Abstract
Inverted repeats in ion-coupled transporters have evolved independently in many unrelated families. It has been suggested that this inverted symmetry is an essential element of the mechanism that allows for the conformational transitions in transporters. We show here that small multidrug transporters offer a model for the evolution of such repeats. This family includes both homodimers and closely related heterodimers. In the former, the topology determinants, evidently identical in each protomer, are weak, and we show that for EmrE, an homodimer from Escherichia coli, the insertion into the membrane is random, and dimers are functional whether they insert into the cytoplasmic membrane with the N- and C-terminal domains facing the inside or the outside of the cell. Also, mutants designed to insert with biased topology are functional regardless of the topology. In the case of EbrAB, a heterodimer homologue supposed to interact antiparallel, we show that one of the subunits, EbrB, can also function as a homodimer, most likely in a parallel mode. In addition, the EmrE homodimer can be forced to an antiparallel topology by fusion of an additional transmembrane segment. The simplicity of the mechanism of coupling ion and substrate transport and the few requirements for substrate recognition provide the robustness necessary to tolerate such a unique and unprecedented ambiguity in the interaction of the subunits and in the dimer topology relative to the membrane. The results suggest that the small multidrug transporters are at an evolutionary junction and provide a model for the evolution of structure of transport proteins.
Keywords: Bacteria, Membrane Proteins, Multidrug Transporters, Protein Structure, Transport Drugs, Ion-coupled Transport, Topology of Membrane Proteins
Introduction
Evolution of large transporter genes is thought to have started from a small single one that duplicated, evolved independently, and then fused to the original one to generate a new gene coding for the large polytopic protein (1). Inverted repeats in ion-coupled transporters have now been observed in transporters from many different families (2, 3). It has been suggested that this inverted symmetry is an essential element of the mechanism that allows for the conformational transitions in transporters (3).
A possible mode for evolution of these inverted repeats has been suggested based on the supposedly inverted topology of EmrE, a small multidrug transporter from Escherichia coli. A claim for an antiparallel topology of the monomers in the EmrE homodimer was made, but this is still a controversial issue and, in our view, is still inconsistent with the biochemical data. The reasoning that the topology of the monomers in the EmrE dimer is parallel and Nin-Cin was supported by data from our laboratory that demonstrated the same topology for all protomers in the intact cell and in membrane vesicles (4), by biochemical studies that established the equivalence of the residues in the protomers (reviewed in Ref. 5 and 6), and by extensive cross-linking studies that suggested the proximity of equivalent residues in the two monomers (7). Moreover, dimers cross-linked at positions not compatible with antiparallel topology were purified to homogeneity and shown to be fully functional based on substrate binding and transport assays (8). To provide an approach that combines in vivo and in vitro studies, we designed a series of genetic fusions (tandems) where the monomers are connected with the C terminus of the first monomer attaching to the N terminus of the second (tail to head). This was achieved by means of defined linkers that are not compatible with an antiparallel topology either because they are too hydrophilic or too short (9).
The suggestion for an antiparallel topology of the monomers was supported by a C-α model of the transmembrane region constructed by considering the evolutionary conservation pattern of each helix (10) and based on a reinterpretation of low resolution electron density maps of two-dimensional crystals of EmrE that showed quasi-symmetry in parts of the structure (11). Although possible, the model is one of many that can be generated with the available data, and unfortunately, no experimental test was provided. An x-ray structure previously retracted (12) has been reanalyzed, and a C-α model of the corrected structure with substrate is very similar to the model proposed based on cryo-EM studies (13). The crystals used to derive the C-α model x-ray structure were obtained with protein solubilized with detergents that inhibit activity by inducing monomerization as shown not only by our data but from the results of the authors as well (8, 13). Monomers arrange in the crystal in a conformation that minimizes the energy needed for crystal formation and do not necessarily reflect the relative topology in the membrane.
In addition, genetic experiments were designed to support the claim for an antiparallel topology. EmrE was fused to the topology reporter proteins alkaline phosphatase and green fluorescent protein, and the results showed that the topology of the EmrE fusion proteins in the membrane is sensitive to the distribution of positive charges in the protein (14). Manipulation of the positive charges generated a set of mutants, some with Nout-Cout (Cout) and others with Nin-Cin (Cin) apparent topology (14, 15). The Cin and Cout mutants did not confer resistance to ethidium, and the authors concluded that this was due to the modified topology. Co-expression of the inactive mutants (Cin and Cout together) restored the ethidium resistance phenotype to the same level as seen with wild type EmrE (15). The suggested interpretation of this finding was that co-expression results in the generation of a functional, antiparallel heterodimer. However, because this conclusion is based solely on the contention that the Cin and Cout mutants are inactive, we decided to re-evaluate these findings that were based only on the phenotypic analysis.
Here we show that small multidrug transporters (SMRs)3 display a remarkable plasticity regarding topology and monomer-monomer interactions. Thus, Cin and Cout mutants are shown to be functional provided they are expressed in a controlled mode and in a strain where the chromosomal wild type emrE was inactivated. Moreover, the wild type protein, when expressed without any tag, inserts into the membrane in a random fashion generating a dual topology; approximately half of the dimers are Nin-Cin, and half are Nout-Cout. These findings are in agreement with the generally accepted functional symmetry of channels, transporters, and ion-coupled transporters where the direction of transport is dictated by the direction of the driving force acting on the transported substrates (except in special cases where there is a kinetic block). However, this is the first demonstration that the directionality of insertion of a transporter in vivo can be random without affecting activity. In the same vein, a detailed analysis of EbrAB, a closely related antiparallel heterodimer from Bacillus subtilis, reveals that in addition to the efficient interaction between the two protomers, one of them can still interact productively with itself to form a functional homodimer, most likely parallel. Finally, in line with the above findings, the homodimeric EmrE can be forced to form an antiparallel dimer by fusion with an additional transmembrane domain. These experiments demonstrate that SMR proteins are in an evolutionary junction and provide an example of how protein fold can evolve while maintaining its function.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Mutagenesis
E. coli DH5α (Invitrogen), JM109 (Novagen), BW25113 (52), HMS174 (DES) (Novagen), and TA15 (53) strains were used throughout this work. BW25113 ΔemrE, Δsmr, ΔmdfA, ΔyjiO, and ΔacrB individual knockouts and combinations were prepared according to Datsenko et al. (52) and described by Tal and Schuldiner (25). For specific labeling with [35S]methionine, BW25113 ΔemrE ΔmdfA cells were transformed with the plasmid pGP1-2, which codes for the T7 polymerase under the inducible control of the λ PL promoter (18). The plasmids used for gene expression of EmrE Cin and Cout and EbrA and B, without any tags are pT7-7 (16, 18) or pACYC184 (54) derivatives. Tagged EmrE is fused at the C terminus to His6 residues and a Myc epitope as described (16). Transfer of EmrE constructs to pACYC184 was performed using NdeI and ClaI restriction sites. GpA-EmrE was constructed by insertion at the BamHI site of E6EMH (9) of a sequence coding for a nondimerizing mutant of human glycophorin A. The DNA sequence was manipulated so that the N terminus of the second EmrE protomer (NP, the first methionine was removed) was connected to the C terminus (RSTPH) of the first EmrE protomer with a linker that codes for the sequence 1GSLEPEITLILFGVMALVIGTILLLLYGIRRLIGSGAAS39. Tandems with linkers with variations of the above sequence displayed similar phenotypes but were not studied further (supplemental Table S1). A “negatively dominated” tandem was built substituting the essential glutamate 14 in the first monomer with glutamine (GpA-EmrE (E14Q)1).
