Abstract
The formation of multiprotein complexes constitutes a key step in determining the function of any translated gene product. Thus, the elucidation of interacting partners for a protein of interest is of fundamental importance to cell biology. Here we describe a simple methodology for the prediction of novel interactors. We have applied this to the developmental transcription factor Brn-3a to predict and verify a novel interaction between Brn-3a and the androgen receptor (AR). We demonstrate that these transcription factors form complexes within the nucleus of ND7 neuroblastoma cells, while in vitro pull-down assays show direct association. As a functional consequence of the Brn-3a-AR interaction, the factors bind cooperatively to multiple elements within the promoter of the voltage-gated sodium channel, Nav1.7, leading to a synergistic increase in its expression. Thus, these data define AR as a direct Brn-3a interactor and verify a simple interacting protein prediction methodology that is likely to be useful for many other proteins.
Keywords: Gene/Transcription, Methods, Neurochemistry, Protein/Protein-Protein Interactions, Transcription, Transcription/Regulation
Introduction
Although the importance of protein-protein interaction is long established, the rise of postgenomic technologies, in particular interactomics in yeast (1), and the growing popularity of systems biology (2) have served to highlight the importance of protein complexes to cell biology. For example, in the field of transcriptional regulation, the discovery of cis-regulatory elements vast distances from the transcriptional start site of a gene requires the formation of protein complexes to bridge often huge expanses of genomic DNA (3). In addition, the concept of promoters and cis-regulatory elements as “coincidence detectors” that integrate and process a diversity of inputs into a decision on the expression of a gene requires the construction of complex molecular machinery to perform these tasks (4). Although the need to study multiprotein complexes in basic research is clear, its relevance to applied disciplines should not be underestimated. In the case of transcriptional complexes, there is a growing body of data supporting the idea that interactions between transcription factors and between transcription factors and co-factors constitute a potent target for therapeutic intervention (5–7). In addition to molecules that block or disrupt protein-protein interactions, the creation of drugs that stabilize a transcriptional complex has been postulated (6), thus suggesting the possibility that, by modulating protein binding events, genes critical to pathological processes may be switched on or off pharmacologically.
The Brn-3a transcription factor (also known as Pou4F1) is a class IV POU (Pit-Oct-Unc) family transcription factor. It is expressed at high levels in the sensory neurons of the trigeminal ganglia and dorsal root ganglia (DRG)2 and in specific structures of the brain (8). Brn-3a is indispensable for normal development because Brn-3a knock-out mice display a severe depletion of sensory neurons. Presumably grossly impaired in nociception and proprioception, these animals are unable to feed, and they die shortly after birth (9, 10). Supporting a key role for Brn-3a in sensory neural development, its overexpression in neuroblastoma cell lines is sufficient to induce differentiation (11–14). Brn-3a has been shown to up-regulate a number of neural genes, including neurofilaments, snap25, the neuropeptide Galanin, and the sodium channel SCN9A as well as the cell cycle inhibitor P21 (12, 14–18). In addition, the predominantly expressed longer splice form of Brn-3a (Brn-3aL) can protect neurons from apoptosis via the up-regulation of survival genes, including HSP27 and BCL-2 (19–23), and the repression of the apoptotic genes BAX and Noxa (18, 24).
The importance of protein-protein interaction to the function of Brn-3a was first identified with the finding that Brn-3a heterodimerizes with Brn-3b in vitro (25). Brn-3a also interacts with a number of unrelated proteins: p53, p73, EWS (Ewing's sarcoma protein), Rin, estrogen receptor (ER), Src-1 (steroid receptor co-activator-1), and HIPK2 (homeodomain-interacting protein kinase-2) (26–32). Co-expression of Brn-3a with either EWS, Rin, Src-1, or HIPK2 has been demonstrated to modify the activation of one or more target genes by Brn-3a (29–32). Moreover, Brn-3a affects the binding of ER to estrogen response elements, whereas the interactions between Brn-3a and p53 or p73 can be synergistic on some promoters and inhibitory on others, indicating that the consequences of binding can be bidirectional (i.e. Brn-3a and the interactor can regulate each other) and promoter-specific (24, 27, 28, 33). Thus, it is clear that the function of Brn-3a is acutely dependent on its interacting partners and can only be understood in terms of the complexes it forms.
Laboratory techniques for identifying interactors, in particular yeast two-hybrid and mass spectrometry, are widely used. These approaches have uncovered many interactions that have greatly advanced our understanding of cellular biology. Nevertheless, the identification of interacting proteins is time-consuming, often requiring a great deal of optimization. More importantly, false positives are commonplace, and data produced by these techniques still need to be verified experimentally. As such, it is perhaps more accurate to consider these techniques strategies to predict candidate interactors rather than to identify interacting proteins directly. With this in mind, we have developed and validated a non-computational approach to successfully predict proteins that associate with Brn-3a. The methodology uses simple mathematics and can be performed by any scientist, without the need for specialized computer programs or training in bioinformatics. While this approach has yielded new insights into how Brn-3a exerts its effects in sensory neuronal differentiation, we anticipate our methodology will be applicable to many other proteins and will allow other laboratories to quickly move their work in new directions.
EXPERIMENTAL PROCEDURES
Non-computational Prediction of Co-complexed Proteins
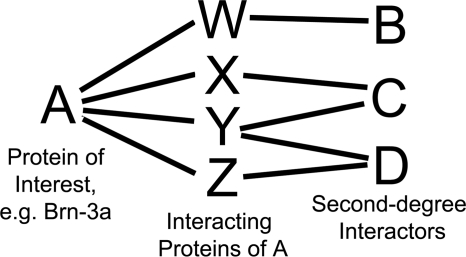
The described non-computational approach to identify potential Brn-3a complex members uses information from the BioGRID protein-protein interaction data base (available on the World Wide Web) (34, 35) and is motivated by the probabilistic network modeling described by Asthana et al. (36). Essentially, the strategy uses the protein-protein interaction records of all known Brn-3a-binding proteins to generate a diagrammatic network of proteins that have been demonstrated to either (a) bind Brn-3a directly or (b) bind more than one of the known Brn-3a-binding proteins. It is this second group of proteins, termed “second degree interactors” that are the novel Brn-3a interactors predicted by this methodology and the focus of the project. Although these proteins have been shown to interact with known Brn-3a-binding proteins, there is no experimental evidence that they form complexes with Brn-3a. A representation of the relationship between a protein of interest, its binding proteins, and its predicted second degree interactors is shown in Fig. 1.
FIGURE 1.
