Abstract
Bone morphogenetic proteins (BMPs) induce ectopic bone formation in muscle tissue in vivo and convert myoblasts such that they differentiate into osteoblastic cells in vitro. We report here that constitutively active Smad1 induced osteoblastic differentiation of C2C12 myoblasts in cooperation with Smad4 or Runx2. In floxed Smad4 mice-derived cells, Smad4 ablation partially suppressed BMP-4-induced osteoblast differentiation. In contrast, the BMP-4-induced inhibition of myogenesis was lost by Smad4 ablation and restored by Smad4 overexpression. A nuclear zinc finger protein, E4F1, was identified as a possible component of the Smad4 complex that suppresses myogenic differentiation in response to BMP signaling. In the presence of Smad4, E4F1 stimulated the expression of Ids. Taken together, these findings suggest that the Smad signaling pathway may play a dual role in the BMP-induced conversion of myoblasts to osteoblastic cells.
Keywords: Cell, Developmental Differentiation, Receptors, Threonine-Serine Kinases, Signal Transduction, Tissue/Organ Systems, Bone, Tissue/Organ Systems, Muscle/Skeletal, Transcription, Smad
Introduction
Bone morphogenetic proteins (BMPs)2 are members of the transforming growth factor-β (TGF-β) superfamily, which regulates the differentiation, proliferation, and death of various types of cells (1). BMPs were originally found in bone matrix as factors responsible for the induction of ectopic bone formation, in which implantation of demineralized bone matrix into muscle tissue induced new bone tissue containing bone marrow (2, 3). Implantation of individual recombinant BMPs, such as BMP-2, BMP-4, BMP-6, and BMP-7, into muscle tissue induces ectopic bone formation in vivo as well (4). This ectopic bone-inducing activity is highly specific to BMPs, because other hormones and cytokines, including TGF-β1 itself, failed to induce ectopic bone formation in muscle tissue in vivo (5). Although many factors, such as BMPs, TGF-βs, fibroblast growth factors, and epidermal growth factor, inhibit myogenic maturation of myoblasts in vitro, only BMPs convert them so that they differentiate to osteoblastic cells, the bone-forming cells in vertebrates (6–10). Thus, the activity of BMPs in myoblast cultures to induce osteoblastic differentiation appears to reflect the ectopic bone-inducing activity of BMPs in vivo (8).
BMP signaling is transduced by two different types of serine/threonine kinase receptors, termed type I and II receptors (1, 11, 12). The BMP-bound type II receptor phosphorylates the type I receptor kinase, and the activated BMP type I receptor in turn phosphorylates downstream substrates such as receptor-regulated Smads (R-Smads), including Smad1, Smad5, and Smad8 and p38 mitogen-activated protein kinase. Phosphorylated R-Smads form heteromeric complexes with Smad4 and translocate into the nucleus to regulate transcription of various target genes, including Id1, which encodes a dominant-negative inhibitor of myogenesis (13–15). Recently, a genetic mutation of ALK2, a BMP type I receptor, was identified in patients with fibrodysplasia ossificans progressiva, an autosomal-dominant disorder characterized by heterotopic bone formation in muscle tissue (16). Our findings indicate that this mutant ALK2 is a constitutively activated and hyper-reactive form of the BMP type I receptor and suggest that downstream signaling of activated receptors play an important role in heterotopic bone formation in muscle tissue under certain pathological conditions (17).
Here, we show that a protein in which the carboxyl-terminal serine residues of Smad1 were substituted with aspartic acids, termed Smad1(DVD), functioned as a constitutively activated Smad1. Overexpression of Smad1(DVD) induced osteoblastic differentiation of C2C12 myoblasts, although the inhibition of myogenic differentiation depended principally on nuclear Smad4 rather than R-Smads. E4F1 was identified as a possible component of the Smad4 complex in the suppression of myogenic differentiation by BMP signaling. R-Smads and Smad4 play important roles in the conversion of myogenic cells to osteoblastic cells by BMPs, data were confirmed with the deletion of Smad4 in mouse embryonic fibroblasts (MEFs).
EXPERIMENTAL PROCEDURES
Expression Vectors
Expression vectors for wild-type mouse Smad1, Smad4, and Smad7 were described previously (13). All of the mutations were introduced by a standard PCR technique using Platinum Pfx DNA polymerase (Invitrogen) and their sequences were confirmed in each expression vector. Smad1 mutants were obtained by substitution of Ser-463 and/or Ser-465 of mouse Smad1 with aspartic acid or alanine residues. A nuclear localization signal derived from an SV40 large T antigen, PPKKKRKV, was inserted between an amino-terminal epitope tag and Smad1 or Smad4. NLS-Smad4(ΔMH1) and NLS-Smad4(ΔMH2) were obtained by deleting amino acids 2–138 and 316–552, respectively, from full-length NLS-Smad4. Both mouse wild-type and dominant-negative Runx2 vectors were kindly provided by Dr. Toshihisa Komori (Nagasaki University) (18). Mouse E4f1 (accession number BC057011) cDNA was obtained by a standard reverse transcription-PCR technique and cloned into pcDEF3 with a FLAG epitope sequence at the 3′ end (19), and E4F1(ΔE3) and the zinc finger mutants (Z1–Z6) were generated by deleting a ubiquitin E3 ligase domain, amino acids 40–84, and substituting two cysteine residues with alanines at positions 195 and 197, 223 and 226, 435 and 438, 519 and 522, and 547 and 550, respectively.
Cell Cultures and Transfections
C2C12 mouse myoblasts, C3H10T1/2 mouse fibroblasts, and COS-7 African green monkey kidney cells were maintained as described (8, 20). The cells were treated with 100 ng/ml of recombinant human BMP-4 (R & D Systems, Inc.) in the presence or absence of chemical inhibitors of mitogen-activated protein kinase, U0126, SB203580, or SP600125 (Merck, Tokyo, Japan) or of the BMP Smad pathway, Dorsomorphin (Calbiochem, San Diego, CA) (20, 21). Lipofectamine 2000 (Invitrogen) was used for plasmid transfections. RNAi Stealth oligonucleotides against murine Smad1 (MSS275578), Smad5 (MSS2755978), Runx2 (MSS202675), Smad4 (MSS206437), and E4f1 (MSS274050) and a scrambled negative control oligonucleotide were purchased from Invitrogen. An expression plasmid for E4f1 microRNA was constructed by subcloning Mmi508063 (Invitrogen) into pcDNA6.2-GW/EmGFP-miR (Invitrogen). MEFs prepared from Smad4floxed/floxed mouse embryos at 11.5 days postcoitum were infected with an adenovirus expressing Cre recombinase (pAxCANCre) or a human histone 2B-GFP unit (pAxH2BGFP) under control of the CAG promoter (22, 23). After being cultured for an additional 48 h, MEFs were transfected with plasmids using Lipofectamine 2000 (Invitrogen) in combinations as described in each figure legend. The cells were further cultured with or without BMP-4 before being examined for BMP activities as described below. A cell line, clone 16, was established by a limiting dilution method from Smad4floxed/floxed MEFs maintained by more than 30-fold serial passage. Bone marrow stromal cells were prepared from tibiae of 8-week-old Smad4floxed/floxed mice.