Resistance to Toxic Compounds
E. coli cells were transformed with the indicated plasmids and were grown overnight at 37 °C in LB medium with the corresponding antibiotic. 5 μl of serial dilutions of the culture were spotted on LB plates containing 30 mm BisTris propane, pH 7.0, with or without the addition of the indicated concentrations of the toxic compounds. When the pACYC184 plasmid was used, IPTG (200 μm) was added for induction of expression. Growth was analyzed after overnight incubation at 37 °C. Each experiment was performed at least twice.
Binding Assays
[3H]Tetraphenylphosphonium (TPP+) binding to His-tagged GpA-EmrE and GpA-EmrE (E14Q)1 was assayed essentially as described (16). The amounts of purified proteins were determined on Coomassie-stained 16% Tricine SDS-PAGE using known amounts of the corresponding protein for calibration. The radioactivity was measured by liquid scintillation. All of the binding reactions were performed in duplicates, and in each experiment the values obtained in a control reaction with 25 μm unlabeled TPP+ were subtracted. All of the experiments were repeated at least twice.
Tagged Construct Topology Determination
HMS174 cells expressing the appropriate single Cys mutant were incubated with the indicated concentrations of MTSES at 30 °C for 20 min. After removal of the MTSES by centrifugation, the cells were rinsed three times and lysed by incubation in a solution containing sucrose (30%), Tris-Cl (30 mm, pH 8.0), 50 μg/ml lysozyme, 10 mm EDTA, and 1.5 μg/ml DNase. The membrane fraction was collected and solubilized in 0.8% dodecyl-β-d-maltoside/sodium buffer (150 mm NaCl and 15 mm Tris-Cl, pH 7.5) for 20 min at 25 °C, allowed to bind to nickel-nitrilotriacetic acid beads for 1 h at 4 °C and rinsed with sodium buffer supplemented with 2.0% SDS and 6 m urea. N-Ethylmaleimide-fluorescein (Pierce) was added to a final concentration of 0.5 mm for 20 min (at 30 °C). The reaction was stopped by dilution with the same buffer containing β-mercaptoethanol at a final concentration of 5 mm. The beads were then washed as described above. The protein was eluted from the beads using a buffer containing 200 mm β-mercaptoethanol, 100 mm Tris-Cl, pH 6.8, 4% SDS, 40% glycerol, 0.2% bromphenol blue (sample buffer), and 450 mm imidazole and analyzed by 16% Tricine SDS-PAGE. Fluorescence labeling analysis of the gel was done using a Fujifilm LAS-1000 imaging system and digitally analyzed with Image Gauge 3.46 Fujifilm software. The results were calibrated to protein amount after staining of the same gels with Coomassie staining, scanning, and quantitation using the same software. Each experiment was performed at least three times.
Untagged Construct Topology Determination
BW25113 ΔemrE ΔmdfA cells bearing pGP1-2 and EmrE and indicated mutants on pT7-7 were metabolically and specifically labeled with [35S]methionine essentially as described in Refs. 17 and 19. The cells were then incubated with MTSES and treated as above except that membrane solubilization was done in 2% SDS, 6 m urea, and 15 mm Tris-Cl, pH 7.5, and 2.5 mm Mal-PEG 5000 (Nektar Transforming Therapeutics, Huntsville, AL). The reaction was stopped after 1 h at 37 °C by the addition of sample buffer. The proteins were analyzed using 16% Tricine SDS-PAGE and visualized with a Fujifilm FLA-3000 imaging system and digitally analyzed with Image Gauge 3.46 Fujifilm software. Each experiment was performed at least three times.
Cross-linking with o-PDM
The membranes prepared from cells expressing EmrE mutants selectively labeled with [35S]methionine were solubilized as described above, and o-PDM was added to a final concentration of 1 mm. The reaction was stopped after 1 h at 45 °C and 10 min at 60 °C by the addition of sample buffer. Samples containing ∼5500 dpm of labeled [35S]EmrE were analyzed on 16% Tricine SDS-PAGE and visualized with a Fujifilm FLA-3000 imaging system and digitally analyzed with Image Gauge 3.46 Fujifilm software. Each experiment was performed twice.
RESULTS
Tags and Reporters May Introduce Bias in Topology Determinations
The topological studies of EmrE in intact cells in our laboratory indicate that the protein assumes a Nin-Cin topology in the membrane (4). In our studies we used a construct in which the wild type gene was fused to a Myc epitope before a six-His tag at the C terminus (16). On the other hand, studies using alkaline phosphatase and green fluorescent protein fusions suggest that the fused wild type protein is mostly in a Nout-Cout configuration (14). We show here that the discrepancy in the topology of the wild type between the two groups stems from the difference in the fusion tags used.
