Abstract
Apical membrane antigen 1 (AMA1) is an essential component of the moving junction complex used by Apicomplexan parasites to invade host cells. We report the 2.0 Å resolution x-ray crystal structure of the full ectodomain (domains I, II, and III) of AMA1 from the pervasive protozoan parasite Toxoplasma gondii. The structure of T. gondii AMA1 (TgAMA1) is the most complete of any AMA1 structure to date, with more than 97.5% of the ectodomain unambiguously modeled. Comparative sequence analysis reveals discrete segments of divergence in TgAMA1 that map to areas of established functional importance in AMA1 from Plasmodium vivax (PvAMA1) and Plasmodium falciparum (PfAMA1). Inspection of the TgAMA1 structure reveals a network of apical surface loops, reorganized in both size and chemistry relative to PvAMA1/PfAMA1, that appear to serve as structural filters restricting access to a central hydrophobic groove. The terminal portion of this groove is formed by an extended loop from DII that is 14 residues shorter in TgAMA1. A pair of tryptophan residues (Trp353 and Trp354) anchor the DII loop in the hydrophobic groove and frame a conserved tyrosine (Tyr230), forming a contiguous surface that may be critical for moving junction assembly. The minimalist DIII structure folds into a cystine knot that probably stabilizes and orients the bulk of the ectodmain without providing excess surface area to which invasion-inhibitory antibodies can be generated. The detailed structural characterization of TgAMA1 provides valuable insight into the mechanism of host cell invasion by T. gondii.
Keywords: Parasitology, Protein Assembly, Protein Domains, Protein Structure, Receptor Structure-Function, X-ray Crystallography, Toxoplasma gondii
Introduction
Toxoplasma gondii, the etiological agent of toxoplasmosis, is a prevalent global pathogen capable of establishing acute and chronic infections in nearly all warm blooded animals (1, 2). Although largely asymptomatic in healthy individuals, T. gondii infections can be lethal to a developing fetus and immunocompromised cancer and AIDS patients (3–6). Toxoplasmosis can also result in severe ocular infections in both children and adults, and encysted forms of the parasite have recently been implicated in neuropsychiatric disorders, such as schizophrenia (7–9).
The success of T. gondii stems from its ability to persist in the environment, utilize several modes of transmission (10), and, importantly, to infect a broad range of host cells (1). A dominant feature that endows T. gondii and, in fact, all Apicomplexan parasites, including Plasmodium, Babesia, Cryptosporidium, and Neospora, with the ability to efficiently invade host cells is a multiprotein complex assembled at the moving junction (MJ)4 (2, 11). The MJ is an electron-dense, ringlike structure formed between the plasma membranes of the apical tip of the motile parasite and the target host cell (12). During invasion, T. gondii is rapidly engulfed within a parasitophorous vacuole (PV) as the MJ traverses in a posterior direction along the length of the parasite (13, 14). As it migrates, the MJ serves as a molecular sieve, selectively filtering host proteins from the PV (12, 15), thereby protecting the parasite from intracellular degradation (16).
Despite the critical role of the MJ in host cell invasion, only limited information exists describing the details of its assembly. This is due, in part, to the absence of structural information for the individual components. Importantly, however, studies with T. gondii have identified rhoptry proteins RON2, -4, -5, and -8 as forming part of the MJ complex targeted to the cytoplasmic face of the host cell membrane (17, 18). Despite an ambiguous orientation of TgRON2 in the membrane, recent studies have demonstrated a clear interaction between TgRON2 and the micronemal protein AMA1 (apical membrane antigen 1) (17–21), a core component of the MJ complex conserved across the phylum. A ligand-receptor model predicts that the parasite is able to provide its own ligand (TgAMA1) to the host cell-embedded RON complex (TgRON2/4/5/8) to promote invasion (18). This feature may explain the ability of Toxoplasma to invade its remarkably extensive cell range from a wide variety of warm blooded animals.
AMA1 was originally identified as an invariant surface antigen on Plasmodium knowlesi merozoites (22, 23), and monovalent Fab fragments of monoclonal antibodies against P. knowlesi AMA1 were sufficient to block in vitro invasion of erythrocytes (24). Subsequent genetic and immunological studies broadly established the importance of AMA1 as a core component of the invasion machinery (21, 25–27). Complete disruption of ama1 results in a lethal phenotype in Plasmodium (28) and T. gondii (26), whereas a conditional ama1 knock-out in T. gondii resulted in tachyzoites severely compromised for invasion (29). Immunological studies have shown that antibodies to both native and recombinant AMA1 recognize a conformational epitope and are protective in animal models of malarial infection (30–35). In addition, anti-AMA1 antibodies extracted from donor sera collected from areas endemic for malaria are both therapeutic and protective (36–38). The importance of AMA1 in both host cell invasion and immune regulation has prompted extensive study (19, 29, 39–43), including testing its potential as a malarial vaccine candidate.
Sequence analysis of AMA1 initially showed it to be a type I integral membrane protein, composed of a small intracellular C-terminal tail, a short trans-membrane region, and a large N-terminal ectodomain (26, 41). The three-domain architecture of the AMA1 ectodomain, originally proposed based on the disulfide bonding pattern (44), was definitively shown in the crystal structure of Plasmodium vivax AMA1 (PvAMA1). This seminal study established that DI and DII adopted a PAN (plasminogen, apple, nematode) motif (45), a module defining a diverse family of adhesins implicated in binding to protein or carbohydrate receptors, while showing little structural homology for DIII. Subsequent structural characterization of a truncated ectodomain of P. falciparum (PfAMA1) incorporating DI and DII (39) allowed for delineation of surface loops disordered in the original PvAMA1 structure (43). Of particular interest was an extended non-polymorphic DII loop that, along with a network of surface loops on DI, formed part of an apical hydrophobic groove. Mutation of a tyrosine (Tyr251-PfAMA1) to an alanine located in the center of this groove was sufficient to abrogate binding to RONs (19), highlighting the importance of this structural feature in formation of the MJ complex (39). A molecular interaction role was also proposed for DIII based on the observations that, when expressed on Chinese hamster ovary cells, DIII was sufficient to bind to the Kx membrane protein on trypsin-treated erythrocytes (46). Although only the original PvAMA1 structure included DIII in the context of DI and DII (43), follow-up structural studies of PfAMA1 DIII alone and in complex with invasion inhibitory antibodies have provided further insight into potential functional roles for this domain (19, 40, 42, 47–49).