Immunohistochemistry, Immunoprecipitation, and Western Blot Analysis
The following antibodies were used for immunohistochemistry, Western blot analysis, and immunoprecipitation: anti-MHC antibody (clone MF-20, Developmental Studies Hybridoma Bank, Iowa City, IA), anti-myogenin (clone F5D, Santa Cruz, Santa Cruz, CA), anti-FLAG antibody (clone M2, Sigma), anti-Myc antibody (clone 9E10, Santa Cruz), anti-Myc polyclonal antibody (Medical & Biological Laboratories Co., Nagoya, Japan), anti-Smad4 antibody (clone B-8, SC-7966, Santa Cruz), anti-V5 antibody (P/N 46–0705, Invitrogen), anti-E4F1 antibody (Bethyl Laboratories, Montgomery, TX), anti-phospho-Smad1/5/8 polyclonal antibody (Cell Signaling, Beverly, MA), anti-Runx2/Cbfa-1 (Medical & Biological Laboratories Co.), and anti-β-actin antibody (I-19, SC-1616, Santa Cruz). For immunohistochemical analysis, target proteins were visualized using a Histofine SimpleStain Kit (Nichirei, Tokyo, Japan) or an Alexa 488- or Alexa 594-conjugated secondary antibody (Invitrogen). A BZ-9000 (Keyence, Tokyo, Japan) microscope was used for fluorescent analysis. Western blot analysis was performed as described (17). The target proteins were immunoprecipitated for 6 h at 4 °C using M2-agarose beads (Sigma). The target proteins were detected using a horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (GE Healthcare).
ALP and Luciferase Assays
ALP activity was stained as a typical marker of osteoblastic differentiation (24). Enzyme activity was measured using p-nitrophenyl phosphate as a substrate (24). Luciferase assays were performed using pGL3MG-185 (25) or IdWT4F-luc reporter plasmids and phRL-SV40 (Promega, Madison, WI) with the Dual-Glo Luciferase Assay System (Promega) as described previously (13).
Dorsoventral Assay in Xenopus Embryos
A dorsoventral assay in Xenopus embryos was performed essentially as described (26). The injected embryos were allowed to develop until stages 34–40 for observation of external appearance and then subjected to histological analysis. The activity of each Smad1 was expressed by a dorso-anterior index (26, 27).
Reverse Transcription-PCR Analysis
Total RNAs were extracted using TRIzol (Invitrogen) and reverse transcribed with SuperScript III (Invitrogen). The PCR was performed using Platinum Pfx DNA polymerase (Invitrogen) as described (28). The primer sets used were previously described (29).
Chromatin Immunoprecipitation Assay
Cells were lysed in ChIP buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1) and sonicated. The chromatin solution was subjected to immunoprecipitation using a OneDay ChIP Kit (Diagenode, Sparta, NJ) according to the manufacturer's instructions. The following antibodies were used: anti-E4F1 (Bethyl), anti-Smad4 (sc-7966, Santa Cruz Biotechnology), anti-MyoD (sc-760, Santa Cruz Biotechnology), and anti-histone H3 (Upstate, Lake Placid, NY). The Myogenin promoter was amplified by PCR using the following primers: 5′-TAATTGAAAGGAGCAGATGAGACGGGG-3′ and 5′-CCATCAGGTCGGAAAAGGCTTGTTC-3′.
Statistical Analysis
Comparisons were made using an unpaired Student's t test. Results are represented as mean ± S.D. Statistical significance is displayed as: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
Induction of the Smad-dependent Pathway by BMP-4 Regulates Both Myogenic and Osteoblastic Differentiation of C2C12 Cells
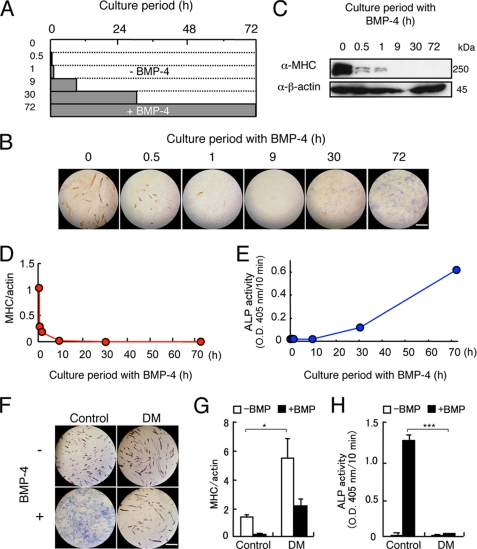
First, we determined the minimal culture periods of C2C12 cells with BMP-4 required for the inhibition of myogenic differentiation and induction of osteoblastic differentiation (Fig. 1A). Treatment of C2C12 cells with 100 ng/ml of BMP-4 for the first 30 min of a 3-day culture markedly suppressed myogenic differentiation on day 3 but did not induce ALP activity (Fig. 1, B–E). ALP-positive cells were detected in cultures treated with BMP-4 for longer than 9 h (Fig. 1, B and E). The BMP-4-induced inhibition of myogenic differentiation and induction of osteoblastic differentiation were blocked by Dorsomorphin, a BMP-Smad specific inhibitor, but not by inhibitors of mitogen-activated protein kinases, suggesting that the Smad-dependent pathway regulates conversion to differentiation (Fig. 1, F–H, and supplemental Fig. S1).
FIGURE 1.
Smad signaling pathway regulates both inhibition of myogenic differentiation and induction of osteoblastic differentiation by BMP-4. A and B, examination of the minimal treatment periods required for inhibition of myogenic differentiation and induction of osteoblastic differentiation by BMP-4 in C2C12 myoblasts. A, scheme of treatment of C2C12 myoblasts with BMP-4 in a window experiment. C2C12 cells were treated for 0, 0.5, 1, 9, 30, or 72 h with 100 ng/ml of BMP-4 (closed bars) and further incubated without BMP-4 (open bars) until 72 h before staining. B, the cells were doubly stained for ALP and MHC in blue and red, respectively, at 72 h. Scale bar, 200 μm. C–E, quantitation of the effects of BMP-4 on myogenic differentiation and osteoblastic differentiation of C2C12 cells. Western blots for MHC (C and D) and measurement of ALP activity (E) were performed on day 3. F–H, effects of Dorsomorphin on BMP-induced differentiation in C2C12 cells. C2C12 cells were preincubated for 1 h with 3 μm Dorsomorphin (DM) and 100 ng/ml of BMP-4 was then added. F, the cells were doubly stained for ALP and MHC on day 3. Scale bar, 400 μm. MHC levels (G) and ALP activity (H) were measured on day 3. Values are mean ± S.D. (n = 3). *, p < 0.05; ***, p < 0.001.