Schematic representation of an interaction network. A schematic interaction network as used in the interacting protein prediction methodology is shown with a simple model system where the protein of interest, A, is linked to three “second degree” interactors, B, C, and D, via four intermediates, W–Z. In this system, all individual interactions have been shown experimentally; however, the idea that A is in complex with B, C, or D is purely theoretical. As such, B, C, and D are predicted, or candidate, interactors of A; these proteins represent the focus of the interacting protein methodology.
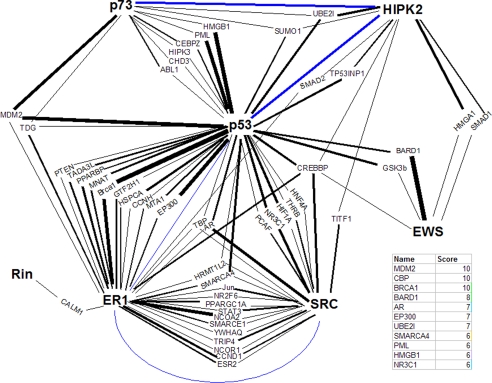
The method used is summarized as follows. 1) The protein-protein interactions of all known Brn-3a-binding proteins were downloaded from BioGRID. 2) Interaction data were cross-referenced in Microsoft Excel for proteins in common (e.g. proteins that have been experimentally determined to bind, for example, p53 and ER). These proteins are the predicted second degree interactors of Brn-3a. 3) To allow for better management of the data, a “nodes and edges” network diagram linking the predicted second degree interactors to the known Brn-3a-binding proteins was drawn in Microsoft Powerpoint, with proteins as nodes and interactions as edges (Fig. 2). 4) Each interaction between Brn-3a-binding proteins and predicted second degree interactors was rated according to experimental evidence supporting it. The amount of data demonstrating an individual interaction is considered a reflection of our confidence that the interaction is real. To do this in a non-subjective manner, interactions were given one point for every method in which the interaction has been shown (e.g. co-immunoprecipitation (co-IP) + yeast two-hybrid = 2). 5) The thicknesses of the edges within the network diagram were adjusted to indicate the score of the interaction they describe (e.g. an interaction demonstrated with three techniques leads to an edge with a three-point weight) (Fig. 2). 6) The score for all predicted second degree interactors was calculated by adding the values for each edge linking it to the rest of the network. Note that for data management purposes, all predicted second degree interactors connected to the network by just two interactions that have each only been shown by one method (e.g. those with the lowest possible score = 2) have been removed from the diagram and excluded from the study.
FIGURE 2.
Network modeling approach for predicting Brn-3a-interacting proteins. The protein-protein interactions of the seven published Brn-3a-binding proteins (shown in boldface type) were downloaded from BioGRID and cross-referenced for common interactors (normal type), and an interaction network was drawn using these proteins. For simplicity, interactions between Brn-3a and its binding proteins and Brn-3a itself are not included. The strength of the data supporting each interaction, scored as the number of methods with which an interaction has been demonstrated, is represented as edge thickness. The total score for a putative Brn-3a interactor is calculated as the sum of all edges to which that protein is connected. To reduce complexity, proteins with the lowest possible score, 2, were excluded. Direct interactions between Brn-3a-binding proteins are depicted in blue and included for reference. The top results of this study are shown in the table.
Underlying this work are two premises. First, the number of experimental methods by which any protein-protein interaction within the databases has been demonstrated is correlated with the likelihood that the interaction is real. Second, the number of connections between a putative second degree interactor and a protein of interest reflects the likelihood that the two proteins will indeed form complexes in cells. In other words, if protein A binds proteins B and C, protein Y binds protein B, and protein Z binds proteins B and C, protein Z can be considered more likely to form complexes with protein A than protein Y.
Experimental Reagents
All consumables were obtained from VWR International Ltd. unless otherwise indicated.
Cell Culture and Transfection
ND7 cells, a mouse neuroblastoma/rat DRG hybridoma cell line (37), were from ATCC. ND7 cells display a sensory neural phenotype and represent the best in vitro model for the study of Brn-3a (37). Cells were cultured in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin at 37 °C and 5% CO2. All transfections were performed using GeneJuice (Merck), following the manufacturer's instructions.
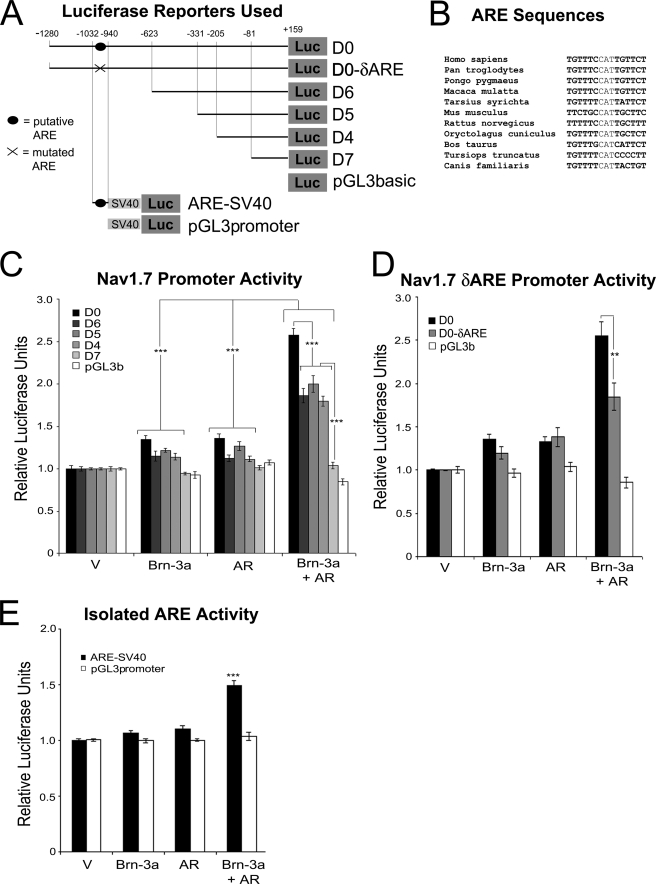
Plasmid encoding FLAG-Brn-3aL has been described previously (13). pSG-AR was provided by Hinrich Gronemeyer (University of Strasbourg, France). pCAN-HA-AR, encoding human AR fused to an N-terminal HA tag, was made as follows. The coding sequence pSG-AR was amplified by PCR using the primers 5′-GGATCCGAAGTGCAGTTAGGGCTG-3′ and 5′-TCTAGATCACTGGGTGTGGAAATAG-3′ to generate a 2.8-kb product consisting of AR, minus the initial ATG, between BamHI and XbaI sites. This PCR product was ligated into pCR-Blunt (Invitrogen) using a Zero Blunt PCR cloning kit. Clones were verified by sequencing, and the AR sequence was excised as a BamHI-XbaI fragment and ligated into the equivalent sites in pCAN-HA. In-frame ligation was confirmed by sequencing, and the expression of a full-length ∼110-kDa product was verified by Western blot. pCR3.1-YFP-Tsg101 (tumor suppressor gene 101) was provided by Jez Carlton and Juan Martin-Serrano (Kings College, London, UK). pMT-ERα has been described previously (27). SCN9A-pGL3-Basic-D0, containing −1280 to +159 of the human SCN9A promoter upstream of luciferase, and the 5′-truncations of this reporter (D6, D5, D4, and D7) plus pRL-TK (Promega) have been described