Abstract
Because the photosynthetic apparatus contains a massive amount of nitrogen in plants, the regulation of its development by sugar signals is important to the maintenance of the carbon-nitrogen balance. In this study we isolated an Arabidopsis mutant (sicy-192) whose cotyledon greening was inhibited by treatments with sugars such as sucrose, glucose, and fructose. In the mutant, the gene encoding plastidic alkaline/neutral invertase (INV-E) was point-mutated at codon 294, with Tyr substituted for Cys (C294Y). Interestingly, the greening of cotyledons in the knock-out INV-E lines was not inhibited by treatment with the sugars. In addition, the knock-out INV-E lines expressing an INV-E:C294Y or INV-E:C294A gene had the same phenotype as sicy-192 mutants, whereas the lines expressing a wild-type INV-E gene had the same phenotype as wild-type plants. A recombinant INV-E:C294Y protein had the same enzymatic activity as a recombinant INV-E protein, suggesting that the Cys-294 residue of INV-E is important for its functions in the chloroplasts. On treatment with sucrose, the expression of photosynthesis-related genes was weaker in seedlings of mutant plants than wild-type seedlings, whereas the activity of nitrate reductase was stronger in the mutant plants than wild-type plants. These findings suggest that Cys-294 of INV-E is associated with the development of the photosynthetic apparatus and the assimilation of nitrogen in Arabidopsis seedlings to control the ratio of sucrose content to hexose content.
Keywords: Carbohydrate/Metabolism, Carbohydrate/Plant, Metabolism/Nitrogen, Photosynthesis, Development, C/N Balance, Invertase
Introduction
Feedback regulation of the development of the photosynthetic apparatus by sugar signals is important to the maintenance of the carbon-nitrogen balance in plants because the apparatus contains half of the nitrogen in plant leaves (1–4). Many researchers have isolated various sugar-hypersensitive mutants of Arabidopsis (5–10). High levels of glucose or sucrose in growth media have been found to prevent the greening of Arabidopsis seedlings (11). Glo (glucose oversensitive) and gss (glucose supersensitive) mutants ceased growing in 222 or 56 mm glucose, whereas sss (sucrose supersensitive) mutant grew in 350 mm sucrose (9, 13). However, why these mutants are hypersensitive to sugars has yet to be elucidated. In contrast, various mutants insensitive to high levels of sugars have been characterized (11, 14, 15). Gin2 mutant plants had a disturbed gene (AtHXK1) encoding hexokinase 1 as a glucose sensor (14, 16–18). The signal transduction pathway via AtHXK1 is associated with physiological functions such as photosynthesis related-gene expression, hormone signaling, senescence, and nitrogen uptake (17, 19, 20). The protein encoded by ABI4 is a regulator to control the expression of the sugar-responsive gene and plays an important role in the development and function of chloroplasts (15, 21–23).
The activity to take up nitrate in Arabidopsis roots is closely linked to levels of sugars as photosynthates (19). The absorbed nitrogen is assimilated by nitrate reductase (NR),3 a key enzyme in the assimilation process, and the transcription of NR is induced by treatment of Arabidopsis with sucrose (24, 25). In many cases, however, these experiments have been carried out under severe carbon starvation conditions. Stitt et al. (26) have indicated that a change in sugar levels is not involved in the regulation of NR expression in tobacco plants under normal growth conditions. Additionally, it remains unclear whether not only sugar but also 2-oxoglutarate and glutamine play a major role in the regulation of NR activity in tobacco (26). Therefore, there is no established theory about a key component regulating the balance between carbon metabolism and nitrogen assimilation in plants.
In this study we isolated an Arabidopsis mutant whose cotyledon greening was inhibited by treatment with a sugar such as sucrose. Map-based cloning revealed that the phenotype of the mutant is caused by the point mutation of an alkaline/neutral (A/N) invertase at codon 294, with a substitution of Tyr for Cys. We analyzed the expression of photosynthesis-related and nitrogen assimilation-related genes and the NR activity in the mutant and discussed the effect of a disturbance of sucrose metabolism on the carbon-nitrogen balance in plastids.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Activation-tagged lines of Arabidopsis thaliana ecotype Colombia were constructed as described previously (27). Sicy-192 mutants were isolated from the T2 lines and crossed with the wild type to remove the tag T-DNA. A. thaliana ecotype Landsberg erecta (Ler), SALK_138953 (KO-INV-E) (28), SALK_015782 (KD-AtHXK1), and abi4 mutant lines were obtained from Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). Sterilized Arabidopsis seeds were placed on 0.8% agar containing Murashige and Skoog's (MS) inorganic salt with or without sugar, incubated in a cold room (4 °C) for 3 days, and moved to a growth chamber (14 h of light, 25 °C/10 h of dark, 22 °C).
Map-based Cloning of sicy-192 Mutant
Ler was crossed with sicy-192 mutants, and F2 progenies showing the yellow cotyledon phenotype on the SUC+ medium were selected. Cleaved amplified polymorphic sequence markers used in the initial mapping analysis were selected from Marker Tracker. Several new markers were developed according to the Monsanto Arabidopsis Polymorphism and Ler Sequence Collections (29), and the two markers closest to the causative gene were KUCAPS5–742 and KUCAPS5–748. Information on these markers is available at The Arabidopsis Information Resource.
Generation of Transgenic Arabidopsis
The genomic DNA fragment containing the INV-E gene region from its promoter (1450 bp from the ATG start site) to its terminator (328 bp from the TGA stop site) was amplified by PCR from genomic DNA of wild-type or sicy-192 mutants using the primers 5′-GGGGAATTCGTTGAGAGAAGGAGACATGA-3′ (the EcoRI site is underlined) and 5′-ACCACCACTGATGGGATCCAGT-3′ (the BamHI site is underlined). After digestion with EcoRI and BamHI, the PCR fragment was inserted between the EcoRI and BamHI cloning sites of the vector pUC19. To convert Cys-294 to Ala, the plasmid was used as a template for PCR with the phosphorylated primers 5′-CTCGCTGATGGTTTCGATATG-3′ and 5′-AGCCAGCTTTAAGATCATCT-3′ by KOD-Plus polymerase (Toyobo), and the PCR product was self-ligated. These plasmids were digested with EcoRI and BamHI, blunted with T4 DNA polymerase, ligated with EcoRI-NotI-BamHI-Adapter (Takara), phosphorylated by T4 polynucleotide kinase, and then inserted between the EcoRI cloning sites of pDH123. The constructs were transferred into Agrobacterium tumefaciens and transformed to KO-INV-E as described by Clough and Bent (30).
