Abstract
Transcription factor IID (TFIID) plays a key role in regulating eukaryotic gene expression by directly binding promoters and enhancer-bound transactivator proteins. However, the precise mechanisms and outcomes of transactivator-TFIID interaction remain unclear. Transcription of yeast ribosomal protein genes requires TFIID and the DNA-binding transactivator Rap1. We have previously shown that Rap1 directly binds to the TFIID complex through interaction with its TATA-binding protein-associated factor (Taf) subunits Taf4, -5, and -12. Here, we identify and characterize the Rap1 binding domains (RBDs) of Taf4 and Taf5. These RBDs are essential for viability but dispensable for Taf-Taf interactions and TFIID stability. Cells expressing altered Rap1 binding domains exhibit conditional growth, synthetic phenotypes when expressed in combination or with altered Rap1, and are selectively defective in ribosomal protein gene transcription. Taf4 and Taf5 proteins with altered RBDs bind Rap1 with reduced affinity. We propose that collectively the Taf4, Taf5, and Taf12 subunits of TFIID represent the physical and functional targets for Rap1 interaction and, furthermore, that these interactions drive ribosomal protein gene transcription.
Keywords: Coregulator Transcription, Gene Transcription, General Transcription Factors, Transcription Coactivators, Transcription Factors, Transcription Regulation, Coactivator, Rap1, TBP-associated Factor, TFIID
Introduction
Activation of eukaryotic RNA polymerase II (pol II)2-transcribed genes requires the action of an ensemble of proteins collectively referred to as coactivators (1, 2). These large multisubunit protein assemblies stimulate mRNA gene transcription at several distinct steps as follows: either by utilizing intrinsic enzymatic activities to alter the biochemical characteristics of transcription proteins and/or chromatin; facilitating accurate formation of preinitiation complexes (PICs) near the transcription start site; stimulating pol II activity; or by serving as scaffolds for the assembly of additional coactivators on target genes. Many variations of these mechanisms have been described, but it is generally accepted that coactivators are targeted by gene-specific enhancer-bound transactivator proteins. Transactivators are minimally composed of distinct DNA binding domains (DBDs) and activation domains (ADs). Classical studies have shown that ADs enhance transcription rates by directly stimulating the formation and/or function of the PIC, which is an assortment of over 40 distinct polypeptides often termed the general transcription factors TFIIA, -B, -D, -E, -F, -H, and RNA pol II (2, 3). However, subsequent work has shown that the ADs of transactivators can also directly bind chromatin-directed coactivators to modulate transcription (1).
TFIID, a complex composed of TATA-binding protein (TBP) and 14 evolutionarily conserved Taf subunits (4–6), is an attractive candidate target for transactivator regulatory interactions within the PIC, both because TFIID promoter binding appears to be rate-limiting in vivo (7) and because transactivators have been shown to bind directly to TFIID subunits (8, 9). Thus, TFIID acts as a general transcription factor and coactivator. The structures of yeast and human TFIID in isolation and in complex with DNA, a subset of general transcription factors, or with activators have been determined using electron microscopy (EM) methods (10–17). The derived structures provide insights into the overall organization of the complex as well as possible modes of interaction of TFIID with a small sampling of possible binding partners (18, 19). However, the functional consequences of these interactions are still not understood. A particular impediment to advancing the understanding of TFIID structure-function relationships is the lack of large numbers of specifically mutated forms of TFIID subunits that can be used to dissect TFIID activity and function in vivo and in vitro.
Budding yeast ribosomal protein genes provide an excellent model for studying the mechanism of TFIID function because RPG transcription is dependent upon TFIID but not SAGA or Mediator, two coactivators implicated in the regulation of many eukaryotic mRNA-encoding genes (20–28). During logarithmic growth, the 137 yeast RPGs are vigorously transcribed and account for ∼50% of all pol II initiation events (29, 30). RPG transcription is coordinately regulated and highly sensitive to diverse environmental stimuli (31). Repressor activator protein 1 (Rap1)-binding sites are found within the majority of RPG enhancers, and Rap1 is the only transactivator absolutely required for RPG transcription, although Rap1 does collaborate with additional transcription factors such as Fhl1/Ifh1/Crf1, Sfp1, Hmo1, and the NuA4 coactivator to modulate RPG expression (21, 24, 30, 32–34). In contrast to the other factors noted above (35–37), Rap1 enhancer occupancy does not change significantly with transcription rates. Importantly, a simple chimeric gene that contains just two RPG Rap1-binding sites fused to a TFIID-independent core promoter exhibits both Rap1- and TFIID-dependent transcription in vivo (21, 24) and in vitro (24). These striking biochemical and genetic interactions have been attributed to direct physical contacts between Rap1 and the TFIID complex (24).
Thus, although the available data clearly indicate that Rap1 and TFIID interact to play essential roles in RPG transcription, exactly how Rap1 physically interacts with the TFIID coactivator and the requirement of these protein-protein interactions for RPG transcription remain unexplored. Here, we have genetically and biochemically dissected the interactions between TFIID and Rap1 and have shown the importance of Rap1-Taf RBD interactions to RPG transcription activation. We propose that collectively Taf4, Taf5, and Taf12, which colocalize within the three-dimensional structure of TFIID, provide the physical and functional interaction targets for Rap1. Finally, our studies have firmly established an important model genetic system with which to dissect the mechanisms of transactivator-TFIID interactions in the context of mRNA gene transcription activation.
MATERIALS AND METHODS
Bacterial Plasmids, Strains, Protein Purification, Yeast Strains, and Molecular Biological Analyses
Deletion mutations in TAF4, TAF5, and RAP1 were generated using PCR-based methods (24) and verified by DNA sequencing. Taf4, Taf5, Taf12, and Rap1 were expressed in Escherichia coli and purified by chromatographic methods that varied depending upon each protein (details available on request). taf4Δ, taf5Δ, taf12Δ, and rap1Δ null mutations were created in yeast strain W303a (supplemental Table S2) carrying appropriate URA3-marked covering plasmids. Centromere/autonomously replicating sequence plasmids containing TAF or RAP1 regulatory sequences upstream of wild type (WT) or mutated open reading frames of test genes (all included HA tag and nuclear localization signal) were then introduced, and the resulting pseudodiploid strains were subjected to plasmid shuffle (5). RBD-targeted Ts+ alleles of TAF4 and TAF5 were generated by error-prone PCR, introduced into shuffling strains, and scored by plasmid shuffle for the ability to support growth at various temperatures. All mutants were then recovered, reintroduced into yeast, reshuffled to ensure that the Ts+ phenotype was plasmid-borne, then passaged through E. coli, and sequence-verified. Whole cell extract preparation, two-hybrid analyses, immunoprecipitation, immunoblotting, RNA preparation, primer extension analyses, and temperature shift studies were performed as described previously (5, 24). Microarray analyses of yeast total RNA were performed using Nimblegen 12-plex yeast oligonucleotide microarrays (Roche Applied Science) by the Vanderbilt University Microarray Shared Resource. Hybridization data were analyzed and organized by hierarchical clustering and displayed as heat maps with ArrayStar version 2.1 software (DNA Star, Inc.). Solution binding assays using optical probe-bound biotinylated Rap1 (see below) and soluble Taf3, Taf5, or Taf4/Taf12 heterodimers were conducted and data analyzed, using an Octet apparatus and software (ForteBio Inc.). Binding was conducted in 25 mm HEPES, pH 7.6, 50 mm (Taf3 and Taf4/12) or 100 mm (Taf5) sodium acetate, 10% glycerol, 1 mm EDTA, 0.1% Nonidet P-40. Taf4 expressed alone was extensively degraded, and thus WT and mutant forms of this protein were coexpressed, purified, and analyzed with Taf12 (38, 39). An N-terminal fragment of Taf5 (amino acids 1–337) was used to allow efficient expression and purification of soluble protein.
