBS twice. EZ-link Sulfo-NHS-LC-Biotin (Thermoscientific) at 0.5 mg/ml was added to cover the surface of the cell layer (1 ml/6 well plate), and the plates were gently shaken at 4 °C for 30 min. Tris-Cl, pH 7.5, was added to a final concentration of 100 nm to stop the biotinylation reaction. The cells were rinsed with ice-cold PBS twice and then lysed with radioimmune precipitation buffer. Part of the lysate was used for Western blots to measure the total level of the targeted protein. To purify surface protein, Neutravidin-agarose beads (Thermoscientific) were added and mixed well with the lysate at 4 °C overnight. The beads were then washed five times with PBS buffer. The cell surface proteins were released by the addition of 30 μl of 1× SDS-PAGE sample loading buffer (Sigma), followed by heating at 100 °C for 5 min.
Fluorescence-activated Cell Sorting (FACS) Analysis
Cells were washed with chilled PBS twice, aliquoted to 1 × 106 cells/tube, and incubated with 10% normal human serum in PBS for 20 min on ice to block Fc receptors on the cell surface. A saturating amount of primary antibody (1 μg) was incubated for 40 min on ice in 100 μl of staining buffer (Dulbecco's PBS, 2% heat-inactivated fetal calf serum, 0.09% sodium azide). After washing with staining buffer twice, secondary antibody (0.5 μg) was added for incubation for 30 min. After completely washing with staining buffer, cells were suspended in ice-cold PBS containing 4′,6-diamidino-2-phenylindole (1 μg/ml; Sigma) and subjected to FACS. The antibodies used were anti-FLAG mouse antibody (Sigma) at 1:100 dilution, phycoerythrin-conjugated anti-mouse IgG (BD Biosciences) at 1:40, anti-human GRP78 rabbit antibody (residues 250–300) (Novus Biologicals, Littleton, CO) at 1:150, anti-His rabbit antibody (Santa Cruz Biotechnology, Inc.) at 1:20, and fluorescein isothiocyanate-conjugated anti-rabbit Ig (Vector Laboratories, Burlingame, CA) at 1:100. FACS data analysis was performed by FACSDiva (BD Biosciences).
Isolation of Secreted GRP78
For isolation of protein secreted in the conditioned medium, 293T cells were transiently transfected with expression vectors for the wild type and mutant proteins. After 6 h, the cells were grown in hybridoma serum-free medium (Invitrogen) for 48 h. The conditioned medium was concentrated with Vivaspin 6 concentrators (10,000 molecular weight cut-off, polyethersulfone membrane) (Sartorius Stedim Biotech, Concord, CA).
For production of secreted GRP78 for cell surface adhesion studies, the GRP78Δ-H expression vector was transiently transfected into 293T cells with Lipofectamine 2000 (Invitrogen). Two days after transfection, the cells were grown in hybridoma serum-free medium (Invitrogen) for up to 10 days (medium was changed every 2 or 3 days). The culture supernatant was collected, and His-tagged protein was purified through a nickel-nitrilotriacetic acid column (Bio-Rad). The purified protein was dialyzed against buffer A (20 mm Tris, pH 8.0, 20 mm NaCl) and loaded onto a Sepharose-Q column (GE Healthcare) equilibrated with buffer A. After extensive wash of the column with buffer A, the elution was performed with buffer (20 mm Tris, pH 8.0) containing increasing concentrations (50–500 mm) of NaCl. The column fractions were run on SDS-PAGE and stained with Coomassie Blue R250.
RESULTS
Construction of Expression Plasmids for Epitope-tagged GRP78 Bearing Specific Mutations
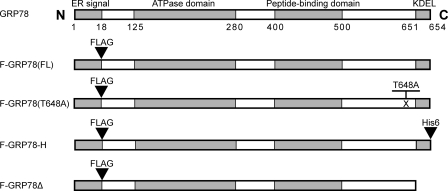
Human GRP78 is a highly conserved protein containing 654 amino acids, which include ER signal peptide (aa 1–18) at the N terminus, an ATPase domain (aa 125–280), a peptide-binding domain (aa 400–500), and a KDEL motif (aa 651–654) at the C terminus (Fig. 1). To facilitate analysis of various forms of GRP78, we first created an expression plasmid for the full-length human GRP78 with a FLAG-tagged epitope inserted immediately after the ER signal peptide (F-GRP78). The vector used was pcDNA3.1, where the Grp78 coding sequence was driven by the cytomegalovirus promoter. This plasmid was used as a template for the construction of the expression plasmid for F-GRP78(T648A), where the threonine (at aa 648) was mutated to alanine; for F-GRP78-H, where the hexahistidine tag epitope was inserted right behind the C terminus KDEL motif; and for F-GRP78Δ, where the KDEL motif was removed (Fig. 1).
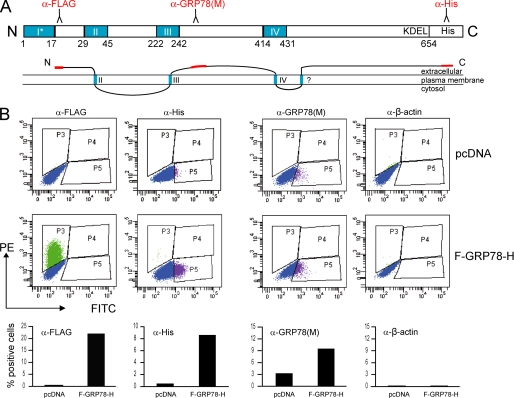
FIGURE 1.
Schematic illustration of the wild type and mutated forms of human GRP78 encoded by the expression plasmids. The ER signal peptide, ATPase domain, peptide-binding domain, and KDEL motif are indicated. The FLAG sequence was inserted immediately after the ER signal peptide. The full-length, wild type protein was denoted as F-GRP78(FL). For glycosylation site mutant F-GRP78(T648A), threonine at position 648 was exchanged to alanine. For F-GRP78-H, six tandem histidine (His6) was added following the KDEL motif of F-GRP78(FL). For F-GRP78Δ, the KDEL motif was deleted from F-GRP78(FL).