AMA1 from P. falciparum and P. vivax are highly homologous with respect to sequence and structure. Comparative sequence analysis, however, reveals significant levels of divergence with AMA1 from T. gondii and other Apicomplexan parasites. Intriguingly, several of these divergent stretches map to sites shown to participate in assembly of the MJ complex, immune regulation, and host cell adhesion in Plasmodium AMA1s. To accurately define the distinctive structural features of TgAMA1, we have solved and refined the crystal structure of the fully processed ectoplasmic region to 2.0 Å resolution. The highly ordered structure provides a nearly complete view of the inter- and intramolecular interactions of DI, DII, and DIII that comprise the ectodomain. The structure of TgAMA1 provides a critical step in defining its elusive role within the MJ and, more broadly, its contribution to the unique invasion characteristics of T. gondii.
EXPERIMENTAL PROCEDURES
Bioinformatics
Boundaries for DI, DII, and DIII were defined based on the paradigm established for PvAMA1 (43). Phylogenic analysis was performed using MEGA 4 (50, 51), and multiple sequences were aligned using Kalign (52, 53). Accession numbers for aligned AMA1 sequences are as follows: T. gondii (ME49_055260), Neospora caninum (BAF45372), P. falciparum (XP_001348015.1), and Babesia bovis (AAS58045.1). The P. vivax (XP_001615447) sequence was modified to reflect the sequence crystallized by Pizarro et al. in 2005 (43).
Cloning, Expression, and Purification
A clone encoding the fully processed ectoplasmic domain of TgAMA1 was generated in a modified pAcGP67b vector (Pharmingen) incorporating a C-terminal hexahistidine tag and thrombin cleavage site. To generate TgAMA1 encoding virus for insect cell protein production, the TgAMA1 clone was transfected with linearized baculovirus DNA into Sf9 cells and amplified to a high titer. Hi-5 cells at 1.8 × 106 cells/ml were infected with amplified virus for 72 h, after which time the supernatant was harvested, concentrated, and applied to a HisTrapFF nickel affinity column. TgAMA1 was eluted with an increasing concentration of imidazole with fractions analyzed by SDS-PAGE and pooled based on purity. The hexahistidine tag was removed by thrombin cleavage, and TgAMA1 was further purified by size exclusion chromatography (Superdex 16/60 200) in HEPES-buffered saline (20 mm HEPES, pH 7.5, 150 mm NaCl). The final yield of purified TgAMA1 was ∼2 mg of purified protein/liter of insect cell culture.
Crystallization and Data Collection
Crystals of TgAMA1 were initially identified in the Index Screen (Hampton Research) and subsequently refined to a final condition of 20% polyethylene glycol 3350, 100 mm HEPES, pH 7.5, and 50 mm NaCl. Small crystals were observed after 2 days and grew to a final size of 0.5 × 0.1 × 0.1 mm within 6 days. The final drops consisted of 1.5 μl of protein (15 mg/ml) with 1.5 μl of reservoir solution and were equilibrated against 100 μl of reservoir solution. Cryoprotection of the TgAMA1 crystal was carried out in mother liquor supplemented with 5% glycerol and 5% ethylene glycol for 20 s and flash-cooled at 100 K directly in the cryostream. Diffraction data were collected on beamline 9-2 at SSRL (Stanford Synchrotron Radiation Laboratory) at a wavelength of 0.9794 Å. A total of 720 images were collected with a 1° oscillation and 2-s exposure.
Data Processing, Structure Solution, and Refinement
Diffraction data to 2.0 Å were processed using Imosflm (54) and Scala (55) in the CCP4 suite of programs (56). Initial phases were obtained by molecular replacement using MOLREP (57) with the individual DI and DII domains of PfAMA1 (Protein Data Bank code 2Q8A) pruned with CHAINSAW (58) to better reflect the TgAMA1 sequence. No molecular replacement solution was obtained for DIII using a pruned or polyserine model. Tracing of the DIII chain was ultimately achieved using 4-fold non-crystallographic symmetry averaging. Solvent molecules were selected using COOT (59), and refinement was carried out using Refmac5 (57). The overall structure of TgAMA1 was refined to an Rcryst of 18.4% and an Rfree of 24.8%. Stereochemical analysis performed with PROCHECK and SFCHECK in CCP4 (56) showed excellent stereochemistry, with more than 95% of the residues in the favored conformations and no residues modeled in disallowed orientations of the Ramachandran plot. Overall, 5% of the reflections were set aside for calculation of Rfree. Data collection and refinement statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell.
| Parameters | Values |
|---|---|
| Data collection | |
| Space group | P1 |
| a, b, c (Å) | 66.15, 76.07, 88.25 |
| α, β, γ (degrees) | 72.19, 71.44, 72.90 |
| Wavelength | 0.9794 |
| Resolution (Å) | 52.24-2.00 |
| Measured reflections | 391,114 (56,271) |
| Unique reflections | 98,629 (14,195) |
| Redundancy | 4.0 (4.0) |
| Completeness (%) | 96.4 (95.3) |
| I/σ(I) | 13.7 (3.3) |
| Rmergea | 0.074 (0.433) |
| Refinement statistics | |
| Resolution range (Å) | 43.90–2.00 (2.05–2.00) |
| Rcrystb | 0.184 (0.228) |
| Rfreec | 0.248 (0.309) |
| No. of atoms | |
| Protein (chain A, B, D, E) | 3225, 3202, 2932, 3149 |
| Solvent | 1177 |
| Glycerol | 6 |
| B-values | |
| Protein (chain A, B, D, E) (Å2) | 25.08, 25.78, 26.28, 25.75 |
| Solvent (Å2) | 35.43 |
| Glycerol (Å2) | 25.89 |
| Root mean square deviation from ideality | |
| Bond lengths (Å) | 0.02 |
| Bond angles (degrees) | 1.97 |
| Ramachandran statistics | |
| Most favored | 95.3% |
| Allowed | 4.7% |
| Disallowed | 0.0% |
a Rmerge = ΣhklΣi|Ihkl,i − [Ihkl]|/ΣhklΣiIhkl,i, where [Ihkl] is the average of symmetry-related observations of a unique reflection.
b Rcryst = Σ|Fobs − Fcalc/ΣFobs, where Fobs and Fcalc are the observed and the calculated structure factors, respectively.
c Rfree is R using 5% of reflections randomly chosen and omitted from refinement.