To test the possible effect of tags, we designed a strategy to assess the topology of untagged proteins that would obviate the need to purify them. Single Cys residues were engineered at the same positions in loops in Myc-His tagged at the C terminus and in untagged proteins. The mutants were specifically labeled metabolically with [35S]methionine as described previously (17–19). The cells were then challenged with the impermeant MTSES reagent, and unreacted thiols were assessed after solubilization and denaturation of the protein with Mal-PEG (20). Reaction with Mal-PEG resulted in a shift in the mobility that can be easily detected after SDS-PAGE analysis (Fig. 1, A and B, bottom panels, compare lanes with and without Mal-PEG). In tagged mutants, cysteine in loop 1 at position 22 (EmrE-K22C) but not at the C terminus, position 110 (EmrE-H110C), reacted quantitatively with MTSES in the intact cell, and therefore only a fraction of ∼30% of the protein reacted with Mal-PEG (Fig. 1A and see also Ref. 4). However, with untagged mutants bearing Cys at the same positions, reactivity of both EmrE-K22C and -H110C was very similar and partial; ∼50–60% of the residues are exposed in each case (Fig. 1B). These results suggest that the Myc-His tag is biasing the protein toward a Nin-Cin topology, whereas the untagged protein displays a dual topology, i.e. approximately half of the protein is Nin-Cin, whereas the other half is Nout-Cout.
FIGURE 1.
Tags introduce bias in topology of undecided proteins. Tagged (A) and untagged (B) EmrE K22C and EmrE H110C were metabolically labeled with [35S]methionine in E. coli BW25113 ΔemrEΔmdfA cells and treated with MTSES at the indicated concentrations as described under “Experimental Procedures.” The unreacted thiols were estimated from the degree of reaction with Mal-PEG. EmrE that reacted with Mal-PEG displays a higher apparent Mr in SDS-PAGE, and the ratios of reacted over nonreacted protein were calculated. The bottom panels in A and B are samples of the types of changes at one concentration of MTSES: 0.3 and 0.1 mm, respectively. C, simplified flow chart of procedure. D, monomers of tagged and untagged EmrE K22C (not shown) and H110C are in a parallel topology relative to each other as judged from the full cross-linking detected with o-PDM.
The Monomers in the Dimer Are Parallel
The term dual topology was previously used to describe the relative topology of the monomers in the dimer (parallel or antiparallel). We use it here to describe the topology of the dimers relative to each other. Previously we showed, using several approaches, that the monomers within each dimer are parallel, i.e. within a given dimer the topology of both monomers is identical (6–9). Because the results described above could also be interpreted as an indication of antiparallel topology and to distinguish between dual and antiparallel topology, we cross-linked the dimers with o-PDM (8), a rigid bifunctional cross-linking agent that can react with Cys residues that are 7–11 Å apart (21). The Cys replacements, H110C at the C terminus and K22C in loop 1, are too far apart to cross-link in an antiparallel structure (∼35–45 Å). Despite this distance and as expected from a parallel topology, efficient cross-linking was observed with o-PDM when either tagged or untagged EmrE was used (Fig. 1D, data for H110C and supplemental Fig. S1, data for K22C as well). Cross-linking with o-PDM was already shown to result from specific interaction within the dimer rather than between dimers (6–9, 22, 23). Further support is provided by an experiment where cross-linking between labeled protomers is observed at the same levels in the presence or absence of excess unlabeled protein (supplemental Fig. S1). In these experiments, membranes with the 35S-labeled EmrE mutant were diluted with the same amount or a 4-fold excess of membranes expressing the same mutant but unlabeled. The fact that the intensity of the cross-linked dimer is not affected by the presence of the unlabeled protein strengthens our contention that the o-PDM-mediated cross-linking is a result of a reaction between Cys residues within the dimer.
We conclude that insertion of untagged EmrE is a random process that results in dual topology when one fraction of the dimer is Nin-Cin and another is Nout-Cout as expected from the very weak topology determinants. However, within the dimer, the preferred relative topology is parallel. If antiparallel dimers exist without experimenter intervention, they are a small minority that cannot be detected with the techniques we developed. The variation in results within the various constructs provides a caveat about the bias that tags and reporters may introduce in topology determinations.
Nin-Cin and Nout-Cout EmrE Constructs Are Functional
The above findings seem to contradict previous reports claiming that expression of EmrE Cin or EmrE Cout individually did not confer resistance to ethidium as shown for wild type EmrE (15). In that report, to push EmrE toward the Nin-Cin or Nout-Cout orientations, the (K+R) bias was systematically modified by substantial mutagenesis (Fig. 2A and Ref. 15). We generated the same mutants and confirm the above results when the expression was tested in the commonly used E. coli DH5α (supplemental Fig. S2). The published conclusion was based only on phenotypic characterization, a powerful tool that provides definite answers when reflecting a positive trait. However, the experiments were done in a wild type strain harboring chromosomal emrE and without induction of expression because of the well known toxic effects of overexpression of membrane proteins. Therefore we carried out a more detailed study of the phenotype of EmrE Cin and Cout under controlled expression conditions. We used the plasmid pACYC184 where a trc promoter controls the expression, and we grew the cells in the presence of 200 μm of the inducer IPTG, conditions under which expression is heightened but not as high as to become toxic (not shown). The increased and controlled expression yielded weak but definite positive phenotypes in several commercially available E. coli strains such as JM109 and HMS174 (supplemental Fig. S2). This is in contrast with the negative phenotypes obtained without induction (Ref. 15 and supplemental Fig. S2, DH5α). In agreement with these findings, weak but distinct phenotypes were also reported in similar experiments with induced BL21 cells (24).
FIGURE 2.
A, schematic representation of the EmrE Cin and EmrE Cout mutations. The mutations were performed as described under “Experimental Procedures.” The sites were selected as in Ref. 15. The squares indicate positive charges in the wild type EmrE and those remaining after mutagenesis; white circles indicate positive charges added. B, phenotype of EmrE Cin and EmrE Cout. E. coli BW25113 ΔemrE, ΔmdfA, or ΔemrEΔmdfA transformed with pACYC184-EmrE (EmrE), pACYC184 (vector), or pACYC184 with each of the EmrE mutants were grown overnight at 37 °C in LB medium containing chloramphenicol. 5 μl of serial dilutions of the culture were spotted on LB plates containing 30 mm BisTris propane, pH 7.0, with 200 μm IPTG and with or without the addition of ethidium (400 μg/ml in top and middle panels and 100 μg/ml in bottom panel), acriflavine (250, 100, and 25 μg/ml, respectively), and methyl viologen (0.6, 0.4, and 0.3 mm, respectively). Growth was analyzed after overnight incubation at 37 °C. In control plates with no toxicants, growth of all strains was similar (not shown). When resistance was tested with the same strains without IPTG, the phenotypes were negative, and with different host strains the results are variable (shown in Fig. S2), stressing again the inadequacy of a negative phenotype to determine the lack of activity of EmrE or similar proteins.