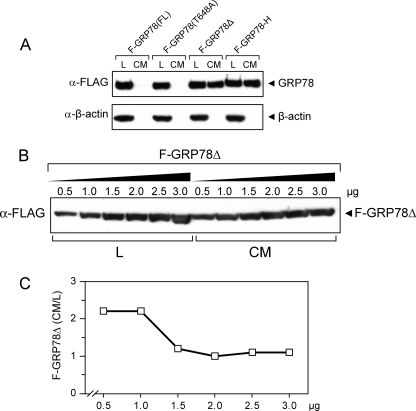
These plasmids, following transfection into 293T cells, produced FLAG-tagged wild type and mutant GRP78 proteins readily detectable in the cell lysates (Fig. 2A). Analysis of the conditioned medium revealed that F-GRP78Δ and F-GRP78-H were secreted into the medium, whereas none was detected for F-GRP78 or F-GRP78(T648A). These results demonstrated that either removal of the KDEL motif or attachment of a peptide at the C terminus adjacent to the KDEL domain would interfere with ER retention of GRP78, leading to secretion. Furthermore, we determined and calculated the ratio of secreted F-GRP78Δ versus the intracellular amount over a range of expression plasmid concentrations. We observed that the ratio is 2-fold greater in the lower dosages as compared with the higher dosages (Fig. 2, B and C). Following these characterizations of expression and secreted levels, the four expression plasmids described above were used for the transfection studies described below for the determination of cell surface relocalization of GRP78.
FIGURE 2.
Measurement of intracellular and secreted GRP78 in wild type and mutant proteins. A, 293T cells were transfected with F-GRP78(FL), F-GRP78(T648A), F-GRP78Δ, or F-GRP78-H. After 6 h, the cells were grown in hybridoma serum-free medium and collected after 48 h. 5% total cell lysate (L) and concentrated conditioned medium (CM) were subjected to Western blot for detection of FLAG-tagged GRP78. β-Actin served as a loading control for the lysates and a test for cell lysis resulting in leakage of intracellular protein into conditioned medium. B, 293T cells were transfected with increasing amounts of F-GRP78Δ DNA in 6-cm dishes. pcDNA was added to equalize the amount of DNA for transfection. 5% total cell lysate and concentrated conditioned medium were subjected to Western blot for detection of F-GRP78Δ. C, after quantitation of B, the ratio of secreted F-GRP78Δ in conditioned medium versus total intracellular F-GRP78Δ in lysate was plotted against the dosage of expression plasmid.
ER Stress Enhances Localization of Endogenous GRP78 to the Cell Surface
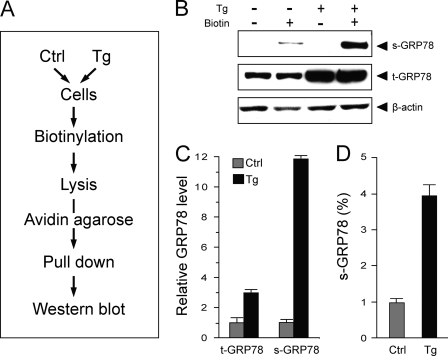
The scheme for monitoring cell surface GRP78 expression is summarized in Fig. 3A. The biotin was charged, such that it could not permeate the cell membrane, and only primary amines exposed on the intact cell surface would be labeled. Following biotinylation and subsequent detergent lysis, the cell lysate was mixed with avidin-agarose resin, which retained biotinylated proteins. Following elution from the resin, the biotinylated proteins, together with the 10% of lysate that served as input control, were subjected to immunoblot to measure GRP78 levels under each experimental condition. By quantitation of the band intensities, the fraction of GRP78 that has redistributed to the cell surface was deduced.
FIGURE 3.
ER stress actively promotes cell surface localization of endogenous GRP78. A, flow chart of the strategy to detect cell surface protein in cells that are either untreated (Ctrl) or subjected to Tg treatment. B, detection of cell surface GRP78 expression in 293T cells either untreated (−) or treated (+) with 300 nm Tg for 16 h. β-Actin served as loading control. Representative Western blots are shown. s-GRP78 and t-GRP78, surface and total intracellular GRP78, respectively. The amount of total lysate was 10% of the amount used for avidin pull-down. C, quantitation of the relative expression level of total intracellular GRP78 and cell surface GRP78 in control and Tg-treated cells. D, the fraction of cell surface versus total intracellular GRP78 in control and Tg-treated cells. The experiments were repeated 3–4 times. The S.D. is shown.
First, we addressed the question of whether cells under regular cell culture conditions exhibit GRP78 on the cell surface and, if so, what is the effect of ER stress on this process. To test this, 293T human embryo kidney fibroblast cells were either cultured under normal conditions or treated for 16 h with thapsigargin (Tg), an inhibitor of the ER ATPase that causes Ca2+ efflux from the ER, leading to ER stress. We observed that in cells under regular culture conditions that were biotinylated, only a very faint band of cell surface GRP78 was detected; upon Tg treatment, this band intensity was increased by 12-fold (Fig. 3, B and C). In contrast, the total amount of intracellular GRP78 only increased by about 3-fold (Fig. 3, B and C). Thorough washing of avidin-agarose beads ensured that surface GRP78 was not contaminated by non-surface GRP78, as indicated by the complete absence of a GRP78 band in the non-biotinylated group (Fig. 3B). Our results showed that prior to ER stress, about 1% of intracellular GRP78 was present on the cell surface, and this increases to about 4% after 16 h of Tg treatment, which is consistent with a 12-fold increase in cell surface GRP78 compared with non-stressed cells because Tg increased the total GRP78 amount by about 3-fold (Fig. 3, C and D). These results suggest that the increase in cell surface GRP78 after ER stress does not simply parallel the increase in total amount of intracellular GRP78; rather, ER stress promotes special mechanism(s) to enhance GRP78 localization to the cell surface.