RESULTS AND DISCUSSION
Domain Divergence; Comparative Sequence Analysis of DI, DII, and DIII
Domain boundaries of the fully processed TgAMA1 ectodomain were defined based on the paradigm established for PvAMA1 and PfAMA1 (Fig. 1). TgAMA1 DI spans residues from Thr67 to Pro287 (residues numbered from initiation methionine in the signal sequence), DII spans from Asn288 to Asn415, and DIII spans from Phe416 to Ala487. TgAMA1 is most closely related to AMA1 from N. caninum, with 75% sequence identity distributed over the entire ectodomain. Increased evolutionary divergence is observed with respect to AMA1s from P. falciparum, P. vivax, and B. babesi, with DIII, in particular, displaying less than 10% sequence identity.
FIGURE 1.
Phylogenic tree and multiple sequence alignments of domains I, II, and III that comprise the ectodomain of AMA1 from T. gondii (TgAMA1), N. caninum (NcAMA1), P. falciparum (PfAMA1), P. vivax (PvAMA1), and B. bovis (BbAMA1). Scale bars on the phylogenetic trees indicate evolutionary distances. Cysteine residues are shown in red and numbered with respect to disulfide bond partner based on the TgAMA1 crystal structure. Residues shown in blue are either invariant or highly conserved in at least four of the five sequences. Domain boundaries were defined based on the paradigm established for Plasmodium AMA1s (39).
Sequence analysis reveals a network of conserved cysteine residues in DI and DII, suggesting a conserved structural core. Several insertions and deletions in the primary sequence, however, map to functionally relevant sites in PvAMA1/PfAMA1 and may be responsible for the unique host cell invasion capabilities of T. gondii. Six invariant cysteines in TgAMA1 DI are supported by an overall moderate level of sequence identity with denoted species (on average 35%) (Fig. 1, top). Several of the insertions are highly polar or proline-rich and map to surface-exposed loops in PfAMA1 predicted to serve as structural filters in governing access to a central groove (39). A single eight-residue deletion at the N terminus of TgAMA1 DI may also carry functional implications as the additional residues in PvAMA1 (43) form an extended strand that connects DI and DIII. DII is approximately two-thirds the size of DI and encodes four invariant cysteines but is less well conserved, with an overall sequence identity of ∼25% (Fig. 1, middle). The most unique feature of TgAMA1 DII is a deletion of 14 residues that maps to the non-polymorphic DII loop of PfAMA1 (39). Recent studies have shown that the PfAMA1 DII loop presents an epitope recognized by an invasion-inhibitory monoclonal antibody and a T cell epitope implicated in the human response to Plasmodium infection (39, 43, 44). The greatest divergence among the AMA1 ectodomains, however, is localized to DIII (Fig. 1, bottom), which is nearly 50% shorter in TgAMA1 relative to Plasmodium AMA1s. Despite comprising only 71 residues, TgAMA1 DIII encodes six cysteines, four of which align with cysteines in PfAMA1/PvAMA1. By contrast, B. bovis DIII encodes just four cysteines, all of which align with Plasmodium AMA1s consistent with a more recent evolutionary divergence.
Protein Production, Crystallization, and Structure Solution of the TgAMA1 Ectodomain
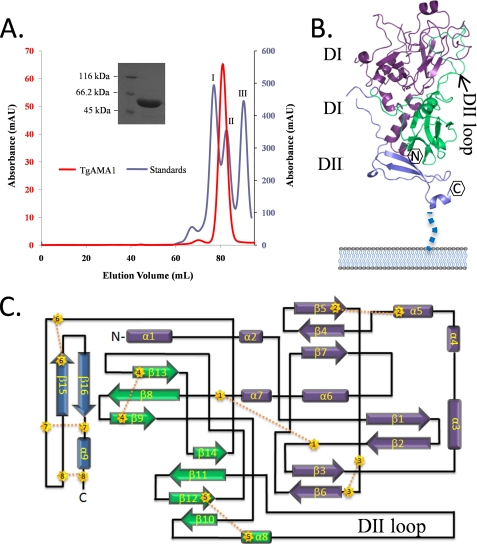
Attempts to produce soluble two-domain (DI and DII) or three-domain (DI, DII, and DIII) versions of TgAMA1 in Escherichia coli were unsuccessful. Ultimately, we were able to recombinantly produce a soluble form of the fully processed TgAMA1 ectodomain (DI, DII, and DIII) using the baculovirus strategy in insect cells. Tangential flow concentration, nickel affinity, and size exclusion chromatography were used to purify TgAMA1 to homogeneity (Fig. 2A). Comparison of the TgAMA1 size exclusion chromatography chromatogram against a series of globular protein standards showed that TgAMA1 eluted as a monomer consistent with Plasmodium AMA1s (39, 43).
FIGURE 2.
Overall structure of the TgAMA1 ectodomain. A, Superdex 200 gel filtration analysis showing that TgAMA1 (red peak) elutes as a monomer of ∼50 kDa. The blue peaks represent protein standards: peak I, conalbumin (75 kDa); peak II, ovalbumin (43 kDa); peak III, carbonic anhydrase (29 kDa). Inset, SDS-PAGE analysis of the column fractions, with TgAMA1 migrating at ∼50 kDa (the expected molecular mass of the TgAMA1 construct is 47,960 Da). B, secondary structure depiction of the TgAMA1 ectodomain displayed in the predicted orientation with respect to the parasite cell surface. Purple, DI; green, DII; slate blue, DIII. C, topology diagram of the TgAMA1 ectodomain (chain A) with cysteine residues depicted by numbered gold stars and disulfide bonds as yellow dotted lines.