Criteria for Clone/Plasmid Selection following Error-prone PCR Mutagenesis
First, multiple rounds of isolation and retransformation into yeast were used to show that the Ts+ phenotype was plasmid-borne. Second, alleles were chosen that conferred a “tight” Ts+ growth phenotype, with approximately WT growth at permissive temperature. Third, loading-adjusted immunoblotting was used to select clones that expressed epitope-tagged proteins of the appropriate molecular weight and steady state protein levels relative to WT. Only those mutant alleles that met all three criteria were included in our analyses; these were then subjected to DNA sequencing to exclude alleles bearing mutations outside the regions targeted for mutagenesis.
Biotinylation of Rap1
Rap1 was biotinylated using the sulfhydryl-reactive EZ-Link biotin-1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido)butane reagent (Thermo Scientific). The biotinylation reaction was carried out at a 2:1 biotin to protein ratio. A 1-ml sample of 1.7 mg/ml Rap1 (in 25 mm HEPES, pH 7.0, 500 mm sodium acetate, 10% glycerol) was incubated with 37 μl of a 5 mg/ml solution of biotin-1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido)butane overnight at 4 °C. After incubation, the Rap1/biotin mixture was dialyzed against 4 liters of 25 mm HEPES, pH 7.6, 500 mm sodium acetate, 10% glycerol (2 liters for 4 h followed by another 2 liters overnight) to remove unreacted biotin.
Gel Shift Assays
A double-stranded oligonucleotide ATATACACCCATACATTGA, containing a single consensus RPG Rap1-binding site (boldface), was end-labeled with [γ-32P]ATP and bacteriophage T4 polynucleotide kinase, desalted, and used for gel shift assays. 20 fmol of labeled double-stranded oligonucleotide (∼10,000 cpm) was incubated with purified recombinant Rap1 (see figure legends) in binding buffer (20 mm HEPES, pH 7.6, 10% v/v glycerol, 100 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, 25 μg/ml bovine serum albumin, and 2.5 μg/ml poly(dG-dC)) for 30 min at 23 °C, loaded, and fractionated on a 6% TBE-buffered polyacrylamide gel (0.15 × 15 × 20 cm) at 15 V/cm for 60 min. Gels were dried, exposed to Kodak K screens, and imaged using a Bio-Rad FX imager. When present, streptavidin and/or biotin was preincubated with Rap1 for 30 min at 23 °C prior to addition to DNA-binding reactions.
RESULTS
We previously demonstrated that yeast Rap1 directly binds TFIID subunits Taf4, Taf5, and Taf12 and also mapped the Taf12 Rap1 binding domain to Taf12 N-terminal sequences that are conserved in sensu strictu yeast strains (24). The Rap1 binding region of Taf12 was shown by others to be dispensable for viability (40) in agreement with our own studies (supplemental Fig. S1, A and B). However, given the colocalization of Taf4, -5, and -12 subunits within TFIID, we reasoned that these three Tafs might collectively represent the functional interaction sites for Rap1 (12, 13). To test this hypothesis, we conducted detailed structure-function analyses of TAF4 and TAF5 to map and characterize the RBDs of these subunits.
Taf4 RBD
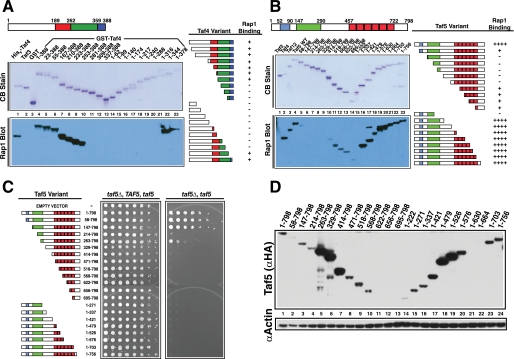
The Taf4 RBD was located by generating a systematic family of N- and C-terminal deletion variants of the protein. Each was expressed in E. coli, purified, and tested by far-Western assay for binding to pure recombinant Rap1. Rap1 bound Taf4 specifically (Fig. 1A, lane 1 versus lanes 2 and 3), and neither deletion of the nonconserved N terminus of Taf4 (amino acids (aa) 1–189; white block of sequences; lanes 5–7), nor the N-terminal portion of the bipartite histone fold domain (HFD) (red-shaded domain; lanes 8–10) had an effect on Rap1 binding. Similarly, C-terminal deletion of aa 345–388, which encode the HFD-related conserved C-terminal domain (Fig. 1A, blue-shaded domain; lanes 22 and 23) had no effect on Rap1 binding. By contrast, removal of residues 253–344 (Fig. 1A, green-shaded domain) completely abolished Rap1 binding (lanes 11–20). To test whether the Taf4 RBD is essential, we scored the ability of the TAF4 deletion family to support viability via a plasmid shuffle assay. Only the evolutionarily conserved residues, including those containing the RBD, were required for viability (supplemental Fig. S1C), results consistent with our previous less extensive mapping studies of TAF4 (38). Note that all mutants were recessive to wild type. Although some variants failed to accumulate to WT levels, complementation did not strictly correlate with intracellular protein concentration (supplemental Fig. S1D, lanes 2 and 8). We conclude from these experiments that the RBD of Taf4 resides between the elements of the conserved bipartite HFD and that this domain plays an important role in yeast cell physiology.
FIGURE 1.
Mapping the RBDs of Taf4 and Taf5. A, schematic of Taf4 nonconserved (white) and conserved residues (colored). N- and C-terminal truncations were expressed, purified, and fractionated by SDS-PAGE and blotted; one blot was stained with Coomassie Blue (CB Stain), and the other was probed with Rap1; Taf4-bound Rap1 was detected with anti-Rap1 IgG (Rap1 Blot). Lane 1 contained positive control His6-Taf4; lanes 2 and 3 contained non-Rap1-binding negative controls, His6-Taf3 and glutathione S-transferase (GST). Taf4 amino acids fused to glutathione S-transferase are indicated above the Coomassie Blue-stained blot, lanes 4–23. Summary of Rap1-Taf4 binding data (right). B, schematic of Taf5 showing nonconserved (white) and conserved sequence elements (colored). N- or C-terminally truncated forms of Taf5 (lanes 4–23) were generated, purified, and tested as in A. Lane 1 contained the negative control, non-Rap1-binding Taf3, and lanes 2 and 3 contained positive control proteins His6-Taf4 and His6-Taf12. Summary of Rap1-Taf5 binding data (right). C, plasmid shuffle complementation assay testing the ability of various truncated forms of Taf5 to support viability. Serial 1:4 dilutions of cells expressing the indicated Taf5 variants were subjected to growth as pseudodiploids (relevant genetic constitution: taf5Δ, TAF5, taf5) or following loss of the URA3-marked TAF5 covering plasmid, scored as 5-fluoroorotic acid resistance (taf5Δ, taf5). Plates were incubated for 3 days at 30 °C. D, steady state protein levels of WT and Taf5 variants measured in whole cell extracts by immunoblotting with anti-HA (Taf5 (αHA)) or anti-actin (αActin) IgGs; actin served as extraction/loading control.