previously (17). SCN9A-pGL3-D0-δARE was made by site-directed mutagenesis to remove the underlined nucleotides from the androgen receptor-response element (ARE) sequence, TGTTTCCATTGTTCT using a QuikChange mutagenesis kit (Stratagene). ARE-SV40, containing an 83-bp sequence of the rat scn9a promoter (equivalent to −1032 to −940 of the human promoter) that includes the ARE, upstream of an SV40 minimal promoter-driven luciferase reporter, was made in the following manner. A 460-bp region of the rat scn9a promoter was amplified from ND7 genomic DNA with the primers 5′-GCCATCTTCTGATTTCTTCC-3′ and 5′-CAAGACACTGTTCCCTGCTATG-3′ and cloned into pGem-T Easy using a TA cloning kit (Promega). A SpeI-XhoI fragment from this plasmid was ligated into the NheI and XhoI sites within the pGL3 promoter (Promega). ARE-SV40 was derived from this vector by excising a PstI-XhoI fragment, followed by blunt-ending and autoligation of the vector backbone. The sequences of ARE-SV40 and all intermediates were confirmed by sequencing. LTRpoly and LTR-3a-as, encoding an inverted 217-nucleotide sequence from the region of Brn-3a mRNA encoding amino acids 41–114 of Brn-3aL has been published before (14). pGL2-HSP27, encoding the human HSP27 proximal promoter upstream of luciferase, has been described previously (19).
Glutathione S-Transferase (GST)-Brn-3a Pull-down Assays
The preparation of GST-Brn-3aL, GST-Brn-3bL, and GST have been described previously (27). In vitro transcribed/translated AR and ERα were produced from pSG-AR and pMT-ERα, respectively, using a TNT Quick Coupled transcription/translation kit (Promega) and 35S-labeled methionine (PerkinElmer Life Sciences). In vitro transcribed/translated luciferase was produced from the control plasmid within the transcription/translation kit. Pull-down assays were performed according to Ref. 27.
Western Blotting and Co-immunoprecipitation
ND7 cells grown in 10-cm plates were transfected with 5 μg of FLAG-tagged Brn-3aL or empty vector in combination with 5 μg of pCAN-HA-AR, pCR3.1-YFP-Tsg101, or empty vector. Cells were harvested 24 h post-transfection.
For Western blots of voltage-gated sodium channel (Nav) expression, cells were lysed in radioimmune precipitation buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 10 mm β-mercaptoethanol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science) in 1× phosphate-buffered saline). Lysates were clarified by centrifugation at 13,000 × g for 10 min, prior to denaturation by the addition of Laemmli sample buffer and boiling. Total Nav expression was detected with pan-Nav antibody (Sigma). Membranes were reprobed with anti-p85 antibody (Upstate) to show protein loading and with anti-FLAG M2 (Sigma) and anti-HA (Roche Applied Science) to show expression of FLAG-Brn-3aL and HA-AR.
For co-IP experiments, cells were extracted using a nuclear extraction protocol. Briefly, cells were lysed in buffer A (10 mm Tris, pH 7.5, 2 mm EDTA, 0.5% Tween 20, and 1 mm phenylmethylsulfonyl fluoride) prior to centrifugation at 2000 × g for 1 min. The resultant nuclear pellet was resuspended in buffer B (buffer A containing 500 mm NaCl), and nuclear proteins were extracted by incubation on ice for 15 min. Finally, extracts were clarified by centrifugation at 13,000 × g for 5 min, and the salt concentration of the final nuclear preparation was adjusted to 150 mm by dilution with buffer A. Anti-FLAG IP of FLAG-Brn-3aL from nuclear extracts was performed by the addition of anti-FLAG M2 antibody at a 1:200 dilution prior to overnight incubation at 4 °C. The next morning, protein G-Sepharose was added to each sample, and after another 1 h of incubation, immunoprecipitates were washed three times in buffer C (buffer A containing 150 mm NaCl) and denatured by the addition of Laemmli sample buffer and boiling. Samples were resolved by SDS-PAGE, transferred to nitrocellulose membrane, and blotted with anti-green fluorescent protein antibody (Sigma) or anti-HA to determine co-precipitation of YFP-Tsg101 or HA-AR, followed by anti-FLAG M2 to verify the IP of FLAG-Brn-3aL from the relevant samples. Expression of YFP-Tsg101 or HA-AR in control samples was shown by blotting the original nuclear extracts.
Promoter Assays
Luciferase assays were performed in 6-well plates, using 0.5 μg of FLAG-Brn-3aL or empty vector in combination with 0.5 μg of pSG-AR or empty vector, together with 0.5 μg of the indicated SCN9A or HSP27 reporter and 0.1 μg of pRL-TK per well. Cells were extracted after 24 h using passive lysis buffer (Promega), and luciferase and Renilla values were measured using a dual luciferase assay kit (Promega) and a Turner Biosystems luminometer (Turner Instruments).
Real-time Reverse Transcription-PCR
RNA was extracted from ND7 cells and used to produce cDNA as published (17). Quantitative real-time PCR was performed using QuantiTect SYBR Green (Qiagen) on an Opticon DNA Engine real-time PCR machine (MJ Research) with the following primers: Nav1.7 forward, 5′-TGACTTGGAAGCTGGGAAAC-3′; Nav1.7 reverse, 5′-TTCCAAGGGTCACGGAGGA-3′; β-actin forward, 5′-AGATGACCCAGATCATGTTTGAG-3′; β-actin reverse, 5′-AGGTCCAGACGCAGGATG-3′; GAPDH forward, 5′-GTGTGAACGGATTTGGCCG-3′; GAPDH reverse, 5′-CCAGTAGACTCCACGACATA-3′. Reactions were performed in duplicate, and to verify product composition, melt curves were carried out from 65 to 95 °C, and reactions were resolved by electrophoresis. Furthermore, experiments performed on dilutions of control cDNA (using the method described in Ref. 38) showed that reverse transcription-PCRs amplifying Nav1.7, β-actin, and glyceraldehyde-3-phosphate dehydrogenase worked with comparable efficiencies (close to 100%; data not shown). Expression of Nav1.7 relative to β-actin or GAPDH was calculated using the 2−ΔΔCt method (38).
