Abstract
During the process of ribosomal assembly, the essential eukaryotic translation initiation factor 6 (eIF6) is known to act as a ribosomal anti-association factor. However, a molecular understanding of the anti-association activity of eIF6 is still missing. Here we present the cryo-electron microscopy reconstruction of a complex of the large ribosomal subunit with eukaryotic eIF6 from Saccharomyces cerevisiae. The structure reveals that the eIF6 binding site involves mainly rpL23 (L14p in Escherichia coli). Based on our structural data, we propose that the mechanism of the anti-association activity of eIF6 is based on steric hindrance of intersubunit bridge formation around the dynamic bridge B6.
Keywords: Electron Microscopy (EM), Ribosome Assembly, Ribosome Function, Ribosome Structure, Ribosomes
Introduction
Eukaryotic translation initiation factor 6 (eIF6,2 p27BBP) binds to large preribosomal subunits during the process of ribosome biogenesis and prevents the premature association with the 40 S ribosomal subunit to form 80 S complexes. eIF6 is localized in the nucleus bound to 66 S preribosomal particles (1) and also in the cytoplasm, where it is associated with free 60 S subunits (2, 3). The nuclear localization of eIF6 is controlled in yeast (4) and mammalian cells (5) via phosphorylation of its two exposed residues, Ser-174 and Ser-175. The depletion of eIF6 leads to a slowdown of translational activity due to a loss of available free 60 S ribosomal subunits and is eventually lethal (6). In yeast, the cytoplasmic release of eIF6 from 60 S subunits is promoted by two factors, the GTPase Efl1p (7) and Sdo1p (Shwachman-Bodian-Diamond syndrome (SBDS) protein of yeast) (8), whereas in mammalian cells, dissociation is mediated by a RACK1-dependent protein kinase C activity (5). Interestingly, it has recently been shown that eIF6 can be part of the mammalian RISC (RNA-induced silencing complex) (9) and that it may play important additional roles in global regulation of translation (10) and cell proliferation (11). The molecular basis of the diverse eIF6 activities, including its interaction with the 60 S subunit and the resulting ribosomal anti-association activity, are poorly understood (11).
EXPERIMENTAL PROCEDURES
Complex Reconstitution
The full-length protein eIF6 from Saccharomyces cerevisiae was overexpressed in Escherichia coli cells and purified via a His6 tag as described previously (6, 12). An anti-association assay with yeast 80 S ribosomes was performed to check for the activity of the purified eIF6 (12). For preparation of the sample, 2 pmol of gradient-purified 60 S subunits were mixed with 10 pmol of eIF6 to reconstitute the 60 S-eIF6 complex in buffer A (20 mm Hepes, 100 mm KCl, 2.5 mm Mg(OAc)2, 1 mm dithiothreitol).
Electron Microscopy and Interpretation
The sample was applied to carbon-coated holey grids. The data set was collected on a Tecnai F30 microscope at 300 kV under low dose conditions and digitized on a Heidelberg drum scanner using a pixel size of 1.23 Å/pixel on object scale. Contrast transfer function was determined with the CTFFIND software (13). After automated particle selection with SIGNATURE (14) followed by visual inspection with WEB (15), 107,000 particles were selected for further processing. The resulting data set was processed using the SPIDER software package (15). Due to the presence of 80 S ribosomal particles in the sample, the data set had to be classified into two subsets (16), representing either 60 S or 80 S ribosomal particles. Around 76,000 particles were used for the final contrast transfer function (CTF)-corrected three-dimensional reconstruction with a resolution of 11.8 Å (8.5 Å, 3 σ criteria) (supplemental Fig. 1S), showing a 60 S ribosome with clear additional extra density for eIF6. The 60 S-eIF6 data set was used for the final CTF-corrected three-dimensional reconstruction with a determined resolution of 11.8 Å, according to 0.5 fourier shell correlation (FSC) criteria (8.5 Å, 3 σ criteria) (supplemental Fig. 1S). The resolution of the cryo-EM reconstruction of the 60 S-eIF6 complex appeared to be limited by preferential orientation of the particles on the grid. Densities for the large ribosomal subunit and for eIF6 were isolated using binary masks. The eIF6 crystal structure of S. cerevisiae (Protein Data Bank (PDB) entry 1G62) was taken as a basis for flexible fitting. First, rigid fitting of the eIF6 structure into the density was performed using a local exhaustive search with the software Mod-EM guided by a cross-correlation coefficient (CCC) between the atomic structure and the density map (17). This search was taking into account both sides and the five possible in-plane rotations of the structure. Next, the best fits (with the highest CCC) were further validated by comparison with the best local fits achieved using the Chimera fit-in-map module (18). Finally, the best eIF6 fit (CCC 0.886) was refined by using the software Flex-EM (19) together with its ribosomal interaction partner rpL23, taking into account the binding interface when eIF6 is bound to rpL23 (supplemental Table 1S). The last C-terminal 20 amino acids of eIF6 were not used for the final fit because they did not influence the fitting result. They are highly likely to be non-structured and, in agreement with that, they could not be localized in the map as density information.