Preparation of Recombinant INV-E
The open reading frame fragment of INV-E or INV-E:C294Y without the coding region of the signal peptide was amplified with KOD-Plus polymerase from the cDNA using 5′-GGGGGTACCTGTACAAACTCGCATGAGGTC-3′ (the KpnI site is underlined) and the phosphorylated primer 5′-TCATACAATAAATGGTTGTTGAGC-3′. The PCR fragment was digested with KpnI and ligated to the KpnI and blunted PstI sites of a pCold2 (Takara). The His-tagged INV-E or His-tagged INV-E:C294Y was expressed in BL21 (DE3) pLysS and purified with TALON Metal Affinity Resin (Clontech) under native conditions, according to the manufacturer's instructions.
Quantitative PCR Analysis
Total RNA was extracted from whole seedlings grown for 5 days using QuickGene RNA cultured cell kit S (Fujifilm). To eliminate any DNA, the RNA was treated with DNase I (Takara) and converted into cDNA using the ReverTra Ace (Toyobo) with the oligo(dT)20 primer. Primer pairs for quantitative reverse transcription-PCR were designed using Primer Express software (Applied Biosystems), and the gene-specific primers are shown in supplemental Table 1. Quantitative PCR was performed with an Applied Biosystems 7300 Real Time PCR system using the SYBR Premix Ex Taq (Takara). The transcript of eIF4A-1 was used as an internal standard in all experiments.
Enzyme Assay
For the invertase assay, plants grown for 5 days were ground to a fine powder in liquid N2, homogenized with a mortar and pestle in the presence of 200 mm Hepes/KOH, pH 7.5, 1 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride, and centrifuged at 20,000 × g for 10 min at 4 °C. The supernatant was desalted with Sephadex G-50 and used to assay A/N-Inv by the method reported by Vargas et al. (28). For the NR assay, plants grown for 5 days were ground with a mortar and pestle in the presence of 50 mm Hepes/KOH, pH 7.5, 25 mm NaF, 5 mm EDTA, 0.014% 2-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride and centrifuged at 20,000 × g for 10 min at 4 °C. NR activity of the supernatants was measured as described by Gibon et al. (31), but at 30 °C. Protein concentrations were determined by the method described by Bradford (32) using bovine serum albumin as the standard.
SDS-PAGE and Immunoblotting
Proteins from the supernatants for the NR assay were separated by SDS-PAGE on 12% polyacrylamide gels and stained with Coomassie Brilliant Blue or blotted onto a polyvinylidene difluoride membrane. The immunoblot analysis was carried out as described previously (33). The membranes were probed with antibodies raised against sedoheptulose-1,7-bisphosphatase (34), against Rubisco large subunit (rbcL), or against glutamine synthetase (GS) (35). The proteins recognized by the primary antibody were revealed with a goat anti-mouse or anti-rabbit IgG coupled to horseradish peroxidase and detected by Chemi-Lumi One L (Nacalai Tesque).
Measurement of Intermediates
Intermediates were measured in the leaves of 5-day-old plants. Levels of sucrose and hexose were determined according to Tamoi et al. (36). The ammonium content of the seedlings was assayed by the method of Bräutigam et al. (37). Glutamic acid and glutamine levels were determined with a l-glutamic acid measuring kit (Roche Applied Science) according to the manufacturer's instructions with some modifications.
RESULTS
Isolation of the sicy-192 Mutant and Cloning of the Causative Gene
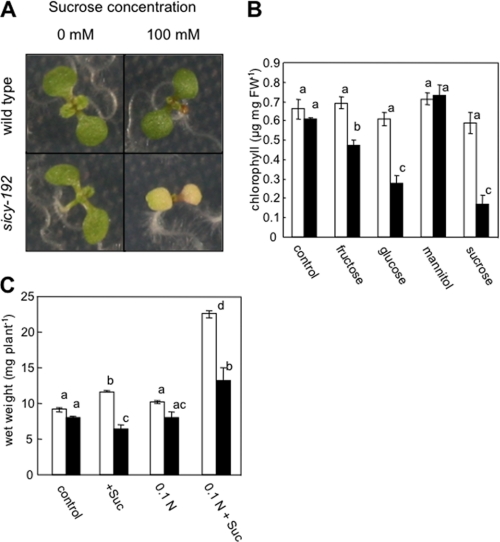
From among ∼10,000 Arabidopsis activation-tagged lines (27), we isolated a mutant line in which the greening of cotyledons was inhibited in medium (SUC+ medium) containing 100 mm sucrose (Fig. 1A). The F2 progeny of a cross between the wild-type and the mutant line had the phenotype without the T-DNA (data not shown), indicating that the insert is not related to the phenotype. The progeny from the mutant without the T-DNA was named sicy (sugar-inducible cotyledon yellow)-192 and used in subsequent experiments. The greening of cotyledons of sicy-192 mutant plants was inhibited by treatment not only with sucrose but also with glucose or fructose (Fig. 1B). But it was not inhibited by treatment with mannitol, indicating that the inhibition is not due to osmotic stress caused by the sugars. In the medium with different C/N balances, the growth of sicy-192 mutants was significantly smaller than that of wild-type plants (Fig. 1C). In particular, in low (6 mm) nitrogen MS medium with 100 mm sucrose, the wet weight of sicy-192 mutants was 41% lower than that of wild-type plants.
FIGURE 1.