Statistical Analysis
Nav1.7 promoter assay data were tested for significance using two-way ANOVA and Bonferroni post hoc tests. mRNA expression data and HSP27 promoter luciferase assays were examined with one-way ANOVA and Student-Newman-Keuls post hoc testing.
RESULTS
Prediction of Brn-3a Interactors
We have developed a method for the prediction of novel interacting proteins of Brn-3a, described in detail under “Experimental Procedures.” In summary, this process combines the construction of a Brn-3a interaction network diagram, consisting of the known Brn-3a-binding proteins and their own published interactors (i.e. direct Brn-3a-interactors and putative “second degree interactors” of Brn-3a; see Fig. 1), with a measure of the strength of the experimental data supporting each individual interaction, namely the number of techniques used to demonstrate that interaction. The likelihood that any second degree interactor will indeed form bona fide complexes with Brn-3a in vivo is then scored as a function of the number of interactions linking it to Brn-3a and of the strength of these interactions.
Using this method, we have produced a list of candidate second degree interactors of Brn-3a, ranked according to the strength of the data linking these molecules to the seven known Brn-3a-binding proteins (Fig. 2). The 11 highest scoring proteins are shown in the accompanying table (Fig. 2). Encouragingly, cyclic AMP-response element-binding protein-binding protein and p300 figure prominently in this list; one would expect this to be the case, given their ubiquity and the key role they play in the regulation of gene expression as transcriptional co-factors and histone acetyltransferases (39).
Also present within the top seven results is the AR. AR is a transcription factor whose classical method of activation involves binding by androgenic steroids, followed by an increase in stability, nuclear accumulation, and subsequent changes to gene expression (40). Nevertheless, the binding of co-factors has been shown to be central to AR biology (41, 42), and an interaction between Brn-3a and another nuclear hormone receptor, ER, has been described previously (27). Thus, we sought to probe the potential interaction between Brn-3a and AR experimentally. Initially, co-IP assays were performed using nuclear extracts from ND7 cells that had been transiently transfected with FLAG-tagged Brn-3a and/or HA-tagged AR. Note that in this and all subsequent experiments, the long isoform of Brn-3a was used (Brn-3aL). As expected, when either protein was expressed individually, HA-AR was not present in anti-FLAG IPs (Fig. 3A). However, when co-expressed, HA-AR was readily detectable, thereby indicating that these proteins form complexes within the nuclei of ND7 cells (Fig. 3A). In addition, we were able to replicate this finding on endogenous protein, as shown by the presence of AR in anti-Brn-3a IPs (Fig. 3B), thus providing important confirmation that these transcription factors interact under normal conditions.
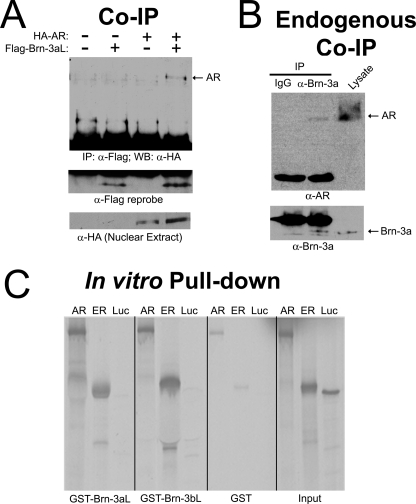
FIGURE 3.
Physical interaction between Brn-3a and the androgen receptor. A, co-IP of transfected protein. ND7 cells were transiently transfected with empty vector, FLAG-Brn-3aL, HA-AR, or HA-AR and FLAG-Brn-3aL in combination. IPs were performed from nuclear extracts with anti-FLAG antibody. IPs were resolved by SDS-PAGE and blotted with anti-HA antibody for the presence of HA-AR (top). IPs were reprobed with anti-FLAG to show FLAG-Brn-3aL (middle). Nuclear extracts were probed with anti-HA to show expression of HA-AR in transfected samples. B, co-IP of endogenous protein. Nuclear extracts from ND7 cells were incubated with anti-Brn-3a antiserum or IgG control. IPs were resolved by SDS-PAGE and blotted with anti-AR antibody for the presence of endogenous AR (top) prior to reprobing for Brn-3a. Lysate from cells overexpressing HA-AR and FLAG-Brn-3a was run alongside as positive control for anti-AR and anti-Brn-3a antibodies. C, in vitro pull-down. [35S]Methionine-labeled, in vitro translated AR, ER (positive control), and luciferase (Luc) (negative control) were incubated with GST-Brn-3aL, GST-Brn-3bL, or GST alone. Precipitates were washed prior to separation by SDS-PAGE. Binding of AR, ER, and Luc to GST-Brn-3aL, GST-Brn-3bL, or GST was visualized by autoradiography. The right-hand panel shows equal quantities of in vitro translation product (Input) as a reference.
Although the prediction methodology has been developed with the objective of determining second degree interactors, we nevertheless sought to determine whether the interaction between Brn-3a and AR can be direct. Such a result would not be unexpected because theoretical studies have shown missing interactions can be “predicted” from partially complete protein-protein interaction data by virtue of the proximity of two proteins within an interaction network (43). To investigate this possibility, in vitro assays were performed using recombinant GST-tagged Brn-3aL, Brn-3bL, or GST alone to pull-down in vitro translated AR. Parallel pull-downs were carried out using in vitro translated ER or luciferase as positive and negative controls, respectively. As expected, ER bound to both GST-Brn-3aL and GST-Brn-3bL to a greater extent than to GST alone, whereas no interactions with luciferase were detected (Fig. 3C). Importantly, and in support of the hypothesis that AR and Brn-3a may interact directly, a greater association was also detected between AR and either recombinant Brn-3 protein than between AR and GST (Fig. 3C). Taken together, these data confirm our bioinformatic prediction of an interaction between Brn-3a and AR in intact cells and demonstrate that the interaction is likely to be direct.
The observed direct interaction between AR and Brn-3a has allowed the position of AR within the network diagram to be “upgraded” from that of a second degree interactor to a direct Brn-3a-binding protein and allowed its own interactors to be integrated into the network (supplemental Fig. 1). Using these new data, we have been able to generate an improved list of putative Brn-3a-interacting proteins (supplemental Table 1). Included in this new list is Tsg101, a protein originally found in screens for tumor suppressor genes and central to a diverse range of processes, including cell cycle arrest and, intriguingly, AR co-activation (41, 44). As a further validation of our interacting protein prediction methodology, we have found Brn-3a and Tsg101 to be physically associated within the nuclei of ND7 cells (supplemental Fig. 2A). In addition, we have independently confirmed the capacity of both Tsg101 and AR to form complexes with Brn-3a via pull-down assays of these proteins from cell lysates with GST-Brn-3a (supplemental Fig. 2B). We are currently validating the remaining predicted Brn-3a interactors in a systematic manner, with priority given to those already documented to be involved in the same biological processes as Brn-3a.