Taf5 RBD
Taf5 contains three evolutionarily conserved domains as follows: N-terminal domains 1 (NTD1; aa 52–90, blue), and 2 (NTD2; aa 147–290, green), and six WD-40 repeats (aa 457–722, red; Fig. 1B). To map the Taf5 RBD, we again generated a family of N- and C-terminally truncated variant proteins and tested each for Rap1 binding as above. Rap1 bound Taf5 specifically (Fig. 1B, cf. lanes 1 and 4) with an apparent affinity similar to Taf4 and Taf12 (lanes 2–4). Removal of Taf5 N-terminal sequences disrupted Rap1-Taf5 interaction (Fig. 1B, lanes 5–10), whereas removal of the C-terminal residues, including the entire WD-40 domain, had no apparent effect on Rap1 binding, although a minor amount of Rap1 binding was observed with a few of the N-terminal deletion variants of Taf5 (lanes 11–13). The significance of this weak Rap1 binding to WD repeats is unknown. The complementation properties of the TAF5 truncation mutants were also scored (Fig. 1C). As reported previously (41), NTD1 was dispensable for viability, and complete removal of NTD2 resulted in lethality. Additional removal of Taf5 N-terminal residues was incompatible with viability, as was C-terminal truncation. These experiments showed that both NTD2 and WD-40 domains contribute essential functions to Taf5 (Fig. 1C); all mutants were recessive to WT, and complementation patterns did not track with Taf5 expression. In fact many noncomplementing Taf5 variants accumulated to levels higher than WT (Fig. 1D). These experiments reveal that both the N-terminal RBD, including NTD2, as well as the WD-40 repeats of Taf5 play essential roles in yeast cell physiology.
Direct Tests of the Contribution of Taf4 and Taf5 RBDs to Viability
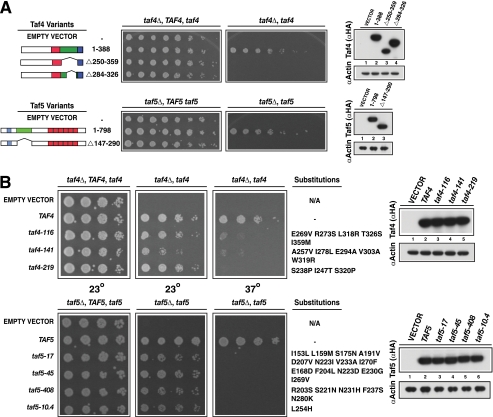
To directly assess the roles of the Taf4 and Taf5 RBDs, we performed two additional genetic experiments. First, the mapped RBDs were deleted in the context of the intact proteins. We reasoned that precise removal of the responsible sequences would, if essential, result in loss of growth. Second, to rule out a potential simple spacer function for the mapped RBDs we tested the sensitivity of these domains to amino acid substitutions, a result that would be predicted if these domains of Taf4 and Taf5 truly contain functional binding sites for Rap1. We tested this hypothesis by subjecting the RBD-encoding sequences of TAF4 and TAF5 to error-prone PCR-mediated mutagenesis. In both experiments, the relevant strains were scored by plasmid shuffle growth tests. Deletion of Taf4 RBD-encoding sequences (i.e. aa 250–350 or aa 284–326) led to loss of viability; WT and variant proteins were expressed comparably (Fig. 2A). Similarly, deletion of TAF5 sequences encoding NTD2 (aa 147–290) led to a conditional growth defect; the Taf5-ΔNTD2 protein accumulated to WT levels at 23 and 37 °C but could not support viability at 37 °C (Fig. 2A, 37 °C, data shown). As seen in Fig. 1, all mutants were recessive. Yeast two-hybrid protein-protein interaction studies showed that the lack of growth evident upon deletion of RBD-encoding sequences is likely not due to a lack of Taf-Taf interactions (supplemental Fig. S2).
FIGURE 2.
Genetic tests of the requirement of Taf4 and Taf5 RBDs for viability. A, internally deleted variants of Taf4 (top) or Taf5 (bottom), along with WT and empty vector controls (left), were tested for the ability to support viability via plasmid shuffle as in Fig. 1 (center). Steady state protein levels of HA-tagged Taf4 and Taf5 proteins (and untagged, empty vector control) were determined by immunoblotting (right). Cells were serially diluted, spotted, and tested for growth at 23 °C (TAF4) or 37 °C (TAF5). B, mutation of either TAF4 or TAF5 RBD-encoding sequences conferred temperature-sensitive growth. Plasmid shuffle was used to test the ability of cells expressing Taf4 (top) or Taf5 (bottom) proteins with altered RBDs to support viability. Cells of the indicated genetic makeup were grown pre- and post-shuffle on plates lacking or containing 5-fluoroorotic acid and tested for growth after 72 h (23 °C) or 48 h (37 °C); N/A indicates not applicable since no second copy of TAF4 (top) or TAF5 (bottom) is present on the test plasmid. Steady state protein levels were scored by immunoblotting as above (right).
We next subjected Taf4 (aa 250–359) and Taf5 (aa 147–290) RBD-encoding sequences to error-prone PCR. RBD-mutagenized TAF4 and TAF5 libraries were generated and used to transform TAF4/taf4Δ or TAF5/taf5Δ strains. The resulting clones were subjected to plasmid shuffle complementation assays that scored growth at various temperatures. Several thousand putative mutants were screened for each gene, and clones that exhibited temperature-sensitive (Ts+) growth were identified and validated using the criteria detailed under “Materials and Methods.” Several unique taf4 and taf5 alleles that met all criteria were isolated, and nearly all encoded multiple amino acid substitutions, and all variant proteins accumulated to WT levels (Fig. 2B). Together, these two additional genetic experiments show that Taf4 and Taf5 RBDs contribute directly to viability.