TgAMA1 crystallized with four molecules in the P1 unit cell. Molecular replacement solutions were independently determined for TgAMA1 DI and DII using PfAMA1 DI and DII as search models with the sequences pruned to reflect TgAMA1 and surface loops removed (39). No molecular replacement solution was obtained for TgAMA1 DIII, and initial electron density maps were inadequate to trace or even manually position a DIII model. Phase improvement strategies incorporating 4-fold non-crystallographic symmetry averaging resulted in maps into which all but three amino acids of DIII were modeled. Each polypeptide chain in the unit cell is largely equivalent with respect to degree of modeled structure and organization as shown by root mean square deviations relative to chain A of 0.61 Å over 361 Cα atoms (chain B), 0.53 Å over 353 Cα atoms (chain D), and 0.44 Å over 386 Cα atoms (chain E). Chain A is the most extensively modeled, yet a small section of loop in chain A (Gln338–Asp352) is reorganized with respect to the analogous regions in the three other NCS-related chains (supplemental Fig. 1). This alternate conformation appears to be a crystallization artifact arising from intermolecular packing. Therefore, structural analysis of this region is based on the loop conformation observed in chains B, D, and E.
Overall Structure
The assembled TgAMA1 ectodomain extends 80 Å in height and, on average, 35 Å in width, as shown in Fig. 2B with respect to its predicted orientation to the membrane. Whereas the bulk of DI (Fig. 2B, purple) is positioned atop DII (Fig. 2B, green), a series of short N-terminal helices span the length of DII, resulting in the N terminus positioned within 21 Å of the C terminus of DIII (Fig. 2B, slate blue). DI and DII are intimately associated and form the bulk of the ectodomain, whereas the majority of the smaller DIII resides at the posterior, membrane-proximal region (Fig. 2B).
The DI domain is composed of small helical bundles, short twisted β-sheets, and an extensive network of random coils. Despite the low secondary structure content, DI is well ordered, due, in part, to the trio of stabilizing disulfide bonds (Fig. 2C). The core of DII is centrally located within the ectodomain with the exception of a 33-residue loop (termed the DII loop) that packs lengthwise against DI, forming an extended interface (Fig. 2B). The base of the DII loop is stabilized by a disulfide bond with a second disulfide bond stabilizing the DII core (Fig. 2C). A 25-residue tether connects the core of DIII to DII, making it possible for the DIII to be positioned at the posterior end of the ectodomain. Of the remaining 46 residues that comprise the DIII core, six are cysteines organized into three disulfide bonds that form a structurally ultrastable cystine knot with disulfide bond 8 (Cys452–Cys479) threading through a ring formed by bond 6 (Cys435–Cys459) and bond 7 (Cys447–Cys471) (Fig. 2C).
To probe the level of structural conservation, a DALI (60) search was individually performed with TgAMA1 DI, DII, and DIII. As expected, TgAMA1 DI shows a high level of structural homology to PfAMA1/PvAMA1 (39, 43), with Z scores ranging from 18 to 22. Intriguingly, however, no structural relationship was identified corresponding to the protein-protein or protein-carbohydrate interacting PAN superfamily (45), as was originally observed for PvAMA1 (43). It is likely that the insertions and deletions in TgAMA1 DI (Fig. 1) may limit its categorization as part of the PAN superfamily. Despite the lower sequence and structural (Z scores from 8 to 12.6) homology for DII resulting from the 14-residue deletion, a clear correlation (Z score of 7.5) is observed with PAN-containing proteins, such as hepatocyte growth factor. This structural feature suggests that DII may participate in ligand recognition. No statistical structural similarity was observed for DIII, consistent with less than 10% sequence identity observed.
Intimate Interfaces; Assembling the TgAMA1 Ectodomain
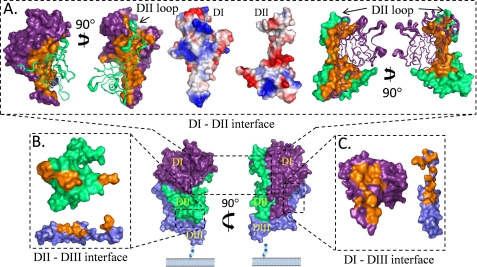
The highly ordered TgAMA1 structure provides an opportunity to thoroughly analyze the inter- and intramolecular interactions that stabilize the structural framework of the ectodomain. Each interdomain interface is formed from numerous non-covalent forces and substantial shape complementarity, as shown by a maximum complexation significance scores of 1.0 (61). To visualize the extensive nature of the interdomain interactions, an “open book” perspective is presented in Fig. 3 with residues contributing to the interfaces displayed in orange.
FIGURE 3.
Assembly of the TgAMA1 ectodomain. Orthogonal surface views of TgAMA1 (chain B) are oriented with respect to the cell membrane; the DI domain is shown in purple, the DII domain in green, and the DIII domain in slate blue (bottom center). An open book perspective provides a view of the interdomain interfaces with buried residues shown in orange. Each interface shows a high degree of structural complementarity as denoted by a complexation significance score of 1.0. A, opposing views of DI and DII are shown with one domain represented as a surface and the other as a secondary structure tube. The diameter of the tube is based on B-factors, with larger diameters representing more flexible regions. The DII loop packs tightly against the DI domain to form a structurally contiguous surface and contributes nearly half of the buried surface area. Electrostatic representations of DI and DII highlight the charge complementarity that promotes assembly. B, the DII/DIII interface is formed by a discontinuous epitope formed primarily by the core cystine knot of DIII. C, the linker region that connects DIII to DII contributes to the buried surface area by packing against the lower portion of DI.