Role for the Brn-3a-AR Interaction in the Regulation of Nav1.7 Expression
The observation of a Brn-3a-AR complex and the fact that these proteins are both transcription factors creates a natural hypothesis that their interaction will result in the altered transcriptional regulation of certain target genes. As such, we sought to determine the effect of co-expressing Brn-3aL and AR on a promoter known to be responsive to both factors.
Nav1.7 (SCN9A) is the predominant sodium channel expressed in the sensory nervous system, and its dysregulation has been implicated in three diseases of peripheral sensory neurons: erythromelalgia, paroxysmal extreme pain disorder, and channelopathy-associated congenital indifference to pain (reviewed in Ref. 45). Importantly, its promoter has been shown to be activated by both Brn-3a (17) and AR3 in prostate cancer models. As such, the SCN9A promoter represented a likely site of action for Brn-3a-AR complexes. Thus we investigated the effects of overexpressing Brn-3aL and AR, alone and in combination, on SCN9A promoter activity in ND7 cells using a series of reporter constructs encoding 5′-truncations of the proximal SCN9A promoter upstream of luciferase. A diagrammatic representation of all SCN9A reporters used in this study is shown in Fig. 4A. Two-way ANOVA revealed a significant effect of all treatments and of promoter truncation and revealed a significant interaction between these variables (Fig. 4C). Transfection of either Brn-3aL or AR alone led to a small increase in promoter activity when compared with vector-transfected controls (Bonferroni post hoc analysis; p < 0.001 versus vector for both treatments). Remarkably, however, and in support of a functional interaction between Brn-3a and AR, the co-expression of these proteins induced synergistic up-regulation of the promoter (p < 0.001 versus all other treatments), although no activation was seen with the shortest SCN9A reporter (D7) or, importantly, with the empty reporter plasmid (Fig. 4C).
FIGURE 4.
Synergistic activation of the SCN9A (Nav1–7) promoter by Brn-3a and AR. A, luciferase reporter constructs used. The SCN9A promoter regions contained within each reporter construct are illustrated. Note that the plasmids used in B encoded progressive truncations of the proximal promoter cloned into the pGL3basic plasmid, whereas the plasmid used in D encoded the putative ARE identified within this promoter cloned into pGL3 promoter, which contains a minimal SV40 promoter. B, alignment of ARE sequences. The putative ARE sequences from all available mammalian genomes in the Ensembl data base are shown. C–E, activation of the SCN9A promoter by Brn-3a and AR. ND7 cells were transfected with the indicated SCN9A promoter reporter constructs and pRL-TK-Renilla plus the indicated combinations of expression plasmids encoding AR, Brn-3aL, or empty vector. After 24 h, SCN9A promoter activity was determined as the ratio of luciferase to Renilla activity. Values shown are the mean values of three independent experiments. Statistical analysis was performed by two-way ANOVA followed by Bonferroni post hoc testing. **, p < 0.01; ***, p < 0.001 between indicated conditions/groups of conditions (C and D) or versus all other conditions (E).
The failure of Brn-3aL and AR to activate the D7 reporter suggested the existence of a discrete DNA element mediating the Brn-3aL- and AR-dependent up-regulation of the SCN9A promoter that lies immediately 5′ to the sequence contained in this plasmid. This observation was supported by the fact that no statistical difference was detected between the activities of the three next longest constructs (D6, D5, and D4), which were all up-regulated between 1.8- and 2-fold by Brn-3aL and AR co-treatment (Fig. 4C). In contrast, the longest reporter construct (D0) was activated ∼2.6-fold, which was significantly greater than all of the other reporters (p < 0.001) and suggested the existence of a second Brn-3a/AR-responsive promoter element within the sequence unique to this construct (Fig. 4B). Interestingly, this promoter region includes a potential ARE at −967 to −953 of the human SCN9A promoter that is largely conserved in all available mammal sequences (TGTTTCcatTGTTCT)3 (see Fig. 4B). To investigate whether this putative ARE may indeed mediate transcriptional activation driven by Brn-3a-AR complexes, we first deleted this element by site-directed mutagenesis and compared the resultant construct to the original D0 reporter. As can be seen in Fig. 4D, this construct was activated by the combination of Brn-3a and AR to a significantly lower degree than the wild-type construct (Bonferroni post hoc analysis; p < 0.01 versus D0). Second, an 83-bp sequence surrounding this ARE was cloned from the rat scn9a promoter into pGL3 promoter, which encodes the luciferase gene under the control of the minimal SV40 promoter (see diagram in Fig. 4A). As expected, the transfection of Brn-3aL or AR alone had little effect on this reporter, but the two factors combined to induce a ∼1.5-fold synergistic activation (Bonferroni post hoc analysis; p < 0.001 versus all other conditions; Fig. 4E). These data confirm the existence of a conserved Brn-3a- and AR-responsive element within this small promoter region and present strong evidence that Brn-3a-AR complexes are able to bind to, and regulate transcription from, classical AREs.
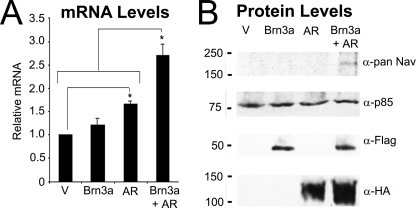
Thus, in confirmation of our bioinformatic prediction, we found Brn-3a and AR to interact directly and within the cell nucleus (Fig. 3), while a synergistic activation of the SCN9A promoter by reporter assay (Fig. 4, B–D) suggested a physiological role for this transcription factor complex. To confirm that our observations are likely to hold true for endogenous Nav1.7 gene expression, we examined the effect of Brn-3aL and AR overexpression on Nav1.7 mRNA levels by quantitative real-time PCR. The overexpression of AR led to a larger increase in Nav1.7 levels than had been seen in luciferase assays (∼1.7-fold; p < 0.001 versus vector), with a trend toward a modest (∼1.2-fold) activation by Brn-3aL. However, a synergistic effect of co-transfection was still observed because the combination of Brn-3aL and AR induced a ∼2.7-fold up-regulation Nav1.7 mRNA levels (p < 0.001 versus all other conditions; Fig. 5A). Finally, and most importantly, we were also able to replicate this result at the level of total Nav protein expression. In Western blots of lysates from empty vector or FLAG-Brn-3aL- or HA-AR-transfected ND7 cells, total Nav expression was below the detectable threshold for the antibody used (Fig. 5B). However, co-expression of FLAG-Brn-3aL and HA-AR led to the detection of a protein species with a molecular mass consistent with that expected for voltage-gated sodium channels (Fig. 5B). These data indicate that the effects of Brn-3a and AR result in corresponding increases in Nav1.7 mRNA and protein levels and thus confirm a functional role for the Brn-3a-AR interaction.