Phenotypes of the wild-type and sicy-192 mutant plants. A, shown are visual phenotypes of the wild-type and sicy-192 mutant grown for 7 days in MS medium with or without 100 mm sucrose. B, shown is the chlorophyll content of the wild-type (open bars) and sicy-192 mutant (solid bars) plants grown for 7 days in MS medium with or without 100 mm of various sugars. C, growth of wild-type (open bars) and sicy-192 mutant (solid bars) plants with various carbon-nitrogen balances is shown. Shown is the wet weight of shoots of the plants grown for 2 weeks in MS medium without sucrose (control), MS medium with sucrose (+Suc), 1/10 n (6 mm nitrogen) MS medium without sucrose (0.1 n), or 1/10 n (6 mm nitrogen) MS medium with sucrose (0.1 n+ Suc). Each value is the mean and S.E. for three replicates. Asterisks indicate that mean values were significantly different compared with those in wild-type plants when analyzed by Student's t test (*, p < 0.05; **, p < 0.01).
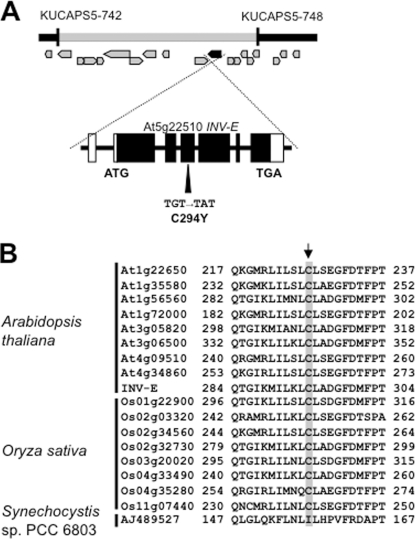
The F1 plants of the cross between sicy-192 and the wild type displayed a normal phenotype, and the F2 progenies showed an ∼3:1 (893:269) rate of segregation (wild type:mutant phenotype) on the SUC+ medium. These findings confirmed that the phenotype of the mutant is caused by a single recessive gene. Map-based cloning indicated the gene causative of the phenotype to lie between the markers KUCAPS5–742 and KUCAPS5–748 on chromosome V and to encode a plastidic A/N invertase INV-E, which had one point mutation in exon 4 at codon 294, a substitution of Tyr for Cys (Fig. 2A). The Cys residue of INV-E is conserved in all A/N invertase proteins in plants including Arabidopsis and rice (Fig. 2B), suggesting that it has an important role in this family. However, some putative A/N invertases in cyanobacteria, for example Synechocystis sp. PCC 6803, do not contain the Cys residue (Fig. 2B).
FIGURE 2.
Map-based cloning of the causative gene of sicy-192 mutant plants and alignment of A/N invertase proteins. A, the locus of the causative gene was mapped to between the markers KUCAPS5–742 and KUCAPS5–748 on chromosome 5. The start codon (ATG) and stop codon (TGA) are indicated. The G to A nucleotide substitution in exon 4 converted Cys to Tyr. B, alignments of A/N invertase proteins of Arabidopsis, rice, and Synechocystis sp. PCC 6803 are shown. An arrow indicates the mutated residue in sicy-192.
Effect of the Mutation of Cys-294 in INV-E on the Greening of Arabidopsis
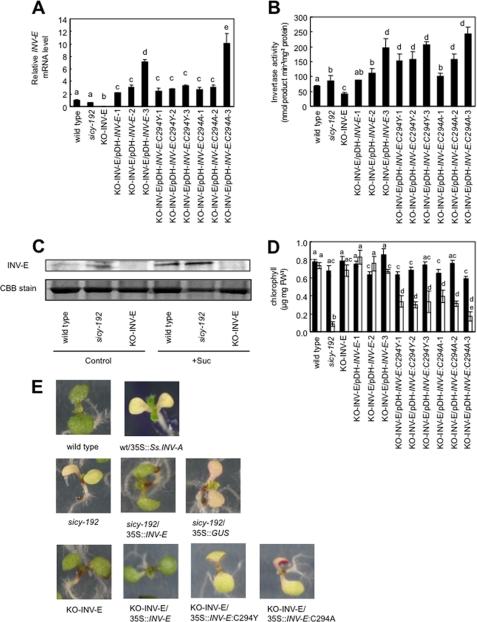
The transcript of INV-E was not detected in the INV-E-knock-out (KO-INV-E) lines (Fig. 3A). The transcript level of INV-E in sicy-192 mutants was almost the same as that in wild-type plants (Fig. 3A). The specific activity of A/N invertase in KO-INV-E lines was also lower than that in wild-type plants. However, in sicy-192 mutants the activity was slightly higher than that in wild-type plants (Fig. 3B). Moreover, the protein levels of INV-E in the mutants were higher than those in the wild-type plants (Fig. 3C). The chlorophyll contents of the KO-INV-E lines in the SUC+ medium were similar to those of the wild-type lines (Fig. 3D). These results suggest that the inhibition of greening of sicy-192 mutant plants is not due to repression of INV-E function. We introduced the expression vector pDH-INV-E:C294Y which retained the mutated INV-E gene region of sicy-192 from the promoter to the terminator into the KO-INV-E lines. The lines obtained expressed the mutated INV-E transcript, resulting in stronger activity of A/N invertase than in the wild-type lines (Fig. 3, A and B). Interestingly, the transgenic lines showed inhibition of greening in the SUC+ medium as did the sicy-192 mutant plants (Fig. 3, D and E). No inhibition of greening was detected in the absence of sucrose (SUC− medium) (Fig. 3D). Transgenic lines into which were introduced the pDH-INV-E vector, which retained the INV-E gene region from wild-type plants, showed almost the same transcript level of INV-E and A/N invertase activity as the lines into which pDH-INV-E:C294Y was introduced. They exhibited no inhibition of greening in the SUC+ medium (Fig. 3, D and E). These findings clearly indicated that the mutation of INV-E in exon 4 at codon 294, a substitution of Tyr for Cys, causes the inhibition of greening of sicy-192 mutants. Transgenic lines into which were introduced a pDH-INV-E:C294A vector, which was modified by replacing the codon for Cys-294 with that for Ala, also showed the same phenotype as sicy-192 mutants (Fig. 3, D and E). Moreover, transgenic lines overexpressing the cyanobacterial invertase gene (INV-A: AJ489527) showed inhibition of greening in the SUC+ medium similar to the sicy-192 mutants (Fig. 3E).
FIGURE 3.