Construction of Constitutively Activated Smad1
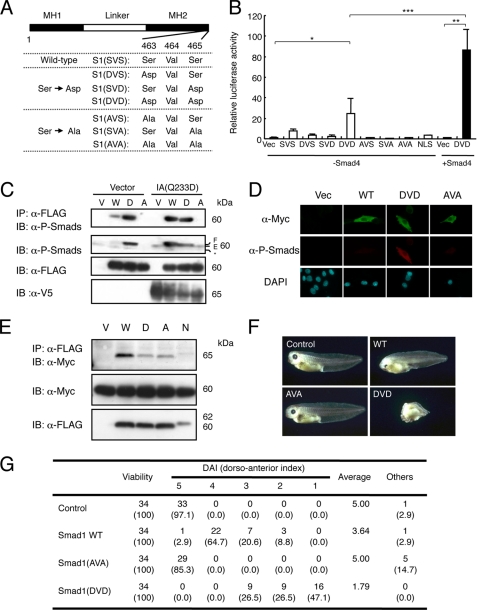
We generated a series of Smad1 mutants, in which one or two serine residues at the carboxyl termini were substituted with aspartic acid or alanine residues (Fig. 2A). Among these Smad1 mutants, Smad1(DVD) exhibited transcriptional activity in a luciferase assay using the ID1 reporter without the addition of BMPs, and this activity was further enhanced by co-transfection with Smad4 (Fig. 2B). NLS-Smad1, in which a nuclear localization signal (NLS) was added to the amino terminus of Smad1, failed to induce luciferase activity. Smad1(DVD) was recognized by the α-phospho-Smad1/5/8 antibody, even in the absence of a constitutively active BMP receptor, BMPR-IA(Q233D), without affecting endogenous phospho-Smad1/5/8 levels (Fig. 2, C and D). Smad1(DVD) did not exhibit changes in cellular localization or interaction with Smad4 that were distinguishable from wild-type or other Smad1 mutants (Fig. 2, D and E). Injection of synthetic Smad1(DVD) mRNA into the dorsal sides of Xenopus embryos induced ventralization, although Smad1(AVA) did not exhibit this activity (Fig. 2F). The average dorso-anterior index values induced by Smad1(DVD) and Smad1(AVA) were 1.79 and 5.00, respectively, indicating that Smad1(DVD) is a constitutively activated Smad1 in Xenopus embryos as well (Fig. 2G).
FIGURE 2.
Establishment of a constitutively activated Smad1. A, construction of mutant Smad1. Serine 463 and/or 465 at the carboxyl terminus of mouse Smad1 was substituted by aspartic acid or alanine. B, transcriptional activities of Smad1 mutants in luciferase assay in C2C12 cells. Wild-type and mutant Smad1 were transfected with IdWT4F-luc in C2C12 cells in the presence or absence of Smad4 (n = 3). Note that Smad1(DVD) activated the reporter activity even in the absence of Smad4. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C and D, Smad1(DVD) was recognized by the α-phospho-Smad1/5/8 antibody. C2C12 cells were co-transfected with empty vector (Vector) or V5-tagged Bmpr-Ia(Q233D) (IA(Q233D)) and an empty vector (V), FLAG-tagged wild-type Smad1 (W), Smad1(DVD) (D), or Smad1(AVA) (A). Whole cell lysates were immunoprecipitated (IP) with anti-FLAG antibody followed by immunoblotting (IB) using α-P-Smads antibody to detect the mutant Smad1 and endogenous Smad1/5/8. D, C2C12 cells transfected with Myc-tagged wild-type or Smad1(DVD), Smad1(AVA), or NLS-Smad1 were stained with α-P-Smads and α-Myc antibodies without BMP stimulation. Note that Smad1(DVD) was detected in the cytoplasm with α-P-Smads antibody. E, interaction with Smad1 and Smad4. C2C12 cells were co-transfected with Myc-tagged Smad4 and an empty vector (V), FLAG-tagged wild-type Smad1 (W), Smad1(DVD) (D), Smad1(AVA) (A), or NLS-Smad1 (N). Whole cell lysates were immunoprecipitated with α-FLAG antibody followed by immunoblotting using α-Myc antibody to detect Smad4 in complex with Smad1. F and G, ventralization inducing activity of Smad1 in Xenopus embryos. F, Xenopus embryos at four-cell stage were injected with 500 pg of synthetic mRNA of wild-type, Smad1(AVA), or Smad1(DVD) into the dorsal marginal region. G, dorsal-anterior index of Xenopus embryos induced by Smad1. Values smaller than 5 indicate degree of ventralization.
Activated Smad1 and Runx2, but Not Smad4, Cooperatively Induce Osteoblastic Differentiation of C2C12 Cells
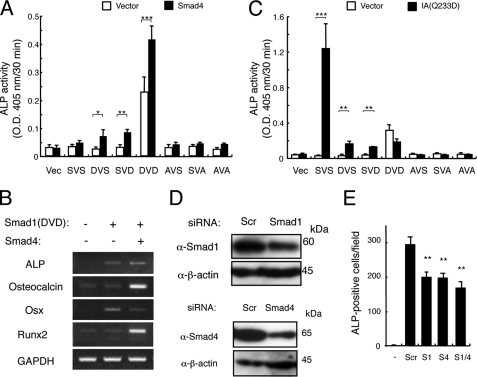
Transient transfection of Smad1(DVD) in C2C12 cells induced expression of osteoblastic differentiation markers such as ALP, Osteocalcin, Runx2, and Osterix; this was further enhanced by the presence of Smad4 for every marker except Osterix, which might be peaked within 3 days before sample preparation (Fig. 3, A and B). BMPR-IA(Q233D) stimulated ALP activity in cooperation with wild-type Smad1, confirming that Smad1 is a critical substrate of the type I receptor for induction of osteoblastic differentiation (Fig. 3C). However, no synergism was observed between BMPR-IA(Q233D) and any Smad1 mutants, including Smad1(DVD), suggesting that Smad1 mutants are not recognized as substrates by the receptor (Figs. 2C and 3C). Transfection of siRNA against Smad1, Smad4, or a combination of the two reduced the ALP activity induced by BMP-4 or BMPR-IA(Q233D) (Fig. 3, D and E). Similar results were obtained using siRNA against Smad5 (data not shown).
FIGURE 3.
Smad1(DVD) induces osteoblastic differentiation of C2C12 myoblasts. A, Smad1(DVD) (DVD) induces ALP activity in C2C12 myoblasts. C2C12 cells were transfected with one of the Smad1 constructs in the presence (closed bars) or absence (open bars) of Smad4, and ALP activity was measured on day 3. Values are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001. B, Smad4 enhances Smad1(DVD) activity. C2C12 cells were transfected with empty vector alone or with Smad1(DVD) in the absence or presence of Smad4. Reverse transcription-PCR was performed on day 3. C, synergism between Smad1 and a constitutively activated BMPR-IA receptor. C2C12 cells were transfected with one of the Smad1 constructs in the absence (open bars) or presence of Bmpr-Ia(Q233D) (closed bars), and ALP activity was determined on day 3. Values are mean ± S.D. (n = 3). D, knockdown of Smad1 and Smad4 in C2C12 cells. C2C12 cells were transfected with 20 nm Smad1 (upper panels) and Smad4 (lower panels) siRNA. Protein levels were determined by Western blots at 24 h. E, knockdown of Smad1 or Smad4 reduced ALP activity induced by BMP-4. The siRNA-transfected C2C12 cells were treated for 3 days with 100 ng/ml of BMP-4 and ALP activity was determined. Values are mean ± S.D. (n = 3). **, p < 0.01 compared with a scrambled siRNA transfection group.