FIGURE 5.
Effect of Brn-3a and AR on endogenous Nav1.7. A, Nav1.7 mRNA expression. ND7 cells were transiently transfected with empty vector, FLAG-Brn-3aL, AR, or AR and FLAG-Brn-3aL in combination. After 24 h, mRNA was extracted, and the levels of Nav1.7 were determined by real-time PCR. Values were normalized to β-actin and GAPDH (n = 6). Statistical analysis was performed using one-way ANOVA with Student-Newman-Keuls post hoc testing; *, p < 0.05 between the indicated conditions/groups of conditions. B, total Nav protein levels. ND7 cells were transiently transfected with empty vector, FLAG-Brn-3aL, HA-AR, or HA-AR and FLAG-Brn-3aL in combination. After 24 h, cells were lysed in radioimmune precipitation buffer, and extracts were resolved by SDS-PAGE. Membranes were blotted with pan-Nav antibody (top panel), anti-p85 to show equivalent protein loading (second panel), anti-FLAG to show FLAG-Brn-3aL (third panel), and anti-HA antibody to show HA-AR (bottom panel).
DISCUSSION
Methodology for the Prediction of Interacting Proteins
Although undeniably simple, our methodology for the prediction of interacting proteins, described here for Brn-3a, clearly works, as evidenced by the experimental verification that both AR (Figs. 3–5) and Tsg101 (supplemental Fig. 2) form complexes with Brn-3a in cells. In addition, we have preliminary data suggesting a direct interaction between Brn-3a and promyelocytic leukemia proteins.4 It is important to note that these are the only predicted Brn-3a interactors to have been tested. Although we would not anticipate all predictions made to be correct (protein-protein interactions are ultimately dependent on specific interaction surfaces and other structural constraints that are not part of our calculations), we still expect our methodology to be equally successful for many other proteins. Indeed, we applied our approach to incomplete protein-protein interaction data from the Parkinson disease-associated protein α-synuclein and were able to predict interactions with four proteins that have been shown experimentally (supplemental Fig. 3), although another two of these proteins have been described as not binding to α-synuclein, thus indicating that our methodology is not infallible (although these proteins may yet bind α-synuclein under different experimental conditions). In support of a wide application for our methodology, it is important to point out that α-synuclein is not a transcription factor. Our technique is easy to perform, requiring no specialized training in mathematics or bioinformatics, and can generate a list of potential interactors in a fraction of the time taken for a standard yeast two-hybrid or mass spectrometry study. We believe this methodology has the potential to be of great use in the future.
Although our prediction strategy has proven successful, there are certainly limitations and improvements that can be made. For example, the methodology is only as good as the protein-protein interaction data used. There are undoubtedly mistakes made in the curation of protein-protein interaction data bases, particularly due to difficulties with protein nomenclature. We would encourage any scientist wishing to use our prediction methodology to be as thorough as possible in checking the original papers from which interaction records used in their predictions were obtained. Another limitation will inevitably come from those proteins for which no existing interactors are known and indeed for those proteins with many previously determined binding proteins; in this latter case, the volumes of data will simply be too great to be managed accurately. In addition, by its very nature, our methodology cannot work for proteins that have just one or no binding partners.
With regard to the methodology itself, it is important to note that we did not take into account the strength of the data linking Brn-3a to its interacting partners, only strength of interactions between these Brn-3a interactors and the putative second degree interactors. In the case of Brn-3a, we have been able to have confidence in its known interactions; however, for many other proteins this will not be the case; future versions of this methodology will need to take into account the strength of all interactions in a network.
A final point about the methodology is the metric that we have used to determine the strength of each interaction, namely the number of methods with which it has been experimentally demonstrated. We consider this a superior metric to the number of publications in which an interaction has been demonstrated because a false positive generated with one technique is less likely to recur using an unrelated technique than when it is repeated with an identical protocol in another laboratory. We have also chosen not to consider certain methods of interaction as “better” than others. Although we do not doubt that some experimental techniques provide more conclusive proof of interaction, we have not made such judgements in order to maintain objectivity. Nevertheless, it is worth noting that for the vast majority of proteins, “weaker” methods of demonstrating protein-protein interaction (e.g. co-localization in confocal microscopy) will only be performed after more direct biochemical studies have been carried out. In other words, there are very few protein-protein interaction records based entirely upon indirect methods. As such, data from these techniques will almost always serve to increase the evidence for an already well supported interaction rather than attempt to make the case on their own.
Our methodology has been designed to be as objective as possible. However, once predictions have been made, the user is still faced with the choice of which predicted interactor to investigate, a decision that will be based on what the user considers to be the protein(s) most likely to function in the same process(es) as the protein of interest. Although we would still recommend the score from the prediction methodology to remain the primary criterion in order to reduce any experimenter bias, we acknowledge that judgments of this nature require a degree of intuition. Nevertheless, prioritization based on other sources of information, in particular function (e.g. gene ontology) and subcellular localization, is not an entirely partisan process because both of these factors have been shown to improve predictions in theoretical studies (46). In light of this, we have applied these criteria to our full list of predicted Brn-3a interactors, which has allowed a subset to be deprioritized (supplemental Table 1). We would also recommend the use of microarray and/or gene expression data (where available) to verify that a transcript encoding the predicted interactor is co-expressed with the protein of interest in a relevant system.
Thus, despite its limitations, we are convinced of the potential of the interacting protein prediction methodology that we have described here. It may not be applicable to all proteins, but the demonstration that the technique works for Brn-3a suggests that it is of great potential as a tool for the study of protein complexes.
Brn-3a-AR Complexes
In this study, we have identified an interaction between AR and Brn-3a (Fig. 3) that appears to promote the binding of both factors to the Nav1.7 promoter, resulting in an increase in the expression of the endogenous gene (Figs. 4 and 5). In the context of our model system, the role of Brn-3a in sensory neuronal differentiation, the association between Brn-3a and AR is intriguing. As mentioned, Nav1.7 is the major voltage-gated sodium channel in sensory neurons and central to nociception (45), and its expression increases during the differentiation of neuroblastoma cell lines (47). In light of this, it is interesting to speculate on what effect the relative expression of Brn-3a and AR in peripheral neurons may have on the sensation of pain. For example, AR is expressed in developing DRG, but its expression drops markedly at birth (48), whereas Brn-3a expression endures into adulthood (9). It may be the case, therefore, that individuals suffering from idiopathic pain have higher AR levels than normal controls, thus making AR (and indeed the Brn-3a-AR interaction) a potential target for the treatment of pain.