RESULTS AND DISCUSSION
Structure of the Ribosomal 60 S-eIF6 Complex
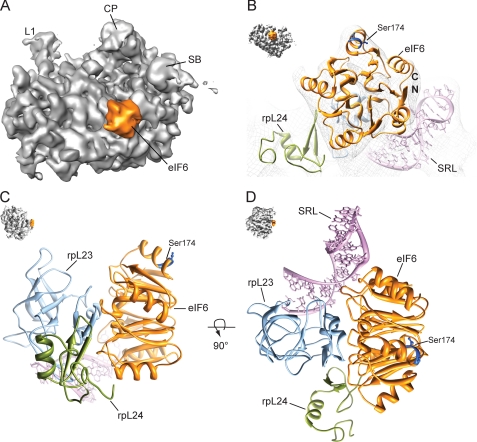
We reconstituted a ribosomal 60 S-eIF6 complex from S. cerevisiae in vitro and used cryo-EM in combination with single particle analysis for a three-dimensional reconstruction. The structure of the 60 S-eIF6 complex reveals an extra density representing eIF6 in the intersubunit space (Fig. 1A). The characteristic five-fold pseudosymmetric structure (12) is resolved in the cryo-EM map (11.8 Å resolution at 0.5 FSC cutoff, supplemental Fig. 1S) and allows the identification of the binding site of eIF6 on the 60 S subunit (Fig. 1B, supplemental Fig. 2S). The main interaction partner of eIF6 is the ribosomal protein rpL23 (L14p in E. coli), which is located at the intersubunit surface of the 60 S subunit (Fig. 1, C and D). This binding site appears to be conserved between kingdoms because it is in agreement with a similar binding site that has recently been suggested for the archaeal aIF6 on the basis of chemical cross-linking and RNA protection experiments (20). The crystal structure of eIF6 from S. cerevisiae (PDB entry 1G62) was taken as a basis for flexible fitting into the assigned EM density for eIF6 using the software Flex-EM (21). The eIF6 binding interface with the 60 S subunit is in good agreement with previously published genetic data. Suppressor mutations in the TIF6 gene which rescue sdo1Δ cells locate to this particular region that has also been suggested to mediate ribosome binding (8). To fit the density, the C-terminal part of rpL23 was adjusted, leading to a small inward movement of the helical C terminus of rpL23 relative to the conformation observed in the 80 S ribosome. The hydrophobic C-terminal interaction surface of rpL23 interacts with the helical binding region of eIF6, both of which show a high degree of conservation. In addition to rpL23, the neighboring ribosomal protein rpL24 (L24e in Haloarcula marismortui) and the highly conserved sarcin-ricin loop (SRL) also contribute to the interaction with eIF6 (Fig. 1D).
FIGURE 1.
Localization of eIF6 in a cryo-EM reconstruction of the 60 S-eIF6 complex. A, the map is shown in the crown view. B, close up of the fitted eIF6 structure in EM-density (mesh). C and D, molecular environment of the eIF6 binding site on the 60 S subunit. Thumbnails show the respective orientation. Colors are: orange, eIF6; magenta, sarcin-ricin loop (SRL)); blue, rpL23; green, rpL24; gray, 60 S ribosomal subunit. Abbreviations are: CP, central protuberance; L1, L1 stalk; SB, stalk base; N/C, N and C terminus of eIF6, respectively).