In total, more than 7350 Å2 of surface area is buried upon assembly of the TgAMA1 ectodomain. The largest interface is formed between DI and DII, resulting in a buried surface area of ∼4849 Å2, with 2319 Å2 contributed from DI and 2530 Å2 from DII (Fig. 3A). The DI/DII interface is stabilized by 21 interdomain hydrogen bonds and three salt bridges (Asn288O-Gln289N; Asp102Oδ1-His357Nδ1, and Arg259Nϵ-Glu330Oϵ1). The role of polar interactions in defining the DI/DII interface is highlighted by an electrostatic surface representation that shows distinct complementary charged surfaces (Fig. 3A, middle). Additional polarity is provided by a small yet well ordered network of buried solvent that may also serve to increase shape complementarity. A major component of the DI/DII interface is contributed by the DII loop that extends from Gly333 to Arg369 and accounts for approximately half of the buried surface area between the two domains. Although the majority of the DII loop is structurally invariant across the four monomers, intramolecular packing results in a contorted segment (Gln338–Asp352) of the DII loop in chain A (Supplemental Fig. 1). As a result, defining the contributions of the DII loop to ectodomain stability is restricted to chains B, D, and E.
The core cystine knot of DIII displays a discontinuous epitope and contributes 637 Å2 of buried surface area, with DII contributing 663 Å2 for a total buried surface area of 1300 Å2 (Fig. 3B). Fourteen hydrogen bonds complement a bifurcated salt bridge between the carboxylate side chain of Asp448 on DIII and the ϵ-amino group of Lys301 and the η-nitrogens of Arg303 on DII. The extended tether that connects DIII to the DI/DII core also contributes to the total buried surface area of 1400 Å2 (740 Å2 from DI and 660 Å2 from DIII) (Fig. 3C). Despite the increased surface area relative to DII/DIII, the DI/DIII interface is stabilized by only six hydrogen bonds and one salt bridge (Arg84NH1-Glu430Oϵ1). Instead, DI/DIII stability relies on complementary hydrophobic surfaces where, for example, DI Phe81 and DIII Phe416 provide the most individual buried surface area of any interface residue. Overall, each domain is intimately associated with the remaining two domains to form a highly stable structure.
Structural Divergence in the Apical Region of TgAMA1; Functional Implications
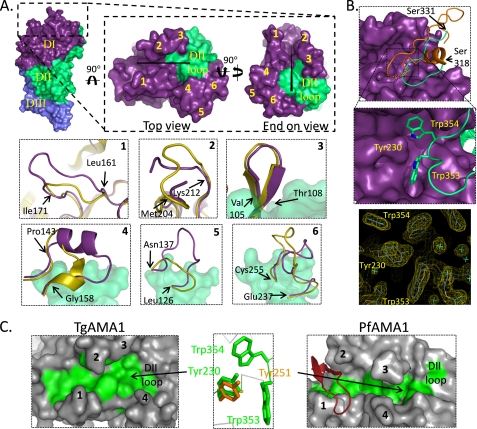
Structural analysis reveals an extended groove at the anterior, or membrane distal tip, of the TgAMA1 ectodomain that extends 30 Å in length and averages ∼10 Å in width (Fig. 4A, inset, horizontal black bar).
FIGURE 4.
Structural and functional implications of the TgAMA1 apical surface. A, surface representations showing the TgAMA1 ectodomain (chain B) with top and end-on views of the membrane distal, apical region with DI and DII colored purple and green, respectively. The black line in the top view represents the surface groove, and the black triangle in the end-on view denotes depth, where the wide part of the triangle is directed toward the viewer. The network of surface loops that define the central groove in TgAMA1 are numbered 1–6 and displayed in yellow on the surface of TgAMA1. Zoomed in views of each loop compared with the analogous loops in PfAMA1 (Protein Data Bank code 2Q8A) (40) are shown in the inset boxes labeled with the appropriate loop number. See “Results and Discussion” for a detailed description of each loop. B, the DII loop (green) is displayed in secondary structure format packed against the surface of DI (purple). The analogous DII loop (orange) from PfAMA1 is shown for comparison. A pair of tryptophan residues (Trp353 and Trp354) on the TgAMA1 DII loop interdigitates into pockets on DI bifurcated by the conserved Tyr230. The ordered structure of Trp353, Trp354, Tyr230, and associated solvent network is shown as a Sigma A-weighted electron density map contoured at 1.3 σ. C, the hydrophobic base of the central grooves in TgAMA1 and PfAMA1 are shown in green with the remainder of the surface shown in gray. Black numbers identify the individual surface loops described above. The red secondary structure represents a portion of the invasion inhibitory antibody 141-1 co-crystallized with PfAMA1 (Protein Data Bank code 2Z8V) (47). Note the structural conservation of the central tyrosine (Tyr230 in TgAMA1 and Tyr251 in PfAMA1) on DI despite the significant reorganization of the DII loop.
Surface loops (identified by yellow numbers in Fig. 4A, inset) span the length of the groove and probably serve as a selectivity filter in mediating access to the base of the groove as proposed for PfAMA1 (39). To assess potential functional implications of these loops, we present structural overlays with the analogous loops in PfAMA1 (40) (Fig. 4A).
Loops 1 and 2 are centrally positioned on opposite sides of the groove, constricting the central segment to ∼6 Å. Despite loop 1 being only three residues (Pro164, Ser165, and Gly166) longer than the analogous loop in PfAMA1, it is significantly reorganized (Fig. 4A, 1) in structure yet well ordered as shown by low B-factors. In PfAMA1, this loop directly coordinates a series of invasion-inhibitory antibodies (40, 47), thereby playing a critical role in pathogenesis. The altered structure of loop 1 in TgAMA1 may, therefore, promote diversity in ligand recognition. A high degree of flexibility is observed in the apical region of loop 2, resulting in three unmodeled residues, yet the overall size is similar between TgAMA1 and PfAMA1 (Fig. 4A, 2). A second set of loops, denoted as loops 3 and 4 in TgAMA1, extend the groove to incorporate the tip of the DII loop. The size of the β hairpin structure in loop 3 is largely conserved (Fig. 4A, 3), although the loop is shifted 1.5 Å toward the central groove in TgAMA1. This displacement is probably due to the reorganized DII loop that is much smaller in TgAMA1. A more striking structural reorganization coupled to the smaller DII loop is observed in loop 4 (Fig. 4A, 4), where the base of the loop provides a hydrophobic backstop with substantial shape complementarity to accommodate the DII loop. The tip of loop 4, however, is highly polar with Glu145, Lys146, Lys149, and Gln150 directed away from the base of the groove, where it may serve as the initial structural filter in defining appropriate ligands. Loops 5 and 6 are positioned at the periphery of the central groove and form a contiguous surface that appears to be critical in promoting correct orientation of the DII loop in the central groove (Fig. 4A, 5 and 6).