INV-E expression level and chlorophyll content in seedlings of KO-INV-E lines expressing mutated INV-E. A, transcript levels of INV-E are shown. These lines were grown in SUC+ medium for 5 days. The values were normalized to the eIF4A-1 expression of each sample and are shown in relation to the RNA level measured in wild-type plants. Each value is the mean and S.E. of three replicates. B, shown is A/N invertase activity in crude extracts of the plants grown in SUC+ medium for 5 days. Each value is the mean and S.E. for three replicates. Bars marked with a different letter were significantly different (p < 0.05) when analyzed by the Student-Newman-Keuls analysis of variance. C, the accumulated levels of INV-E in seedlings of the wild-type, sicy-192 mutants, and KO-INV-E lines are shown. Seedlings were grown in the SUC+ medium (Suc) or SUC− medium (control) for 5 days, and their soluble proteins (10 μg lane−1) were separated by 12% SDS-PAGE and analyzed by immunoblotting using antibody against INV-E. Molecular masses of marker proteins are indicated on the left. CBB, Coomassie Brilliant Blue. D, chlorophyll content of the plants grown in the SUC+ medium (solid bars) or SUC− medium (open bars) is shown. Each value is the mean and S.E. for three replicates. Asterisks indicate that mean values were significantly different compared with those for the control treatment when analyzed by Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). FW, fresh weight. E, shown is the phenotype of 5-day-old seedlings in 100 mm sucrose-containing medium.
Effects of the Mutation of INV-E on Enzymatic Properties
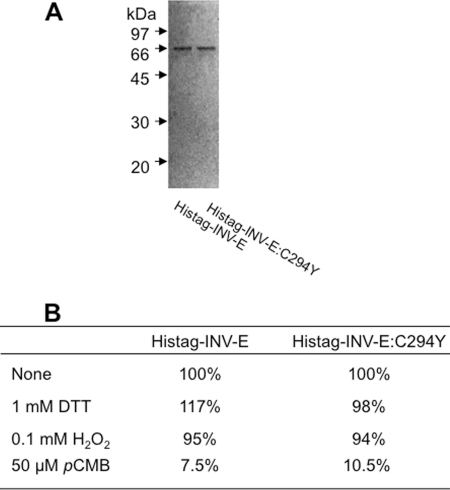
To assess effects of the mutation of INV-E in sicy-192 mutants on the enzymatic properties of INV-E, we characterized the recombinant His-tagged INV-E:C294Y and recombinant His-tagged INV-E. Each INV-E cDNA fragment missing the transit peptide region was PCR-amplified, cloned downstream of the His-tag sequence, and expressed in Escherichia coli. Both recombinant His-tagged proteins were purified (Fig. 4A) and then used for the A/N invertase assay. The Vmax values of the recombinant INV-E and the recombinant INV-E:C294Y proteins for sucrose were calculated to be 40.1 ± 2.5 and 50.0 ± 3.2 mmol of product min−1 mg−1 of protein, respectively, and their Km values for sucrose were 15.5 ± 2.1 and 17.5 ± 2.9 mm, respectively. The degree of inhibition of INV-E:C294Y by each thiol-modifying reagent was almost the same as that of INV-E (Fig. 4B). Neither INV-E nor INV-E:C294Y could hydrolyze other disaccharides (maltose, lactose, trehalose, cellobiose, and galactinol) (data not shown). Thus, there was no remarkable difference in kinetic parameters between the two recombinant proteins.
FIGURE 4.
Enzymatic properties of recombinant INV-E and INV-E:C294Y proteins. A, purified His-tagged INV-E and His-tagged INV-E:C294Y proteins (0.5 μg of protein lane−1) were separated on a12% SDS-PAGE gel and detected by silver staining. B, shown is the effect of thiol-modifying reagents on the invertase activity of recombinant INV-E and INV-E:C294Y. The specific activities of the recombinant INV-E and INV-E:C294Y proteins without the reagent (none) were 32.8 and 40.8 mmol of product min−1 mg−1 protein, respectively, and were normalized (= 100%). DTT, dithiothreitol; pCMB, p-chloromercuribenzoate.
AtHXK1 and ABI4 Are Not Essential for the Greening Inhibition of sicy-192
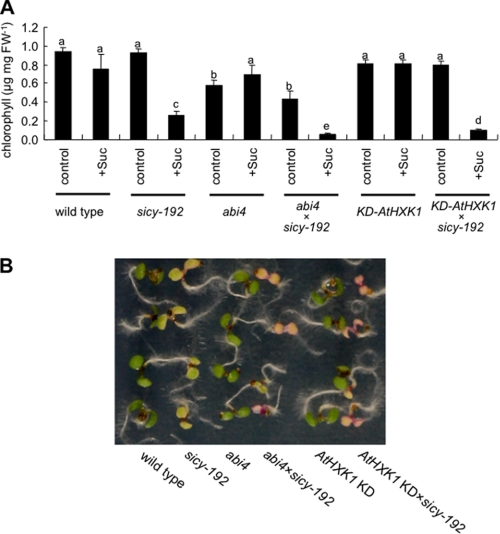
According to the data described above, it is assumed that sicy-192 mutants are associated with a disturbance of signal transduction between sugar metabolism and chlorophyll biosynthesis. It has been reported that gin6 (ABI4) and gin2 (AtHXK1) are insensitive to high concentrations of glucose (16, 21). AtHXK1 and ABI4 are known to be involved in the inhibition of photosynthesis-related gene expression by high levels of sugar (17, 21). To identify whether AtHXK1 or ABI4 is essential for the inhibition of greening of sicy-192 mutants on treatment with sugar, we generated double mutants harboring mutations in INV-E:C294Y and AtHXK1 or ABI4 and analyzed their phenotypes. In the AtHXK1 knockdown line (KD-AtHXK1), the expression of AtHXK1 mRNA in the mutants was significantly repressed (38). The ABI4 mutant expressed a frameshifted ABI4 gene that disrupted the C-terminal half of the protein (39). The greening of the double mutant lines was also inhibited in the SUC+ medium (Fig. 5). These findings indicated that the signal transduction pathways involved in AtHXK1 and ABI4 are not essential for the phenotype of sicy-192 mutants.
FIGURE 5.