Overexpression of the ER Form of GRP78 Results in Cell Surface Localization Independent of ER Stress
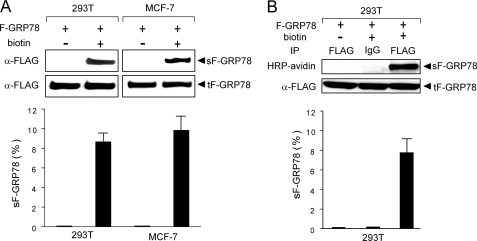
To test whether overexpression of the ER form of GRP78 in cells maintained in regular culture conditions is sufficient for cell surface localization of GRP78, we selected 293T due to high transfection efficiency in this cell line. 293T cells were transfected with expression plasmid for FLAG-GRP78 (F-GRP78) and then subjected to analysis of cell surface GRP78 by protein biotinylation and Western blot. Because F-GRP78 contains the ER signal peptide, it is expected to be synthesized as an ER protein, and then subsequently a fraction translocates to the cell surface. We observed that in cells cultured under regular conditions, about 8.5% of F-GRP78 was detected on the cell surface (Fig. 4A). The lack of a protein band in cells that were not biotinylated confirmed specific labeling of cell surface protein. To examine whether this applies to other cell types, the same transfection and biotinylation experiments were performed in the MCF-7 breast adenocarcinoma cells, which showed about 10% of F-GRP78 was expressed at the cell surface (Fig. 4A).
FIGURE 4.
Detection of cell surface GRP78 following ectopic expression in 293T and MCF-7 cells. The cells were transfected with 2.0 μg of F-GRP78 in 6-well dishes, and either biotinylated or non-biotinylated. The cell lysates were subjected to avidin pull-down (A) or immunoprecipitation (IP) with anti-FLAG antibody (B), followed by Western blots with anti-FLAG antibody or developed with horseradish peroxidase-avidin, as indicated. The amount of total lysate was 10% of the amount used for avidin pull-down or immunoprecipitation. Representative Western blots are shown. sF-GRP78 and tF-GRP78, surface and total intracellular F-GRP78, respectively. After quantitation, the percentage of cell surface F-GRP78 was calculated, and it is presented below the autoradiogram. The experiments were repeated 3–4 times. The S.D. is shown.
As a complementary measure to analyze GRP78 surface expression, 293T cells were transfected with expression plasmid for F-GRP78 and biotinylated, and the cell lysate was subjected to immunoprecipitation with antibody against the FLAG epitope, which specifically recognized F-GRP78. Cell surface F-GRP78 was detected by horseradish peroxidase-conjugated avidin and total F-GRP78 by immunoblot with anti-FLAG antibody. Our results showed that about 7.5% of F-GRP78 was detected by this method (Fig. 4B). Thus, two independent methods confirm that about 7–10% of ectopically expressed GRP78 can locate to the cell surface.
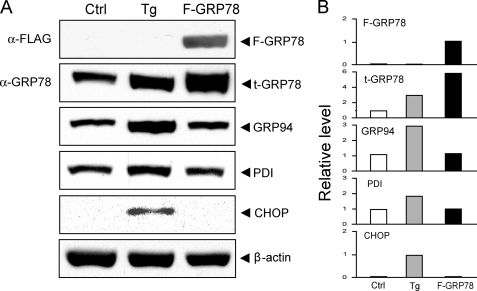
Next we determined whether the localization of ectopically expressed GRP78 to the cell surface is a consequence of ER stress resulting from overexpression of GRP78. For this purpose, we examined the ER stress marker proteins by Western blot using cell lysates prepared from F-GRP78-transfected 293T cells with or without Tg treatment (Fig. 5A). The protein band intensities were quantitated, and after normalization to the loading control, β-actin, the fold changes were summarized (Fig. 5B). As expected, F-GRP78 was only detected in the transfected cells. Using anti-GRP78 antibody, which could detect both F-GRP78 and endogenous GRP78, we observed a 3-fold increase in Tg-treated cells and a 6-fold increase in the transfected cells. In Tg-treated cells, the GRP94 level was elevated 3-fold, PDI was elevated 1.8-fold, and CHOP was induced. In the cells transfected with F-GRP78, the levels of GRP94 and PDI were minimally affected, and there was no induction of CHOP, suggesting that overexpression of GRP78 alone does not lead to ER stress. Hence, under conditions of overexpression, GRP78 can relocalize to the cell surface independent of ER stress.
FIGURE 5.
Ectopic expression of GRP78 does not induce ER stress. A, 293T cells were either treated with Tg (300 nm) for 16 h or transfected with full-length FLAG-tagged GRP78 for 48 h. Cells without Tg treatment and transfection served as control (Ctrl). Total cell lysate was prepared and subjected to Western blot analysis. The protein levels of exogenous GRP78 (F-GRP78); total intracellular GRP78 (t-GRP78); endogenous GRP94, PDI, and CHOP; and β-actin, which served as loading control, are shown. B, quantitation of the protein band intensities shown in A, with the level of the control cells set as 1.
Deletion of the C-terminal ER Retention Motif of GRP78 Alters Cell Surface Expression
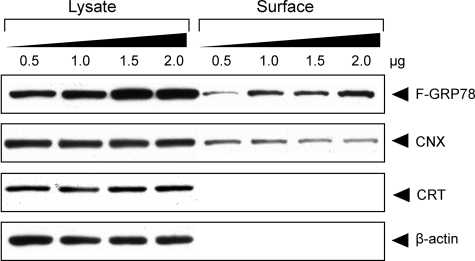
One explanation for cell surface localization of GRP78 is that when there is a high level of GRP78, the capacity of the KDEL retrieval system may be exceeded. To test this, we first established the assay system where increasing amounts of full-length F-GRP78 were expressed in 293T cells. The surface proteins were labeled by biotinylation and pulled down by avidin-agarose resin as outlined in Fig. 3A, and the amount of cell surface and total F-GRP78 was analyzed by Western blots. As shown in Fig. 6, cell surface F-GRP78 increased in parallel with the increase in intracellular F-GRP78. As reported previously (37), calnexin (CNX), an ER transmembrane protein, was also detected on the cell surface (Fig. 6). In contrast, cell surface expression of calreticulin, an ER lumenal protein, was minimal even upon long exposure, and β-actin, a cytosolic protein, was not detected on the cell surface. Because the level of endogenous cell surface CNX is relatively constant in the transfected cells, it was used as an internal cell surface protein loading control to normalize the level of cell surface GRP78 and β-actin as an internal control for contamination by cytosolic protein.