As well as being expressed in developing DRG (48), the involvement of AR and androgens in inducing neurite outgrowth in other neural subtypes is well established, most notably in motoneurons (49, 50). Indeed, as in the case of Brn-3a (11, 12, 14), the overexpression of AR in ND7 cells is sufficient to induce neurite outgrowth (data not shown). However, in contrast to SCN9A promoter activation, no additive effect on neurite outgrowth is seen when Brn-3a and AR are co-expressed (data not shown). As such, it is more likely that Brn-3a-AR complexes only regulate a subset of genes involved in neural differentiation, a hypothesis that is supported by preliminary studies on other Brn-3a target promoters; e.g. P21 and Galanin do not display an additive up-regulation by the co-expression of AR and Brn-3a (data not shown), whereas the HSP27 promoter is activated synergistically by these factors (supplemental Fig. 4).
Finally, although our studies of the Brn-3a-AR interaction have been confined to sensory neurons, it would be wrong to overlook the potential role of this interaction in other systems, most notably prostate cancer. The involvement of AR in prostate cancer is well documented, although its precise function and requirements remain elusive. Certainly, at early stages, prostate tumors require androgens to proliferate, and androgen ablation remains a successful treatment (51). At later stages, the disease becomes refractory to androgen ablation; however, curiously, knockdown of AR is still sufficient to inhibit the growth of androgen-independent cancers, indicating that although androgens may no longer be required, AR remains critical for survival (52, 53). Intriguingly, relative to age-matched controls, Brn-3a expression is elevated in prostate cancer tissue, and its expression increases with disease progression (54). Thus, in light of the interaction between Brn-3a and AR described in this work, it may be the case that late stage prostate cancers are able to use Brn-3a to increase activation of AR-regulated genes, thereby reducing the requirement for androgens. Although this notion requires testing, the idea that Brn-3a-AR complexes may be at work in prostate cancer represents an intriguing hypothesis for future work.
Acknowledgments
We thank Dr. Scott Fraser and Prof. Mustafa Djamgoz (Imperial College, London, UK) for provision of anti-pan-Nav antibody; Drs. Jez Carlton and Juan Martin-Serrano (King's College) and Prof. Hinrich Gronemeyer (University of Strasbourg, France) for provision of Tsg101 and AR expression plasmids, respectively; and Dr. Peter Morris for technical assistance. We are particularly grateful to Dr. Sascha Ott (University of Warwick, UK) for helpful discussions and critical reading of the manuscript. We also thank Drs. Sean Barry, Mattia Calissano, Patricia Cogram, and Sam Ounzain (University College London Institute of Child Health), Dr. Ana Antunes-Martins (University of Warwick), and John Reid and Prof. Lorenz Wernisch (Medical Research Council Biostatistics Unit) for input.
This work was supported in part by the Biotechnology and Biological Science Research Center and Prostate UK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
J. K. J. Diss, M. Djamgoz, and D. S. Latchman, unpublished data.
D. C. Berwick and D. S. Latchman, unpublished data.
- DRG
- dorsal root ganglia
- AR
- androgen receptor
- Nav
- voltage-gated sodium channel
- ER
- estrogen receptor
- YFP
- yellow fluorescent protein
- HA
- hemagglutinin
- IP
- immunoprecipitation
- GST
- glutathione S-transferase
- ARE
- AR response element
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance.
REFERENCES
- 1.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 2.Koentges G. (2008) Nature 451, 658–663 [DOI] [PubMed] [Google Scholar]
- 3.Cook P. R. (2003) J. Cell Sci. 116, 4483–4491 [DOI] [PubMed] [Google Scholar]
- 4.Panne D. (2008) Curr. Opin. Struct. Biol. 18, 236–242 [DOI] [PubMed] [Google Scholar]
- 5.Klein C., Vassilev L. T. (2004) Br. J. Cancer 91, 1415–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arndt H. D. (2006) Angew. Chem. Int. Ed. Engl. 45, 4552–4560 [DOI] [PubMed] [Google Scholar]
- 7.Emami K. H., Nguyen C., Ma H., Kim D. H., Jeong K. W., Eguchi M., Moon R. T., Teo J. L., Oh S. W., Kim H. Y., Moon S. H., Ha J. R., Kahn M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12682–12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latchman D. S. (1998) Int. J. Biochem. Cell Biol. 30, 1153–1157 [DOI] [PubMed] [Google Scholar]
- 9.McEvilly R. J., Erkman L., Luo L., Sawchenko P. E., Ryan A. F., Rosenfeld M. G. (1996) Nature 384, 574–577 [DOI] [PubMed] [Google Scholar]
- 10.Eng S. R., Gratwick K., Rhee J. M., Fedtsova N., Gan L., Turner E. E. (2001) J. Neurosci. 21, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakin N. D., Morris P. J., Theil T., Sato T. N., Möröy T., Wilson M. C., Latchman D. S. (1995) J. Biol. Chem. 270, 15858–15863 [DOI] [PubMed] [Google Scholar]
- 12.Smith M. D., Dawson S. J., Latchman D. S. (1997) Mol. Cell. Biol. 17, 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calissano M., Faulkes D., Latchman D. S. (2005) Brain Res. Mol. Brain Res. 141, 10–18 [DOI] [PubMed] [Google Scholar]
- 14.Berwick D. C., Calissano M., Corness J. D., Cook S. J., Latchman D. S. (2009) Brain Res. 1256, 8–18 [DOI] [PubMed] [Google Scholar]
- 15.Smith M. D., Morris P. J., Dawson S. J., Schwartz M. L., Schlaepfer W. W., Latchman D. S. (1997) J. Biol. Chem. 272, 21325–21333 [DOI] [PubMed] [Google Scholar]