An even more dramatic factor influencing the phenotype was identified when using strains well defined genetically and functionally (Fig. 2B). In these strains we have inactivated one or multiple multidrug transporters (25). Although not related in amino acid sequence, several multiple multidrug transporters are functionally related and even share some of the substrates. We show that the ability to confer resistance is independent of the sensitivity of the host cell because even in very sensitive strains where the genes coding for the functionally related transporters MdfA, yjiO, or AcrAB were inactivated, either individually or together, the Cin and Cout mutants still provide only a feeble resistance to ethidium and acriflavine as shown for the other strains tested (data shown only for MdfA; Fig. 2B). Strikingly, however, when the chromosomal gene coding for EmrE is inactivated, the phenotype for the three substrates tested is distinct and comparable with that shown by wild type EmrE, somewhat weaker for Cout but very clear (Fig. 2B).
The results described above imply that the presence of EmrE is detrimental to the phenotypic expression of the Cin and Cout mutants. This may be due to a nonproductive interaction between wild type EmrE and the mutants. This reasoning is further supported by a series of experiments where negative dominance was observed when co-expressing the functional Cin and Cout mutants with E14C EmrE, an inactive mutant (supplemental Fig. S3). In these experiments we had to balance the need for relatively high levels of expression of the Cin and Cout mutants with low expression levels of E14C EmrE that were obtained from the constitutive leak of T7 promoter that control the plasmidic gene. The best results were obtained at low (25 μm IPTG) or no induction when the phenotype conferred by the Cin and Cout mutants is not as strong as shown under full induction (compare supplemental Fig. S3 with Fig. 2B, bottom panel). Under these conditions the phenotype conferred by the Cin and Cout mutants is significantly impaired when co-expressed with the inactive E14C EmrE mutant (supplemental Fig. S3).
We conclude that Cin and Cout mutants confer a robust resistance, provided they are expressed in a cell where the chromosomal gene coding for EmrE has been inactivated and the expression levels are enhanced by induction. The results also explain the variability in the phenotype conferred by the Cin and Cout mutants in different E. coli strains that could be correlated with different levels of expression of chromosomal emrE in the various strains (Fig. 2, supplemental Fig. S2, and Ref. 24).
When Is Heterodimerization Obligatory?
The finding that expression of chromosomal emrE may have a deleterious effect on specific mutants led us also to investigate the case of EbrAB, a heterodimer from the SMR family considered obligatory solely on the basis of phenotype (26, 27) (Fig. 3A). We confirm previous findings that expression of each one of the protomers, EbrA or EbrB, alone does not confer resistance to the toxicants when tested in a wild type strain such as DH5α (supplemental Fig. S4). The same phenotype is observed in strains where either mdfA, a gene coding for an MFS multidrug transporter, or acrA, coding for a subunit of the major multidrug transporter AcrAB, has been inactivated (Fig. 3B). However, EbrB but not EbrA confers robust resistance to ethidium and acriflavine (not shown) when expressed in any one of three strains where the chromosomal emrE gene was inactivated alone or in combination with other SMRs (Δsmr) or with mdfA (Fig. 3B, bottom panel). The results imply that EbrB has not completely crossed the point where it can interact only in an antiparallel mode with EbrA. It can also interact with itself (with a lower affinity), forming a homodimer most likely with a parallel topology. In the presence of EmrE, it interacts in a nonproductive way similar to what we showed above for the EmrE Cin and Cout mutant proteins.
FIGURE 3.
EbrAB, a heterodimer that can still function as a homodimer. A, schematic representation of EbrAB. The squares represent the positive charges in each monomer. B, phenotype. E. coli BW25113 ΔacrA, ΔmdfA, ΔemrE, Δsmr, or ΔemrEΔmdfA transformed with pACYC184-EmrE (EmrE), pACYC184 (vector), or pACYC184 with EbrA, EbrB, or both genes together were grown overnight at 37 °C in LB medium containing chloramphenicol. 5 μl of serial dilutions of the culture were spotted on LB plates containing 30 mm BisTris propane, pH 7.0, with 200 μm IPTG and with or without the addition of ethidium (ΔacrA, 12.5 μg/ml; ΔmdfA, ΔemrE, and Δsmr, 400 μg/ml; and ΔemrEΔmdfA, 100 μg/ml). Growth was analyzed after overnight incubation at 37 °C. In control plates with no toxicants, growth of all strains was similar (not shown).
Forcing a Homodimer with Identical Protomers into an Antiparallel Configuration
A wide range of biochemical experiments from our laboratory supports the contention that the topology of the monomers in the native EmrE dimer is parallel and Nin-Cin (4–9). To provide an approach that combines in vivo and in vitro studies, we designed a series of genetic fusions (tandems) where the monomers are connected C terminus of the first to N terminus of the second monomer (tail to head). This was achieved by means of defined linkers that are not compatible with an antiparallel topology either because they are too hydrophilic or too short (9). In these studies we showed that all the constructs are functional in vivo because they confer resistance to EmrE substrates, and they catalyze energy-dependent ethidium efflux. In addition, the proteins were purified and reconstituted into proteoliposomes and shown to catalyze transport with kinetic constants similar to the wild type EmrE. Moreover, we showed that the functional unit is the dimer itself and does not result from interdimeric interaction.
Because of the high plasticity documented above, where the dimers can insert in both topologies and the antiparallel heterodimer can also function as a parallel homodimer, we asked whether also in the EmrE homodimer there is a potential for an antiparallel interaction without introducing further mutations. For this purpose, we generated a series of tandem EmrE constructs with nine transmembrane domains (TM) by genetic fusion of the monomers (N terminus of the second EmrE protomer to the C terminus of the first one) with an additional transmembrane segment in between them (Fig. 4A). The segment inserted was based on the sequence of a mutant of human glycophorin A (GpA) that does not dimerize (28). Such a fusion, with an odd number of TMs, would have the N and C termini at opposite sides of the membrane, contrasting with the previously generated eight TM proteins with hydrophilic linkers of variable length (9). Fusions with several sequences of GpA and several linkers were constructed (supplemental Table S1), and they all yielded functional proteins. Only the first one will be described in depth and will be named GpA-EmrE.
FIGURE 4.