RPG Transcription in taf4 and taf5 Cells
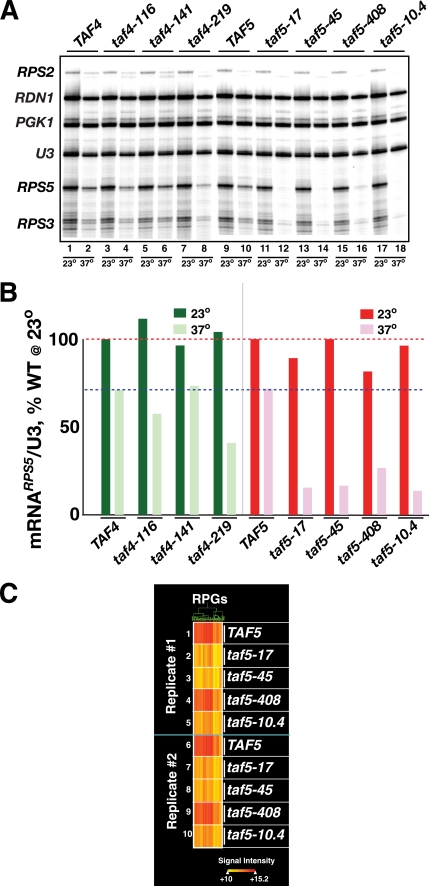
Given the growth defects associated with alteration of Taf4 and Taf5 RBDs, the known roles of Rap1 and TFIID in RPG transcription, and the importance of ongoing ribosome synthesis to cellular growth, we examined the effect of temperature up-shift on RPG transcription in WT and taf RBD− Ts+ cells. Log phase cultures were grown at 23 °C; half the culture shifted to 37 °C for 2 h, and total RNA was extracted from the control 23 °C culture and the 37 °C-shifted culture. Specific transcripts were detected using a multiplex primer extension assay. Three TFIID-dependent RPG transcripts (RPS2, RPS3, and RPS5), two TFIID-independent genes (PGK1 and rRNA-encoding RDN1), and U3 small nucleolar RNA (loading control) were scored simultaneously (Fig. 3, A, gel image, and B, quantification for RPS5). RPG transcript levels were reduced in two of three taf4 mutants at 37 °C, whereas all taf5 mutants displayed a dramatic reduction upon shift to 37 °C. The temperature-dependent reduction in transcript levels was specific to the RPGs because the levels of the TFIID-independent PGK1 mRNA were not reduced at 37 °C (21), and as expected, RDN1 transcript levels were only moderately reduced (31). The transcriptional defects seen with the various mutants were quantitatively similar, regardless of whether RPS5 (Fig. 3B), RPS2, or RPS3 were quantified (data not shown). Measurements of total poly(A)+ mRNA abundance, as well as additional RPG-specific primer extension assays, further supported the hypothesis that transcription of the RPG regulon requires Taf-RBD function (data not shown).
FIGURE 3.
Effect of temperature shift of taf4 Ts+ and taf5 Ts+ strains on RPG transcription. A, log phase cells expressing the indicated TAF4 and TAF5 alleles (top) were grown at 23 or 37 °C for 2 h and harvested, and total RNA was extracted. Equal amounts of RNA were subjected to multiplex primer extension analyses using a mixture of 5′-32P-labeled gene-specific oligonucleotide primers (RPS2, RDN1, PGK1, U3, RPS5, and RPS3). Extension products were fractionated on a sequencing gel that was dried, exposed to a Kodak K-screen, and scanned with a Bio-Rad FX imager; relevant portion of image is shown. B, scan was analyzed using Bio-Rad QuantityOne software. RPS5 mRNA-specific signals were normalized to WT and U3 signals and plotted; dashed lines represent signals of WT at 23 °C (top line, red) and 37 °C (bottom line, blue). C, effect of taf5 mutations on expression of the RPG regulon. The 37 °C TAF5 and taf5 Ts+ total RNA samples analyzed in A and B above (Replicate #1, 1–5), as well as the equivalent RNA from an independent biological replicate (Replicate #2, 6–10) were Cy5-labeled and hybridized to Nimblegen oligonucleotide yeast genome arrays. The hybridization signals of the RPGs whose expression changed by 2-fold (up- or down) in the taf5-17-derived sample relative to TAF5 are plotted in heat map format following hierarchical clustering. Hybridization signal intensity is indicated by the “heat” scale shown (bottom).
Effect of taf5 Mutations on Transcription of the RPG Regulon
Given the dramatic effect on RPG expression caused by disrupting Taf5 RBD function, we tested if temperature-induced inactivation of the Taf5 RBD caused a general RPG transcriptional defect. WT and taf5 Ts+ strains were grown at 23 °C and shifted to 37 °C, and replicate RNA samples from two independent experiments were prepared (Fig. 3, A and B). RNA from each strain grown at 37 °C was labeled and hybridized to oligonucleotide expression arrays (23 °C RNA hybridization profiles were all essentially identical; data not shown). Fig. 3C shows the results of these analyses for the majority of RPGs. The data are presented in the form of clustered heat maps comparing specific transcript abundance (see signal intensity scale Fig. 3C, lower right) in the taf5-17 mutant cells to TAF5 cells; transcripts in the other strains were clustered accordingly. Expression of 1347 genes varied up or down 2-fold in this comparison. For ease of comprehension, only the RPG data from the 37 °C samples are presented. This presentation format facilitates direct comparison of signals from column to column (Fig. 3C). The entire data set, including results for all 1347 genes, is presented in supplemental Fig. S3. Essentially the entire RPG family behaved similarly (110 of 137 RPGs; Fig. 3C); RPG transcripts were down-regulated 2–8-fold upon shift to nonpermissive temperature (Fig. 3C and supplemental Table S1). The extent of loss of RPG transcription varied among the different taf5 mutants, with taf5-17 being the most affected and taf5-408 the least affected, a pattern observed in both biological replicates and consistent with the gene-by-gene primer extension analyses (Fig. 3, A and B). Collectively, the data of Fig. 3 indicate that the RBD function of both Taf4 and Taf5 is required for RPG transcription, and as directly shown in the case of Taf5, RBD function is necessary to activate expression of essentially the entire RPG family.
Altered Forms of Taf4 and Taf5 Bind Rap1 with Reduced Affinity
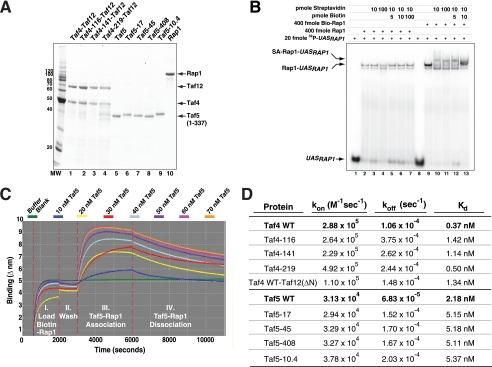
To test whether the taf4- and taf5-encoded proteins exhibited a biochemical loss of function phenotype (i.e. reduced Rap1 binding affinities) as predicted by the recessive nature of the corresponding alleles, we performed biolayer interferometry binding assays to determine rate constants for association (kon) and dissociation (koff) between Tafs and Rap1. This approach allowed us to calculate equilibrium dissociation constants (i.e. KD = koff/ kon) for Taf-Rap1 interactions. The integrity and purity of the recombinant proteins used are shown in Fig. 4A. Biotinylated Rap1 retained DNA binding activity (supplemental Fig. S4) even when bound to streptavidin (Fig. 4B). Biotinylated Rap1 was loaded on streptavidin-derivatized microfiber optical probes and incubated with increasing concentrations of Tafs. Formation and dissociation of Rap1-Taf complexes were monitored optically in real time. Typical Taf5-Rap1 kinetic binding curves are presented in Fig. 4C. Specific binding (Fig. 1, A and B, and supplemental Fig. S5) was concentration-dependent and reached equilibrium rapidly, whereas the resulting Rap1-Taf complexes dissociated slowly. A summary of binding assays with WT and altered forms of Taf4 (Taf4/Taf12 heterodimers) and Taf5 is presented in Fig. 4D. The affinity of interaction between WT forms of the proteins is high, all in the nanomolar range (cf. Fig. 1, A and B, with Fig. 4D) as follows: Taf4/12-Rap1 KD ∼ 0.4 × 10−9 m; Taf5-Rap1 KD ∼ 2 × 10−9 m. As predicted, the variant Taf forms bound Rap1 with a lower affinity (3–5-fold) than WT; dissociation rates were most often affected by RBD mutation. To test if the N-terminal Taf12 RBD present in the Taf4/Taf12 heterodimer contributed to binding as predicted by our previous studies (24), we assayed the binding of Rap1 to WT-Taf4/ΔNTaf12. This heterodimer bound Rap1 ∼3.5-fold less well than WT-Taf4/Taf12. Together, these in vitro binding data confirm and extend the molecular genetic analyses presented in Figs. 1–3 and support the idea that the RBDs of Taf4, Taf5, and Taf12 are all direct in vivo targets of Rap1.