A, chlorophyll contents of single and double mutant lines grown in the SUC+ medium (Suc) or SUC− medium (control) for 7 days are shown. Each value is the mean and S.E. for three replicates. Bars marked with a different letter were significantly different (p < 0.05) when analyzed by the Student-Newman-Keuls analysis of variance. B, shown is the phenotype of 5-day-old seedlings in 100 mm sucrose-containing medium. FW, fresh weight.
Photosynthetic Carbon Metabolism and Nitrate Assimilation in sicy-192 Mutants
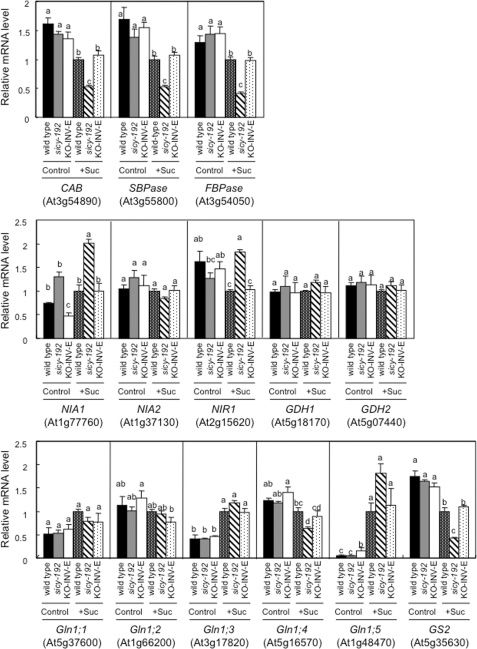
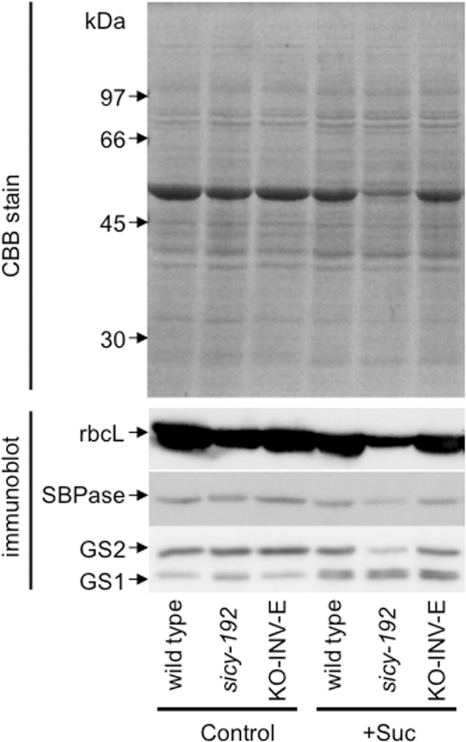
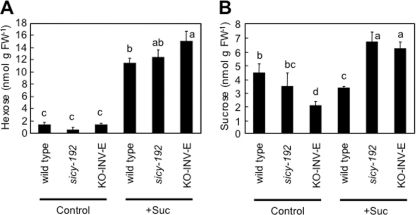
The transcript levels of the photosynthesis-related chlorophyll a/b-binding protein, sedoheptulose-1,7-bisphosphatase, and plastidic fructose-1,6-bisphosphatase genes were lower in the 5-day-old sicy-192 plants than wild-type plants in the SUC+ medium (Fig. 6). The protein levels of rbcL and sedoheptulose-1,7-bisphosphatase were also lower in the sicy-192 seedlings (Fig. 7). However, these decreases were not observed in the SUC− medium (Figs. 6 and 7). With the SUC− medium, there was no significant difference in hexose or sucrose content between the sicy-192 mutants and wild-type plants (Fig. 8). In the mutants, however, sucrose content was significantly larger than that in the wild-type plants in the SUC+ medium (Fig. 8B).
FIGURE 6.
The transcript levels of photosynthesis and nitrate assimilation-related genes in seedlings of the wild type and sicy-192 mutants and KO-INV-E lines. Seedlings were grown in the SUC+ medium (Suc) or SUC− medium (Control) for 5 days, and their RNA was analyzed by real time reverse transcription-PCR. The values were normalized to the eIF4A-1 (At3g13920) expression of each sample and are shown in relation to the RNA level measured in sucrose-treated wild-type plants. Each value is the mean and S.E. for three or four replicates. Bars marked with a different letter were significantly different (p < 0.05) when analyzed by the Student-Newman-Keuls analysis of variance. CAB, chlorophyll a/b-binding protein; SBPase, sedoheptulose-1,7- bisphosphatase; FBPase, fructose-1,6-bisphosphatase.
FIGURE 7.
The accumulated levels of rbcL, sedoheptulose-1,7-bisphosphatase (SBPase), and GSs in seedlings of the wild type and sicy-192 mutants and KO-INV-E lines. Seedlings were grown in the SUC+ medium (Suc) or SUC− medium (Control) for 5 days, and their soluble proteins (10 μg lane−1) were separated by 12% SDS-PAGE and analyzed by immunoblotting with antibody against rbcL, sedoheptulose-1,7-bisphosphatase (SBPase), or GS. Molecular masses of marker proteins are indicated on the left. CBB, Coomassie Brilliant Blue.
FIGURE 8.
Hexose (A) and sucrose (B) levels in seedlings of the wild type and sicy-192 mutants and KO-INV-E lines. The seedlings were grown in the SUC+ medium (+Suc) or SUC− medium (Control) for 5 days, and then hexose and sucrose levels in the seedlings were determined. Each value is the mean and S.E. for four replicates. Bars marked with a different letter were significantly different (p < 0.05) when analyzed by the Student-Newman-Keuls analysis of variance. FW, fresh weight.
