Abstract
The phosphatidylinositol 3-kinase (PI3K)-protein kinase B (PKB)/Akt-PTEN signal transduction pathway orchestrates a variety of fundamental cell processes and its deregulation is implicated in many human diseases. Although the importance of this pathway to many cellular functions is well established, the mechanisms by which it achieves context-specific physiological outcomes in response to a variety of stimuli, using a relatively limited pool of effectors, remain largely unknown. Spatial restriction of signaling events is one means by which cells coordinate specific responses using common molecules. To investigate the subcellular location-specific roles of the major PI3K effector PKB/Akt in various cell processes, we have developed a novel experimental system employing cellular compartment-directed PKB/Akt pseudosubstrate inhibitors. Subcellular location-restricted PKB/Akt inhibition in the 3T3L1 adipocyte differentiation model revealed that nuclear and plasma membrane, but not cytoplasmic, PKB/Akt activity is required for terminal adipocyte differentiation. Nuclear and plasma membrane pools of PKB/Akt were found to contribute to distinct stages of adipocyte differentiation, revealing that PKB/Akt activity impacts multiple points of this program. Our work establishes the use of localized pseudosubstrate PKB/Akt inhibitors as an effective method for the dissection of PKB/Akt signaling.
Keywords: Adipocyte, Akt PKB, Differentiation, Protein Kinases, Subcellular Organelles, Localized Kinase Inhibitors, Pseudosubstrate
Introduction
The phosphatidylinositol 3-kinase (PI3K)2-protein kinase B (PKB)/Akt-phosphatase and tensin homolog (PTEN) pathway is an evolutionarily conserved signaling cascade implicated in the regulation of several fundamental cell processes including cell proliferation, survival, motility and size, glucose metabolism, and differentiation. Appropriate execution of PI3K-PKB/Akt-PTEN signaling is of critical importance to the proper functioning of cells and organisms, exemplified by the variety of developmental defects associated with disruption of its constituents in model organisms (1). Moreover, the components of the pathway are frequent targets of mutations associated with cancer and other human diseases (2).
Upon activation in response to a variety of cellular stimuli, PI3K phosphorylates the D3 position of the phosphoinositide PI(4,5)P2, leading to the generation of the second messenger PI(3,4,5)P3 at the plasma membrane. PI(3,4,5)P3 acts as a docking site for pleckstrin homology (PH) domain containing proteins such as PKB/Akt and its activating kinase, 3-phosphoinositide-dependent kinase 1 (3). Upon recruitment to PI(3,4,5)P3 and activation-specific phosphorylation by 3-phosphoinositide-dependent kinase 1, as well as additional phosphorylation by mammalian target of rapamycin complex 2 (mTORC2) (4), fully activated PKB/Akt executes a wide spectrum of cellular responses acting through a variety of intracellular substrates (5). The role of this signaling pathway in the process of adipocyte differentiation has been demonstrated by genetic disruption of PKB/Akt in mice, which results in impaired adipogenesis (6), as well as cell culture models of adipocyte differentiation, in which PI3K inhibition or PKB/Akt knockdown blocks the ability of cells to differentiate into adipocytes (7, 8).
Cellular localization of PKB/Akt contributes to its tight regulation. The requirement for PKB/Akt translocation to the plasma membrane for its activation by 3-phosphoinositide-dependent kinase 1 (3, 9) ensures that the kinase is not aberrantly activated in the cytoplasm in the absence of cell stimulation. Following activation at the plasma membrane, PKB/Akt translocates to various cellular locations to phosphorylate its targets. Remarkably, PKB/Akt substrates have been identified at a variety of locations within the cell, such as TSC2 (10) and PRAS40 (11) in the cytoplasm, FOXO transcription factors in the nucleus (12), AS160 at cellular membranes (13), Bad at the mitochondria (14), and Girdin/APE at the cytoskeleton (15). Despite a significant understanding of the functions of the growing group of PKB/Akt substrates, little is known of the mechanisms that govern access of PKB/Akt to its distinctly located targets. Recent discovery that the adaptor protein containing PH domain, PTB domain, and leucine zipper motif 1 (Appl1) interacts with PKB/Akt and directs its activity toward glycogen synthase kinase 3 but not TSC2, although requiring endosomal localization, establishes distinct subcellular localization of PKB/Akt as a determinant of its substrate specificity (16).
Here, we describe the design and implementation of a novel experimental system allowing subcellular location-specific inhibition of PKB/Akt. Using this system we investigated the effects of compartment-restricted PKB/Akt inhibition on adipocyte differentiation and found that nuclear and plasma membrane, but not cytoplasmic, PKB/Akt activity is required for this process. Interestingly, nuclear and plasma membrane pools of PKB/Akt were found to contribute to different stages of adipocyte differentiation. Our work establishes localized PKB/Akt inhibition as a powerful system for prioritization of PKB/Akt substrates.
EXPERIMENTAL PROCEDURES
Materials
All materials were from Sigma unless otherwise stated. Monoclonal antibodies against the HA epitope (clone 12CA5) and myc epitope (clone 9E10) were prepared from hybridomas according to standard protocol. α-FLAG (M2) antibody was purchased from Sigma, α-58K Golgi protein, α-calnexin, and α-glyceraldehyde-3-phosphate dehydrogenase from Abcam, α-c/EBPβ, α-c/EBPα, and α-cyclin A from Santa Cruz, α-P-TSC2, α-P-Akt substrate, and α-PKB/Akt from Cell Signaling, α-P-PRAS40 and α-glucose transporter 4 from Millipore, α-p27Kip1 from BD Transduction Laboratories, α-glutathione S-transferase (GST) from GE Healthcare, and α-HA-fluorescein (clone 3F10) from Roche Applied Science.
Generation of Localized PKB/Akt Inhibitors
Myc-tagged PKB/Akt inhibitors (PKBis) localized to the nucleus (via SV40 large T antigen amino acids 126–132 (17)) and cytoplasm (via influenza B NS2/NEP amino acids 10–19 (18)) were obtained from Intrexon Corp. (Blacksburg, VA). Subsequently, cDNA encoding GST was cloned in-frame with inhibitor cDNA to generate GST fusion proteins. GST-fused inhibitor cDNAs were then cloned into vectors encoding localization signals directed to the Golgi (N-terminal 81 amino acids of human β1,4-galactosyltransferase), ER (C-terminal 10 amino acids from cytochrome b5), and plasma membrane (N-terminal 14 amino acids of human Src). cDNAs encoding localization sequence-tagged, GST-fused, myc-tagged PKBis were then subcloned into the pTRE2-Hyg tetracycline-inducible vector (Clontech) and pBMN-I-GFP retroviral vector (Garry Nolan laboratory; Addgene plasmid 1736).