FIGURE 6.
Dosage-dependent increase of surface expression of GRP78. 293T cells were transfected with the indicated amount of F-GRP78 expression plasmid. F-GRP78 and CNX were detected on the cell surface by Western blot, whereas calreticulin (CRT) was barely detected on the cell surface. β-Actin served as loading control for the input lysate, which represents 10% of the amount used for avidin pull-down to detect surface protein.
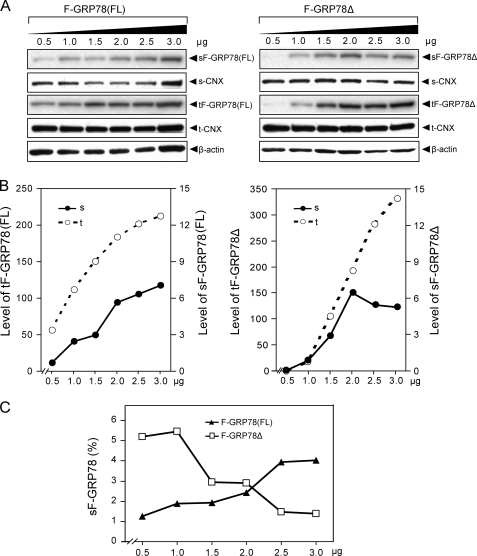
To test for the importance of the KDEL sequence in regulating GRP78 cell surface expression, we created F-GRP78Δ, which is identical to F-GRP78 but with the deletion of the KDEL sequence motif (Fig. 1). In principle, F-GRP78Δ would not be subjected to the KDEL retrieval system. In this set of experiments, the wild type F-GRP78 was referred to as full-length F-GRP78 (F-GRP78(FL)) to distinguish it from the KDEL deletion mutant. 293T cells were transfected with increasing amounts of the F-GRP78(FL) and F-GRP78Δ in parallel. The Western blots of cell surface and total lysate for detection of F-GRP78, CNX, and β-actin are shown in Fig. 7A. The protein band intensities were quantitated and normalized against CNX and β-actin for cell surface protein and lysate loading, respectively. The levels of surface and total F-GRP78 in relation to the different doses of transfected expression vector are presented in Fig. 7B (note different scales for total and surface protein), and surface F-GRP78 as a percentage of total transfected protein for each dosage is presented in Fig. 7C. Our results showed similarities and differences between the two forms of GRP78. For F-GRP78(FL), the level of cell surface protein increased, correlating with increasing amounts of intracellular F-GRP78(FL), with the percentage of cell surface GRP78 increasing from 1% at the low dosage to 4% at the higher doses (Fig. 7, A–C). The 2.5% value at the 2.0-μg dosage was about 3-fold lower than that observed in Fig. 4A because these experiments were performed in larger culture dishes with a 2.5-fold lower ratio of transfected DNA/cell. For F-GRP78Δ, at the lower dosages, there was a substantially lesser intracellular amount of F-GRP78Δ compared with F-GRP78 (Fig. 7, A and B). This is consistent with F-GRP78Δ being secreted outside the cell (Fig. 2). Interestingly, at higher dosages, the intracellular level of F-GRP78Δ was even slightly higher than F-GRP78(FL). For cell surface F-GRP78Δ, at the lower dosages, the level of surface F-GRP78Δ was very low, correlating with the very low amount of F-GRP78Δ retained inside the cell (Fig. 7, A and B). Nonetheless, the signal of hybridization was readily detectable by the Fluor-S Max MultiImager and could be quantitated. At higher doses, the level of surface F-GRP78Δ increased in parallel to the increase in the intracellular form; however, it reached a plateau, or even decreased at the highest doses (Fig. 7, A and B). After normalization with the total intracellular amount of F-GRP78Δ, our results showed that in the low dosages, despite the lower overall level, about 5% of intracellular F-GRP78Δ exists as surface protein, compared with 1–2% of F-GRP78(FL), suggesting that deletion of the KDEL motif could promote surface expression (Fig. 7C). However, at higher dosages, the trend was reversed with cell surface F-GRP78Δ at a 1% level compared with F-GRP78(FL) at 4%. Collectively, these results suggest that deletion of the C-terminal KDEL motif affects cell surface presentation of GRP78; however, the effects are dosage-dependent.
FIGURE 7.
Deletion of the ER retrieval signal, KDEL, affects cell surface localization of GRP78. A, 293T cells in 6-cm dishes were transfected with increasing amounts of F-GRP78(FL) or F-GRP78Δ, as indicated, and pcDNA vector was added to equalize total DNA amount in each transfection. Cell surface proteins were purified and analyzed as described in the legend to Fig. 6. s, surface proteins; t, lysate input. B, the relative level of F-GRP78(FL) and F-GRP78Δ on cell surface or in total lysate. After quantitation of protein band intensity, surface F-GRP78(FL) and surface F-GRP78-Δ were normalized against surface CNX, whereas total intracellular F-GRP78(FL) and F-GRP78-Δ were normalized against β-actin. The levels of surface and total intracellular F-GRP78(FL) or F-GRP78Δ are plotted against the dosage of the expression plasmid. C, the percentages of cell surface F-GRP78(FL) and F-GRP78Δ were calculated and plotted against the dosage of the expression plasmid.