To investigate the balance between the demand for nitrogen for the development of the photosynthetic apparatus and the supply of utilizable nitrogen to the cell by nitrate assimilation in sugar-treated sicy-192 mutant plants, the transcript levels of nitrate assimilation-related genes and the activity of NR were analyzed. In the SUC+ medium, transcript levels of the NR gene (NIA1) and nitrite reductase gene (NIR1) were higher in the sicy-192 plants than wild-type plants. But there were no changes in the transcript levels of another NR gene (NIA2) in normal or SUC+ medium (Fig. 6). It has been reported that free NR and phosphorylated NR are active, whereas phosphorylated NR in a complex with 14-3-3 protein is inactive (40). Total NR activity can be measured in the presence of EDTA, whereas only active NR can be measured in the presence of Mg2+ (40). The activities of total and active NR were higher in sicy-192 mutants than wild-type plants in the SUC+ medium (supplemental Fig. S1, A and B). In sicy-192 mutants in the SUC+ medium, the NR activity state, which accounted for the NR activity of the active types as a percentage of total NR activity, was higher than that in the wild-type plants (supplemental Fig. S1C). Increases in the transcript level, activity, and the activity state of NR were not detected in the SUC− medium (Figs. 6 and 7).
The transcript and protein levels of GS2 were suppressed in sicy-192 plants in the SUC+ medium (Figs. 6 and 7). On the other hand, the protein levels of GS1 in sicy-192 were almost the same as those in the wild-type plants in the SUC+ medium (Fig. 7). The transcript levels of two glutamate dehydrogenase genes, GDH1 and GDH2, in sicy-192 were similar to those in the wild-type plants (Fig. 6). The levels of ammonium, nitrate, glutamate, and glutamine in sicy-192 in the SUC− medium were almost the same as those in wild-type plants. However, the levels of ammonium, nitrate, and glutamate in sicy-192 mutants were higher than those in the wild-type plants in the SUC+ medium, whereas the levels of glutamine in the mutants were lower than those in the wild-type plants in the SUC+ medium (supplemental Fig. S2). These phenotypes were not observed in the KO-INV-E lines.
DISCUSSION
The photosynthetic apparatus contains a massive amount of nitrogen, and thus, its development closely depends on the distribution of nitrogen in plants (1–4). The regulatory system for the distribution of nitrogen by sugar signals appears to be important for the adjustment of primary metabolism to ensure growth, survival, and completion of the life cycle in plants (4). To uncover the regulatory system, we isolated an Arabidopsis sicy-192 mutant whose greening was hypersensitive to treatment with sugar (Fig. 1). Under high C/low N conditions, the growth of sicy-192 mutants was remarkably inhibited compared with that of wild-type plants (Fig. 1C), suggesting that the phenotypes of the sicy-192 mutants depend on the carbon to nitrogen ratio rather than carbohydrate status alone. In the mutant plants, not only the chlorophyll content but also the expression of photosynthesis-related genes was inhibited in the SUC+ medium (Figs. 6 and 7). These findings indicated that the development of the photosynthetic apparatus in sicy-192 mutants was repressed by the sugar treatment. Thus, this mutant seemed to be a suitable tool to investigate the regulation of the carbon-nitrogen balance in plants.
The causative mutation of the phenotype was due to the conversion of Cys-294 to Tyr of INV-E (Figs. 2 and 3). Although phenotypes of the sicy-192 mutants were caused by a single recessive gene, the greening of KO-INV-E was not inhibited in SUC+ medium. In addition to the finding that the KO-INV-E lines overexpressing the mutated INV-E showed the same phenotype as sicy-192 plants (Fig. 3B), transgenic lines overexpressing a cyanobacterial invertase gene (INV-A: AJ489527) lacking the Cys residue corresponding to Cys-294 in INV-E showed inhibition of greening in SUC+ medium similar to the sicy-192 mutant (Fig. 3, D and E). These results suggest that the increased A/N-INV activity in plastids causes inhibition of greening in the sicy-192 mutant. The kinetic parameters of the His-tagged INV-E:C294Y were almost the same as those of the His-tagged INV-E in vitro (Fig. 4). Although the transcriptional level of INV-E was not altered, the protein and activity levels of INV-E were higher in the mutants than in the wild-type plants (Fig. 3, B and C). These findings indicated that the mutant INV-E is more stable than the wild-type INV-E, and Cys-294 is necessary to regulate the stability of INV-E in vivo during greening. Furthermore, there is a possibility that the Cys-294 of INV-E is involved in interacting with proteins to regulate INV-E activity during germination. Lou et al. (42) have reported that the cytosolic invertase (CINV1: At1g35580) interacts with phosphatidylinositol monophosphate 5-kinase and then regulates sugar-mediated root growth of Arabidopsis. Although CINV1 had the Cys residue corresponding to Cys-294 of INV-E, it was unclear that this residue was necessary for interacting with phosphatidylinositol monophosphate 5-kinase. Moreover, Hothorn et al. (43) reported that the activity of cell-wall invertase was inhibited by interaction with an inhibitor protein.
The sicy-192 plants are recessive but gain-of-function mutants. At present, just why the sicy-192 mutant is recessive cannot be explained, although there is the following possibility. It has been reported that A/N invertase acts as homo-octamer or homo-tetramer in higher plants (41). Only when the holoenzyme consists of mutated subunits, will INV-E:C294Y become more stable. That is, the holoenzymes consisting of hetero-oligomers containing at least one subunit of INV-E may show the same stability as the native INV-E. On the other hand, the subunit of INV-A may not have the ability to assemble with the subunit of the native INV-E to form the holoenzyme, and thus, INV-A exists as a homo-oligomer in vivo.
It is well known that A/N invertase catalyzes the hydrolysis of sucrose, which is restricted to the cytosol in higher plants (44). However, Vargas et al. (28) have recently reported that INV-E was located in chloroplasts of A. thaliana, and the starch content of the KO-INV-E lines was lower than that of the wild-type plants. Moreover, Gerrits et al. (45) genetically expressed a plastid-targeted levansucrase that converts sucrose into fructan in plastids of tobacco and potato plants and observed that chloroplasts of the transgenic tobacco plants and amyloplasts of the transgenic potato accumulated high levels of fructan. Although direct evidence that sucrose exists in chloroplasts and that the sucrose transporter is located on chloroplast membranes is still lacking, these reports suggest that sucrose is metabolized in chloroplasts. The hexoses produced from sucrose by A/N invertase in plastids are phosphorylated by plastidic hexokinase and plastidic fructokinase and then used for the metabolism of carbon in plastids including starch synthesis, the oxidative pentose-phosphate pathway, and glycolysis (46–49). Accordingly, the catalytic activity of plastidic invertase affected carbon metabolism in plastids and really was needed to inhibit development of the photosynthesis apparatus in sugar-treated seedlings. The double mutants of sicy-192;KD-AtHXK1 and sicy-192;abi4 were as sugar-sensitive as the sicy-192 mutants (Fig. 5), indicating that the changes of carbon metabolism in plastids of sicy-192 mutants are independent of the alteration of the expression of photosynthesis-related genes through ABI4- and AtHXK1-independent signal transduction pathways.