Inhibitor-Kinase Interaction Assay
HEK 293 cells were co-transfected with non-localized GST-fused inhibitor and various AGC kinases by the calcium phosphate method. 48 h post-transfection cells were lysed in CHAPS lysis buffer (10 mm Tris-HCl, pH 7.4, 1 mm MgCl2, 1 mm EGTA, 0.5% CHAPS, 10% glycerol, 50 mm NaF, 100 μm sodium orthovanadate and protease inhibitor mixture (Calbiochem)) and normalized for protein content. Equal amounts of cell lysate were incubated with CHAPS lysis buffer-equilibrated glutathione-Sepharose (GE Healthcare) at 4 °C with rotation overnight. Glutathione-Sepharose-purified proteins were washed 3 times in CHAPS lysis buffer containing 0.2 m NaCl and dissociated from beads using 2× SDS loading buffer (100 mm Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 15% glycerol, 0.25% bromphenol blue) prior to separation by SDS-PAGE, transfer to PVDF membrane (Millipore), and immunoblotting.
In Vitro Kinase Assay
Bacterially purified GST or GST-PKBi (125 pmol/reaction) was preincubated with epitope-tagged kinases immunoprecipitated from transiently transfected HEK 293 cells for 10 min at 30 °C. Kinase assays were then initiated by the addition of the respective substrate (250 pmol/reaction), 500 μm cold ATP and 5 μCi of [γ-32P]ATP diluted in the respective kinase assay buffers. Kinase assay buffers were: PKB/Akt and p70S6K (10 mm MOPS, pH 7.2, 20 mm MgCl2, 2.5 mm EGTA, 12.5 mm β-glycerol phosphate, 0.5 mm sodium orthovanadate, 0.5 mm dithiothreitol) and p90RSK (50 mm MOPS, pH 7.4, 10 mm MgCl2, 10 mm MnCl2, 2 mm EGTA, 20 mm β-glycerol phosphate, 1 mm dithiothreitol). The substrate for PKB/Akt and p70S6K was Crosstide (GRPRTSSFAEG) and for p90RSK, RSK substrate (KKRNRTLTV).
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and permeabilized with 0.2% Triton X-100 in PBS for 2 min at room temperature. Nonspecific protein binding was blocked using blocking buffer (1.5% bovine serum albumin, 1.5% goat serum in PBS) for 1 h at room temperature. Primary antibody was diluted in blocking buffer (9E10 α-myc, 1:100; α-58K Golgi protein, 1:100; α-calnexin, 1:100; α-PKB/Akt, 1:100) and used to stain blocked cells overnight at 4 °C. After 3 washes with PBS cells were incubated with secondary antibodies (Molecular Probes) diluted 1:400 in blocking buffer for 1 h at 4 °C. After a further 3 washes with PBS, coverslips were mounted onto slides with Mowiol 4-88 reagent (Calbiochem) containing 1 μg/ml of 4′,6-diamidino-2-phenylindole dihydrochloride and imaged on a Zeiss Axiovert 200M microscope equipped with a Roper Scientific Coolsnap HQ camera. For HA-FOXO3a localization assay, the α-HA-fluorescein clone 3F10 (Roche Applied Science) was added (2 μg/ml) in combination with secondary antibody. α-c/EBPβ immunofluorescence was performed as described (19).
Preparation of the Plasma Membrane Fraction
3T3L1 cells were retrovirally transduced, starved overnight in Dulbecco's modified Eagle's medium (DMEM) containing 0.2% FBS, then stimulated with 10% FBS for either 5 min or 2 h. Plasma membrane preparation was as described (20). Briefly, cells were washed twice with ice-cold PBS and homogenized in HES buffer (20 mm HEPES, pH 7.5, 250 mm sucrose, and 1 mm EDTA) containing protease inhibitors (Calbiochem) with a 27½-gauge needle. Homogenized cells were centrifuged at 3,000 × g and the supernatant collected and centrifuged at 20,000 × g to produce a crude plasma membrane fraction. The resulting plasma membrane containing pellet was resuspended in 2× SDS loading buffer and separated by SDS-PAGE.
Preparation of Nuclear Lysates
Retrovirally transduced 3T3L1 cells were washed with ice-cold PBS and resuspended in sucrose buffer (0.32 m sucrose, 10 mm Tris-HCl, pH 8.0, 3 mm CaCl2, 2 mm MgOAc, 0.1 mm EDTA, 0.5% Nonidet P-40, 1 mm dithiothreitol, and protease inhibitors). Nuclei were collected by centrifugation at 500 × g and washed with sucrose buffer without Nonidet P-40. Nuclei were then resuspended in low salt buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 20 mm KCl, 0.2 mm EDTA, 25% glycerol, 0.5 mm dithiothreitol, and protease inhibitors), and high salt buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 800 mm KCl, 0.2 mm EDTA, 25% glycerol, 1% Nonidet P-40, 0.5 mm dithiothreitol, and protease inhibitors) was added slowly with gentle mixing. Samples were incubated for 45 min at 4 °C with rotation, and centrifuged 15 min to clear the nuclear extracts.
FOXO3a Luciferase Assay
COS7 cells were transiently co-transfected with the pGL3-FOXO luciferase reporter, pCMV-β-galactosidase, and the indicated constructs. Luciferase activity was assayed (Promega Luciferase Assay System) and normalized to β-galactosidase activity, which was assayed using Tropix Emerald reagent according to the manufacturer's instructions.
Adipocyte Differentiation
3T3L1 preadipocytes were cultured in DMEM containing 10% FBS until confluence and maintained for 48 h (day 0). Cells were induced to differentiate using MDI induction medium (DMEM containing 10% FBS and 0.5 mm 3-isobutyl-1-methylxanthine, 1 μm dexamethasone, and 1 μg/ml of insulin) for 2 days, followed by insulin medium (DMEM containing 10% FBS and 1 μg/ml insulin) for 2 days, at which time the cells were fed with DMEM containing 10% FBS every 2 days. Terminal differentiation was assayed by Oil Red O staining: cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, stained with Oil Red O (3 μg/ml in 60% isopropyl alcohol) for 1 h at room temperature, washed twice with distilled water, and plates were dried and scanned for images. The integrated density of each sample was calculated using ImageJ (NIH). Oil Red O dye was extracted using 100% isopropyl alcohol and measured by spectrophotometry at 510 nm.
Mitotic Clonal Expansion Analysis
For enumeration experiments, 3T3L1 preadipocytes were seeded in quadruplicate, retrovirally transduced, and stimulated with MDI induction medium for the indicated times. Cells were rinsed, trypsinized, and counted using the Z2 Coulter cell and particle counter. For cell cycle analysis, 3T3L1 preadipocytes were seeded in triplicate, retrovirally transduced, and stimulated with MDI induction medium for 20 h. Cells were trypsinized, fixed with 2% paraformaldehyde and 2% FBS in PBS, permeabilized with 0.2% Triton X-100, then stained with 50 μg/ml of propidium iodide in 3.8 mm sodium citrate supplemented with 0.5 μg/ml of RNase A. DNA content was analyzed by flow cytometry (BD Biosciences FACSCalibur using CellQuest software).