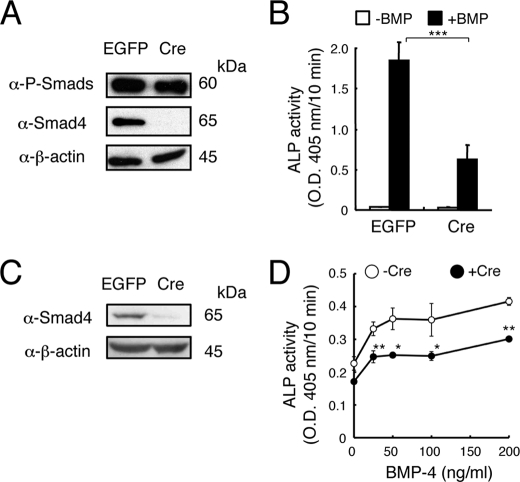
The role of Smad proteins in osteoblastic differentiation was further examined using MEFs prepared from Smad4floxed/floxed mice. The MEFs had been infected with an adenovirus expressing Cre recombinase or EGFP in vitro before being treated with BMP-4. Western blot analysis revealed that the phosphorylation of Smad1/5/8 in response to BMP-4 was independent of Smad4 (Fig. 4A). In these MEF cultures, the ALP activity induced by BMP-4 was reduced but not eliminated in the Cre-expressing MEFs (Fig. 4B). We further examined the role of Smad4 in osteoblastic differentiation using bone marrow stromal/osteoblastic cells prepared from Smad4floxed/floxed mice. Again, Smad4 was deleted in vitro by infection with an adenovirus expressing Cre, but expression of ALP was not eliminated in these cells; in fact, ALP expression was still induced by BMP-4 in a dose-dependent fashion in Smad4-deleted cultures (Fig. 4, C and D). These results suggested that Smad4 is not essential for BMP-induced osteoblastic differentiation but that it may enhance BMP signaling.
FIGURE 4.
Smad4 is not essential for the osteoblastic differentiation induced by BMP-4. A, MEFs prepared from Smad4floxed/floxed were infected with adenovirus expressing Cre recombinase or EGFP. The cells were stimulated for 30 min with 100 ng/ml of BMP-4 to induce phosphorylation of Smad1/5/8. B, adenovirus-infected MEFs were treated for 3 days with 100 ng/ml of BMP-4, and ALP activity was determined on day 3. Values are mean ± S.D. (n = 5). ***, p < 0.001. C, Western blot analysis of Smad4-ablated bone marrow stromal/osteoblastic cells prepared from Smad4floxed/floxed mice. D, dose-dependent induction of ALP activity by BMP-4 in Smad4floxed/floxed-derived bone marrow stromal/osteoblastic cells infected with Cre-expressing adenovirus or uninfected cells. Values are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01.
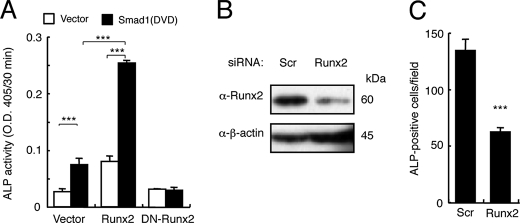
Runx2 is essential for osteoblast differentiation and also interacts with R-Smads (30–32). Overexpression of Smad1(DVD) or Runx2 alone induced ALP activity in C2C12 cells, and co-expression of Smad1(DVD) and Runx2 further increased ALP activity (Fig. 5A). In contrast, a dominant-negative form of Runx2 blocked ALP induction by Smad1(DVD) (Fig. 5A). RNAi knockdown of Runx2 reduced numbers of ALP-positive cells in C2C12 cultures induced by BMPR-IA(Q233D) (Fig. 5, B and C). Taken together, these findings suggest that phosphorylated R-Smads and Runx2 may cooperatively induce osteoblast differentiation in response to BMPs.
FIGURE 5.
Runx2 and Smad1(DVD) cooperatively induce osteoblastic differentiation of C2C12 cells. A, C2C12 cells were co-transfected with Smad1(DVD) and Runx2 or dominant-negative Runx2, and ALP activity was determined on day 3. Values are mean ± S.D. (n = 3). ***, p < 0.001. B, 20 nmol of Runx2 siRNA or scrambled oligonucleotide was transfected in C2C12 cells, and protein levels were determined by Western blots at 24 h post-transfection. C, the siRNA-transfected C2C12 cells were co-transfected with Bmpr-Ia(Q233D) and Smad1, and ALP activity was determined on day 3. Values are mean ± S.D. (n = 3). ***, p < 0.001 compared with a scrambled siRNA transfection group.
Smad4 Is Involved in Inhibition of Myogenic Differentiation by BMPs
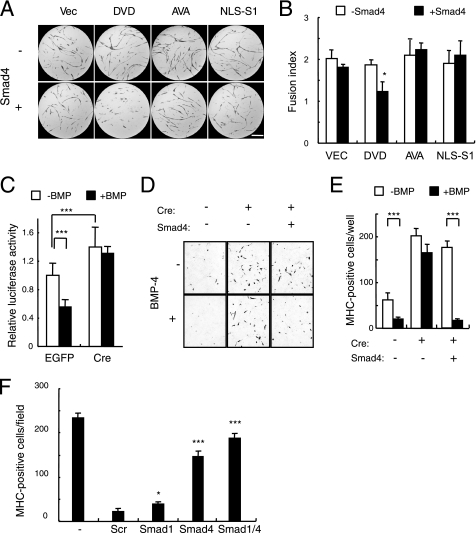
We next examined the roles that the Smad signaling pathway plays in the inhibition of myogenic differentiation by BMPs. Overexpression of Smad1 mutants, including Smad1(DVD), did not inhibit the myogenic differentiation induced by MyoD. Co-transfection of Smad4 with Smad1(DVD) was required to reduce the number of MHC-positive cells (Fig. 6, A and B). This role of Smad4 in myogenic differentiation was confirmed in Smad4floxed/floxed MEFs. The transcriptional activity of MyoD was suppressed by BMP-4 in MEF cultures infected with a control virus (Fig. 6C). In contrast, the basal transcriptional activity of MyoD was increased ∼1.4-fold in MEFs infected with the Cre-expressing adenovirus. This increase was not suppressed by BMP-4, suggesting that Smad4 is essential for the suppression of myogenic differentiation by BMPs (Fig. 6C). This hypothesis was further confirmed using the floxed Smad4 MEF cell line, clone 16. Transient transfection of MyoD in clone 16 induced a small number of MHC-positive cells, which were reduced in number by BMP-4 (Fig. 6, D and E). Infection by Cre-expressing adenovirus not only increased the number of MHC-positive cells, but also maintained this increase in the presence of BMP-4 (Fig. 6, D and E). Transient transfection of Smad4 in Cre-adenovirus-infected cultures restored the suppression of myogenic differentiation in response to BMP-4 (Fig. 6, D and E). Moreover, RNAi knockdown of Smad4 increased the number of MHC-positive cells in C2C12 cell cultures treated with BMP-4 (Fig. 6F). In contrast, knockdown of Smad1 had minimal effects on these cultures (Fig. 6F).
FIGURE 6.