A, schematic representation of GpA-EmrE, an EmrE tandem with nine transmembrane helices. GpA-EmrE is shown schematically; transmembrane helices are drawn as boxes. The N terminus of the second EmrE protomer was connected to the C terminus of the first EmrE protomer with a linker derived from a mutant of human GpA (dark box) that does not dimerize (28) and six additional hydrophilic amino acids as described under “Experimental Procedures.” The Myc-His tag is fused to the C terminus of the second monomer. B, growth phenotype of cells expressing GpA-EmrE. E. coli BW25113 ΔemrEΔmdfA cells transformed with either pT7-7-EmrE (EmrE), pT7-7 (vector), or pT7-7-GpA-EmrE (GpA-EmrE) were grown overnight at 37 °C in LB medium with ampicillin. 5 μl of serial dilutions of the culture were spotted on the LB plates containing 30 mm BisTris propane, pH 7.0, with or without 100 μg/ml ethidium, 25 μg/ml acriflavine, or 0.3 mm methyl viologen. Growth was analyzed after overnight incubation at 37 °C.
The activity was tested in vivo by assessment of the resistance it confers to E. coli cells. This was achieved in solid medium containing either ethidium (100 μg/ml), acriflavine (25 μg/ml), or methyl viologen (0.3 mm) (Fig. 4B). Cells expressing either EmrE or GpA-EmrE displayed substantial and similar resistance to the three compounds.
To further characterize the protein activity and to rule out the possibility that the transport observed in vivo is due to proteolytic products, GpA-EmrE was purified and assayed for binding activity. GpA-EmrE bound the high affinity substrate Tetraphenyl phosphonium (TPP+) with a KD of 2.4 ± 0.5 nm, very similar to that of the wild type protein or the tandem E6EMH (9, 29) (supplemental Fig. S5A). Purified GpA-EmrE reconstituted into proteoliposomes transports both substrates: the monovalent 1-methyl-4-phenylpyridinium and the divalent methyl viologen (supplemental Fig. S5, B and C).
GpA-EmrE Is the Functional Unit
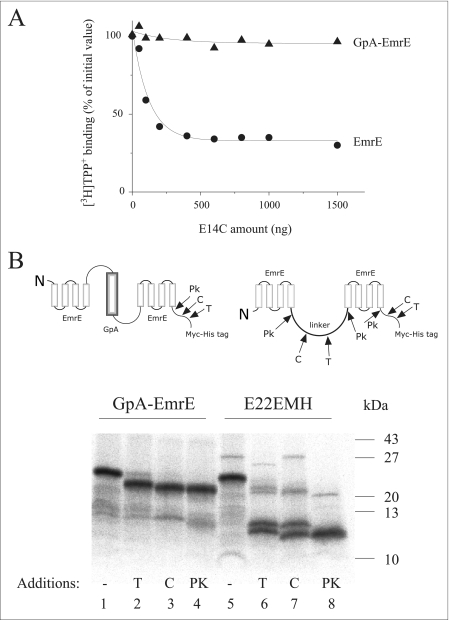
The possibility that the functional unit may be formed by the interaction of two dimers was ruled out by several experimental approaches. First, we used negative dominance analysis. Specifically, replacement of glutamate 14 in EmrE with an uncharged amino acid completely abolishes binding, but the mutants can form mixed dimers with other functional EmrE protomers. The mixed dimers display at least a 20-fold lower affinity toward TPP+, a phenomenon resulting from negative dominance of the inactive mutant on the functional one (23, 30). In these experiments, increasing amounts of the well characterized mutant EmrE E14C were added to wild type EmrE and to GpA-EmrE, and subsequently the mixtures were subjected to the monomer swapping procedure and assayed for TPP+ binding activity (Fig. 5A). The TPP+ concentration used in this assay, 2.5 nm, equals the KD of EmrE (9, 29) and GpA-EmrE. Therefore, inhibition of binding reflects the formation of mixed dimers because the contribution of heterodimers EmrE/E14C is insignificant because of their lower affinity to TPP+. After monomer swapping with a 50-fold excess of the EmrE E14C mutant, the binding activity of the wild type protein was inhibited by more than 60% (Fig. 5A, circles). Under the same conditions, the binding activity of GpA-EmrE was hardly affected (Fig. 5A, triangles).
FIGURE 5.
A, GpA-EmrE does not take part in the intermolecular interactions with other EmrE molecules. Negative dominance of an inactive mutant (EmrE E14C) on EmrE activity and the lack of effect on GpA-EmrE activity were demonstrated as follows. Increasing amounts of membranes containing the indicated amounts of inactive EmrE mutant E14C were solubilized in 1% dodecyl-β-d-maltoside/sodium buffer and mixed with solubilized membranes containing 40 ng of EmrE (●) or GpA-EmrE (▴). After incubation at 80 °C (10 min), the mixture was transferred to 4 °C and assayed for [3H]TPP+ binding as described under “Experimental Procedures.” B, probing the packing of GpA-EmrE. Membranes bearing 35S-radiolabeled GpA-EmrE (lanes 1–4) and E22EMH (lanes 5–8) were incubated with the corresponding proteases (T, trypsin; C, chymotrypsin; PK, proteinase K) as described in Ref. 9. A cartoon is shown indicating the proteolysis sites of the different enzymes: E22EMH is digested by chymotrypsin (after Phe112 in the linker after monomer 1 and the corresponding residue after monomer 2) and trypsin (after Lys119 as above) to produce two polypeptides with a similar molecular mass (11–12-kDa apparent mass in SDS-PAGE) as a result of the digestion of the linker. Proteinase K digests all the linker and the tag. Under the conditions used, the released tag was not detectable; only a minor uncut and tag-less fraction of E22EMH is detectable. In the case of GpA-EmrE only the tag is digested producing a polypeptide of ∼22–24 kDa. The very minor low molecular weight bands observed are present also in the absence of added proteases and may represent small amounts of unrelated proteins.
To further substantiate the contention that GpA-EmrE is the functional unit, we mutated one of the two essential glutamates (one in each protomer) that correspond to the membrane embedded glutamate 14 in wild type EmrE. GpA-EmrE-(E14Q)1 where one of the essential glutamates was changed to glutamine, confers only a very low but significant resistance to ethidium and acriflavine (not shown) and displays binding affinity to TPP+ (120 ± 59 nm) almost 2 orders of magnitude lower than the wild type protein (9). GpA-EmrE-(E14Q)1 reconstituted in proteoliposomes transports monovalent (1-methyl-4-phenylpyridinium) but not divalent (methyl viologen) substrates (supplemental Fig. S5, B and C). Transport of the monovalent 1-methyl-4-phenylpyridinium is inhibited by another monovalent (TPP+) but not by a divalent (methyl viologen) substrate (supplemental Fig. S5C). A parallel tandem protein with eight TMs bearing only one Glu14 was also shown to display similar phenotype and affinities and lost the ability to transport divalent substrates. The findings are explained by the fact that the mutated transporters bearing only one Glu14 residue exchange only 1H+/substrate as opposed to 2H+/substrate for the wild type proteins (9).