RESULTS
Development of a Novel Experimental System to Study Subcellular Compartment-specific PKB/Akt Signaling
We hypothesized that specificity within the PI3K signaling pathway is, at least in part, achieved by compartmentalization of its constituents resulting in compartment-specific outcomes. To explore the locus-specific aspects of signaling via PKB/Akt, we developed agents allowing interference with PKB/Akt function within distinct areas of the cell. The interference strategy was founded on the principles of substrate recognition by protein kinases, which interact with unphosphorylated substrates with high affinity and catalyze phosphate transfer to the target residue, creating a lower affinity interaction moiety and leading to their dissociation from the substrate. Substitution of the target residue within the substrate recognition sequence with a non-phosphorylatable amino acid has been found to result in “kinase trapping” and inhibition of kinase activity akin to the mechanism naturally employed by the pseudosubstrate regulatory regions of proteins (21).
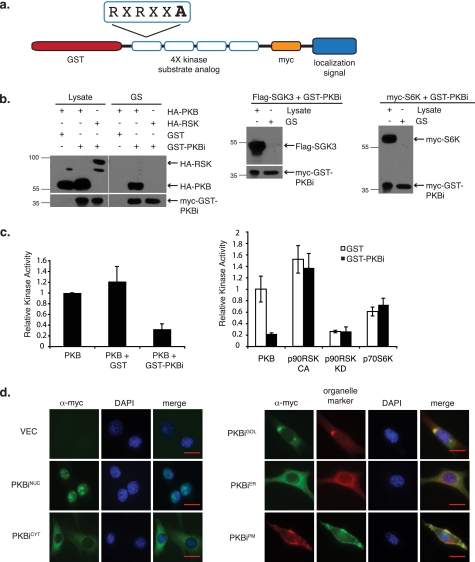
To develop PKBis, four variations of the PKB/Akt substrate recognition sequence RXRXX(S/T), containing alanines in place of the target serines or threonines (Intrexon Corp.), were cloned in-frame with the cDNA encoding GST (Fig. 1a). To facilitate immunodetection, a myc epitope tag was also included. PKB/Akt is a member of the AGC family of kinases, which share similar substrate recognition sequences. We therefore examined the specificity of PKBi by testing its ability to physically interact with a panel of AGC kinases. As shown in Fig. 1b, affinity purified PKBi readily co-precipitated with PKB/Akt, but not p90RSK, p70S6K, or SGK3, indicating that even under conditions of their overexpression, PKBi did not interact with the other AGC kinase family members. Consistently, purified PKBi interfered with PKB/Akt kinase activity toward the Crosstide substrate in vitro, decreasing it to less than 30% of the activity achieved under control conditions (Fig. 1c). In contrast, PKBi did not impact the kinase activities of p90RSK or p70S6K toward their preferred peptide substrates (Fig. 1c).
FIGURE 1.
Design and characterization of compartment-restricted PKBis. a, schematic representation of cDNAs encoding PKBi. b, HEK 293 cells were co-transfected with cDNA encoding a GST-PKBi fusion protein and HA-tagged PKB/Akt (HA-PKB), HA-tagged p90RSK2 (HA-RSK), myc-tagged p70S6K (myc-S6K), or FLAG-tagged SGK3 (FLAG-SGK3). Cell lysates were subjected to glutathione-Sepharose (GS) purification and purified proteins were washed, separated by SDS-PAGE, transferred to PVDF membrane, and immunoblotted with the indicated antibodies. c, bacterially purified GST or GST-PKBi was incubated with epitope-tagged kinases immunoprecipitated from transiently transfected HEK 293 cells. In vitro kinase assays were carried out in triplicate with the immobilized kinases and their respective substrates in the presence of [γ-32P]ATP and the supernatant was transferred to P81 phosphocellulose paper. Samples were washed and the resulting counts per minute recorded by scintillation. Values were plotted relative to PKB/Akt activity for three experiments (error bar = S.E.). d, NIH 3T3 cells stably transfected with plasmids encoding doxycyclin-inducible localized PKB/Akt inhibitors were treated overnight with 1 μg/ml of doxycyclin and immunofluorescence was performed against the myc tag. PKBiNUC- and PKBiCYT-expressing cells were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). PKBiGOL- and PKBiER-expressing cells were co-stained with antibody against 58K Golgi protein and calnexin, respectively. PKBiPM-expressing cells were transiently transfected with cDNA encoding GFP-fused PH domain of PKB/Akt. Scale bar = 20 μm.
We next sought to apply PKBis toward PKB/Akt in a subcellular compartment-specific manner. For this, signal peptides directing specific subcellular localization sequences were engineered in-frame with the PKBi (Fig. 1a). Namely, influenza B NS2/NEP amino acids 10–19 were used to achieve cytoplasmic localization (PKBiCYT) (18), N-terminal 14 amino acids of human Src were used for localization to the plasma membrane (PKBiPM), SV40 large T antigen amino acids 126–132 were used to direct nuclear localization (PKBiNUC) (17), N-terminal 81 amino acids of human β1,4-galactosyltransferase for localization in the Golgi lumen (PKBiGOL) (22) and C-terminal 10 amino acids from cytochrome b5 for localization to the outer surface of the ER (PKBiER) (23). The subcellular localization of compartment-directed PKBis was tested by immunofluorescence against the myc tag and co-localization with known organelle markers (Fig. 1d). True to their localization tags, each PKBi was found to co-localize with its compartment-specific marker.
Given the observed interaction between PKBi and PKB/Akt, we asked whether localized PKBis might sequester PKB/Akt to the targeted loci, which could confound interpretation of the consequences of localized PKB/Akt inhibition. In vector-transfected, as well as nuclear- (PKBiNUC), cytoplasm- (PKBiCYT), Golgi- (PKBiGOL), and ER- (PKBiER) localized PKBi-transfected cells, endogenous PKB/Akt displayed predominantly nuclear localization with varying degrees of cytoplasmic and plasma membrane staining among different cells (supplemental Fig. S1). Although there was a modest increase in plasma membrane-localized PKB/Akt upon expression of plasma membrane-localized PKBi (PKBiPM), ample amounts of PKB/Akt remained distributed throughout the cell (supplemental Fig. S1). To assess the temporal dynamics of PKB/Akt translocation and phosphorylation in the presence of PKBiPM, we compared the PKB/Akt recovered from plasma membrane fractions of control and PKBiPM-expressing cells. Consistent with previous reports, we observed increased phosphorylation of PKB/Akt upon its inhibition, an effect attributed to the engagement of the negative feedback through p70S6K toward insulin receptor substrates 1/2 (24–26). Nevertheless, the temporal pattern of PKB/Akt activation was unchanged in response to PKBiPM, as were the total PKB/Akt levels at the plasma membrane of PKBiPM-expressing cells (supplemental Fig. S1). Thus, PKB/Akt sequestration by the localized PKBis is not predicted to contribute to the biological effects of localized PKB/Akt inhibition.