- 16.Budhram-Mahadeo V., Morris P. J., Lakin N. D., Theil T., Ching G. Y., Lillycrop K. A., Möröy T., Liem R. K., Latchman D. S. (1995) J. Biol. Chem. 270, 2853–2858 [DOI] [PubMed] [Google Scholar]
- 17.Diss J. K., Calissano M., Gascoyne D., Djamgoz M. B., Latchman D. S. (2008) Mol. Cell. Neurosci. 37, 537–547 [DOI] [PubMed] [Google Scholar]
- 18.Budram-Mahadeo V., Morris P. J., Latchman D. S. (2002) Oncogene 21, 6123–6131 [DOI] [PubMed] [Google Scholar]
- 19.Farooqui-Kabir S. R., Budhram-Mahadeo V., Lewis H., Latchman D. S., Marber M. S., Heads R. J. (2004) Cell Death Differ. 11, 1242–1244 [DOI] [PubMed] [Google Scholar]
- 20.Smith M. D., Dawson S. J., Boxer L. M., Latchman D. S. (1998) Nucleic Acids Res. 26, 4100–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ensor E., Smith M. D., Latchman D. S. (2001) J. Biol. Chem. 276, 5204–5212 [DOI] [PubMed] [Google Scholar]
- 22.Smith M. D., Ensor E. A., Coffin R. S., Boxer L. M., Latchman D. S. (1998) J. Biol. Chem. 273, 16715–16722 [DOI] [PubMed] [Google Scholar]
- 23.Smith M. D., Melton L. A., Ensor E. A., Packham G., Anderson P., Kinloch R. A., Latchman D. S. (2001) Mol. Cell. Neurosci. 17, 460–470 [DOI] [PubMed] [Google Scholar]
- 24.Hudson C. D., Morris P. J., Latchman D. S., Budhram-Mahadeo V. S. (2005) J. Biol. Chem. 280, 11851–11858 [DOI] [PubMed] [Google Scholar]
- 25.Theil T., Rödel B., Spiegelhalter F., Möröy T. (1995) J. Biol. Chem. 270, 30958–30964 [DOI] [PubMed] [Google Scholar]
- 26.Budhram-Mahadeo V., Morris P. J., Smith M. D., Midgley C. A., Boxer L. M., Latchman D. S. (1999) J. Biol. Chem. 274, 15237–15244 [DOI] [PubMed] [Google Scholar]
- 27.Budhram-Mahadeo V., Parker M., Latchman D. S. (1998) Mol. Cell. Biol. 18, 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson C. D., Sayan A. E., Melino G., Knight R. A., Latchman D. S., Budhram-Mahadeo V. (2008) Cell Death Differ. 15, 1266–1278 [DOI] [PubMed] [Google Scholar]
- 29.Calissano M., Latchman D. S. (2003) Oncogene 22, 5408–5414 [DOI] [PubMed] [Google Scholar]
- 30.Gascoyne D. M., Thomas G. R., Latchman D. S. (2004) Oncogene 23, 3830–3840 [DOI] [PubMed] [Google Scholar]
- 31.Dennis J. H., Budhram-Mahadeo V., Latchman D. S. (2002) Biochem. Biophys. Res. Commun. 294, 487–495 [DOI] [PubMed] [Google Scholar]
- 32.Wiggins A. K., Wei G., Doxakis E., Wong C., Tang A. A., Zang K., Luo E. J., Neve R. L., Reichardt L. F., Huang E. J. (2004) J. Cell Biol. 167, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Sanchez C., Budhram-Mahadeo V. S., Latchman D. S. (2002) Nucleic Acids Res. 30, 4872–4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breitkreutz B. J., Stark C., Reguly T., Boucher L., Breitkreutz A., Livstone M., Oughtred R., Lackner D. H., Bähler J., Wood V., Dolinski K., Tyers M. (2008) Nucleic Acids Res. 36, D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006) Nucleic Acids Res. 34, D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asthana S., King O. D., Gibbons F. D., Roth F. P. (2004) Genome Res. 14, 1170–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood J. N., Bevan S. J., Coote P. R., Dunn P. M., Harmar A., Hogan P., Latchman D. S., Morrison C., Rougon G., Theveniau M. (1990) Proc. Biol. Sci. 241, 187–194 [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen T. D., Livak K. J. (2008) Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 39.Chan H. M., La Thangue N. B. (2001) J. Cell Sci. 114, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 40.Kemppainen J. A., Lane M. V., Sar M., Wilson E. M. (1992) J. Biol. Chem. 267, 968–974 [PubMed] [Google Scholar]
- 41.Burgdorf S., Leister P., Scheidtmann K. H. (2004) J. Biol. Chem. 279, 17524–17534 [DOI] [PubMed] [Google Scholar]
- 42.Lin D. Y., Fang H. I., Ma A. H., Huang Y. S., Pu Y. S., Jenster G., Kung H. J., Shih H. M. (2004) Mol. Cell. Biol. 24, 10529–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H., Paccanaro A., Trifonov V., Gerstein M. (2006) Bioinformatics 22, 823–829 [DOI] [PubMed] [Google Scholar]
- 44.Ruland J., Sirard C., Elia A., MacPherson D., Wakeham A., Li L., de la Pompa J. L., Cohen S. N., Mak T. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dib-Hajj S. D., Cummins T. R., Black J. A., Waxman S. G. (2007) Trends Neurosci. 30, 555–563 [DOI] [PubMed] [Google Scholar]
- 46.Ben-Hur A., Noble W. S. (2005) Bioinformatics 21, Suppl. 1, i38–46 [DOI] [PubMed] [Google Scholar]
- 47.Kawaguchi A., Asano H., Matsushima K., Wada T., Yoshida S., Ichida S. (2007) Neurochem. Res. 32, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 48.Luo H., Liu J., Kang D., Cui S. (2008) Histochem. Cell Biol. 129, 525–533 [DOI] [PubMed] [Google Scholar]
- 49.Marron T. U., Guerini V., Rusmini P., Sau D., Brevini T. A., Martini L., Poletti A. (2005) J. Neurochem. 92, 10–20 [DOI] [PubMed] [Google Scholar]
- 50.Estrada M., Uhlen P., Ehrlich B. E. (2006) J. Cell Sci. 119, 733–743 [DOI] [PubMed] [Google Scholar]
- 51.Dehm S. M., Tindall D. J. (2007) Mol. Endocrinol. 21, 2855–2863 [DOI] [PubMed] [Google Scholar]
- 52.Eder I. E., Culig Z., Ramoner R., Thurnher M., Putz T., Nessler-Menardi C., Tiefenthaler M., Bartsch G., Klocker H. (2000) Cancer Gene Ther. 7, 997–1007 [DOI] [PubMed] [Google Scholar]
- 53.Hååg P., Bektic J., Bartsch G., Klocker H., Eder I. E. (2005) J. Steroid Biochem. Mol. Biol. 96, 251–258 [DOI] [PubMed] [Google Scholar]
- 54.Diss J. K., Faulkes D. J., Walker M. M., Patel A., Foster C. S., Budhram-Mahadeo V., Djamgoz M. B., Latchman D. S. (2006) Prostate Cancer Prostatic Dis. 9, 83–91 [DOI] [PubMed] [Google Scholar]