FIGURE 4.
Binding affinity of Rap1 to WT and mutated Taf4 and Taf5. A, purified Taf5 (WT and variants) or Taf4/Taf12 heterodimers (composed of WT Taf12, WT Taf4, and indicated variants) were fractionated by SDS-PAGE and visualized with Sypro Ruby staining using a Bio-Rad FX Imager. B, biotinylated-Rap1 is active for DNA binding in the absence and presence of streptavidin. DNA binding assays were performed in the presence of the indicated components (top); relevant species indicated (UASRAP1; Rap1-UASRAP1; Streptavidin (SA)-Rap1-UASRAP1). C, representative kinetic binding traces monitoring the binding of varying concentrations of WT Taf5 (residues 1–337) to optical probe-bound biotinylated Rap1. Binding, expressed as Δnm (y axis), was monitored in real time (time/s; x axis) using a ForteBio Octet apparatus. Binding was conducted in four phases shown: I, II, III, and IV. D, kinetic binding constants for WT and RBD-altered Taf4, Taf5, and Taf12 (Taf12-ΔN; residues 286–539). All proteins were analyzed in >3 independent experiments with multiple preparations of proteins, and kon, koff, and KD kinetic binding constants were calculated following global fit of the data. Values for the mutant proteins were compared with WT (bold); all data were significantly different from WT (p < 0.05) as determined using a two-tailed t test.
Genetic Interactions between TAF4, TAF5, and RAP1
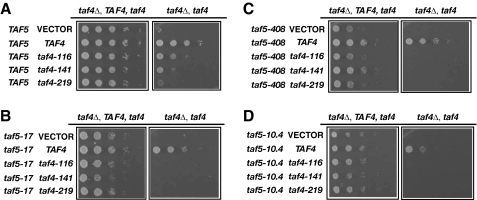
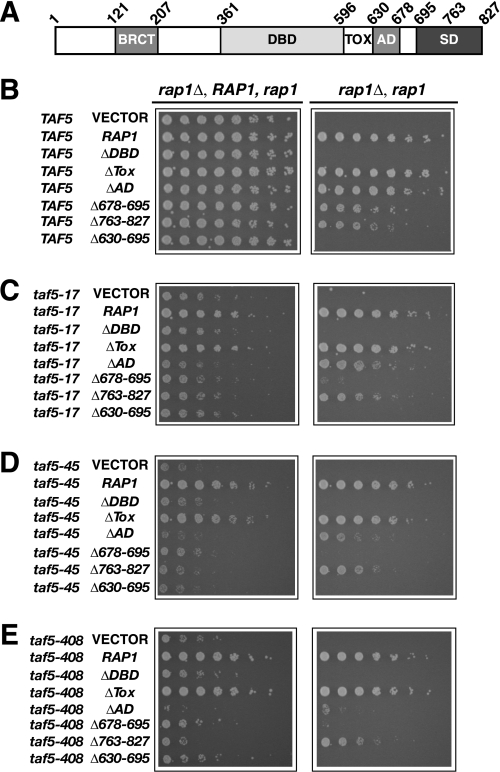
Finally, we tested whether combining mutant alleles of the Taf-encoding genes together, or with various mutant variants of RAP1, would lead to synthetic sick or lethal (SSL) phenotypes as expected if these proteins directly contribute to the same process. We constructed strains in which taf4 alleles were combined with taf5-17, taf5-408, or taf5-10.4 alleles (Fig. 5). Synthetic lethality occurred whenever a taf4 allele was combined with a taf5 allele (Fig. 5, B–D). Similarly, we constructed strains carrying both taf5 and rap1 mutant alleles, focusing on RAP1 sequences encoding the DBD and C terminus of the protein (see Fig. 6A) because we have previously shown that these domains contribute to TFIID binding (24). As expected, deletion of the Rap1 DBD caused lethality, regardless of TAF5 constitution (Fig. 6, B–E, ΔDBD). Surprisingly, simultaneous removal of Rap1 sequences encoding aa 630–695 containing the AD (630–678) plus additional residues with as yet unknown function (678–695) were also lethal regardless of TAF5 status (Fig. 6, B–E, Δ630–695). Removal of residues 678–695, although viable in an otherwise WT background, resulted in a strong SSL phenotype when combined with any of the tested taf5 alleles (Fig. 6, C–E). Collectively, the results of these genetic tests support a model of direct Rap1-Taf interactions in vivo.
FIGURE 5.
taf4 Ts+ and taf5 Ts+ mutants exhibit synthetic lethality when combined. Cells carrying the indicated alleles of taf4 and taf5 were tested for synthetic interactions by plasmid shuffle. Serial ¼ dilutions of cells carry the following: A, TAF5 plus empty vector, TAF4, or taf4 Ts+ alleles; B, taf5-17 + vector, TAF4, or taf4 Ts+ alleles; C, taf5-408 + vector, TAF4, or taf4 Ts+ alleles; D, taf5-10.4 + vector, TAF4, or taf4 Ts+ alleles, were grown on plates lacking (left, each panel) or containing 5-fluoroorotic acid (right, each panel) for 72 h at 30 °C.
FIGURE 6.
taf5 Ts+ mutants display synthetic genetic interactions with rap1 mutants. A, schematic of Rap1 indicating the C-terminal BRAC1 protein (BRCT), DBD, toxicity (Tox), AD, and silencing (SD) domains, and amino acid coordinates delineating the domains. B–E, cells carrying the indicated TAF5 and RAP1 alleles were tested by plasmid shuffle for SSL phenotypes as in Fig. 5.
DISCUSSION
In this study, we have identified and characterized the biochemical and genetic interactions between the DNA binding transactivator Rap1 and subunits of the TFIID coactivator. Our study represents the first description of direct, specific, mutationally sensitive, high affinity transactivator-Taf interactions in the yeast system. We have identified the RBDs within two essential TFIID subunits, Taf4 and Taf5, studies that complement our previous work that defined an RBD in Taf12. These three RBDs lack any significant sequence conservation, and the properties of each are unique; the Taf5 RBD is acidic (calculated pI 6.5), whereas both Taf4 and Taf12 RBDs are basic (pI 9.4 and 9.2, respectively) suggesting that each interacts with distinct portions, domains, or isoforms of Rap1, a hypothesis consistent with our previous identification of multiple TFIID binding domains within the Rap1 DBD and C terminus (24) and the fact that Rap1 has been reported to be subject to post-translational modification (42).