Compartment-specific Reduction in Phosphorylation of PKB/Akt Targets Using Localized PKBis
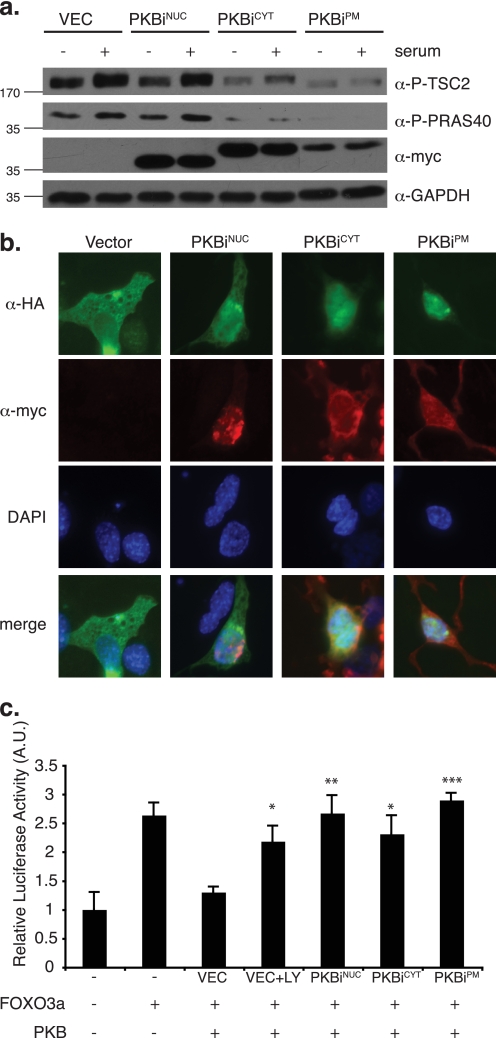
We examined the effect of localized PKBis on two well documented cytoplasmic PKB/Akt substrates, TSC2 (10) and PRAS40 (11). Under conditions of starvation and subsequent serum stimulation, TSC2 and PRAS40 phosphorylation by PKB/Akt was impaired in NIH 3T3 cells expressing the cytoplasmic and plasma membrane-localized PKBis, whereas PKBiNUC expression had no effect on these targets (Fig. 2a). Similarly, PKBiCYT and PKBiPM specifically affected phosphorylation of the downstream cytoplasmic targets p70S6K and ribosomal S6 protein (supplemental Fig. S2). Expression levels of PKBiGOL and PKBiER were insufficient to monitor their effects using lysis of whole cell populations (data not shown). To test whether PKBis could inhibit the nuclear PKB/Akt pool, we investigated subcellular localization of the FOXO family member, the transcription factor FOXO3a, a nuclear target of PKB/Akt (12). Upon PKB/Akt-mediated phosphorylation, FOXO3a is exported from the nucleus, making this property a readout for nuclear PKB/Akt activity (12). Surprisingly, in addition to PKBiNUC, expression of PKBiCYT and PKBiPM also led to the nuclear accumulation of FOXO3a (Fig. 2b). In contrast, sequestration of PKBi to either the Golgi or the ER impeded its ability to down-regulate nuclear PKB/Akt kinase activity toward FOXO3a, resulting in cytoplasmic FOXO3a localization (supplemental Fig. S3).
FIGURE 2.
Subcellular compartment-restricted PKB/Akt inhibition. a, NIH 3T3 cells transduced with retrovirus encoding localized myc-tagged PKBis were starved in medium containing 0.2% FBS overnight. Starved cells were either left untreated or stimulated with 10% FBS for 15 min. Total cell lysates were normalized for protein content and 25 μg was separated by SDS-PAGE followed by transfer to PVDF membrane and immunoblotting with the indicated antibodies against cytoplasmic PKB/Akt substrates (P-TSC2, P-PRAS40), epitope-tagged inhibitors (myc), and loading control (GAPDH, glyceraldehyde-3-phosphate dehydrogenase). b, 3T3L1 cells were transiently co-transfected with cDNA encoding HA-tagged FOXO3a and myc-tagged localized PKBis. Co-staining was performed using fluorescein-conjugated α-HA antibody (green) and indirect immunofluorescence against the myc tag (red). c, COS7 cells were transiently co-transfected with the pGL3-FOXO luciferase reporter of FOXO activity, FOXO3a, PKB/Akt, the indicated localized PKBi constructs, and equal amounts of β-galactosidase cDNA. LY294002 (LY) treatment was 12.5 μm overnight. Luciferase activity was assayed and normalized to β-galactosidase activity to account for differences in transfection efficiency. Three experiments were performed in triplicate and values were plotted relative to basal pGL3-FOXO luciferase activity (error bars = S.E.; *, p < 0.02; **, p < 0.01; ***, p < 0.001). DAPI, 4′,6-diamidino-2-phenylindole.
Phosphorylation of FOXO3a by PKB/Akt followed by its nuclear export leads to down-regulation of its transcriptional activity (12). We monitored the effect of localized PKBis on the transcriptional activity of FOXO3a using a FOXO3a-responsive luciferase reporter (27). As anticipated, transfection of PKB/Akt led to a decrease in FOXO3a activity, which could be restored by cell treatment with the PI3K inhibitor LY294002 (Fig. 2c). Consistent with their effect on FOXO3a localization, the nucleus-, cytoplasm-, and plasma membrane-specific inhibitors of PKB/Akt rescued the negative effect of PKB/Akt on FOXO3a transcriptional activity (Fig. 2c).
To decipher whether the effect on FOXO3a was unique, or whether PKBiCYT and PKBiPM may affect other nuclear PKB/Akt substrates, we performed immunoblot analysis with the anti-phospho-Akt substrate antibody on nuclear lysates from PKBiNUC-, PKBiCYT-, and PKBiPM-expressing cells (supplemental Fig. S4). Upon serum stimulation of control cells, increased phosphorylation of several nuclear proteins can be detected (supplemental Fig. S4, arrows). Apart from a ∼100 kDa protein species whose phosphorylation was reduced by all PKBis, Akt substrate phospho-reactivity of other nuclear targets was specifically reduced only by PKBiNUC, consistent with a high degree of specificity of this reagent toward nuclear PKB/Akt. Thus, compartment-specific PKB/Akt inhibition can be achieved by directing PKBis to specific subcellular loci, and PKB/Akt inhibition at the plasma membrane and the cytoplasm may impair a subset of subsequent downstream signaling events, even at other locations of action, such as the nucleus.