Anti-association Activity of eIF6
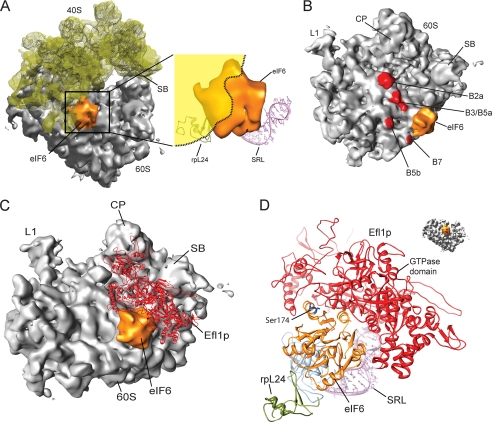
The anti-association activity of eIF6 prevents binding of the 40 S ribosomal subunit to the 60 S subunit. When the 40 S subunit in its 80 S-bound conformation is superimposed with the 60 S-eIF6 complex, it is evident that both eIF6 and the 40 S subunit share a common binding region on the 60 S subunit and cannot be present simultaneously (Fig. 2A). A large part of the eIF6 density would clash with the rRNA of the 40 S subunit, explaining the mutually exclusive binding by simple steric hindrance (Fig. 2B). The position of eIF6 at the 60 S subunit surface coincides exactly with the dynamic intersubunit bridge B6 formed by the C-terminal region of the ribosomal protein rpL23 (22). Therefore, in the presence of eIF6, the bridge B6 and the nearby located bridge B5a, both formed by rpL23, are directly affected. In addition, the surrounding intersubunit contacts B7, B3, and B5b also can no longer be formed, explaining the observed highly efficient prevention of subunit joining (Fig. 2B). The phosphorylation sites of eIF6, Ser-174 and Ser-175, are located at the accessible surface of the bound eIF6, which is not involved in the interaction with the 60 S subunit (Fig. 1B). Also, the C-terminal nuclear localization sequence (and Ser-235), which was not visualized in the crystal structure, would be located at the accessible outer surface of eIF6.
FIGURE 2.
Mechanism of the anti-association activity of eIF6. A, cryo-EM map of the 60 S-eIF6 complex superimposed with the 40 S subunit (semitransparent yellow) in its 60 S-bound conformation. A close-up view of the interfering region of the 40 S subunit is indicated as yellow shadow. B, intersubunit bridges (red) in the vicinity of eIF6. The binding position of eIF6 at the 60 S subunit coincides directly with bridge B6. C, the binding of Efl1p would allow the release eIF6. Homology model of the eEF2-like GTPase Efl1p (red) in its putative ribosome-bound conformation together with eIF6. D, model of the putative Efl1p binding environment on the 60 S ribosomal subunit. Colors are: orange, eIF6; magenta, sarcin-ricin loop (SRL)); blue, rpL23; green, rpL24; gray, 60 S ribosomal subunit. Abbreviations are: SB, stalk base; CP, central protuberance; L1, L1 stalk.
Dissociation of eIF6 from the 60 S Ribosomal Subunit
The elongation factor-like GTPase Efl1p, which is required for the release of eIF6 in yeast, is highly homologous to eEF2. It has been shown that Efl1p can compete with eEF2 for ribosome binding, resulting in inhibition of the ribosome-associated GTPase activity of eEF2 (23). Therefore, it is safe to assume that they share a similar binding site, which is known for eEF2 to be the canonical translation factor binding site (24). We modeled Efl1p using HHpred (25) and MODELLER (26) with the ribosome-bound eEF2 (PDB entry 1S1H) as a template, resulting in a model for the 60 S ribosomal subunit-bound Efl1p. Strikingly, in this position, Efl1p could bind concomitantly with eIF6 and would perfectly embrace eIF6 to facilitate an interaction with its GTPase domain (Fig. 2C). A small conformational change of Efl1p in this position would be sufficient to trigger GTP-dependent release of eIF6 from the 60 S subunit (Fig. 2D).
Our structural information provides the basis for a molecular understanding of the conserved activity of eIF6 as a ribosomal anti-association factor. It remains to be seen to what extent the ribosomal binding mode of eIF6 is also functionally involved in its additional activities in higher eukaryotes.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (Grants SFB594 and SFB646) and the Center for Integrated Protein Science Munich (to R. B.), by a grant from the Deutsche Forschungsgemeinschaft SFB740 (to T. M.), and by grants from the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (Anwenderzentrum).