Smad4 is involved in the inhibition of myogenic differentiation. A and B, Smad1(DVD) inhibits myogenic differentiation only in the presence of Smad4. C3H10T1/2 fibroblasts were co-transfected with MyoD and empty vector (Vec), Smad1(DVD), Smad1(AVA), or NLS-Smad1 (NLS-S1) in the absence (upper panels) or presence (lower panels) of Smad4 (A). Myogenic cells were immunostained for MHC (red) on day 5. Scale bar, 400 μm. B, fusion index was determined in the presence (closed bars) or absence (open bars) of Smad4. Values are mean ± S.D. (n = 3). *, p < 0.05. C, Smad4 is required for suppression of transcriptional activity of MyoD by BMP-4. MEFs prepared from Smad4floxed/floxed were infected with adenovirus expressing EGFP or Cre recombinase, and then the transcriptional activity of MyoD was determined in the presence or absence of 100 ng/ml of BMP-4. Values are mean ± S.D. (n = 10). ***, p < 0.001. D and E, Smad4 is essential for inhibition of myogenic differentiation by BMPs. Clonal cell line 16 infected with Cre-expressing or EGFP-expressing control adenovirus was transfected with MyoD and stained for MHC on day 5 (D). E, the numbers of MHC-positive cells were counted in cultures prepared as in D. Values are mean ± S.D. (n = 6). ***, p < 0.001. F, 20 nmol of Smad1 or Smad4 siRNA or a scrambled oligonucleotide was transfected into C2C12 cells. These cells were treated with 100 ng/ml of BMP-4, and MHC-positive cells were counted on day 5. Values are mean ± S.D. (n = 3). *, p < 0.05; ***, p < 0.001 compared with a scrambled siRNA transfection group.
Inhibition of Myogenic Differentiation by Nuclear Smad4
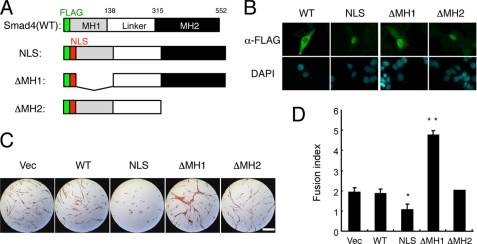
Because Smad4 may translocate into the nucleus as a complex with Smad1/5/8 in response to BMP stimulation, we generated NLS-Smad4 and deletion mutants lacking amino-terminal MH1 (NLS-Smad4(ΔMH1)) and carboxyl-terminal MH2 (NLS-Smad4(ΔMH2)) domains, respectively (Fig. 7A). Nuclear localization of these NLS-Smad4 mutants was confirmed, although wild-type Smad4 was mainly detected in the cytoplasm (Fig. 7B). Overexpression of full-length NLS-Smad4 suppressed the myogenic differentiation of C3H10T1/2 cells induced by MyoD in a dose-dependent manner (Fig. 7, C and D, and data not shown). Unexpectedly, NLS-Smad4(ΔMH1), but not NLS-Smad4(ΔMH2), stimulated myogenic differentiation, suggesting that NLS-Smad4(ΔMH1) behaved in a dominant-negative fashion (Fig. 7, C and D). The Smad4 MH2 domain may thus interact with other molecules essential for inhibition of myogenic differentiation.
FIGURE 7.
Nuclear Smad4 inhibits myogenic differentiation. A, scheme of construction of FLAG-tagged Smad4 mutants. B, cellular localization of FLAG-tagged Smad4 mutants. The cells were immunostained with α-FLAG antibody. Scale bar, 25 μm. C, effects of Smad4 mutants on myogenic differentiation. C3H10T1/2 cells were co-transfected with MyoD and one of the Smad4 constructs and immunostained for MHC on day 5. Scale bar, 400 μm. D, fusion index was determined from cultures prepared as in C. Values are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole.
Involvement of E4F1 in the Inhibition of Myogenic Differentiation by BMP Signaling
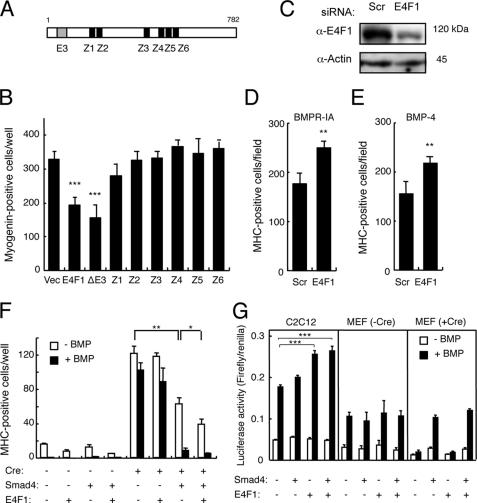
We searched a protein-protein interaction data base that was constructed based on the mammalian two-hybrid method established by the RIKEN group (33) and found several proteins that formed complexes with Smad4. Among these proteins, we focused on E4F1, which contains six zinc fingers and a ubiquitin E3 ligase domain (Fig. 9A), because it appeared to be one of the transcription factors principally responsible for inhibition of myogenic differentiation by Smad4.
FIGURE 9.
E4F1 is involved in the BMP-induced inhibition of myogenesis. A, schematic structure of E4F1. E3, an E3 ubiquitin ligase domain; Z, zinc fingers. B, overexpression of E4F1 inhibits myogenic differentiation. C3H10T1/2 cells were co-transfected with MyoD and wild-type E4F1, E4F1(ΔE3), or a zinc finger mutant E4F1 and immunostained for myogenin on day 3. Values are mean ± S.D. (n = 3). ***, p < 0.001. C, 20 nmol of E4F1 siRNA or scrambled oligonucleotide was transfected in C2C12 cells, and protein levels were determined by Western blots at 24 h post-transfection. D and E, the siRNA-transfected C2C12 cells were co-transfected with Bmpr-Ia(Q233D) (D) or treated with 100 ng/ml of BMP-4 (E), and MHC-positive cells were counted on day 5. Values are mean ± S.D. (n = 3). **, p < 0.01 compared with each scrambled siRNA transfection group. F, E4F1 suppresses myogenesis cooperatively with Smad4. Smad4floxed/floxed-derived clonal cell line 16 was infected with or without a Cre-expressing adenovirus. The cells were transfected with or without Smad4 in the presence or absence of Smad4, cultured with or without 100 ng/ml of BMP-4, and stained for MHC on day 3. Values are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01. G, E4F1 stimulates Id1 expression. C2C12 cells and MEF clone 16 infected with Cre adenovirus were transfected with Id985-luc, E4F1, and Smad4 and were then cultured in the presence or absence of 50 ng/ml of BMP-4. Luciferase activity was determined on day 1. Values are mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
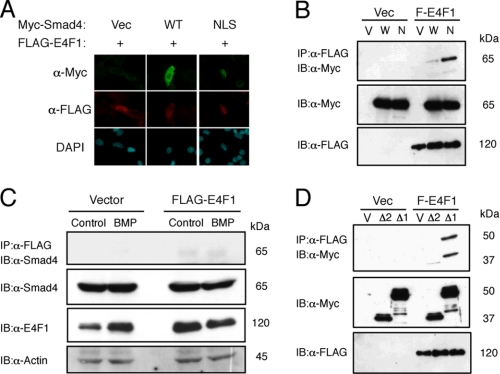
FLAG-tagged E4F1 was expressed in nuclei, co-localized with NLS-Smad4, and bound to larger amounts of NLS-Smad4 than wild-type Smad4 (Fig. 8, A and B). Interaction between endogenous Smad4 and FLAG-E4F1 was also detected in C2C12 cells (Fig. 8C). NLS-Smad4(ΔMH1), but not NLS-Smad4(ΔMH2), bound to E4F1, confirming that the complex is formed via the MH2 domain of Smad4 (Fig. 8D).