Subcellular-specific Roles for PKB/Akt in the Regulation of Adipocyte Differentiation
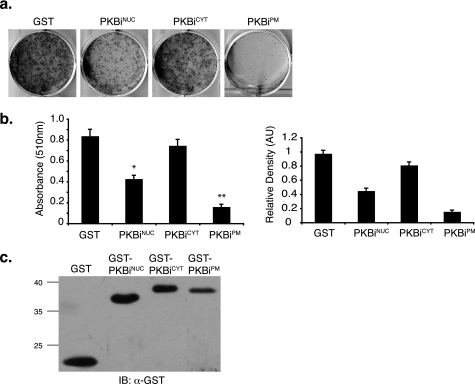
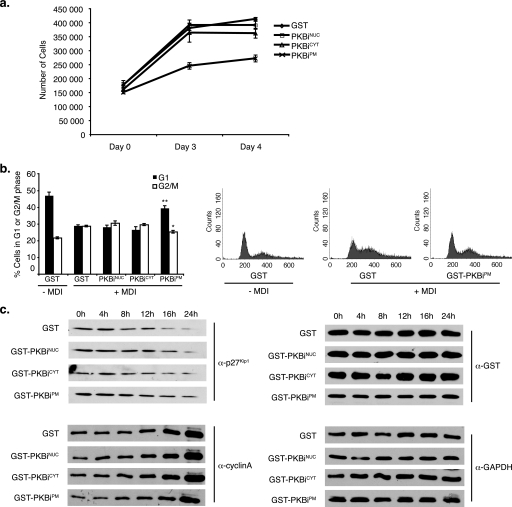
One of the many physiological outputs of PI3K-PKB/Akt signaling is adipose tissue differentiation (28). We sought to investigate whether spatially distinct pools of PKB/Akt may differentially contribute to the process of adipocyte differentiation using 3T3L1 pre-adipocytes as a model system. PKBis were retrovirally transduced into 3T3L1 cells and the cells were differentiated. Judged by the Oil red O accumulation and the densitometric image analysis 8 days following induction of differentiation, expression of either PKBiNUC or PKBiPM significantly impaired adipocyte differentiation, whereas PKBiCYT had no effect (Fig. 3, a and b). Reduced differentiation in PKBiNUC- and PKBiPM-expressing cells was not due to gross reduction in cell number, because these cells reached and maintained confluence throughout the differentiation process (supplemental Fig. S5).
FIGURE 3.
Subcellular compartment-specific regulation of adipocyte differentiation by PKB/Akt. 3T3L1 cells were transduced with retrovirus encoding localized GST-PKBi fusion proteins and induced to undergo adipocyte differentiation as described under “Experimental Procedures.” Differentiation was assayed by staining with Oil red O and quantified by measuring absorbance of isopropyl alcohol-extracted dye at 510 nm and by densitometry of Oil red O images. a, representative image of Oil red O staining. b, absorbance values of isopropyl alcohol-extracted Oil red O (left panel) and densitometry values of Oil red O images (right panel). Three experiments were performed in triplicate (error bars = S.E.; *, p < 1.4 × 10−5; **, p < 2.7 × 10−6). c, representative Western blot against GST showing inhibitor expression at day 8.
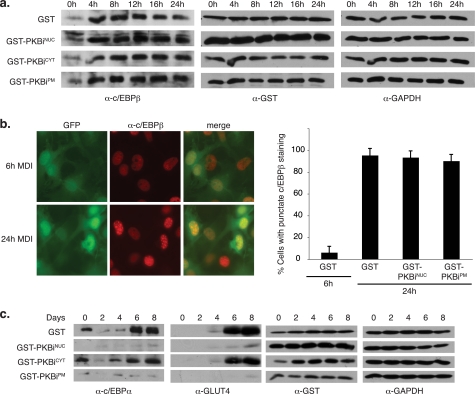
To gain further insight into the impact of localized PKBis on adipocyte differentiation, we tested the expression of CCAAT/enhancer-binding protein (c/EBP) β, a marker of the early stages of differentiation (29). Differentiation-specific induction of c/EBPβ remained unaffected upon PKBiNUC, PKBiCYT, or PKBiPM expression (Fig. 4a), consistent with previous findings that PKB/Akt is not required for c/EBPβ expression (6). Although c/EBPβ levels increase by 4 h after stimulation with differentiation medium, its ability to bind DNA and exert its transcriptional activity emerged 24 h after stimulation and can be measured by monitoring a distinct pattern of nuclear c/EBPβ staining (30). Immunofluorescence against endogenous c/EBPβ in 3T3L1 cells expressing PKBiNUC, PKBiPM, or controls revealed no difference in the punctate nuclear c/EBPβ staining, a surrogate marker for its association with DNA, indicative of progression through the differentiation program (Fig. 4b). Thus, PKB/Akt activity is not required for the induction or DNA-binding activity of c/EBPβ and the early stages of adipocyte differentiation.
FIGURE 4.
PKBiNUC and PKBiPM inhibit expression of late differentiation markers. a, 3T3L1 cells were transduced with retrovirus encoding localized GST-PKBi fusion proteins and induced to undergo adipocyte differentiation for the indicated time periods. Total protein lysates were normalized for protein content and separated by SDS-PAGE followed by transfer to PVDF membrane and immunoblotting with the indicated antibodies. b, 3T3L1 cells seeded on coverslips were transduced with retrovirus encoding GST or GST-PKBiPM and induced to undergo adipocyte differentiation. Cells were fixed at 6 and 24 h after induction and immunofluorescence was performed against endogenous c/EBPβ. Representative images of GST-expressing cells are shown. Green fluorescent protein (GFP)-positive (therefore retrovirally infected) cells exhibiting punctate nuclear c/EBPβ staining were counted from 10 fields per condition (error bars = S.E.). c, 3T3L1 cells were treated as in a. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We next examined endogenous mid- and late-stage adipocyte differentiation markers upon localized PKB/Akt inhibition. Expression of c/EBPα, which emerges midway through the differentiation program and precedes the transcription of most adipocyte-specific genes (29), was significantly impaired upon expression of PKBiNUC and PKBiPM, but not PKBiCYT (Fig. 4c). Consistent with this, expression of the terminal differentiation marker glucose transporter 4 was also reduced upon PKBiNUC and PKBiPM, but not PKBiCYT expression (Fig. 4c). Taken together, our data reveal that PKB/Akt activity at the plasma membrane and in the nucleus, but not in the cytoplasm, affects expression of mid- and late-stage adipocyte differentiation markers and is required for terminal differentiation.
Regulation of Mitotic Clonal Expansion by Plasma Membrane-localized PKB/Akt
As part of the adipocyte differentiation program, pre-adipocytes undergo a phase of growth arrest followed by mitotic clonal expansion (MCE) (29). Although cells expressing PKBiNUC proliferated at a rate equivalent to that of control cells, MCE of PKBiPM-expressing cells was significantly reduced (Fig. 5a). Cell cycle profile monitoring of differentiating cells expressing PKBis revealed that expression of PKBiPM blocked the G1 exit induced upon mitogenic stimulation (Fig. 5b). Consistently, levels of the cell cycle inhibitor p27Kip1, found to decrease 16–24 h following stimulation with differentiation medium, were sustained upon PKBiPM expression (Fig. 5c). Finally, levels of cyclin A, an indicator of S-phase entry normally induced 16–24 h into clonal expansion, lagged behind that of control cells in PKBiPM-expressing cells (Fig. 5c). Thus, our results demonstrate that PKB/Akt activities in the nucleus and at the plasma membrane are required for adipocyte differentiation, and uncover a specific role for plasma membrane PKB/Akt activity in progression through the MCE phase of this program.