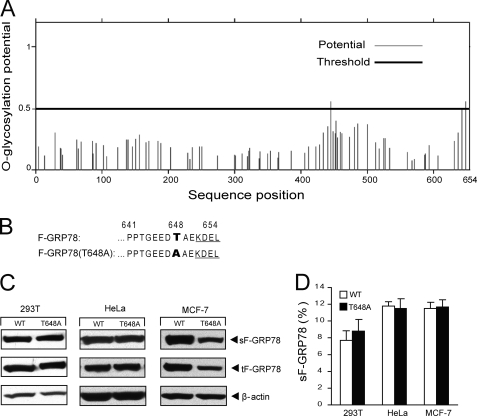
Mutation of the Putative O-Linked Glycosylation Site at the C Terminus of GRP78 Does Not Affect Its Cell Surface Localization
Recent reports suggest the existence of an O-linked glycosylated form of GRP78 at the cell surface, and the site was implicated at the C terminus of GRP78 (34, 35). Analysis of potential O-linked glycosylation sites on human GRP78 by the Net OGly 3.1 program revealed the strongest site at threonine 648 with close proximity to the KDEL motif at the C terminus of GRP78 (Fig. 8A). One possibility is that upon modification of this site, it may mask or interfere with the KDEL retrieval system, leading to GRP78 escape from the ER to the cell surface. To test this, F-GRP78(T648A) was constructed where threonine at aa 648 was mutated to alanine, thus destroying the putative O-linked glycosylation site (Fig. 8B). This, in principle, will result in more efficient KDEL retrieval and less cell surface expression. Following transfection of F-GRP78(T648A) and the wild type control (F-GRP78) into 293T cells, surface GRP78 protein was monitored by biotinylation, avidin purification, and immunoblotting. Our results showed a minimal difference in cell surface GRP78 expression between the wild type and T648A mutant in 293T cells at the dose shown or at other dosages (Fig. 8C) (data not shown). Similar results were observed in other cell types, including HeLa and MCF-7 cells (Fig. 8C). In all three cell lines, the level of surface expression of F-GRP78 ranges from about 8 to 12%, and this is not affected by the T648A mutation (Fig. 8D).
FIGURE 8.
Mutation of O-linked glycosylation site (T648A) does not affect cell surface translocation of GRP78. A, schematic diagram of O-linked glycosylation sites predicted by the Net OGly 3.1 program for human GRP78. The threshold line of glycosylation potential is indicated as a black boldface line. The amino acid sequence position is shown below. B, the amino acid sequences surrounding the putative O-linked glycosylation site at aa 648 of human GRP78, with the mutated site indicated in boldface type. C, detection of surface and total intracellular F-GRP78 (wild type and mutant) in MCF7, HeLa, and 293T cells. β-Actin served as the loading control for the lysate input. Representative Western blots are shown. D, after quantitation of protein band intensity, the percentages of cell surface F-GRP78 (sF-GRP78; wild type (WT) and mutant) in the different cell lines were calculated and are presented. The experiments were repeated 3–4 times. The S.D. is shown.
Multiple Domains of GRP78 Are Exposed on the Cell Surface
Although GRP78 is generally a hydrophilic protein, it contains several hydrophobic regions, and a subfraction exhibits properties of a transmembrane protein (6). Analysis of the human GRP78 amino acid sequence by the TMpred predication program revealed four potential transmembrane domains, I (aa 1–17), II (aa 29–45), III (aa 222–242), and IV (aa 414–431), with prediction scores of 806, 379, 260, and 493, respectively (Fig. 9A). Thus, considering that the ER membrane is the source for the plasma membrane, one mechanism for cell surface localization of GRP78 is that it is in part derived from the subfraction of GRP78 existing as an ER transmembrane protein.
FIGURE 9.
Multiple domains of GRP78 are exposed on cell surface. HeLa cells transfected with F-GRP78-H containing N-terminal FLAG tag and C-terminal His tag were subjected to FACS analysis. Cells transfected with pcDNA vector were used as negative control. A, schematic drawing of human GRP78 with the potential transmembrane domains (I–IV) predicted by TMPred (ExPasy tools) indicated in blue. The ER signal sequence is indicated as an asterisk. The numbers below refer to the amino acid residues. The domains recognized by the antibodies used for FACS analysis are indicated in red. The α-GRP78 (M) antibody recognizes the middle domain of human GRP78 spanning aa 250–300. Below is a proposed topology model of cell surface GRP78, with the putative transmembrane domains indicated in blue and the extracellular domains identified by FACS in red. B, exposure of the N terminus, C terminus, and middle domain (aa 250–300) of GRP78 at the cell surface was detected by FACS analysis using the antibodies as indicated. β-Actin was used as control for cell integrity. The percentage of positive cells is shown under each FACS profile. PE, phycoerythrin; FITC, fluorescein isothiocyanate.
To test this, we first examined which domain(s) of GRP78 is exposed to the cell surface, using FACS analysis of native cells. In order to use high affinity antibody with specificity for GRP78 and not other related proteins, we created F-GRP78-H, where the N and C termini of GRP78 were tagged by the FLAG and His epitope, respectively (Fig. 1). The plasmids, F-GRP78-H, and the empty vector pcDNA, which served as negative control, were transfected into HeLa cells, followed by FACS analysis. Our results showed that both the FLAG and the His epitopes were detected on the cell surface, implying that both the N and C termini of GRP78 are exposed on the cell surface (Fig. 9B). Furthermore, using an antibody against the middle domain of GRP78 (spanning aa 250–300), we also detected exposure of this domain to the cell surface (Fig. 9B). Because this antibody is not able to differentiate between endogenous and exogenous GRP78, a positive signal was detected in cells transfected with pcDNA due to the endogenous surface GRP78, and this signal was higher in cells overexpressing F-GRP78-H. As negative controls, β-actin was not detected on the surface, and omission of either the primary or secondary antibody eliminated any cell surface detection (Fig. 9B) (data not shown). These results show that at least three domains (N, C, and middle) of GRP78 are exposed on the cell surface. Combining these observations with the TMpred plot and earlier biochemical analysis of GRP78 in microsomes (6), a potential transmembrane configuration for cell surface GRP78 is proposed (Fig. 9A).