FIGURE 8.
E4F1 inhibits myogenic differentiation in cooperation with nuclear Smad4. A, E4F1 and Smad4 overlapped in nuclei in C2C12 cells. FLAG-tagged E4F1 and empty vector (Vec), Myc-Smad4 (WT), or Myc-NLS-Smad4 (NLS) were cotransfected and stained with α-Myc and α-FLAG antibodies. B and C, E4F1 interacts with Smad4 in vivo. B, COS-7 cells were co-transfected with FLAG-E4F1 and empty vector (Vec), Myc-Smad4 (WT), or Myc-NLS-Smad4 (NLS). Whole cell lysates were immunoprecipitated (IP) with α-FLAG antibody followed by immunoblotting (IB) using α-Myc antibody. C, C2C12 cells transfected with Flag-E4F1 were treated for 1 h without or with 100 ng/ml of BMP-4. Whole cell lysates were immunoprecipitated with M2-agarose beads followed by immunoblotting with antibodies for E4F1, Smad4, and actin. D, the MH2 domain of Smad4 is required for interaction with E4F1. The interaction between E4F1 and Smad4 was determined by cotransfection of Flag-E4F1 and Myc-NLS-Smad4(ΔMH2) or Myc-NLS-Smad4(ΔMH1) in COS-7 cells. DAPI, 4′,6-diamidino-2-phenylindole.
Both wild-type E4F1 and E4F1(ΔE3), but not zinc finger mutants, suppressed myogenic differentiation, suggesting that E4F1 may inhibit myogenesis as a transcription factor rather than as a ubiquitin ligase (Fig. 9B). Although Myogenin is one of the targets of MyoD and is markedly suppressed by BMP signaling, we could not detect Smad4 or E4F1 binding to the Myogenin promoter in response to BMP-4 (supplemental Fig. S2). RNAi knockdown of E4F1 increased the number of MHC-positive C2C12 cells in the presence of BMP signaling (Fig. 9, D and E). Similar results were obtained using a plasmid-based microRNA expression vector for E4F1 (supplemental Fig. S3). We further examined the role of E4F1 in myogenesis in cell line clone 16 established from Smad4floxed/floxed MEFs. Again, deletion of Smad4 by Cre-adenovirus infection increased the number of MyoD-induced MHC- and myogenin-positive myogenic cells (Fig. 9, F and G). Co-transfection of Smad4 and E4F1 markedly reduced the number of myogenic cells, suggesting that E4F1 acts cooperatively with Smad4 (Fig. 9F).
Id1–3 suppress myogenesis and are targets of BMP signaling. Transfection of E4f1 increased Id1-, Id2-, and Id3-luc activities in C2C12 cells treated with and without BMP-4 (Fig. 9G, and data not shown). This stimulation by E4F1 seemed to be Smad4 dependent because the activity was lost by Smad4 ablation and restored by Smad4 overexpression in MEF clone 16 (Fig. 9G).
DISCUSSION
In the present study, we examined the molecular mechanisms underlying the conversion of myoblasts by BMPs, allowing their differentiation into osteoblastic cells. It has been suggested that a unique type of intracellular BMP signaling is involved in this conversion, because other inhibitors of myogenic differentiation, such as TGF-β and fibroblast growth factors, do not induce ectopic bone formation in vivo or osteoblastic differentiation in vitro (12). We found that the inhibition of myogenic differentiation by BMP-4 required treatment for less than 1 h, although induction of osteoblastic differentiation required treatment for more than 9 h. Both activities of BMPs were dependent on the Smad pathway, suggesting that related but distinct mechanisms regulate the conversion of myoblasts into osteoblastic cells. Because we failed to detect cells positive for both MHC and ALP in C2C12 cell cultures treated with BMPs (9, 12), it appeared that osteoblastic differentiation is activated only in immature myoblasts that have not yet initiated myogenic differentiation (34). This hypothesis was confirmed by our preliminary observation that BMPs did not induce ALP activity in mature multinucleated myotubes.3
BMP treatment can convert the differentiation pathway of myoblasts into osteoblastic cells and overexpression of constitutively activated BMP type I receptors such as BMPR-IA, BMPR-IB, and ALK2 can have the same effect without requiring the addition of BMPs (35, 36). However, we found that levels of endogenous Smad1 and Smad5 were low in C2C12 cells and that overexpression of wild-type Smad1 was required for induction of osteoblastic differentiation by BMPR-IA(Q233D). These findings suggested that downstream signaling of BMP type I receptors, rather than BMP type II and co-receptors, plays an important role in the conversion of myoblast differentiation. We established a constitutively activated Smad1, Smad1(DVD), to directly examine the role of the Smad pathway without activation of other signaling pathways induced by BMP receptors, such as the mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways. Serine residues were substituted with aspartic residues in Smad1(DVD) to introduce negative charges in the SVS motif at the carboxyl terminus, the site of phosphorylation by type I BMP receptors. These substitutions may induce conformational changes and stimulate interaction with coactivators such as p300, OAZ, and Runx2 (11). Indeed, Smad1(DVD) was directly recognized by the α-phospho-Smad1/5/8 antibody, suggesting that its three-dimensional structure is similar to that of native Smad1 phosphorylated by the receptors. We found that co-transfection of Smad1(DVD) with Smad4 was capable of inducing osteoblastic differentiation of C2C12 myoblasts, and this induction was not inhibited by co-expression with Smad7, suggesting that Smad1(DVD) induces activation downstream of BMP type I receptors without activating endogenous BMPs or receptors. However, the ALP activity induced by Smad1(DVD) and Smad4 was lower than that induced by cotransfection of a constitutively activated BMPR-IA receptor and wild-type Smad1, although it was higher than that induced by BMPR-IA(Q233D) alone or Smad1(EVE), in which serine residues had been substituted with glutamic acids instead of aspartic acids.3 These findings suggested that native phosphorylated Smad1 may have higher affinity for the coactivators required for osteoblastic differentiation than Smad1(DVD) or Smad1(EVE). This hypothesis will require further testing.
The Smad signaling pathway was also involved in the inhibition of myogenic differentiation. In contrast to osteoblast differentiation, however, this inhibitory activity of the Smad pathway appeared to be mainly dependent on Smad4 rather than R-Smads. In particular, the nuclear-targeted Smad4 markedly suppressed myogenic differentiation, although overexpression of NLS-Smad4 did not induce osteoblastic differentiation,3 suggesting that Smad4 in complex with R-Smads inhibits myogenic differentiation after translocation from the cytoplasm to the nucleus in response to BMP stimulation. Because Smad4 is a common Smad among the TGF-β superfamily members, it may also be involved in mediating the effects of other myogenic inhibitors, such as TGF-βs, myostatin, and activin (37, 38).