FIGURE 5.
PKBiPM inhibits mitotic clonal expansion. a, 3T3L1 cells were transduced with retrovirus encoding localized GST-PKBi fusion proteins and stimulated with differentiation medium (MDI). Cell number was assayed using a Z2 Coulter Counter on the indicated days. Three experiments were performed in quadruplicate (error bars = S.E.). b, propidium iodide staining of cells was performed 20 h after induction of differentiation. The proportion of cells in G1 and G2/M phases was measured by flow cytometry in three experiments performed in triplicate (error bars = S.E.). Representative propidium iodide profiles are shown. c, retrovirally transduced 3T3L1 cells were stimulated with MDI for the indicated times. Total protein lysates were normalized for protein content and separated by SDS-PAGE followed by transfer to PVDF membrane and immunoblotting with the indicated antibodies.
DISCUSSION
Subcellular Localization Imparts Specificity during Signal Transduction
Spatial regulation of signaling molecules is a well documented mode of attaining specificity in signal transduction. Many cytoplasmic kinases translocate to the plasma membrane as part of their activation, whereas transcription factors are commonly regulated by their reversible translocation to the nucleus. Restricted localization can govern entire transcriptional programs by controlling access to target genes, as is the case with the nuclear PKB/Akt substrates, members of the FOXO family of transcription factors (12). Multimolecular signaling complexes can also be regulated by subcellular localization. For example, protein kinase A anchoring proteins direct protein kinase A toward specifically localized targets (31). Similarly, JNK-interacting protein scaffolds the components of the JNK signaling pathway to control their activation and cellular localization, such as at the tips of neuronal cell processes and at synaptic junctions (32). Protein kinase C (PKC) isozymes are also subject to distinct subcellular targeting, such as PKCα to the ER, PKCδ to the Golgi, and PKCϵ to the punctate structures throughout the cytoplasm (33).
Compartmentalization of PI3K pathway components is apparent during cell migration and chemotaxis, when PI(3,4,5)P3 concentrations locally increase at the leading edge of cells due to PI3K localization there and PTEN accumulation at the trailing edge (reviewed in Ref. 34). During phagocytosis, macrophages exhibit sharply confined PI(3,4,5)P3 accumulation at the phagosomal cup (35). PKB/Akt localization is also dynamically regulated. Following PI3K activation, PKB/Akt is recruited to the plasma membrane via interaction of its PH domain with PI(3,4,5)P3, which facilitates a conformational change and access to its activating kinases 3-phosphoinositide-dependent kinase 1 (3) and mTORC2 (4). Once fully activated, PKB/Akt translocates to a range of subcellular loci to phosphorylate its numerous substrates, including the cytoplasm (10, 11), nucleus (12), cellular membranes (13), mitochondria (14), and cytoskeleton (15).
Significantly, preferential localization of PKB/Akt to the plasma membrane due to a mutation in its PH domain has been associated with breast, colorectal, and ovarian cancers (36). Moreover, the extent of PKB/Akt nuclear localization was found to increase during the progression from normal tissue to low grade prostatic intraepithelial neoplasia, high grade prostatic intraepithelial neoplasia, and invasive carcinomas (reviewed in Ref. 37). Predominantly nuclear PKB/Akt has also been reported in certain lung and breast cancers, as well as in acute myeloid leukemia, and has been associated with invasion and metastasis in human thyroid cancer tissue (37). These findings illustrate the importance of the spatial organization of PKB/Akt in the normal functioning of cells. In this study, we demonstrate that distinct pools of PKB/Akt activity in the nucleus and at the plasma membrane, but not in the cytoplasm, govern the process of adipocyte differentiation. Moreover, our work establishes the use of PKBis to probe the importance of the spatial organization of PKB/Akt in the maintenance of cellular homeostasis and its deregulation in disease.
Novel Experimental Method for the Dissection of Subcellular-specific PKB/Akt Signaling
The strategy to use subcellular-targeted PKBis to probe compartment-specific roles of PKB/Akt hinges on the efficient and specific inhibition of PKB/Akt by these agents. Members of the AGC family of kinases share similar substrate recognition determinants. For example, the consensus recognition sequence for PKB/Akt is RXRXX(S/T) (where S/T represents the serine or threonine targeted for phosphorylation) (38), whereas p90RSK and p70S6K can also tolerate lysine residues at the −5 and −3 positions: (R/K)X(R/K)XX(S/T) (5). Despite these similarities, PKBi (composed of four variations of the consensus target sequence derived from natural PKB/Akt substrates) efficiently inhibited PKB/Akt but did not interact with other AGC kinases, nor did it inhibit their kinase activity in vitro (Fig. 1, b and c). This is consistent with the ability of pseudosubstrate inhibitors to inhibit other kinases with high specificity, including JNK (39), p60c-Src protein-tyrosine kinase (40), and PKC (41).
Subcellular-specific Roles for PKB/Akt in Adipocyte Differentiation
Application of localized PKBis in the 3T3L1 model system of adipocyte differentiation identified locus-specific roles for PKB/Akt in this cellular process. Specifically, we observed impaired differentiation upon expression of PKBiNUC and PKBiPM, but not PKBiCYT, despite the ability of PKBiCYT to inhibit phosphorylation of cytoplasmic PKB/Akt targets TSC2 and PRAS40 (Fig. 2a). This suggests that nuclear and plasma membrane, but not cytoplasmic pools of PKB/Akt are required for adipocyte differentiation.
Although this is the first report of subcellular-specific PKB/Akt signaling in the regulation of adipocyte differentiation, the role of PKB/Akt in this process has been previously established. The combined targeted disruption of two of the three PKB/Akt isoforms in mice, namely PKBα/Akt1 and PKBβ/Akt2, results in impaired adipogenesis (6). Furthermore, unlike PKBβ/Akt2−/− MEFs, the same cells from PKBα/Akt1−/− animals fail to undergo adipocyte differentiation (6, 8). Significantly, only re-expression of PKBα/Akt1 can rescue this deficiency, indicating that there is no redundancy of PKB/Akt isoforms in regulating this process. Consistently, knockdown of PKBα/Akt1 in 3T3L1 cells also blocks their ability to differentiate (7). In contrast, the PKBβ/Akt2 isoform is responsible for the maintenance of glucose homeostasis (reviewed in Ref. 28). Interestingly, preferential localization of PKBβ/Akt2 to the plasma membrane has been implicated as a means of isoform-specific regulation of glucose transport (42). Cytoplasmic linker protein (Clip) R-59, a scaffolding protein, was found responsible for tethering PKB/Akt to the plasma membrane and affecting access to its substrates in adipocytes (43). This protein displays preferential affinity for PKBβ/Akt2, presumably leading to its distinct localization to the plasma membrane and resulting in isoform-specific regulation of glucose transport. Whether a similar mechanism of isoform-specific subcellular localization may contribute to the specific role of PKBα/Akt1 in adipogenesis, however, remains to be determined.