Extracellular GRP78 Is Not Able to Stably Attach to Cell Surface
Our observation that multiple domains of GRP78 are exposed to the cell surface raises the possibility that another mechanism for cell surface localization of GRP78 is via the recapture of secreted GRP78 onto the cell surface. Because GRP78 could be detected in plasma (38), it is possible that a fraction of GRP78 is actively or passively secreted outside the cell and subsequently reattached to the cell surface through interactions with cell surface client proteins. To test this possibility, we created expression plasmid encoding GRP78Δ-H for large scale preparation of secreted GRP78 (Fig. 10A). GRP78Δ-H does not contain the FLAG tag and is also devoid of the KDEL sequence; however, it contains a hexahistidine tag epitope at the C terminus to facilitate purification of the secreted protein. GRP78Δ-H is expected to be synthesized in the ER, transit through the Golgi, and be secreted outside the cell. Upon transfection into 293T cells, GRP78Δ-H was secreted into the culture medium and purified by nickel-agarose chromatography followed by Q column chromatography (Fig. 10B). The Q column fractions were subjected to gel electrophoresis, and Coomassie Blue staining confirmed the purity of GRP78Δ-H (Fig. 10C). This form of secreted GRP78 and recombinant full-length GRP78 (rGRP78) containing the KDEL motif were used to test whether extracellular GRP78 can bind to the cell surface of viable cells, with bovine serum albumin serving as a negative control. Following incubation, cell surface proteins were detected by biotinylation and purified by avidin-agarose beads. Total cell lysate was also prepared to test whether GRP78Δ-H or rGRP78 was being internalized inside the cells. Using the anti-KDEL antibody in the Western blot assays, we detected endogenous GRP78 and GRP94, both of which contain the KDEL motif, in the cell lysate input; however, no rGRP78 was detected on the cell surface following biotinylation (Fig. 10D). Using anti-His antibody in the Western blot assays, GRP78Δ-H was detected in the purified protein control, but none was detected in the cell lysate or cell surface protein fraction (Fig. 10D). As positive controls, Na,K-ATPase (NKA α1) was detected on the cell surface following biotinylation, and this enzyme as well as β-actin were detected in the cell lysate (Fig. 10D). Collectively, these experiments suggest that it is unlikely that cell surface GRP78 results from reattachment of extracellular GRP78.
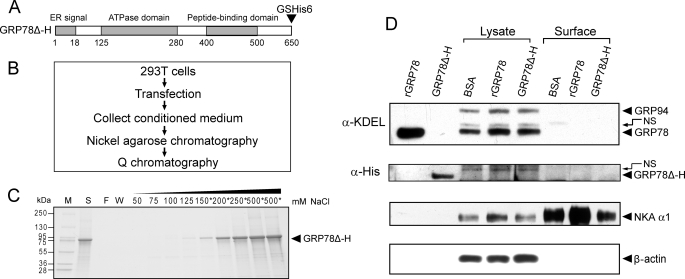
FIGURE 10.
Extracellular GRP78 does not stably bind to cell surface. A, schematic illustration of GRP78Δ-H mutant encoded by expression plasmid, in which KDEL motif was deleted and glycine (G) and serine (S), followed by six tandem histidine (GSHis6), were added at the C terminus of GRP78. B, methodological flow chart of expression and purification of secreted GRP78Δ-H. C, Q chromatography purification for GRP78Δ-H. Protein purified by nickel-agarose affinity chromatography was loaded onto a Q column, and after extensive wash of the column, elution was performed with buffer containing the indicated concentration (50–500 mm) of NaCl. All fractions were run on SDS-PAGE followed by Coomassie Blue staining. The fractions marked with an asterisk were collected and saved. The molecular weight of protein marker (M) is indicated on the left. S, protein initially loaded onto the Q column; F, flow-through fraction; W, wash fraction. D, detection of extracellular GRP78 entering or binding to surface of 293T cells. 10 μg of rGRP78, purified GRP78Δ-H, or bovine serum albumin (BSA) were added in the medium of 293T cells in 6-well dishes, respectively, and incubated for 24 h. The cell surface and 20% lysate input of GRP78 and GRP78Δ-H were detected by Western blot against either anti-KDEL or anti-His antibody. 1 μg of rGRP78 and purified GRP78Δ-H were loaded as positive control. The upper band above the positive control is a nonspecific (NS) band. Sodium-potassium-ATPase (NKA α1) served as cell surface protein loading control. β-Actin was used as lysate input loading control.
DISCUSSION
In view of the emerging importance of cell surface GRP78 in controlling cell signaling and viability, it is important to understand to what extent GRP78 is presented on the cell surface and the mechanism by which that is achieved. Studies on cell surface GRP78 expression performed in tissue culture show much variability, ranging from expression of GRP78 in various cancer cell lines to no expression in normal fibroblast cell lines (18) to positive reactivity with GRP78 binding peptide motifs whether or not they are tumor cells (17). There are also conflicting reports of whether GRP78 is expressed on specific cell lines, such as the PC-3 prostate cancer cells (33, 39). The discrepancies are probably due to different techniques used to detect cell surface GRP78, which is present at very low amounts and in only a subpopulation of cells, and the divergent results could also represent intrinsic variabilities among cultured cell lines. Here we used cell surface biotinylation as the primary method for identification and isolation of cell surface GRP78, coupled with FACS analysis of living cells. Whereas previous studies primarily examined endogenous GRP78, using both polyclonal and monoclonal antibodies against GRP78, we examined in addition tagged versions of wild type and mutated GRP78, using epitope antibodies that recognize GRP78 with specificity and high affinity. Due to their high transfection efficiency, we have selected the 293T cells as the model system, with additional analysis performed in HeLa and MCF-7 cells for independent confirmation. Our studies reveal several novel findings toward understanding how GRP78, an ER lumen protein, is localized to the cell surface.
We report here that in cells maintained under normal culture conditions, the amount of surface GRP78 is extremely low but nonetheless detectable with a high sensitivity assay. Substantially higher levels of GRP78 localize to the cell surface following ER stress. This is consistent with the notion that as the amount of intracellular GRP78 and other ER chaperones also bearing KDEL increases, the KDEL retrieval system may be overwhelmed such that a fraction of GRP78 escapes to the cell surface. We report here that the surface GRP78 does not simply increase in parallel with intracellular GRP78; rather, it is about 4-fold higher than the increase in intracellular GRP78, suggesting that ER stress may activate specific mechanisms for GRP78 surface localization and/or inactivate mechanisms for its ER retention.