The TgAMA1 DII loop (Fig. 4B) is composed of random coil, yet it is reasonably well ordered due to the tight packing against DI. A pair of tryptophan residues (Trp353 and Trp354) forms the tip of the DII loop interdigitating into discrete hydrophobic pockets on DI (Fig. 4B). The first pocket is formed on one side by Leu155, Tyr148, Val142, and Pro143 from the base of loop 4 and on the opposite side by Tyr230. The second pocket, which accommodates Trp354 from the DII loop, also makes use of Tyr230 and is completed by Tyr215, Val105, Ala203, Tyr213, and Tyr110 derived from loops 2 and 3. Note that despite the reorganization of Gln338 to Asp352 from the DII loop in chain A, the region encompassing Trp353 and Trp354 is well anchored and structurally conserved across each of the four monomers (supplemental Fig. 1). Although the presentation of the tryptophan pair in TgAMA1 is not conserved in PfAMA1, the central tyrosine (Tyr230 in TgAMA1; Tyr251 in PfAMA1) is invariant, with a root mean square deviation of less than 0.2 Å (Fig. 4B, middle). The conserved location of this tyrosine suggests a key role in defining the function of the central groove, and, indeed, mutation of Tyr251 in PfAMA1 to alanine was sufficient to abrogate formation of the MJ complex (19). Based on these observations and the recent evidence of a direct interaction between TgAMA1 and RON2 in the absence of RON4, -5, and -8 (18), we predict that this central tyrosine may serve as a hot spot residue in mediating AMA1-RON2 complex formation.
In addition to the central tyrosine, the residues that form the base of the central groove are primarily hydrophobic, leading to the initial description as a hydrophobic trough in the PvAMA1 (43) and PfAMA1 (39) crystal structures. In TgAMA1, 15 residues (Ile185, Leu179, Phe174, Phe197, Ile171, Phe163, Ile161, Met203, Tyr230, Val142, Trp354, Val231, Trp353, Trp253, and Leu155) form the hydrophobic trough. Of these, 10 are spatially conserved with PfAMA1, where the original nine-residue trough (39) was recently expanded to 12 residues (40). The additional hydrophobic residues in TgAMA1 form a second layer of hydrophobicity near the constricted region between loops 1 and 2 and compensate for the polar substitution of Thr201 in TgAMA1 loop 2 for the structurally analogous Met224 in PfAMA1, thereby maintaining a contiguous hydrophobic surface. An additional noteworthy feature is a small yet well defined solvent network incorporating the hydroxyl group of Tyr230 (Fig. 4B, bottom). Overall, the hydrophobic trough in TgAMA1 is shorter and wider relative to the analogous region in PfAMA1 (Fig. 4C, left and right). This, in conjunction with the reorganized network of surface loops, may define the divergent repertoire of host cells infected by Toxoplasma and Plasmodium.
Global Structural Rearrangement in TgAMA1 DIII
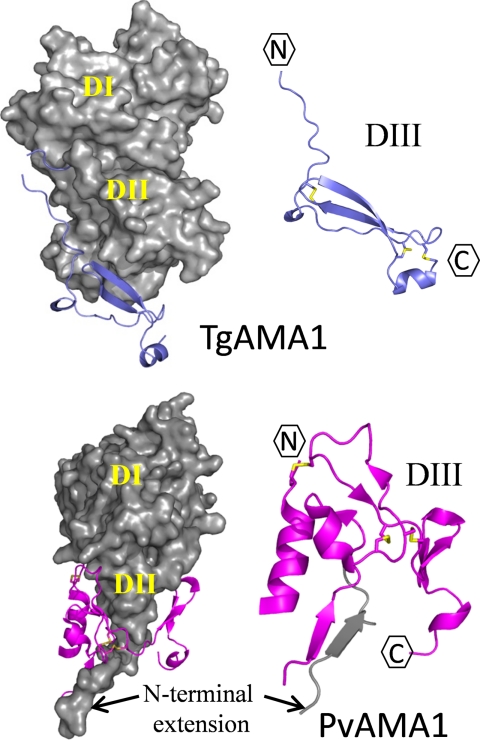
The structural reorganization of the TgAMA1 apical region (DI/DII) is likely to have a profound impact on assembly of the MJ complex, yet it is DIII that displays the most divergence between TgAMA1 and PvAMA1/PfAMA1. This observation is especially interesting because DIII has been directly implicated in mediating erythrocyte adhesion with PfAMA1 (46). Thus, the reorganization of TgAMA1 DIII may also contribute to the broad infectivity of T. gondii.
A comparative structural analysis revealed a rationale for why TgAMA1 DIII was substantially smaller than PvAMA1/PfAMA1 DIII (Fig. 5). As described above, the N terminus of PvAMA1 DI is eight residues longer than in TgAMA1 with the additional residues adopting a single β strand that extends away from the DI core toward DIII. PvAMA1 DIII forms a saddle-like structure with a central groove to accommodate and stabilize this N-terminal extension. The shorter N-terminal region of TgAMA1 DI does not extend to DIII and therefore obviates the need for additional stabilizing features contributed by DIII. Interestingly, the minimalist structure of TgAMA1 DIII is sufficiently large to adopt the structurally ultrastable cystine knot that may serve as a foundation to properly orient the DI/DII core with respect to the parasite cell membrane. In addition to a base structural role, the smaller TgAMA1 DIII provides little excess surface area to which growth inhibitory antibodies might be generated as recently suggested for PfAMA1 DIII (48). In this study, engineered peptodomimetics of PfAMA1 DIII were used to identify two immunodominant epitopes comprising the linear sequences KRIKLN and DEGNKKII capable of generating a protective antibody response (48). With the exception of the two terminal isoleucine residues, the residues that comprise these epitopes are located in the divergent region of PfAMA1 DIII not represented in the smaller TgAMA1 DIII (Fig. 1).