RBD-encoding regions of TAF4 and TAF5 are both essential for viability and sensitive to targeted mutagenesis; mutation leads to temperature conditional growth without affecting Taf-Taf interactions or TFIID stability (Figs. 1 and 2, and supplemental Figs. S2, S6, and S7). Importantly, the recessive taf4 and taf5 RBD− Ts+ alleles produce proteins that cause defects in RPG transcription at nonpermissive temperatures (Fig. 3), have reduced Rap1 binding capacity in vitro (Fig. 4), and exhibit SSL interactions with each other and RAP1 (Figs. 5 and 6). Collectively, these results support the hypothesis that direct Rap1-TFIID interactions drive ribosomal protein gene transcription (21, 24) and that the RBDs contained within the Taf4, Taf5, and Taf12 subunits of TFIID are key physical and functional in vivo targets for Rap1-driven RPG transactivation.
Taf5 RBD
Our data show that the N-terminal 337 aa of Taf5 contain the RBD of this TFIID subunit. Although both NTD1 and NTD2 of Taf5 are evolutionarily conserved (41, 43), NTD1 is dispensable for viability. By contrast, precise removal of NTD2 confers a Ts+ phenotype, as does alteration of specific amino acid residues within NTD2. We have previously shown that the N termini of each Taf5 molecule localizes to the C-lobe of TFIID, whereas Taf5 C termini extend through lobes A and B, results that lead us to propose that Taf5 dimerizes via N-terminal sequences (Fig. 7) (13). However, conflicting data regarding the ability of NTD2 to dimerize have been reported (41, 43), and additional work will be needed to resolve this issue. Our results do indicate that if the Taf5-Taf5 interaction is mediated through N-terminal sequences, the dimerization domain is likely distinct from the RBD because Rap1 efficiently binds holo-TFIID (supplemental Figs. S6 and S7), the physiological context for any potential Taf5 dimerization (24). Taf5 is subject to PTMs, some of which map to NTD2 (44, 45). Thus, it is reasonable to propose that Taf5 PTMs influence Rap1-TFIID interactions, perhaps in response to the several signaling pathways that integrate nutrient availability with RPG transcription. Dimerization and PTM status notwithstanding, our data demonstrate that Rap1 readily interacts with both full-length Taf5 and the isolated Taf5 N terminus and that NTD2-altered forms of Taf5 bind Rap1 with ∼2.5-fold reduced affinity compared with WT. In vivo, mutants expressing these forms of Taf5 display significant reductions in RPG expression relative to WT when mutant cells are shifted to 37 °C. Without additional detailed molecular information regarding the precise mechanism(s) of Rap1-mediated transactivation, it is impossible to predict the magnitude of the decrease in RPG transcription expected for any of the individual RBD-mutated Ts+ taf mutants. This challenge is made all the more difficult given the fact that multiple RBDs exist within TFIID. It is possible that each RBD, when bound by Rap1, acts in a unique mechanistic fashion by affecting different aspects of PIC formation and/or function. Nevertheless, our recessive loss-of-function taf5 mutants displayed decreased transcription of the RPG regulon, a result consistent with the fact that all of the altered Taf5 proteins are defective for Rap1 binding. Altogether, our data shed new light on the physiological importance of the Taf5 N terminus, reveal the mechanistic basis of its interaction with Rap1, and clearly show that regions of Taf5 distinct from the WD-40 repeats (46) are also critical to Taf5 function.
FIGURE 7.
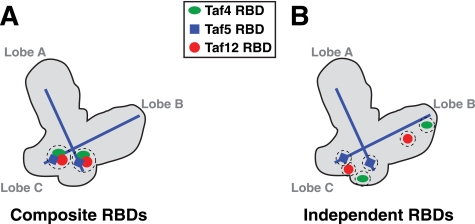
Possible organization of Taf4, -5, and -12 RBDs within the TFIID complex. Shown is a schematic of yeast TFIID with A, B, and C lobes labeled (13). TFIID contains two copies of both Taf5 and the Taf4/Taf12 heterodimer (5), and all three subunits have been immunomapped within the structure. The N termini of the two molecules of Taf5 map to lobe C, whereas the two Taf5 C termini map to lobes A and B. The localization and orientation of the Taf5 subunits are indicated by the blue lines, whereas Taf4, -5, and -12 RBDs are indicated by the green ovals, blue squares, and red circles, respectively. A, model depicting two tightly localized Rap1-binding sites (RBDs, two dashed circles) composed of combined Taf4, -5, and -12; Composite RBDs. B, model depicting six independent Rap1-binding sites within Taf4, -5, and -12 (six dashed circles); Independent RBDs. Note that the currently available resolution of S. cerevisiae TFIID subunits is too low to assign precise locations for the Taf5 N termini and Taf4/12 heterodimer pairs (17).
Do Taf4/Taf12 Heterodimers Provide Independent RBDs?
Our data show that distinct RBDs exist in the Taf4/Taf12 heterodimer. Like Taf5, 2 mol of Taf4/Taf12 heterodimer are present per mol of TFIID and Taf4 and -12 colocalize within the B- and C-lobes of yeast TFIID (5, 12). The Taf4-12 complex contributes importantly to the assembly and/or stability of TFIID (47), and recently it was reported that TBP binding to the TFIID-Taf complex induces a large conformational change in TFIID involving Taf4-12 (15). Taf4 is a TFIID-specific subunit, whereas Taf12, like Taf5, is shared with SAGA (20). Importantly, shared Taf function in the context of RPG transcription derives entirely from TFIID because SAGA, despite containing five shared Tafs (i.e. Taf5, -6, -9, -10, and -12), makes no apparent functional contribution to RPG transcription under the conditions of our study (supplemental Fig. S8) (21). However, it is likely that Rap1 collaborates with SAGA to regulate transcription of other genes, and our data support this notion (supplemental Fig. S8) (20, 32). As with SAGA, we also failed to detect any significant contribution of Mediator to RPG transcription (supplemental Fig. S8), results consistent with recent studies of genome-wide Mediator promoter occupancy (22, 23) (although this point remains controversial (26–28)). Regardless, it is clear that TFIID serves as a Rap1 coactivator during RPG transcription by making direct contacts with RBD-containing Tafs.
Interestingly, Taf4 is the TFIID subunit for which the greatest number of direct regulatory protein-Taf interactions has been identified. For example, the transcription factors Sp1, c-Jun, CREB, TCF/Pan, HP1α/β, and Caenorhabditis elegans differentiation modulators OMA-1/2 all bind Taf4 (8, 16, 48). In addition, cell type-specific isoforms of Taf4 have been described in metazoans that contribute to the diversity of regulatory inputs impinging on TFIID (49–51). The so-called TafH domain within metazoan Taf4 is responsible for mediating interactions with numerous activators (52); however, yeast Taf4 lacks an obvious TafH domain. We speculate that the yeast Taf4/Taf12 heterodimer, with at least two independent activator-binding sites (for Rap1 and possibly other transactivators), collectively fulfills the roles of the metazoan Taf homology domain. Such distribution of function among multiple yeast proteins has been seen in the case of the TFIID-associated protein Bdf1, which supplies bromodomain functions absent from yeast Taf1 but present in metazoan Taf1, resulting in comparable conservation of overall TFIID function from yeast to man (33).