Nonetheless, ER stress is not obligatory for cell surface localization of GRP78. GRP78 surface localization is readily detected in non-stressed cells with ectopic expression of GRP78. Examination of unfolded protein response markers in the transfected cells confirmed that ER stress is not triggered. Thus, this offers an experimental system to study mechanisms for GRP78 cell localization without the complication of other factors influenced by ER stress. Here, using specific mutations of GRP78, we tested several hypotheses that could in principle account for cell surface localization of GRP78. First, we examined whether deletion of the C-terminal KDEL motif, which is known to cause secretion of GRP78 outside the cell, affects surface localization. Our results revealed that at low dosages, the fraction of surface to intracellular protein was higher for F-GRP78Δ compared with the full-length protein, suggesting that escape from the KDEL retrieval mechanism could enhance surface expression. However, why this trend was reversed remains to be determined. One speculation is that cell surface presentation of F-GRP78Δ is linked to secretion because at the lower dosages the fraction of secreted to intracellular F-GRP78Δ was also higher.
The discovery of patient antisera specifically recognizing O-linked GRP78 on the surface of malignant cells (34) and the discovery that C terminus modification of GRP78 accounts for cancer-specific antibody specificity (35) prompted us to test the hypothesis that the putative O-linked glycosylation site of GRP78 is within close proximity of the KDEL motif and that modification of this site could mask the KDEL motif and allow GRP78 to escape the KDEL retrieval system. Our results showed no significant change in the level or the electrophoretic mobility of surface GRP78 regardless of whether the putative glycosylation site was intact or mutated in three different cell types. It is possible that none of the cell lines that we tested possesses this O-linked glycosylation modification mechanism. It is noted that the O-linked glycosylated form of GRP78 represents a very minor fraction of total GRP78 (34). Thus, if cell surface GRP78 also contains a very low amount of the glycosylated form, this could be a minor pathway below the detection sensibility of our assays.
Previously, we reported the existence of a subpopulation of GRP78 that exists as an ER transmembrane protein with its N-terminal region exposed to the cytosol (6). This was demonstrated by limited trypsin digestion of isolated microsomes, yielding a major resistant carboxyl band of about 35 kDa and a minor band of about 50 kDa. This was further confirmed with sodium carbonate extraction of the microsome membrane fractions, showing that GRP78 was present in both the membrane and the lumenal fractions (6). Since ER membrane is a source of the plasma membrane, this form of GRP78 could be cycled to the cell surface. Consistent with this, we observed that calnexin, another ER transmembrane protein, is exposed to the cell surface, in agreement with an earlier report (37). In this model, the C-terminal region of GRP78 is expected to be exposed extracellularly. In support of this, studies using antibodies against the C terminus of GRP78 in FACS analysis detected surface GRP78 expression (18, 33). Here we observed that the His epitope tagged at the C terminus of GRP78 is also exposed. However, evidence is accumulating that the N terminus of GRP78 is also exposed on the cell surface. Thus, using antibody that targets the N-terminal region of GRP78, it was demonstrated that the interaction between surface GRP78 and extracellular Par-4 and GPI-anchored Cripto and T-cadherin could be negatively affected (29, 30, 33). Here using FACS analysis of ectopically expressed GRP78 bearing the FLAG epitope at the N terminus, we demonstrated directly that the N terminus of GRP78 is exposed. Furthermore, we discovered here that a middle domain of GRP78 is also exposed. We found no evidence that extracellular GRP78 binds stably to cell surface. Recent studies suggest that specific cell types may utilize different proteins for transporting GRP78 to the cell surface. For example, the ER transmembrane protein, MTJ-1, is implicated as the GRP78 carrier protein in macrophages (40). Recently, the tumor suppressor Par-4 is reported to be required for GRP78 cell surface localization in PC-3 cells, although the molecular basis for translocation of Par-4 inside the ER remains to be determined (33, 41). In conclusion, our studies address fundamental mechanisms for GRP78 cell surface localization and open up new areas of investigations for partner protein complexes for its surface localization because it is evident that surface GRP78 plays critical roles in cell signaling, proliferation, and survival and has great potential for therapeutic interventions.
Acknowledgments
We thank Dr. James Ou for helpful discussion. We also thank Dr. Hui Zhou and the USC Norris Comprehensive Cancer Center and Stem Cell Institute Cell Sorting Core Facility for assistance in FACS analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA027607 and CA111700 (to A. S. L.).
- ER
- endoplasmic reticulum
- aa
- amino acid(s)
- CNX
- calnexin
- FACS
- fluorescence-activated cell sorting
- F-GRP78
- FLAG-GRP78
- Tg
- thapsigargin
- PDI
- protein-disulfide isomerase
- PBS
- phosphate-buffered saline
- rGRP78
- recombinant full-length GRP78.