The atomic coordinates and structure factors (code 2x7n) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The cryo-electron microscopic map has been deposited in the 3D-EM database under accession number EMD-1705.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S and 2S and Table 1S.
- eIF6
- eukaryotic initiation factor 6
- eEF2
- eukaryotic translation elongation factor 2
- cryo-EM
- cryo electron microscopy
- Efl1p
- elongation factor-like protein 1 of yeast
- CCC
- cross-correlation coefficient.
REFERENCES
- 1.Horsey E. W., Jakovljevic J., Miles T. D., Harnpicharnchai P., Woolford J. L., Jr. (2004) RNA 10, 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valenzuela D. M., Chaudhuri A., Maitra U. (1982) J. Biol. Chem. 257, 7712–7719 [PubMed] [Google Scholar]
- 3.Russell D. W., Spremulli L. L. (1979) J. Biol. Chem. 254, 8796–8800 [PubMed] [Google Scholar]
- 4.Basu U., Si K., Deng H., Maitra U. (2003) Mol. Cell. Biol. 23, 6187–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhäuser N., Marchisio P. C., Biffo S. (2003) Nature 426, 579–584 [DOI] [PubMed] [Google Scholar]
- 6.Si K., Maitra U. (1999) Mol. Cell. Biol. 19, 1416–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senger B., Lafontaine D. L., Graindorge J. S., Gadal O., Camasses A., Sanni A., Garnier J. M., Breitenbach M., Hurt E., Fasiolo F. (2001) Mol. Cell 8, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 8.Menne T. F., Goyenechea B., Sánchez-Puig N., Wong C. C., Tonkin L. M., Ancliff P. J., Brost R. L., Costanzo M., Boone C., Warren A. J. (2007) Nat. Genet. 39, 486–495 [DOI] [PubMed] [Google Scholar]
- 9.Chendrimada T. P., Finn K. J., Ji X., Baillat D., Gregory R. I., Liebhaber S. A., Pasquinelli A. E., Shiekhattar R. (2007) Nature 447, 823–828 [DOI] [PubMed] [Google Scholar]
- 10.Gandin V., Miluzio A., Barbieri A. M., Beugnet A., Kiyokawa H., Marchisio P. C., Biffo S. (2008) Nature 455, 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miluzio A., Beugnet A., Volta V., Biffo S. (2009) EMBO Rep. 10, 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groft C. M., Beckmann R., Sali A., Burley S. K. (2000) Nat. Struct. Biol. 7, 1156–1164 [DOI] [PubMed] [Google Scholar]
- 13.Mindell J. A., Grigorieff N. (2003) J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 14.Chen J. Z., Grigorieff N. (2007) J. Struct. Biol. 157, 168–173 [DOI] [PubMed] [Google Scholar]
- 15.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 16.Penczek P. A., Frank J., Spahn C. M. (2006) J. Struct. Biol. 154, 184–194 [DOI] [PubMed] [Google Scholar]
- 17.Topf M., Baker M. L., John B., Chiu W., Sali A. (2005) J. Struct. Biol. 149, 191–203 [DOI] [PubMed] [Google Scholar]
- 18.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 19.Topf M., Lasker K., Webb B., Wolfson H., Chiu W., Sali A. (2008) Structure 16, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benelli D., Marzi S., Mancone C., Alonzi T., la Teana A., Londei P. (2009) Nucleic Acids Res. 37, 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topf M., Baker M. L., Marti-Renom M. A., Chiu W., Sali A. (2006) J. Mol. Biol. 357, 1655–1668 [DOI] [PubMed] [Google Scholar]
- 22.Spahn C. M., Beckmann R., Eswar N., Penczek P. A., Sali A., Blobel G., Frank J. (2001) Cell 107, 373–386 [DOI] [PubMed] [Google Scholar]
- 23.Graindorge J. S., Rousselle J. C., Senger B., Lenormand P., Namane A., Lacroute F., Fasiolo F. (2005) J. Mol. Biol. 352, 355–369 [DOI] [PubMed] [Google Scholar]
- 24.Spahn C. M., Gomez-Lorenzo M. G., Grassucci R. A., Jørgensen R., Andersen G. R., Beckmann R., Penczek P. A., Ballesta J. P., Frank J. (2004) EMBO J. 23, 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Söding J., Biegert A., Lupas A. N. (2005) Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995) Proteins 23, 318–326 [DOI] [PubMed] [Google Scholar]