The region we have identified in yeast Taf4 as an RBD was previously referred to as the Taf4 “spacer domain” because it separates the elements of its noncanonical bipartite HFD (38, 39). Taf4 from various organisms binds DNA directly, and binding requires residues within the yeast Taf4 RBD/spacer domain (53). The human Taf4 spacer domain also binds TFIIA in vitro, thus providing important contributions to both TFIID promoter binding and PIC function, directly illustrating a biochemical contribution of this conserved domain to transcriptional activation (54). Further emphasizing the importance of this region to TFIID function, in Drosophila a repressor protein competes with TFIIA for Taf4 binding to inhibit transcription (55). These data, combined with our work, indicate that the so-called Taf4 spacer domain is in fact an ancient domain that plays central roles in transcriptional activation. Our demonstration that yeast Taf4, -5, and -12 contain distinct functional RBDs adds importantly to the existing body of work on the coactivator functions of TFIID. We are currently constructing additional RBD site-directed taf4 mutants and expect that this will allow us to identify a broader range of RPG transcription activation defects compared with the relatively mild transcriptional phenotypes observed in the taf4 RBD mutants described here.
Distinct or Composite RBDs?
The presence of multiple functional RBDs within TFIID (Taf4, -5, and -12) is reminiscent of activator interactions with SWI/SNF, SAGA, SPT3-TAFII31-GCN5L acetylase, and Mediator complexes where activators have been shown to interact with several subunits of each of these coactivators (56–59). These data suggest that redundancy of transactivator targets within coactivators is likely to be a widespread phenomenon. However, to date, direct transactivator-coactivator interactions have only been described for TFIID-independent yeast genes. Our findings therefore allow for new insights into transactivator-TFIID coactivator interactions.
Taf4, -5, and -12 are all present in 2 mol/mol TFIID complex (5), and the location of each has been mapped within the yeast structure (12, 13). TFIID therefore contains at a maximum six RBDs. We propose two (limit) models for the organization of TFIID RBDs (Fig. 7). Either there are two composite Rap1 interaction sites within TFIID (Fig. 7A) or, alternatively, there are six independent RBDs (Fig. 7B). Each model offers unique advantages. Composite RBDs would allow for a stronger interaction with Rap1 by providing a larger/higher local concentration of binding surfaces with increased affinity/avidity, perhaps specializing this specific domain of TFIID for this particular transactivator. Either one or both composite high affinity sites could then serve to functionally interact with Rap1 to drive high level RPG transcription (Fig. 7A). This model is attractive given that 50% of all RNA pol II initiation events in log phase cells occur on Rap1-driven RPGs (29). Alternatively, if each RBD is independent (Fig. 7B), such distributed RBDs would allow for increased conformational interaction flexibility between enhancer-bound Rap1 (or any other transactivator that may work by binding these surfaces) and TFIID. A recent cryo-EM study reported a high resolution structure of Schizosaccharomyces pombe TFIID (15). These authors performed both Taf4 immunomapping and docking of a partial (Taf4-12)2 HFD heterotetramer structure (39) on TFIID. They conclude that both copies of Taf4-12 heterodimer reside together within a small domain of TFIID. Unfortunately, the S. pombe TFIID structure was not compared with the equivalent EM-derived Saccharomyces cerevisiae structures, so it is currently not possible to extrapolate from one structure to the other. If the S. pombe TFIID Taf4-12 mapping data hold true for S. cerevisiae TFIID, this would support the composite RBD model. Future studies will resolve these issues. Finally, it should be noted that most ribosomal protein gene enhancers have two or more variably oriented binding sites for Rap1. Genome-wide, many Rap1-binding sites are located ∼200 bp upstream of RPG transcription start sites (30, 33), and the ∼160 bp of DNA between RAP1 enhancers and RPG promoters is nucleosome-deficient and thus presumably quite conformationally flexible, a feature that would facilitate Rap1-RBD interaction regardless of the mode of RBD organization within TFIID.
In conclusion, our data provide new insights into the organization and function of the multisubunit TFIID coactivator complex and its interactions with the essential transactivator Rap1, while simultaneously providing key information that will enable future dissection of activator-coactivator interactions. The interactions between Rap1 and the TFIID coactivator play essential roles in mRNA gene transcription control. Elucidating the mechanisms of this process remains a major, yet poorly understood problem in eukaryotic biology. Our work adds importantly to the existing body of knowledge.
Acknowledgments
We thank Drs. M. K. Tripathi, M. B. Chandrasekharan, M. A. Bendjennat, M. V. Singh, and R. L. Mernaugh for fruitful discussions during the course of this study. J. H. L. acknowledges a generous gift by Dr. Harold P. Erickson.
This work was supported, in whole or in part, by National Institutes of Health Grant GM52461.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8, Tables S1 and S2, and additional references.
- pol II
- RNA polymerase II
- TFIID
- transcription factor IID
- RPG
- ribosomal protein gene
- RBD
- Rap1 binding domain
- TBP
- TATA box-binding protein
- Taf
- TBP-associated factor
- PIC
- preinitiation complex
- DBD
- DNA binding domain
- AD
- activation domain
- HA
- hemagglutinin
- WT
- wild type
- Ts
- temperature-sensitive
- HFD
- histone fold domain
- CCTD
- conserved C-terminal domain
- NTD1/2
- N-terminal domains
- WD-40
- Trp-Asp (W-D) dipeptide or β-transducin repeats
- SSL
- synthetic sick, lethal
- PTM
- post-translational modification
- SAGA
- Spt-Ada-Gcn-acetylase complex
- aa
- amino acid.