One would expect repression of the development of the photosynthetic apparatus to cause a down-regulation of nitrate assimilation. To assess this, we examined the activity of NR as a key component of the nitrate assimilation pathway. Unexpectedly, the transcript level of NIA1 and the NR activity were higher in the sicy-192 mutant plants than wild-type plants in the SUC+ medium (Fig. 6 and supplemental Fig. S1). However, it has been reported that NR activity was regulated not only at the transcriptional level but also at the posttranslational level via phosphorylation and subsequent binding to 14-3-3 protein, leading to its degradation (40). In fact, the activity of NR was higher in the sicy-192 mutant plants than wild-type plants in the SUC+ medium (supplemental Fig. S1). Thus, the suppression of NR activity at the posttranslational level may be affected by the change of sucrose-hydrolytic activity via the mutation of INV-E in plastids.
To elucidate the change in metabolism downstream of nitrate assimilation in sugar-treated sicy-192 mutants, we determined the ammonium levels and the expression of ammonium-metabolizing enzymes. The seedlings of sicy-192 mutants accumulated significantly more ammonium in the SUC+ medium than did the wild-type seedlings (supplemental Fig. S2). It has been reported that high levels of ammonium were toxic to plant cells, leading to cell death (50). The level of ammonium in sicy-192 mutants was lower in the SUC+ medium than SUC− medium, indicating that the inhibition of the greening of sicy-192 mutants was not caused by the toxic effect of ammonium (supplemental Fig. S2). Plant leaves generate ammonium during photorespiration at up to 10 times the rate at which they assimilate nitrate (51), and then ammonium is mainly eliminated by GS2 located in plastids and mitochondria (52–54). Although the decrease in the rbcL level suggested that photorespiration declined in the seedlings (Fig. 7), the ammonium level was higher in sicy-192 mutants than wild-type plants in the SUC+ medium (supplemental Fig. S2). The transcripts and/or protein levels of other possible ammonium-assimilatory enzymes, GS1s (35) and glutamate dehydrogenases (12), in sicy-192 mutants were almost the same as those in the wild type (Figs. 6 and 7), indicating the accumulation of ammonium and glutamate is due to the decline in the GS2 protein level and the acceleration in the assimilation of nitrate, and thus, the decrease in glutamine might lead to down-regulation of the biosynthesis of chlorophyll and amino acids (supplemental Fig. S2).
In sicy-192 mutants, the development of the photosynthetic apparatus was inhibited in the SUC+ medium, whereas the assimilation of nitrate was enhanced. These phenotypes of sicy-192 were observed only during the greening of seedlings. Accordingly, INV-E is regulated for maintenance of the level of sucrose or its metabolites in plastids, preventing an imbalance between the supply of nitrogen to cells via nitrate assimilation and the demand for nitrogen for development of the photosynthetic apparatus. Thus, Cys-294 is necessary to regulate the activity of INV-E under conditions where the balance of carbon and nitrogen sources is disrupted.
Acknowledgments
We thank Dr. Akiho Yokota (Nara Institute of Science and Technology), Dr. Tomoyuki Yamaya (Tohoku University), and Dr. Toshiharu Shikanai (Kyoto University) for kindly providing the antibodies (Rubisco and GS) and pDH123 vector.
This work was supported by the Core Research for Evolutional Science and Technology Project of the Japan Science and Technology Agency (Grant 2005-2010; to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
- NR
- nitrate reductase
- A/N invertase
- alkaline/neutral invertase
- GS
- glutamine synthase
- rbcL
- Rubisco large subunit;
- KO-INV-E
- INV-E-knock-out
- MS
- Murashige and Skoog's.
REFERENCES
- 1.Evans J. R., Seemann J. R. (1989) Photosynthesis, pp. 183–205, Alan R. Liss, New York [Google Scholar]
- 2.Hikosaka K., Terashima I. (1996) Funct. Ecol. 10, 335–343 [Google Scholar]
- 3.Martin T., Oswald O., Graham I. A. (2002) Plant Physiol. 128, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul M. J., Pellny T. K. (2003) J. Exp. Bot. 54, 539–547 [DOI] [PubMed] [Google Scholar]
- 5.Castle L. A., Meinke D. W. (1994) Plant Cell 6, 25–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hauser M. T., Morikami A., Benfey P. N. (1995) Development 121, 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mita S., Hirano H., Nakamura K. (1997) Plant Physiol. 114, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Németh K., Salchert K., Putnoky P., Bhalerao R., Koncz-Kálmán Z., Stankovic-Stangeland B., Bakó L., Mathur J., Okrész L., Stabel S., Geigenberger P., Stitt M., Rédei G. P., Schell J., Koncz C. (1998) Genes Dev. 12, 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pego J. V., Kortstee A. J., Huijser C., Smeekens S. C. M. (2000) J. Exp. Bot. 51, 407–416 [DOI] [PubMed] [Google Scholar]
- 10.Sheen J., Zhou L., Jang J. C. (1999) Curr. Opin. Plant Biol. 2, 410–418 [DOI] [PubMed] [Google Scholar]
- 11.Rognoni S., Teng S., Arru L., Smeekens S. C. M., Perata P. (2007) Plant Growth Regul. 52, 217–228 [Google Scholar]