GpA-EmrE Is Properly Packed
The glycophorin A linker was designed to be long and hydrophobic enough to cross the membrane plane (supplemental Table S1 and Fig. 4A). The results described above demonstrate that the additional TM has no significant effect on the activity of the protein and therefore most likely does not affect the overall packing of the dimer, a prerequisite for function. However, because manipulations of this type may potentially affect integration of transmembrane segments and may induce generation of semi-inverted topologies, we tested whether the insertion of the linker affected the packing of the protomers in GpA-EmrE. The membrane domain of EmrE is completely resistant to a battery of proteases. After exposure of EmrE to either chymotrypsin or proteinase K, only the C-terminal tag is digested even at prolonged overnight digestions (9). When a parallel tandem with an exposed hydrophilic linker (E22EMH) is digested by three proteases: trypsin, chymotrypsin, and proteinase K (Ref. 9 and Fig. 5B, lanes 5–8), the digestion results in the production of two polypeptides with similar apparent mass (11–12 kDa). GpA-EmrE, on the other hand, is resistant to treatment with any of the three enzymes (Fig. 5B, lanes 1–4). Only the C-terminal tag is digested, producing a polypeptide with correspondingly lower apparent mass (22–24 kDa) as predicted from the sites of digestion of the three enzymes. The results support the contention that both the glycophorin A linker and the rest of the protein are well packed, embedded in the membrane, and protected from digestion.
The Protomers in GpA-EmrE Display Antiparallel Configuration
To test whether the relative topology of the protomers in GpA-EmrE is as proposed, we assessed accessibility to impermeant thiol reagents of single Cys residues engineered in Cys-less GpA-EmrE. As a control we used EmrE mutants with a single residue engineered at positions previously shown as accessible (K22C) or inaccessible (H110C) (4). After challenging the cells with the impermeant MTSES reagent at the indicated concentrations, the membranes were prepared, and the proteins were purified. To assess reactivity with MTSES, free thiols were reacted after solubilization and denaturation with N-ethylmaleimide-fluorescein, and the fluorescence was estimated after SDS-PAGE. As previously shown, in EmrE, the accessible residue K22C reacts with MTSES in intact cells, and therefore only a small fraction (∼ 25%) of unreacted thiols is detected (Fig. 6A). On the other hand when a mutant bearing residue H110C is used, most (∼80%) of the Cys residues are unreacted (Fig. 6A). The GpA-EmrE mutants used bear a single Cys residue engineered in the first loop (K22C) of either the first or the second monomer. The K22C in the second monomer is practically fully accessible to the impermeant MTSES reagent because only a small fraction (∼15%) of unreacted thiols is detected after exposure (Fig. 6B). On the other hand K22C in the first monomer is only partly accessible, suggesting that the majority of the protein is organized with the first monomer in a Nout configuration and the second in a Cin (Fig. 6B).
FIGURE 6.
Topology of GpA-EmrE. BW25113 ΔemrEΔmdfA cells bearing (A) EmrE K22C (♦) and EmrE H110C (•) or (B) GpA-EmrE K22C in the first (■) or the second (▴) monomer were treated with MTSES at the indicated concentrations, and the unreacted thiols were estimated from the degree of reaction with N-ethylmaleimide-fluorescein as described under “Experimental Procedures.” A simplified flow chart is shown in the right panel.
DISCUSSION
The results described in this paper document a unique plasticity in the interaction of protomers within the EmrE dimer and of the dimer relative to the membrane. Thus, dimers can insert with either Nin-Cin or Nout-Cout topology and monomers that usually interact in a parallel mode can be forced to interact productively in an antiparallel mode. In addition, in the heterodimer EbrAB, EbrB can interact productively with EbrA or with itself.
It was previously proposed that antiparallel topology could be obtained by co-expression of inactive EmrE mutants where the (K+R) bias of the EmrE protomers was changed in opposite directions by mutagenesis (15). This conclusion necessitates further experimental support because the reassessment of the EmrE mutants that were designed to insert into the membrane with the Nin-Cin or Nout-Cout topology shows that they are both functional and capable of removing substrate from the cell. A similar conclusion was reached when expressing the same mutants in E. coli BL21 IPTG-induced cells and following growth continuously in liquid medium (24). The results described here emphasize the need for caution in the analysis of negative phenotypes. We have shown previously examples of negative phenotypes even though the protein expressed is active (31). The ability of cells to carry a given function (in this case growth in a toxic environment) depends on the levels of the expressed protein, its affinity to the substrate, and the rate at which the substrate can be removed relative to the leaks, parameters that may well differ from strain to strain (25). Furthermore, the activity of other transporters that share the same substrate and the presence of proteins that interfere with the function may confuse interpretation (25). Here we also show that the mutants interact with the wild type protein produced by the chromosomal emrE gene in a nonproductive interaction that causes confusion in the interpretation of the phenotypes.
In the SMR family of multidrug transporters homodimers (e.g. EmrE, Hsmr, BPsmr, PAsmr, and TBsmr) and heterodimers (e.g. EbrAB) have been identified (26, 27, 32, 33). Membrane protein topology is largely governed by the positive-inside rule, i.e. loops rich in Lys and Arg residues tend to orient toward the cytoplasm (34). Analysis of the (K+R) bias suggests a very low bias for the homodimers and an antiparallel arrangement of the protomers in the heterodimers (14). EmrE and EbrAB confer resistance to ethidium and, therefore, SMR homodimers and heterodimers with a supposed different relative topology of the protomers perform very similar, if not identical, functions. Moreover, modification of the (K+R) bias of the heterodimer EbrAB has resulted in mutants of EbrA and EbrB that can function as homodimers (35). In other words, a number of mutations introduced by design may revert the traits acquired during evolution and change a heterodimer to a functional homodimer. Here we show that even without any further mutations, EbrB is capable of interacting with itself, provided the chromosomal gene coding for EmrE is inactivated. The findings are in agreement with a model where EbrB can interact with EbrA, EmrE, and itself in decreasing order of affinities. The interaction with EmrE is a nonproductive one that may result in the formation of a heterodimer with low or no activity. We have previously shown that EmrE can interact with a variety of homologues forming heterodimers with various affinities (32).