Our data indicate that nuclear and plasma membrane pools of PKB/Akt do not contribute to the induction and DNA-binding activity of the early adipogenesis marker c/EBPβ, but to subsequent steps of the differentiation program. Although expression of either PKBiNUC or PKBiPM ultimately resulted in impaired differentiation, analysis of MCE revealed the requirement for plasma membrane-, but not nuclear-localized PKB/Akt activity for this phase of the differentiation process (Fig. 5, a and b). During MCE, growth-arrested preadipocytes synchronously traverse the G1/S checkpoint, which is characterized by the expression of cyclin A and the degradation of p27Kip1 (44). Expression of PKBiPM, but not PKBiNUC or PKBiCYT, blocked the changes in these cell cycle regulators (Fig. 5c), suggesting that PKB/Akt substrates at the plasma membrane alone are capable of impacting the cell cycle machinery. The fact that cytoplasmic PKB/Akt inhibition has no effect in this regard highlights the requirement for further investigation of subcellular localization as a regulatory mechanism for molecules implicated in control of the cell cycle.
The downstream target of PKB/Akt signaling, mTORC1, has been shown to play a role in adipogenesis by regulating peroxisome proliferator-activated receptor-γ expression (45). Furthermore, the mTORC1 inhibitor rapamycin disrupts the positive feedback transcriptional effects of c/EBPα and peroxisome proliferator-activated receptor-γ, and amino acids are required for peroxisome proliferator-activated receptor-γ activity (46). To monitor mTORC1 activity in our system, we analyzed the phosphorylation of p70S6K and its downstream target, ribosomal protein S6, following induction of various PKBis (supplemental Fig. S2). Phosphorylation of both proteins was reduced upon induction of PKBiCYT and PKBiPM, but not PKBiNUC. Thus, the impact of nuclear PKB/Akt inhibition on adipogenesis is likely not mediated by mTORC1. On the other hand, based on its impact on TSC2 phosphorylation (Fig. 2), PKBiPM may, at least in part, act through mTORC1. Taken together, our results reveal that discrete subcellular pools of PKB/Akt activity can regulate adipocyte differentiation via distinct molecular mechanisms.
Localized Inhibition of PKB/Akt and Nuclear Targets
Both PKBiNUC and PKBiPM affected the localization and transcriptional activity of FOXO3a, a member of the FOXO family of transcription factors (Fig. 2, b and c), implicating FOXO down-regulation as one mechanism by which nuclear and plasma membrane PKB/Akt pools regulate adipocyte differentiation. PKB/Akt-mediated inhibitory phosphorylation of FOXO1, a closely related member of this family, has been linked to the induction of peroxisome proliferator-activated receptor-γ expression during adipogenesis (6, 7). FOXO1 can also regulate the cell cycle through transcriptional regulation of p27Kip1 (47), and consistently, FOXO1 inhibition by PKBα/Akt1, accompanied by p27Kip1 degradation, has been shown to contribute to the MCE phase of adipocyte differentiation (8). PKB/Akt activity toward additional locus-specific targets, such as GATA2, a nuclear anti-adipogenic factor that is excluded from the nucleus following PKB/Akt-mediated phosphorylation (48), may also contribute to this process.
The fact that PKBiPM inhibits PKB/Akt at the site of its activation at the plasma membrane raises the possibility that this inhibitor may affect downstream PKB/Akt targets at other subcellular locations. Indeed, PKBiPM displayed prominent effects on FOXO3a localization and transcriptional activity (Fig. 2, b and c). Considering that PKBiCYT expression failed to affect terminal adipocyte differentiation (Fig. 3) while affecting phosphorylation of known cytoplasmic targets (Fig. 2a) implies that translocation of activated PKB/Akt may include yet to be discovered molecules, whose interaction with this kinase may protect it from the activity of specific PKBis. Interestingly, PKB/Akt has also been found to translocate to the nucleus prior to its activation (49), indicating that spatially distinct PKB/Akt pools impervious to the action of other localized PKBis may exist.
It is worth noting that we unexpectedly observed nuclear FOXO3a accumulation upon PKBiCYT expression in a number of cell lines (Fig. 2b and supplemental Fig. S3), suggesting that this occurrence is not specific to 3T3L1s. The effect could be explained by a role for cytoplasmic PKB/Akt in the retention of FOXOs in the cytoplasm. The nucleo-cytoplasmic shuttling of FOXOs involves their phosphorylation-dependent interaction with 14-3-3 proteins in the nucleus and their nuclear export (50). It is possible that cytoplasmic PKB/Akt activity is necessary to counter the activity of cytoplasmic phosphatases and maintain FOXOs in their phosphorylated and thus the 14-3-3-bound state in the cytoplasm.
Outlook
As spatial regulation continues to emerge as a mechanism of fine-tuning major signaling pathways, the challenge lies in developing efficient experimental strategies to elucidate the specific roles of distinct subcellular pools of signaling molecules. We have developed and characterized a novel method for the subcellular-localized inhibition of PKB/Akt and employed it to uncover nuclear- and plasma membrane-specific roles for PKB/Akt in the regulation of adipocyte differentiation. Due to their mechanisms of action, previous strategies for the investigation of PKB/Akt function such as genetic manipulation, RNA interference, and pharmacological inhibition were unable to address its subcellular-specific properties. The experimental system presented here both compliments and improves on these strategies and will be critical for the elucidation of the roles of PKB/Akt in the processes of cell survival, proliferation, growth, metabolism, and differentiation. This and future work with localized PKB/Akt inhibitors will facilitate the molecular dissection of the PI3K-PKB/Akt pathway and may aid in the design and development of peptide-mimetic drugs targeting PKB/Akt.
Acknowledgment
We thank K. MacAulay for critical reading of the manuscript.
This work was supported by funding from the Canadian Breast Cancer Foundation (Ontario branch) (to V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PI3K
- phosphatidylinositol 3-kinase
- PKB/Akt
- protein kinase B/Akt
- PTEN
- phosphatase and tensin homolog
- TSC2
- tuberous sclerosis 2 protein
- PRAS40
- proline-rich Akt substrate of 40 kDa
- FOXO
- forkhead box O
- p90RSK
- p90 ribosomal S6 kinase
- p70S6K
- p70 ribosomal S6 kinase
- SGK3
- serum/glucocorticoid-regulated kinase 3
- ER
- endoplasmic reticulum
- PH
- pleckstrin homology
- mTORC2
- mammalian target of rapamycin complex 2
- PKBis
- PKB/Akt pseudosubstrate inhibitors
- GST
- glutathione S-transferase
- c/EBP
- CCAAT/enhancer-binding protein
- MCE
- mitotic clonal expansion
- JNK
- c-Jun N-terminal kinase
- HA
- hemagglutinin
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MOPS
- 4-morpholinepropanesulfonic acid
- PVDF
- polyvinylidene difluoride
- PBS
- phosphate-buffered saline
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- PKC
- protein kinase C
- PI
- phosphoinositide.