REFERENCES
- 1.Taatjes D. J., Marr M. T., Tjian R. (2004) Nat. Rev. Mol. Cell Biol. 5, 403–410 [DOI] [PubMed] [Google Scholar]
- 2.Thomas M. C., Chiang C. M. (2006) Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M., Gann A. (1997) Nature 386, 569–577 [DOI] [PubMed] [Google Scholar]
- 4.Poon D., Bai Y., Campbell A. M., Bjorklund S., Kim Y. J., Zhou S., Kornberg R. D., Weil P. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8224–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders S. L., Garbett K. A., Weil P. A. (2002) Mol. Cell. Biol. 22, 6000–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tora L. (2002) Genes Dev. 16, 673–675 [DOI] [PubMed] [Google Scholar]
- 7.Kuras L., Struhl K. (1999) Nature 399, 609–613 [DOI] [PubMed] [Google Scholar]
- 8.Albright S. R., Tjian R. (2000) Gene 242, 1–13 [DOI] [PubMed] [Google Scholar]
- 9.Cler E., Papai G., Schultz P., Davidson I. (2009) Cell. Mol. Life Sci. 66, 2123–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andel F., 3rd, Ladurner A. G., Inouye C., Tjian R., Nogales E. (1999) Science 286, 2153–2156 [DOI] [PubMed] [Google Scholar]
- 11.Brand M., Leurent C., Mallouh V., Tora L., Schultz P. (1999) Science 286, 2151–2153 [DOI] [PubMed] [Google Scholar]
- 12.Leurent C., Sanders S., Ruhlmann C., Mallouh V., Weil P. A., Kirschner D. B., Tora L., Schultz P. (2002) EMBO J. 21, 3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leurent C., Sanders S. L., Demény M. A., Garbett K. A., Ruhlmann C., Weil P. A., Tora L., Schultz P. (2004) EMBO J. 23, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grob P., Cruse M. J., Inouye C., Peris M., Penczek P. A., Tjian R., Nogales E. (2006) Structure 14, 511–520 [DOI] [PubMed] [Google Scholar]
- 15.Elmlund H., Baraznenok V., Linder T., Szilagyi Z., Rofougaran R., Hofer A., Hebert H., Lindahl M., Gustafsson C. M. (2009) Structure 17, 1442–1452 [DOI] [PubMed] [Google Scholar]
- 16.Liu W. L., Coleman R. A., Ma E., Grob P., Yang J. L., Zhang Y., Dailey G., Nogales E., Tjian R. (2009) Genes Dev. 23, 1510–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papai G., Tripathi M. K., Ruhlmann C., Werten S., Crucifix C., Weil P. A., Schultz P. (2009) Structure 17, 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders S. L., Jennings J., Canutescu A., Link A. J., Weil P. A. (2002) Mol. Cell. Biol. 22, 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi H., Martin-Brown S., Washburn M. P., Florens L., Conaway J. W., Conaway R. C. (2009) J. Biol. Chem. 284, 32405–32412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X. Y., Bhaumik S. R., Green M. R. (2000) Science 288, 1242–1244 [DOI] [PubMed] [Google Scholar]
- 21.Mencía M., Moqtaderi Z., Geisberg J. V., Kuras L., Struhl K. (2002) Mol. Cell 9, 823–833 [DOI] [PubMed] [Google Scholar]
- 22.Fan X., Chou D. M., Struhl K. (2006) Nat. Struct. Mol. Biol. 13, 117–120 [DOI] [PubMed] [Google Scholar]
- 23.Fan X., Struhl K. (2009) PLoS One 4, e5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garbett K. A., Tripathi M. K., Cencki B., Layer J. H., Weil P. A. (2007) Mol. Cell. Biol. 27, 297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huisinga K. L., Pugh B. F. (2007) Genome Biol. 8, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrau J. C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M. G., van de Peppel J., Werner M., Holstege F. C. (2006) Mol. Cell 22, 179–192 [DOI] [PubMed] [Google Scholar]
- 27.Zhu X., Wirén M., Sinha I., Rasmussen N. N., Linder T., Holmberg S., Ekwall K., Gustafsson C. M. (2006) Mol. Cell 22, 169–178 [DOI] [PubMed] [Google Scholar]
- 28.Ansari S. A., He Q., Morse R. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16734–16739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner J. R. (1999) Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 30.Venters B. J., Pugh B. F. (2009) Genome Res. 19, 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudra D., Warner J. R. (2004) Genes Dev. 18, 2431–2436 [DOI] [PubMed] [Google Scholar]
- 32.Lieb J. D., Liu X., Botstein D., Brown P. O. (2001) Nat. Genet. 28, 327–334 [DOI] [PubMed] [Google Scholar]
- 33.Koerber R. T., Rhee H. S., Jiang C., Pugh B. F. (2009) Mol. Cell 35, 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohde J. R., Cardenas M. E. (2003) Mol. Cell. Biol. 23, 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lempiäinen H., Uotila A., Urban J., Dohnal I., Ammerer G., Loewith R., Shore D. (2009) Mol. Cell 33, 704–716 [DOI] [PubMed] [Google Scholar]
- 36.Schawalder S. B., Kabani M., Howald I., Choudhury U., Werner M., Shore D. (2004) Nature 432, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 37.Zanton S. J., Pugh B. F. (2006) Genes Dev. 20, 2250–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thuault S., Gangloff Y. G., Kirchner J., Sanders S., Werten S., Romier C., Weil P. A., Davidson I. (2002) J. Biol. Chem. 277, 45510–45517 [DOI] [PubMed] [Google Scholar]
- 39.Werten S., Mitschler A., Romier C., Gangloff Y. G., Thuault S., Davidson I., Moras D. (2002) J. Biol. Chem. 277, 45502–45509 [DOI] [PubMed] [Google Scholar]
- 40.Moqtaderi Z., Yale J. D., Struhl K., Buratowski S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14654–14658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romier C., James N., Birck C., Cavarelli J., Vivarès C., Collart M. A., Moras D. (2007) J. Mol. Biol. 368, 1292–1306 [DOI] [PubMed] [Google Scholar]
- 42.Tsang J. S., Henry Y. A., Chambers A., Kingsman A. J., Kingsman S. M. (1990) Nucleic Acids Res. 18, 7331–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharya S., Takada S., Jacobson R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer-Guittaut M., Birsoy K., Potel C., Elliott G., Jaffray E., Desterro J. M., Hay R. T., Oelgeschläger T. (2005) J. Biol. Chem. 280, 9937–9945 [DOI] [PubMed] [Google Scholar]
- 45.Mischerikow N., Spedale G., Altelaar A. F., Timmers H. T., Pijnappel W. W., Heck A. J. (2009) J. Proteome Res. 8, 5020–5030 [DOI] [PubMed] [Google Scholar]
- 46.Durso R. J., Fisher A. K., Albright-Frey T. J., Reese J. C. (2001) Mol. Cell. Biol. 21, 7331–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright K. J., Marr M. T., 2nd, Tjian R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12347–12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R. (2008) Cell 135, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W. L., Coleman R. A., Grob P., King D. S., Florens L., Washburn M. P., Geles K. G., Yang J. L., Ramey V., Nogales E., Tjian R. (2008) Mol. Cell 29, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mengus G., Fadloun A., Kobi D., Thibault C., Perletti L., Michel I., Davidson I. (2005) EMBO J. 24, 2753–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fadloun A., Kobi D., Delacroix L., Dembélé D., Michel I., Lardenois A., Tisserand J., Losson R., Mengus G., Davidson I. (2008) Oncogene 27, 477–489 [DOI] [PubMed] [Google Scholar]
- 52.Wang X., Truckses D. M., Takada S., Matsumura T., Tanese N., Jacobson R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7839–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao H., Revach M., Moshonov S., Tzuman Y., Gazit K., Albeck S., Unger T., Dikstein R. (2005) Mol. Cell. Biol. 25, 206–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guermah M., Tao Y., Roeder R. G. (2001) Mol. Cell. Biol. 21, 6882–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olave I., Reinberg D., Vales L. D. (1998) Genes Dev. 12, 1621–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein J., Nolden M., Sanders S. L., Kirchner J., Weil P. A., Melcher K. (2003) J. Biol. Chem. 278, 6779–6786 [DOI] [PubMed] [Google Scholar]
- 57.Prochasson P., Neely K. E., Hassan A. H., Li B., Workman J. L. (2003) Mol. Cell 12, 983–990 [DOI] [PubMed] [Google Scholar]
- 58.Reeves W. M., Hahn S. (2005) Mol. Cell. Biol. 25, 9092–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gamper A. M., Roeder R. G. (2008) Mol. Cell. Biol. 28, 2517–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]