FIGURE 5.
Structural divergence in DIII. The DI and DII domains of TgAMA1 (chain A) and PvAMA1 (Protein Data Bank code 1W81) (43) are displayed as gray surfaces in the same orientation. The TgAMA1 and PvAMA1 DIII domains are shown in slate blue and magenta secondary structures, respectively. Cysteine residues that define the cystine knot configuration are shown as yellow side chains.
CONCLUSIONS
T. gondii is one of the most successful parasites, yet a detailed molecular mechanism describing assembly and function of the MJ complex remains elusive. The highly ordered crystal structure of TgAMA1 presented herein reveals an intriguing level of divergence from its Plasmodium counterparts. While maintaining a conserved structural core in DI and DII, reorganized structural elements in TgAMA1 map to areas of established functional importance in PfAMA1, including a network of surface loops that frame a central hydrophobic groove. Because AMA1 is vital for parasitic invasion and has been previously shown to interact with a variety of other proteins during MJ formation, the implications of novel features leading to altered ligand binding sites are profoundly significant. More specifically, we predict that the hydrophobic groove (and in particular Tyr230) plays a key role in engaging RON2 during assembly of the MJ complex. Our structure of the complete TgAMA1 ectodomain will help catalyze a better understanding of the role AMA1 plays in host cell invasion by T. gondii and, indeed, all Apicomplexan parasites. The structural details provided here will also be useful to refine AMA1 vaccine development efforts for both Plasmodium and Toxoplasma.
This work was supported by Canadian Institutes of Health Research (CIHR) Grant MOP82915 (to M. J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The atomic coordinates and structure factors (code 2x2z) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MJ
- moving junction
- TgAMA1
- PvAMA1, and PfAMA1, T. gondii, P. vivax, and P. falciparum AMA1, respectively
- PV
- parasitophorous vacuole
- RON
- rhoptry neck protein.
REFERENCES
- 1.Tenter A. M., Heckeroth A. R., Weiss L. M. (2000) Int. J. Parasitol. 30, 1217–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill D., Dubey J. P. (2002) Clin. Microbiol. Infect. 8, 634–640 [DOI] [PubMed] [Google Scholar]
- 3.Jackson M. H., Hutchison W. M. (1989) Adv. Parasitol. 28, 55–105 [DOI] [PubMed] [Google Scholar]
- 4.Luft B. J., Brooks R. G., Conley F. K., McCabe R. E., Remington J. S. (1984) JAMA 252, 913–917 [PubMed] [Google Scholar]
- 5.Luft B. J., Remington J. S. (1992) Clin. Infect. Dis. 15, 211–222 [DOI] [PubMed] [Google Scholar]
- 6.McDonald J. C., Gyorkos T. W., Alberton B., MacLean J. D., Richer G., Juranek D. (1990) J. Infect. Dis. 161, 769–774 [DOI] [PubMed] [Google Scholar]
- 7.Flegr J., Havlícek J., Kodym P., Malý M., Smahel Z. (2002) BMC Infect. Dis. 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henriquez S. A., Brett R., Alexander J., Pratt J., Roberts C. W. (2009) Neuroimmunomodulation 16, 122–133 [DOI] [PubMed] [Google Scholar]
- 9.Torrey E. F., Yolken R. H. (2003) Emerg. Infect. Dis. 9, 1375–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones J. L., Kruszon-Moran D., Wilson M. (2003) Emerg. Infect. Dis. 9, 1371–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibley L. D. (2004) Science 304, 248–253 [DOI] [PubMed] [Google Scholar]
- 12.Aikawa M., Miller L. H., Johnson J., Rabbege J. (1978) J. Cell Biol. 77, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel R., Schupp K., Raether W., Bierther F. W. (1980) Int. J. Parasitol. 10, 309–313 [DOI] [PubMed] [Google Scholar]
- 14.Suss-Toby E., Zimmerberg J., Ward G. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8413–8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carruthers V., Boothroyd J. C. (2007) Curr. Opin. Microbiol. 10, 83–89 [DOI] [PubMed] [Google Scholar]
- 16.Mordue D. G., Monroy F., La Regina M., Dinarello C. A., Sibley L. D. (2001) J. Immunol. 167, 4574–4584 [DOI] [PubMed] [Google Scholar]
- 17.Alexander D. L., Mital J., Ward G. E., Bradley P., Boothroyd J. C. (2005) PLoS Pathog. 1, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besteiro S., Michelin A., Poncet J., Dubremetz J. F., Lebrun M. (2009) PLoS Pathog. 5, e1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins C. R., Withers-Martinez C., Hackett F., Blackman M. J. (2009) PLoS Pathog. 5, e1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Straub K. W., Cheng S. J., Sohn C. S., Bradley P. J. (2009) Cell Microbiol. 11, 590–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H., Compaore M. K., Lee E. G., Liao M., Zhang G., Sugimoto C., Fujisaki K., Nishikawa Y., Xuan X. (2007) Mol. Biochem. Parasitol. 151, 205–212 [DOI] [PubMed] [Google Scholar]
- 22.Deans J. A., Alderson T., Thomas A. W., Mitchell G. H., Lennox E. S., Cohen S. (1982) Clin. Exp. Immunol. 49, 297–309 [PMC free article] [PubMed] [Google Scholar]
- 23.Deans J. A., Thomas A. W., Alderson T., Cohen S. (1984) Mol. Biochem. Parasitol. 11, 189–204 [DOI] [PubMed] [Google Scholar]
- 24.Thomas A. W., Deans J. A., Mitchell G. H., Alderson T., Cohen S. (1984) Mol. Biochem. Parasitol. 13, 187–199 [DOI] [PubMed] [Google Scholar]