Antiparallel topology of a homodimer presents many intriguing implications regarding biogenesis and insertion. It would be expected that topology determinants in identical protomers would dictate a similar mode of insertion, resulting in a parallel arrangement. If a special mechanism that can direct insertion only of antiparallel homodimers exists, it is unprecedented and remains to be recognized. In the case of a protein with ambiguous topology determinants, one could expect a mixed population of dimers with parallel (a fraction with Nin-Cin as well as a fraction with Nout-Cout topology) and antiparallel protomers. However, even with a random insertion, the proportion of each species would be determined by the relative affinity of interaction. With the tools we have developed, we were able to detect dual topology dimers but not antiparallel ones. We suggest that in the case of native EmrE, the parallel interaction is preferred, but it can be imposed by proper manipulations as shown here or by mutations accumulated during evolution. Usually it is assumed that a very accurate design of protein-protein interaction is essential for the function of an oligomeric protein. However, protein domain promiscuity seems to play a major role in the creation and modulation of molecular functionality, especially for signal transduction. Such domains connect components of signal transduction networks through specific protein-protein interactions and delivering effectors to the sites of their action (36, 37). In other cases, the composition of oligomeric channels has been shown to be dependent on the specific cell types with the same subunits able to generate homo-oligomers as well as hetero-oligomers of varying composition (38).
We propose that the plasticity described here could provide an example of how inverted repeats in modern large polytopic transporters may have developed. The interaction interfaces must be appropriate for generation of a stable oligomer, and a substrate binding cavity must supply interaction points at given and fixed locations. We suggest that the promiscuity among interacting protomers reported here is tolerated also functionally because in EmrE, as in other proteins interacting with multiple substrates, the size of the binding pocket must be large enough to allow different molecules to reside in it in different orientations and to establish interactions with different sets of residues on the pocket walls (39–41). Therefore, the minimal structure where we find the necessary elements for substrate recognition and coupling may be provided by a dimer with the protomers in either one of the possible topologies.
In light of the plasticity described here, it will be important to describe experiments to test the proposed models of EmrE structure (10, 13). Both models have been obtained from low resolution data, and in the case of the x-ray data derived model, the crystals were obtained under conditions that inactivate the protein because the detergents used disturb dimerization (8, 13).
An additional aspect that contributes to the plasticity described in this report stems from the generally accepted functional symmetry of channels, transporters, and ion-coupled transporters where the direction of transport is dictated by the direction of the driving force acting on the transported substrates. Nevertheless, this is the first demonstration that the directionality of insertion of a transporter in vivo can be random without affecting activity. We suggest that this is possible because of the simplicity of the coupling mechanism of EmrE that allows also for the flexibility needed to allow function in a parallel as well as in an antiparallel dimer. In the proposed catalytic mechanism of EmrE, the binding sites of substrate and protons overlap, and occupancy is mutually exclusive. In other words, either molecule binds to the transporter only when the binding site is not occupied by the other and induces a shift in the equilibrium. The suggested mechanism necessitates the fine-tuning of the pKa of the glutamyl carboxyls at position 14, and indeed this pKa was estimated as between 7.5 to 8.0, depending on the technique used (29). In a protein with an Asp replacement that displays a lower pKa (5.8–6.3), coupling of the two fluxes is impaired (42). Aromatic residues from three TMs have also been identified as playing a role in proton and substrate binding (31, 43). We speculate that in the absence of substrates, the carboxyls of Glu14, located approximately in the middle of the binding cavity, are stabilized by interaction with protons or with at least part of aromatic residues contributed by TM1, TM2, and TM3. The aromatic residues provide an environment that may explain the unusually high pKa of these carboxyls (44) and allow for interaction with the hydrophobic substrates as has been documented in other proteins that bind similar substrates (45, 46). Such a mechanism would be rather insensitive to the specific nature and distribution of the aromatic residues around the carboxyls and could therefore provide the necessary stepping stone for the evolution of the topological inverted repeats observed in modern transporters.
The evolutionary challenge of recognition and transport of a wide spectrum of substrates may have selected for SMR heterodimers that originated from gene duplication of the more ancient homodimers. After gene duplication, a relatively small number of mutations in the hydrophilic domains may be sufficient to convert homodimers into heterodimers and vice versa. In this manner, one protein with an only slightly modified sequence may extend the range of the substrate specificity (5). The number of new combinations (and therefore new and wider spectrum of substrates) possible with heterodimers is undoubtedly higher than one can obtain with a homodimer (5). Topology evolution of larger proteins can now be envisaged starting from the SMR heterodimers, which can then fuse and give rise to larger proteins, some of them such as LacY, GlpT, and AcrB (39, 40, 47, 48) with domains in parallel orientation and others such as aquaporins, ClC channel, and a number of transporters with two oppositely oriented membrane domains (2, 3, 49–51).
This work was supported, in whole or in part, by National Institutes of Health Grant NS16708. This work was also supported by Grant 119/04 from the Israel Science Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- SMR
- small multidrug transporter
- IPTG
- isopropyl β-d-thiogalactopyranoside
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- MTSES
- (2-sulfonatoethyl) methane-thiosulfonate
- Mal-PEG
- methyl-polyethylene glycol-maleimide-MR 5000
- o-PDM
- o-phenylenedimaleimide
- TM
- transmembrane domain
- GpA
- glycophorin A
- TPP+
- tetraphenylphosphonium.