The MH1 and MH2 domains of Smads have been shown to be involved in DNA binding and interaction with other proteins, respectively (39). Our deletion analysis suggested that nuclear Smad4 may interact with other transcriptional factor(s) and recruit them to the target DNA sequences via the MH2 and MH1 domains, respectively, to suppress myogenesis. This hypothesis was further supported by the finding of stimulation of myogenic differentiation by NLS-Smad4(ΔMH1); this mutant Smad4 lacking DNA-binding activity may quench the transcriptional activity of the complex via the MH2 domain. It also appeared that a component of the Smad4 complex, interacting through the MH2 domain, is critical for inhibition of myogenic differentiation in response to BMPs. In the present study, we identified E4F1 as one of the components of the Smad4 complex in the nucleus, interacting through the MH2 domain. E4F1 is a zinc finger DNA-binding protein, identified as a cellular target of viral oncoproteins and shown to regulate the cell cycle (40–42). Our findings indicated that overexpression of E4F1 inhibited myogenic differentiation cooperatively with Smad4. Moreover, RNAi knockdown of E4F1 prevented the inhibition of myogenic differentiation by BMP signaling. Although E4F1 was recently shown to act as a ubiquitin E3 ligase of p53 (43), our findings indicated that deletion of the ubiquitin E3 ligase domain from E4F1 still allowed inhibition of myogenic differentiation. However, all of the zinc finger structures of E4F1 seemed to be important for this inhibitory activity. Taken together, these findings suggest that Smad4, which undergoes nuclear translocation in response to BMP stimulation, may interact with E4F1 in the nucleus to suppress myogenic differentiation as a transcription factor, independent of its ubiquitin E3 ligase activity. Recently, it was reported that Smad4 regulates the processing of pri-microRNA into mature microRNA in response to BMP-2 treatment (44). The direct target gene(s) of the complex still needs to be identified. It is interesting to note that loss-of-function mutations of p53 and Smad4 were identified in some tumors, suggesting that mutations in the Smad4-E4F1-p53 axis might play a role in tumorigenesis (45, 46).
We found that E4F1 stimulated the expression of Id1–3 in the presence of Smad4. Id proteins inhibit myogenesis and are targets of BMP signaling. Recently, insufficient skeletal muscle repair was reported in Id1+/−Id3−/− mice after muscle injury (47). BMP signaling may also up-regulate Id expression in healing muscle tissue (47). Because expression of Ids leads to cell cycle progression, the E4F1-induced Ids may suppress myogenic differentiation and maintain myoblast proliferation. Further studies are needed to elucidate the physiological roles of Smads and E4F1 in muscle development and regeneration in vivo.
In the present study, we obtained an unexpected finding related to Smads. BMP-induced osteoblastic differentiation was not completely blocked in the Smad4-deleted MEFs. There are some possible explanations for this finding: 1) undetectable levels of Smad4 still remained in the MEFs expressing Cre recombinase, 2) an alternative pathway, including a novel Co-Smad, transduced BMP signaling, or 3) Smad4 is not essential for the osteoblastic differentiation induced by BMPs. Recently, evidence has been presented that bone and cartilage tissues were formed during development in the absence of functional Smad4 in mice, although such mice exhibited abnormalities (48). Deletion of Smad4 in mouse mature osteoblasts using a Cre-loxP system significantly reduced bone volume and osteoblast function in vivo, but they still had bone tissues and osteoblasts (48). Further study will be required to elucidate the roles of Smad4 in bone metabolism.
In conclusion, we found that the Smad-dependent pathway regulates both the inhibition of myogenic differentiation and the induction of osteoblastic differentiation induced by BMPs. The introduction of negative charges at the carboxyl terminus of Smad1 may play an important role in the induction of osteoblast differentiation in response to BMPs. In contrast, nuclear Smad4, rather than R-Smad, and E4F1, a novel partner of nuclear Smad4, are responsible for the inhibition of myogenic differentiation by BMPs.
Acknowledgments
We thank Drs. Naoyuki Takahashi, Tatsuo Suda, Ken Yagi, and Masami Muramatsu and members of the Division of Pathophysiology, Research Center for Genomic Medicine, Saitama Medical University, and the Department of Biochemistry, School of Dentistry, Showa University, for valuable comments, discussions, and encouragement. We are grateful to Drs. J. A. Langer, T. Komori, K. Kawakami, and C. Deng for kindly providing constructs, reagents, and mice. We thank Kiyoshiro Imawano for encouragement.
This work was supported in part by Health and Labor Sciences Research Grants for Research on Measures for Intractable Research from the Ministry of Health, Labor and Welfare of Japan (to T. K.), grant-in-aids from Saitama Medical University Internal Grants (to T. F. and T. K.), grant-in-aids from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T. F. and T. K.), a grant-in-aid for “Support Project of Strategic Research Center in Private Universities” from the Ministry of Education, Culture, Sports, Science and Technology of Japan to Saitama Medical University Research Center for Genomic Medicine (to T. K.), a grant-in-aid from the Sankyo Foundation of Life Science (to T. K.), a grant-in-aid from the Kawano Masanori Memorial Foundation for Promotion of Pediatrics (to T. K.), a grant-in-aid from the Novo Nordisk Award for Growth and Development (to T. K.), a grant-in-aid from Japan Intractable Diseases Research Foundation (to T. F.), and a grant-in-aid from the Takeda Science Foundation (to T. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
J. Nojima, T. Takada, and T. Katagiri, unpublished data.
- BMP
- bone morphogenetic protein
- ALP
- alkaline phosphatase
- BMPR-IA
- bone morphogenetic protein receptor type IA
- MHC
- myosin heavy chain
- TGF-β
- transforming growth factor-β
- ChIP
- chromatin immunoprecipitation
- NLS
- nuclear localization signal
- EGFP
- enhanced green fluorescent protein
- MEF
- mouse embryonic fibroblast
- RNAi
- RNA interference
- siRNA
- small interfering RNA
- R-Smad
- receptor-regulated Smad
- luc
- luciferase.