- 25.Gaffar F. R., Yatsuda A. P., Franssen F. F., de Vries E. (2004) Infect. Immun. 72, 2947–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hehl A. B., Lekutis C., Grigg M. E., Bradley P. J., Dubremetz J. F., Ortega-Barria E., Boothroyd J. C. (2000) Infect. Immun. 68, 7078–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silvie O., Franetich J. F., Charrin S., Mueller M. S., Siau A., Bodescot M., Rubinstein E., Hannoun L., Charoenvit Y., Kocken C. H., Thomas A. W., Van Gemert G. J., Sauerwein R. W., Blackman M. J., Anders R. F., Pluschke G., Mazier D. (2004) J. Biol. Chem. 279, 9490–9496 [DOI] [PubMed] [Google Scholar]
- 28.Triglia T., Healer J., Caruana S. R., Hodder A. N., Anders R. F., Crabb B. S., Cowman A. F. (2000) Mol. Microbiol. 38, 706–718 [DOI] [PubMed] [Google Scholar]
- 29.Mital J., Meissner M., Soldati D., Ward G. E. (2005) Mol. Biol. Cell 16, 4341–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders R. F., Crewther P. E., Edwards S., Margetts M., Matthew M. L., Pollock B., Pye D. (1998) Vaccine 16, 240–247 [DOI] [PubMed] [Google Scholar]
- 31.Cohen S., McGregor I. A., Carrington S. (1961) Nature 192, 733–737 [DOI] [PubMed] [Google Scholar]
- 32.Deans J. A., Knight A. M., Jean W. C., Waters A. P., Cohen S., Mitchell G. H. (1988) Parasite Immunol. 10, 535–552 [DOI] [PubMed] [Google Scholar]
- 33.Narum D. L., Ogun S. A., Batchelor A. H., Holder A. A. (2006) Infect. Immun. 74, 5529–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narum D. L., Ogun S. A., Thomas A. W., Holder A. A. (2000) Infect. Immun. 68, 2899–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stowers A. W., Kennedy M. C., Keegan B. P., Saul A., Long C. A., Miller L. H. (2002) Infect. Immun. 70, 6961–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodder A. N., Crewther P. E., Anders R. F. (2001) Infect. Immun. 69, 3286–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polley S. D., Mwangi T., Kocken C. H., Thomas A. W., Dutta S., Lanar D. E., Remarque E., Ross A., Williams T. N., Mwambingu G., Lowe B., Conway D. J., Marsh K. (2004) Vaccine 23, 718–728 [DOI] [PubMed] [Google Scholar]
- 38.Sabchareon A., Burnouf T., Ouattara D., Attanath P., Bouharoun-Tayoun H., Chantavanich P., Foucault C., Chongsuphajaisiddhi T., Druilhe P. (1991) Am. J. Trop. Med. Hyg. 45, 297–308 [DOI] [PubMed] [Google Scholar]
- 39.Bai T., Becker M., Gupta A., Strike P., Murphy V. J., Anders R. F., Batchelor A. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coley A. M., Gupta A., Murphy V. J., Bai T., Kim H., Foley M., Anders R. F., Batchelor A. H. (2007) PLoS Pathog. 3, 1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donahue C. G., Carruthers V. B., Gilk S. D., Ward G. E. (2000) Mol. Biochem. Parasitol. 111, 15–30 [DOI] [PubMed] [Google Scholar]
- 42.Igonet S., Vulliez-Le Normand B., Faure G., Riottot M. M., Kocken C. H., Thomas A. W., Bentley G. A. (2007) J. Mol. Biol. 366, 1523–1537 [DOI] [PubMed] [Google Scholar]
- 43.Pizarro J. C., Vulliez-Le Normand B., Chesne-Seck M. L., Collins C. R., Withers-Martinez C., Hackett F., Blackman M. J., Faber B. W., Remarque E. J., Kocken C. H., Thomas A. W., Bentley G. A. (2005) Science 308, 408–411 [DOI] [PubMed] [Google Scholar]
- 44.Hodder A. N., Crewther P. E., Matthew M. L., Reid G. E., Moritz R. L., Simpson R. J., Anders R. F. (1996) J. Biol. Chem. 271, 29446–29452 [DOI] [PubMed] [Google Scholar]
- 45.Tordai H., Banyai L., Patthy L. (1999) FEBS Lett. 461, 63–67 [DOI] [PubMed] [Google Scholar]
- 46.Kato K., Mayer D. C., Singh S., Reid M., Miller L. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5552–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson K. A., Streltsov V. A., Coley A. M., Dolezal O., Hudson P. J., Batchelor A. H., Gupta A., Bai T., Murphy V. J., Anders R. F., Foley M., Nuttall S. D. (2007) Structure 15, 1452–1466 [DOI] [PubMed] [Google Scholar]
- 48.Mueller M. S., Renard A., Boato F., Vogel D., Naegeli M., Zurbriggen R., Robinson J. A., Pluschke G. (2003) Infect. Immun. 71, 4749–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nair M., Hinds M. G., Coley A. M., Hodder A. N., Foley M., Anders R. F., Norton R. S. (2002) J. Mol. Biol. 322, 741–753 [DOI] [PubMed] [Google Scholar]
- 50.Kumar S., Nei M., Dudley J., Tamura K. (2008) Brief Bioinform. 9, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 52.Lassmann T., Sonnhammer E. L. (2005) BMC Bioinformatics 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lassmann T., Sonnhammer E. L. (2006) Nucleic Acids Res. 34, W596–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- 55.Evans P. R. (2005) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 56.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 57.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 58.Schwarzenbacher R., Godzik A., Grzechnik S. K., Jaroszewski L. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1229–1236 [DOI] [PubMed] [Google Scholar]
- 59.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 60.Holm L., Sander C. (1996) Science 273, 595–603 [DOI] [PubMed] [Google Scholar]
- 61.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]