REFERENCES
- 1.Saier M. H., Jr. (1994) Microbiol. Rev. 58, 71–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramson J., Wright E. M. (2009) Curr. Opin. Struct. Biol. 19, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrest L. R., Rudnick G. (2009) Physiology 24, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ninio S., Elbaz Y., Schuldiner S. (2004) FEBS Lett. 562, 193–196 [DOI] [PubMed] [Google Scholar]
- 5.Schuldiner S. (2007) Trends Biochem. Sci. 32, 252–258 [DOI] [PubMed] [Google Scholar]
- 6.Schuldiner S. (2009) Biochim. Biophys. Acta 1794, 748–762 [DOI] [PubMed] [Google Scholar]
- 7.Soskine M., Steiner-Mordoch S., Schuldiner S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soskine M., Mark S., Tayer N., Mizrachi R., Schuldiner S. (2006) J. Biol. Chem. 281, 36205–36212 [DOI] [PubMed] [Google Scholar]
- 9.Steiner-Mordoch S., Soskine M., Solomon D., Rotem D., Gold A., Yechieli M., Adam Y., Schuldiner S. (2008) EMBO J. 27, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleishman S. J., Harrington S. E., Enosh A., Halperin D., Tate C. G., Ben-Tal N. (2006) J. Mol. Biol. 364, 54–67 [DOI] [PubMed] [Google Scholar]
- 11.Ubarretxena-Belandia I., Baldwin J. M., Schuldiner S., Tate C. G. (2003) EMBO J. 22, 6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang G., Roth C. B., Reyes C. L., Pornillos O., Chen Y. J., Chen A. P. (2006) Science 314, 1875. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y. J., Pornillos O., Lieu S., Ma C., Chen A. P., Chang G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18999–19004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rapp M., Granseth E., Seppälä S., von Heijne G. (2006) Nat. Struct. Mol. Biol. 13, 112–116 [DOI] [PubMed] [Google Scholar]
- 15.Rapp M., Seppälä S., Granseth E., von Heijne G. (2007) Science 315, 1282–1284 [DOI] [PubMed] [Google Scholar]
- 16.Muth T. R., Schuldiner S. (2000) EMBO J. 19, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yerushalmi H., Lebendiker M., Schuldiner S. (1995) J. Biol. Chem. 270, 6856–6863 [DOI] [PubMed] [Google Scholar]
- 18.Tabor S., Richardson C. C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taglicht D., Padan E., Schuldiner S. (1991) J. Biol. Chem. 266, 11289–11294 [PubMed] [Google Scholar]
- 20.Li J., Xu Q., Cortes D. M., Perozo E., Laskey A., Karlin A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11605–11610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green N. S., Reisler E., Houk K. N. (2001) Protein Sci. 10, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbaz Y., Salomon T., Schuldiner S. (2008) J. Biol. Chem. 283, 12276–12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbaz Y., Steiner-Mordoch S., Danieli T., Schuldiner S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHaourab H. S., Mishra S., Koteiche H. A., Amadi S. H. (2008) Biochemistry 47, 7980–7982 [DOI] [PubMed] [Google Scholar]
- 25.Tal N., Schuldiner S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9051–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Ma C., Pornillos O., Xiu X., Chang G., Saier M. H., Jr. (2007) Biochemistry 46, 5218–5225 [DOI] [PubMed] [Google Scholar]
- 27.Masaoka Y., Ueno Y., Morita Y., Kuroda T., Mizushima T., Tsuchiya T. (2000) J. Bacteriol. 182, 2307–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmon M. A., Flanagan J. M., Treutlein H. R., Zhang J., Engelman D. M. (1992) Biochemistry 31, 12719–12725 [DOI] [PubMed] [Google Scholar]
- 29.Adam Y., Tayer N., Rotem D., Schreiber G., Schuldiner S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17989–17994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotem D., Sal-man N., Schuldiner S. (2001) J. Biol. Chem. 276, 48243–48249 [DOI] [PubMed] [Google Scholar]
- 31.Elbaz Y., Tayer N., Steinfels E., Steiner-Mordoch S., Schuldiner S. (2005) Biochemistry 44, 7369–7377 [DOI] [PubMed] [Google Scholar]
- 32.Ninio S., Rotem D., Schuldiner S. (2001) J. Biol. Chem. 276, 48250–48256 [DOI] [PubMed] [Google Scholar]
- 33.Ninio S., Schuldiner S. (2003) J. Biol. Chem. 278, 12000–12005 [DOI] [PubMed] [Google Scholar]
- 34.Heijne G. V. (1986) EMBO J. 5, 3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikukawa T., Nara T., Araiso T., Miyauchi S., Kamo N. (2006) Biochim. Biophys. Acta 1758, 673–679 [DOI] [PubMed] [Google Scholar]
- 36.Basu M. K., Carmel L., Rogozin I. B., Koonin E. V. (2008) Genome Res. 18, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcotte E. M., Pellegrini M., Ng H. L., Rice D. W., Yeates T. O., Eisenberg D. (1999) Science 285, 751–753 [DOI] [PubMed] [Google Scholar]
- 38.Cull-Candy S., Kelly L., Farrant M. (2006) Curr. Opin. Neurobiol. 16, 288–297 [DOI] [PubMed] [Google Scholar]
- 39.Murakami S., Nakashima R., Yamashita E., Matsumoto T., Yamaguchi A. (2006) Nature 443, 173–179 [DOI] [PubMed] [Google Scholar]
- 40.Seeger M. A., Schiefner A., Eicher T., Verrey F., Diederichs K., Pos K. M. (2006) Science 313, 1295–1298 [DOI] [PubMed] [Google Scholar]
- 41.Godsey M. H., Zheleznova Heldwein E. E., Brennan R. G. (2002) J. Biol. Chem. 277, 40169–40172 [DOI] [PubMed] [Google Scholar]
- 42.Yerushalmi H., Schuldiner S. (2000) Biochemistry 39, 14711–14719 [DOI] [PubMed] [Google Scholar]
- 43.Rotem D., Steiner-Mordoch S., Schuldiner S. (2006) J. Biol. Chem. 281, 18715–18722 [DOI] [PubMed] [Google Scholar]
- 44.Soskine M., Adam Y., Schuldiner S. (2004) J. Biol. Chem. 279, 9951–9955 [DOI] [PubMed] [Google Scholar]
- 45.Zheleznova E. E., Markham P. N., Neyfakh A. A., Brennan R. G. (1999) Cell 96, 353–362 [DOI] [PubMed] [Google Scholar]
- 46.Schumacher M. A., Miller M. C., Grkovic S., Brown M. H., Skurray R. A., Brennan R. G. (2001) Science 294, 2158–2163 [DOI] [PubMed] [Google Scholar]
- 47.Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 48.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 49.Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J. B., Engel A., Fujiyoshi Y. (2000) Nature 407, 599–605 [DOI] [PubMed] [Google Scholar]
- 50.Dutzler R., Campbell E. B., Cadene M., Chait B. T., MacKinnon R. (2002) Nature 415, 287–294 [DOI] [PubMed] [Google Scholar]
- 51.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 52.Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg E. B., Arbel T., Chen J., Karpel R., Mackie G. A., Schuldiner S., Padan E. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2615–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang A. C., Cohen S. N. (1978) J. Bacteriol. 134, 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]