REFERENCES
- 1.Ni M., Lee A. S. (2007) FEBS Lett. 581, 3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendershot L. M. (2004) Mt. Sinai J. Med. 71, 289–297 [PubMed] [Google Scholar]
- 3.Lee A. S. (2005) Methods 35, 373–381 [DOI] [PubMed] [Google Scholar]
- 4.Fu Y., Lee A. S. (2006) Cancer Biol. Ther. 5, 741–744 [DOI] [PubMed] [Google Scholar]
- 5.Lee A. S. (2007) Cancer Res. 67, 3496–3499 [DOI] [PubMed] [Google Scholar]
- 6.Reddy R. K., Mao C., Baumeister P., Austin R. C., Kaufman R. J., Lee A. S. (2003) J. Biol. Chem. 278, 20915–20924 [DOI] [PubMed] [Google Scholar]
- 7.Li J., Lee B., Lee A. S. (2006) J. Biol. Chem. 281, 7260–7270 [DOI] [PubMed] [Google Scholar]
- 8.Ranganathan A. C., Zhang L., Adam A. P., Aguirre-Ghiso J. A. (2006) Cancer Res. 66, 1702–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyrko P., Schönthal A. H., Hofman F. M., Chen T. C., Lee A. S. (2007) Cancer Res. 67, 9809–9816 [DOI] [PubMed] [Google Scholar]
- 10.Virrey J. J., Dong D., Stiles C., Patterson J. B., Pen L., Ni M., Schönthal A. H., Chen T. C., Hofman F. M., Lee A. S. (2008) Mol. Cancer Res. 6, 1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumeister P., Dong D., Fu Y., Lee A. S. (2009) Mol. Cancer Ther. 8, 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro S., Pelham H. R. (1986) Cell 46, 291–300 [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto A., Hanawalt P. C. (2000) Cancer Res. 60, 3921–3926 [PubMed] [Google Scholar]
- 14.Sun F. C., Wei S., Li C. W., Chang Y. S., Chao C. C., Lai Y. K. (2006) Biochem. J. 396, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni M., Zhou H., Wey S., Baumeister P., Lee A. S. (2009) PLoS ONE 4, e6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C., Bhattacharjee G., Boisvert W., Dilley R., Edgington T. (2003) Am. J. Pathol. 163, 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arap M. A., Lahdenranta J., Mintz P. J., Hajitou A., Sarkis A. S., Arap W., Pasqualini R. (2004) Cancer Cell 6, 275–284 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Steiniger S. C., Kim Y., Kaufmann G. F., Felding-Habermann B., Janda K. D. (2007) Mol. Pharmacol. 4, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Gronow M., Selim M. A., Papalas J., Pizzo S. V. (2009) Antioxid. Redox Signal. 11, 2299–2306 [DOI] [PubMed] [Google Scholar]
- 20.Wang M., Wey S., Zhang Y., Ye R., Lee A. S. (2009) Antioxid. Redox Signal. 11, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin B. K., Wang H., Yim A. M., Le Naour F., Brichory F., Jang J. H., Zhao R., Puravs E., Tra J., Michael C. W., Misek D. E., Hanash S. M. (2003) J. Biol. Chem. 278, 7607–7616 [DOI] [PubMed] [Google Scholar]
- 22.Kim Y., Lillo A. M., Steiniger S. C., Liu Y., Ballatore C., Anichini A., Mortarini R., Kaufmann G. F., Zhou B., Felding-Habermann B., Janda K. D. (2006) Biochemistry 45, 9434–9444 [DOI] [PubMed] [Google Scholar]
- 23.Davidson D. J., Haskell C., Majest S., Kherzai A., Egan D. A., Walter K. A., Schneider A., Gubbins E. F., Solomon L., Chen Z., Lesniewski R., Henkin J. (2005) Cancer Res. 65, 4663–4672 [DOI] [PubMed] [Google Scholar]
- 24.McFarland B. C., Stewart J., Jr., Hamza A., Nordal R., Davidson D. J., Henkin J., Gladson C. L. (2009) Cancer Res. 69, 5537–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misra U. K., Gonzalez-Gronow M., Gawdi G., Hart J. P., Johnson C. E., Pizzo S. V. (2002) J. Biol. Chem. 277, 42082–42087 [DOI] [PubMed] [Google Scholar]
- 26.Misra U. K., Gonzalez-Gronow M., Gawdi G., Wang F., Pizzo S. V. (2004) Cell. Signal. 16, 929–938 [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Gronow M., Cuchacovich M., Llanos C., Urzua C., Gawdi G., Pizzo S. V. (2006) Cancer Res. 66, 11424–11431 [DOI] [PubMed] [Google Scholar]
- 28.Shani G., Fischer W. H., Justice N. J., Kelber J. A., Vale W., Gray P. C. (2008) Mol. Cell. Biol. 28, 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelber J. A., Panopoulos A. D., Shani G., Booker E. C., Belmonte J. C., Vale W. W., Gray P. C. (2009) Oncogene 28, 2324–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippova M., Ivanov D., Joshi M. B., Kyriakakis E., Rupp K., Afonyushkin T., Bochkov V., Erne P., Resink T. J. (2008) Mol. Cell. Biol. 28, 4004–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triantafilou K., Fradelizi D., Wilson K., Triantafilou M. (2002) J. Virol. 76, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jindadamrongwech S., Thepparit C., Smith D. R. (2004) Arch. Virol. 149, 915–927 [DOI] [PubMed] [Google Scholar]
- 33.Burikhanov R., Zhao Y., Goswami A., Qiu S., Schwarze S. R., Rangnekar V. M. (2009) Cell 138, 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauschert N., Brändlein S., Holzinger E., Hensel F., Müller-Hermelink H. K., Vollmers H. P. (2008) Lab. Investig. 88, 375–386 [DOI] [PubMed] [Google Scholar]
- 35.Jakobsen C. G., Rasmussen N., Laenkholm A. V., Ditzel H. J. (2007) Cancer Res. 67, 9507–9517 [DOI] [PubMed] [Google Scholar]
- 36.Li J., Ni M., Lee B., Barron E., Hinton D. R., Lee A. S. (2008) Cell Death Differ. 15, 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okazaki Y., Ohno H., Takase K., Ochiai T., Saito T. (2000) J. Biol. Chem. 275, 35751–35758 [DOI] [PubMed] [Google Scholar]
- 38.Laverrière A., Landau R., Charvet I., Irion O., Bischof P., Morales M., Cohen M. (2009) Mol. Hum. Reprod. 15, 569–574 [DOI] [PubMed] [Google Scholar]
- 39.Misra U. K., Deedwania R., Pizzo S. V. (2006) J. Biol. Chem. 281, 13694–13707 [DOI] [PubMed] [Google Scholar]
- 40.Misra U. K., Gonzalez-Gronow M., Gawdi G., Pizzo S. V. (2005) J. Immunol. 174, 2092–2097 [DOI] [PubMed] [Google Scholar]
- 41.Lee A. S. (2009) Cancer Biol. Ther. 8, 2103–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]