REFERENCES
- 1.Katagiri T., Suda T., Miyazono K. (2008) The Bone Morphogenetic Proteins, pp. 121–149, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 2.Urist M. R. (1965) Science 150, 893–899 [DOI] [PubMed] [Google Scholar]
- 3.Wozney J. M., Rosen V., Celeste A. J., Mitsock L. M., Whitters M. J., Kriz R. W., Hewick R. M., Wang E. A. (1988) Science 242, 1528–1534 [DOI] [PubMed] [Google Scholar]
- 4.Wozney J. M., Rosen V. (1998) Clin. Orthop. Relat. Res. 346, 26–37 [PubMed] [Google Scholar]
- 5.Sampath T. K., Muthukumaran N., Reddi A. H. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7109–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salminen A., Braun T., Buchberger A., Jürs S., Winter B., Arnold H. H. (1991) J. Cell Biol. 115, 905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida S., Fujisawa-Sehara A., Taki T., Arai K., Nabeshima Y. (1996) J. Cell Biol. 132, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A., Suda T. (1994) J. Cell Biol. 127, 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massagué J., Cheifetz S., Endo T., Nadal-Ginard B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8206–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D., Black B. L., Derynck R. (2001) Genes Dev. 15, 2950–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyazono K., Maeda S., Imamura T. (2005) Cytokine Growth Factor Rev. 16, 251–263 [DOI] [PubMed] [Google Scholar]
- 12.Wan M., Cao X. (2005) Biochem. Biophys. Res. Commun. 328, 651–657 [DOI] [PubMed] [Google Scholar]
- 13.Katagiri T., Imada M., Yanai T., Suda T., Takahashi N., Kamijo R. (2002) Genes Cells 7, 949–960 [DOI] [PubMed] [Google Scholar]
- 14.Liu C. J., Ding B., Wang H., Lengyel P. (2002) Mol. Cell. Biol. 22, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Rovira T., Chalaux E., Massagué J., Rosa J. L., Ventura F. (2002) J. Biol. Chem. 277, 3176–3185 [DOI] [PubMed] [Google Scholar]
- 16.Shore E. M., Xu M., Feldman G. J., Fenstermacher D. A., Cho T. J., Choi I. H., Connor J. M., Delai P., Glaser D. L., LeMerrer M., Morhart R., Rogers J. G., Smith R., Triffitt J. T., Urtizberea J. A., Zasloff M., Brown M. A., Kaplan F. S. (2006) Nat. Genet. 38, 525–527 [DOI] [PubMed] [Google Scholar]
- 17.Fukuda T., Kohda M., Kanomata K., Nojima J., Nakamura A., Kamizono J., Noguchi Y., Iwakiri K., Kondo T., Kurose J., Endo K., Awakura T., Fukushi J., Nakashima Y., Chiyonobu T., Kawara A., Nishida Y., Wada I., Akita M., Komori T., Nakayama K., Nanba A., Maruki Y., Yoda T., Tomoda H., Yu P. B., Shore E. M., Kaplan F. S., Miyazono K., Matsuoka M., Ikebuchi K., Ohtake A., Oda H., Jimi E., Owan I., Okazaki Y., Katagiri T. (2009) J. Biol. Chem. 284, 7149–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama Z., Yoshida C. A., Furuichi T., Amizuka N., Ito M., Fukuyama R., Miyazaki T., Kitaura H., Nakamura K., Fujita T., Kanatani N., Moriishi T., Yamana K., Liu W., Kawaguchi H., Nakamura K., Komori T. (2007) Dev. Dyn. 236, 1876–1890 [DOI] [PubMed] [Google Scholar]
- 19.Goldman L. A., Cutrone E. C., Kotenko S. V., Krause C. D., Langer J. A. (1996) BioTechniques 21, 1013–1015 [DOI] [PubMed] [Google Scholar]
- 20.Katagiri T., Akiyama S., Namiki M., Komaki M., Yamaguchi A., Rosen V., Wozney J. M., Fujisawa-Sehara A., Suda T. (1997) Exp. Cell Res. 230, 342–351 [DOI] [PubMed] [Google Scholar]
- 21.Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008) Nat. Chem. Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X., Li C., Herrera P. L., Deng C. X. (2002) Genesis 32, 80–81 [DOI] [PubMed] [Google Scholar]
- 23.Kanegae Y., Lee G., Sato Y., Tanaka M., Nakai M., Sakaki T., Sugano S., Saito I. (1995) Nucleic Acids Res. 23, 3816–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodaira K., Imada M., Goto M., Tomoyasu A., Fukuda T., Kamijo R., Suda T., Higashio K., Katagiri T. (2006) Biochem. Biophys. Res. Commun. 345, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 25.Ohto H., Kamada S., Tago K., Tominaga S. I., Ozaki H., Sato S., Kawakami K. (1999) Mol. Cell. Biol. 19, 6815–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki A., Thies R. S., Yamaji N., Song J. J., Wozney J. M., Murakami K., Ueno N. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10255–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao K. R., Elinson R. P. (1988) Dev. Biol. 127, 64–77 [DOI] [PubMed] [Google Scholar]
- 28.Hattori H., Ishihara M., Fukuda T., Suda T., Katagiri T. (2006) Biochem. Biophys. Res. Commun. 343, 1118–1123 [DOI] [PubMed] [Google Scholar]
- 29.Zhao B., Katagiri T., Toyoda H., Takada T., Yanai T., Fukuda T., Chung U. I., Koike T., Takaoka K., Kamijo R. (2006) J. Biol. Chem. 281, 23246–23253 [DOI] [PubMed] [Google Scholar]
- 30.Hanai J., Chen L. F., Kanno T., Ohtani-Fujita N., Kim W. Y., Guo W. H., Imamura T., Ishidou Y., Fukuchi M., Shi M. J., Stavnezer J., Kawabata M., Miyazono K., Ito Y. (1999) J. Biol. Chem. 274, 31577–31582 [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y. W., Yasui N., Ito K., Huang G., Fujii M., Hanai J., Nogami H., Ochi T., Miyazono K., Ito Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10549–10554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito Y., Miyazono K. (2003) Curr. Opin. Genet. Dev. 13, 43–47 [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H., Fukunishi Y., Kagawa I., Saito R., Oda H., Endo T., Kondo S., Bono H., Okazaki Y., Hayashizaki Y. (2001) Genome Res. 11, 1758–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pownall M. E., Gustafsson M. K., Emerson C. P., Jr. (2002) Annu. Rev. Cell Dev. Biol. 18, 747–783 [DOI] [PubMed] [Google Scholar]
- 35.Akiyama S., Katagiri T., Namiki M., Yamaji N., Yamamoto N., Miyama K., Shibuya H., Ueno N., Wozney J. M., Suda T. (1997) Exp. Cell Res. 235, 362–369 [DOI] [PubMed] [Google Scholar]
- 36.Fujii M., Takeda K., Imamura T., Aoki H., Sampath T. K., Enomoto S., Kawabata M., Kato M., Ichijo H., Miyazono K. (1999) Mol. Biol. Cell 10, 3801–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McPherron A. C., Lawler A. M., Lee S. J. (1997) Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- 38.He L., Vichev K., Macharia R., Huang R., Christ B., Patel K., Amthor H. (2005) Anat. Embryol. 209, 401–407 [DOI] [PubMed] [Google Scholar]
- 39.Massagué J., Wotton D. (2000) EMBO J. 19, 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Cam L., Lacroix M., Ciemerych M. A., Sardet C., Sicinski P. (2004) Mol. Cell. Biol. 24, 6467–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney R. J., Rothammer K., Fernandes E. R. (1998) Nucleic Acids Res. 26, 1681–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee K. A., Green M. R. (1987) EMBO J. 6, 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Cam L., Linares L. K., Paul C., Julien E., Lacroix M., Hatchi E., Triboulet R., Bossis G., Shmueli A., Rodriguez M. S., Coux O., Sardet C. (2006) Cell 127, 775–788 [DOI] [PubMed] [Google Scholar]
- 44.Sato M. M., Nashimoto M., Katagiri T., Yawaka Y., Tamura M. (2009) Biochem. Biophys. Res. Commun. 383, 125–129 [DOI] [PubMed] [Google Scholar]
- 45.Levine A. J., Momand J., Finlay C. A. (1991) Nature 351, 453–456 [DOI] [PubMed] [Google Scholar]
- 46.Hahn S. A., Schutte M., Hoque A. T., Moskaluk C. A., da Costa L. T., Rozenblum E., Weinstein C. L., Fischer A., Yeo C. J., Hruban R. H., Kern S. E. (1996) Science 271, 350–353 [DOI] [PubMed] [Google Scholar]
- 47.Clever J. L., Sakai Y., Wang R. A., Schneider D. R. (2010) Am. J. Physiol. Cell Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan X., Weng T., Zhang J., Wang J., Li W., Wan H., Lan Y., Cheng X., Hou N., Liu H., Ding J., Lin F., Yang R., Gao X., Chen D., Yang X. (2007) J. Cell Sci. 120, 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]