REFERENCES
- 1.Scheid M. P., Woodgett J. R. (2001) Nat. Rev. Mol. Cell Biol. 2, 760–768 [DOI] [PubMed] [Google Scholar]
- 2.Yuan T. L., Cantley L. C. (2008) Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 5.Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng X. D., Xu P. Z., Chen M. L., Hahn-Windgassen A., Skeen J., Jacobs J., Sundararajan D., Chen W. S., Crawford S. E., Coleman K. G., Hay N. (2003) Genes Dev. 17, 1352–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J., Liao K. (2004) J. Biol. Chem. 279, 35914–35922 [DOI] [PubMed] [Google Scholar]
- 8.Yun S. J., Kim E. K., Tucker D. F., Kim C. D., Birnbaum M. J., Bae S. S. (2008) Biochem. Biophys. Res. Commun. 371, 138–143 [DOI] [PubMed] [Google Scholar]
- 9.Andjelković M., Alessi D. R., Meier R., Fernandez A., Lamb N. J., Frech M., Cron P., Cohen P., Lucocq J. M., Hemmings B. A. (1997) J. Biol. Chem. 272, 31515–31524 [DOI] [PubMed] [Google Scholar]
- 10.Inoki K., Li Y., Zhu T., Wu J., Guan K. L. (2002) Nat. Cell Biol. 4, 648–657 [DOI] [PubMed] [Google Scholar]
- 11.Kovacina K. S., Park G. Y., Bae S. S., Guzzetta A. W., Schaefer E., Birnbaum M. J., Roth R. A. (2003) J. Biol. Chem. 278, 10189–10194 [DOI] [PubMed] [Google Scholar]
- 12.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 13.Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. (2002) J. Biol. Chem. 277, 22115–22118 [DOI] [PubMed] [Google Scholar]
- 14.Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M. E. (1997) Cell 91, 231–241 [DOI] [PubMed] [Google Scholar]
- 15.Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., Takahashi M. (2005) Dev. Cell 9, 389–402 [DOI] [PubMed] [Google Scholar]
- 16.Schenck A., Goto-Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., Zerial M. (2008) Cell 133, 486–497 [DOI] [PubMed] [Google Scholar]
- 17.Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984) Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 18.O'Neill R. E., Talon J., Palese P. (1998) EMBO J. 17, 288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarmo M. N., Landry A., Molgat A. S., Gagnon A., Sorisky A. (2009) Exp. Cell Res. 315, 411–418 [DOI] [PubMed] [Google Scholar]
- 20.Chang L., Chiang S. H., Saltiel A. R. (2007) Endocrinology 148, 27–33 [DOI] [PubMed] [Google Scholar]
- 21.House C., Kemp B. E. (1987) Science 238, 1726–1728 [DOI] [PubMed] [Google Scholar]
- 22.Geshi N., Jørgensen B., Ulvskov P. (2004) Planta 218, 862–868 [DOI] [PubMed] [Google Scholar]
- 23.Mitoma J., Ito A. (1992) EMBO J. 11, 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah O. J., Wang Z., Hunter T. (2004) Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 26.Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 27.Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dummler B., Hemmings B. A. (2007) Biochem. Soc. Trans. 35, 231–235 [DOI] [PubMed] [Google Scholar]
- 29.Gregoire F. M., Smas C. M., Sul H. S. (1998) Physiol. Rev. 78, 783–809 [DOI] [PubMed] [Google Scholar]
- 30.Tang Q. Q., Lane M. D. (1999) Genes Dev. 13, 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 32.Whitmarsh A. J. (2006) Biochem. Soc. Trans. 34, 828–832 [DOI] [PubMed] [Google Scholar]
- 33.Jaken S. (1996) Curr. Opin. Cell Biol. 8, 168–173 [DOI] [PubMed] [Google Scholar]
- 34.Kölsch V., Charest P. G., Firtel R. A. (2008) J. Cell Sci. 121, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall J. G., Booth J. W., Stambolic V., Mak T., Balla T., Schreiber A. D., Meyer T., Grinstein S. (2001) J. Cell Biol. 153, 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpten J. D., Faber A. L., Horn C., Donoho G. P., Briggs S. L., Robbins C. M., Hostetter G., Boguslawski S., Moses T. Y., Savage S., Uhlik M., Lin A., Du J., Qian Y. W., Zeckner D. J., Tucker-Kellogg G., Touchman J., Patel K., Mousses S., Bittner M., Schevitz R., Lai M. H., Blanchard K. L., Thomas J. E. (2007) Nature 448, 439–444 [DOI] [PubMed] [Google Scholar]
- 37.Martelli A. M., Faenza I., Billi A. M., Manzoli L., Evangelisti C., Falà F., Cocco L. (2006) Cell. Signal. 18, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 38.Obata T., Yaffe M. B., Leparc G. G., Piro E. T., Maegawa H., Kashiwagi A., Kikkawa R., Cantley L. C. (2000) J. Biol. Chem. 275, 36108–36115 [DOI] [PubMed] [Google Scholar]
- 39.Bonny C., Oberson A., Negri S., Sauser C., Schorderet D. F. (2001) Diabetes 50, 77–82 [DOI] [PubMed] [Google Scholar]
- 40.Alfaro-Lopez J., Yuan W., Phan B. C., Kamath J., Lou Q., Lam K. S., Hruby V. J. (1998) J. Med. Chem. 41, 2252–2260 [DOI] [PubMed] [Google Scholar]
- 41.Harris T. E., Persaud S. J., Saermark T., Jones P. M. (1995) Biochem. Soc. Trans. 23, 187S. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez E., McGraw T. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding J., Du K. (2009) Mol. Cell. Biol. 29, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Q. Q., Otto T. C., Lane M. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H. H., Huang J., Düvel K., Boback B., Wu S., Squillace R. M., Wu C. L., Manning B. D. (2009) PloS One 4, e6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J. E., Chen J. (2004) Diabetes 53, 2748–2756 [DOI] [PubMed] [Google Scholar]
- 47.Dijkers P. F., Medema R. H., Pals C., Banerji L., Thomas N. S., Lam E. W., Burgering B. M., Raaijmakers J. A., Lammers J. W., Koenderman L., Coffer P. J. (2000) Mol. Cell. Biol. 20, 9138–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menghini R., Marchetti V., Cardellini M., Hribal M. L., Mauriello A., Lauro D., Sbraccia P., Lauro R., Federici M. (2005) Circulation 111, 1946–1953 [DOI] [PubMed] [Google Scholar]
- 49.Wang R., Brattain M. G. (2006) Cell. Signal. 18, 1722–1731 [DOI] [PubMed] [Google Scholar]
- 50.Greer E. L., Brunet A. (2005) Oncogene 24, 7410–7425 [DOI] [PubMed] [Google Scholar]