- 12.Skopelitis D. S., Paranychianakis N. V., Paschalidis K. A., Pliakonis E. D., Delis I. D., Yakoumakis D. I., Kouvarakis A., Papadakis A. K., Stephanou E. G., Roubelakis-Angelakis K. A. (2006) Plant Cell 18, 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolland F., Moore B., Sheen J. (2002) Plant Cell Suppl.185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolland F., Baena-Gonzalez E., Sheen J. (2006) Annu. Rev. Plant Biol. 57, 675–709 [DOI] [PubMed] [Google Scholar]
- 15.Rook F., Hadingham S. A., Li Y., Bevan M. W. (2006) Plant Cell Environ. 29, 426–434 [DOI] [PubMed] [Google Scholar]
- 16.Jang J. C., León P., Zhou L., Sheen J. (1997) Plant Cell 9, 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., Hwang I., Jones T., Sheen J. (2003) Science 300, 332–336 [DOI] [PubMed] [Google Scholar]
- 18.Cho Y. H., Yoo S. D., Sheen J. (2006) Cell 127, 579–589 [DOI] [PubMed] [Google Scholar]
- 19.Lejay L., Gansel X., Cerezo M., Tillard P., Müller C., Krapp A., von Wirén N., Daniel-Vedele F., Gojon A. (2003) Plant Cell 15, 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pourtau N., Jennings R., Pelzer E., Pallas J., Wingler A. (2006) Planta 224, 556–568 [DOI] [PubMed] [Google Scholar]
- 21.Acevedo-Hernández G. J., León P., Herrera-Estrella L. R. (2005) Plant J. 43, 506–519 [DOI] [PubMed] [Google Scholar]
- 22.Arenas-Huertero F., Arroyo A., Zhou L., Sheen J., León P. (2000) Genes Dev. 14, 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D. P. (2007) Science 316, 700–701 [DOI] [PubMed] [Google Scholar]
- 24.Cheng C. L., Acedo G. N., Cristinsin M., Conkling M. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1861–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincentz M., Moureaux T., Leydecker M. T., Vaucheret H., Caboche M. (1993) Plant J. 3, 315–324 [DOI] [PubMed] [Google Scholar]
- 26.Stitt M., Müller C., Matt P., Gibon Y., Carillo P., Morcuende R., Scheible W. R., Krapp A. (2002) J. Exp. Bot. 53, 959–970 [DOI] [PubMed] [Google Scholar]
- 27.Ogawa T., Ishikawa K., Harada K., Fukusaki E., Yoshimura K., Shigeoka S. (2009) Plant J. 57, 289–301 [DOI] [PubMed] [Google Scholar]
- 28.Vargas W. A., Pontis H. G., Salerno G. L. (2008) Planta 227, 795–807 [DOI] [PubMed] [Google Scholar]
- 29.Jander G., Norris S. R., Rounsley S. D., Bush D. F., Levin I. M., Last R. L. (2002) Plant Physiol. 129, 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 31.Gibon Y., Blaesing O. E., Hannemann J., Carillo P., Höhne M., Hendriks J. H., Palacios N., Cross J., Selbig J., Stitt M. (2004) Plant Cell 16, 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 33.Yoshimura K., Miyao K., Gaber A., Takeda T., Kanaboshi H., Miyasaka H., Shigeoka S. (2004) Plant J. 37, 21–33 [DOI] [PubMed] [Google Scholar]
- 34.Tamoi M., Nagaoka M., Shigeoka S. (2005) Biosci. Biotechnol. Biochem. 69, 848–851 [DOI] [PubMed] [Google Scholar]
- 35.Ishiyama K., Inoue E., Watanabe-Takahashi A., Obara M., Yamaya T., Takahashi H. (2004) J. Biol. Chem. 279, 16598–16605 [DOI] [PubMed] [Google Scholar]
- 36.Tamoi M., Nagaoka M., Miyagawa Y., Shigeoka S. (2006) Plant Cell Physiol. 47, 380–390 [DOI] [PubMed] [Google Scholar]
- 37.Bräutigam A., Gagneul D., Weber A. P. (2007) Anal. Biochem. 362, 151–153 [DOI] [PubMed] [Google Scholar]
- 38.Aki T., Konishi M., Kikuchi T., Fujimori T., Yoneyama T., Yanagisawa S. (2007) J. Exp. Bot. 58, 3239–3248 [DOI] [PubMed] [Google Scholar]
- 39.Finkelstein R. R., Wang M. L., Lynch T. J., Rao S., Goodman H. M. (1998) Plant Cell 10, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser W. M., Huber S. C. (2001) J. Exp Bot. 52, 1981–1989 [DOI] [PubMed] [Google Scholar]
- 41.Lee H. S., Sturm A. (1996) Plant Physiol. 112, 1513–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lou Y., Gou J. Y., Xue H. W. (2007) Plant Cell 19, 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hothorn M., Wolf S., Aloy P., Greiner S., Scheffzek K. (2004) Plant Cell 16, 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J. Q., Black C. C. (1992) Arch. Biochem. Biophys. 295, 61–69 [DOI] [PubMed] [Google Scholar]
- 45.Gerrits N., Turk S. C., van Dun K. P., Hulleman S. H., Visser R. G., Weisbeek P. J., Smeekens S. C. (2001) Plant Physiol. 125, 926–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giese J. O., Herbers K., Hoffmann M., Klösgen R. B., Sonnewald U. (2005) FEBS Lett. 579, 827–831 [DOI] [PubMed] [Google Scholar]
- 47.Damari-Weissler H., Kandel-Kfir M., Gidoni D., Mett A., Belausov E., Granot D. (2006) Planta 224, 1495–1502 [DOI] [PubMed] [Google Scholar]
- 48.Claeyssen E., Rivoal J. (2007) Phytochemistry 68, 709–731 [DOI] [PubMed] [Google Scholar]
- 49.Lunn J. E. (2007) J. Exp. Bot. 58, 35–47 [DOI] [PubMed] [Google Scholar]
- 50.Block M. D., Botterman J., Vandewiele M., Dockx J., Thoen C., Gosselé V., Movva N. R., Thompson C., Montagu M. V., Leemans J. (1987) EMBO J. 6, 2513–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallsgrove R. M., Key A. J., Lea P. J., Miflin B. J. (1983) Plant Cell Environ. 6, 301–309 [Google Scholar]
- 52.Kozaki A., Takeba G. (1996) Nature 384, 557–560 [Google Scholar]
- 53.Hodges M. (2002) J. Exp. Bot. 53, 905–916 [DOI] [PubMed] [Google Scholar]
- 54.Taira M., Valtersson U., Burkhardt B., Ludwig R. A. (2004) Plant Cell 16, 2048–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]