Abstract
The formation of heteromeric tetramers is a common feature of voltage-gated potassium (Kv) channels. This results in the generation of a variety of tetrameric Kv channels that exhibit distinct biophysical and biochemical characteristics. Kv2 delayed rectifier channels are, however, unique exceptions. It has been previously shown that mammalian Kv2.1 and Kv2.2 are localized in distinct domains of neuronal membranes and are not capable of forming heteromeric channels with each other (Hwang, P. M., Glatt, C. E., Bredt, D. S., Yellen, G., and Snyder, S. H. (1992) Neuron 8, 473–481). In this study, we report a novel form of rat Kv2.2, Kv2.2long, which has not been previously recognized. Our data indicate that Kv2.2long is the predominant form of Kv2.2 expressed in cortical pyramidal neurons. In contrast to the previous findings, we also found that rat Kv2.1 and Kv2.2long are colocalized in the somata and proximal dendrites of cortical pyramidal neurons and are capable of forming functional heteromeric delayed rectifier channels. Our results suggest that the delayed rectifier currents, which regulate action potential firing, are encoded by heteromeric Kv2 channels in cortical neurons.
Keywords: Antibodies, Channels/Potassium, Methods/Confocal Microscopy, Methods/Immunochemistry, Neurobiology/Neuroscience, Protein/Assembly
Introduction
Voltage-gated potassium (Kv) channels play pivotal roles in regulating neuronal excitability, shaping action potentials, and modulating spike patterns (1). Mammalian neurons express a number of Kv channel subunits (>20), which are assembled into functional tetrameric channels (2). These subunits are classified into 12 subfamilies based on their primary structures (3). One remarkable aspect of Kv channels is that subunits in the same subfamily can form heteromeric channels, which dramatically increases the molecular and functional diversity of Kv channels (2, 4–7). By doing so, neurons express a wide variety of K+ conductances from the limited number of Kv subunit genes. However, the Kv2 subfamily, composed of Kv2.1 and Kv2.2 subunits, is a unique exception.
The cDNA sequences of Kv2.1 (8) and Kv2.2 (9) predict proteins that are similar in N-terminal and membrane-spanning domains (∼90% homology), but they differ substantially across the large cytoplasmic C-terminal region (∼50%). Although they can assemble with modulatory α-subunits, Kv5, Kv6, Kv8, and Kv9 (10–13), it has been shown that mammalian Kv2.1 and Kv2.2 do not form heteromeric channels. This has been well supported by two major findings. First, Kv2.1 and Kv2.2 show very distinct subcellular distributions. It has been reported that Kv2.2 is diffusely localized in the dendrites of cortical and hippocampal pyramidal neurons (14–16), whereas Kv2.1 is localized in large clusters in the somata and proximal dendrites (17, 18). Second, studies using subtype-specific dominant-negative pore mutants clearly showed that rat Kv2.1 and Kv2.2 in sensory neurons and HEK293 cells do not form functional heteromeric channels (19).
However, we found that the originally reported cDNA sequence of rat Kv2.2 (9) differs from the rat genomic sequence of Kv2.2. This results in two different protein sequences, the original form (802 amino acids) and a novel longer form (907 amino acids) of Kv2.2 (called Kv2.2long hereafter). In this study, we provide evidence that, unlike the original form of Kv2.2 (called Kv2.2short hereafter), Kv2.1 and Kv2.2long are colocalized in large clusters in the somata and proximal dendrites of cortical pyramidal neurons, are physically associated, and form heteromeric delayed rectifier channels.
EXPERIMENTAL PROCEDURES
Immunohistochemistry
Immunohistochemistry was performed as described (20). Briefly, rats were anesthetized by the injection of 50 mg/kg sodium pentobarbital (intraperitoneal). Animals were then fixed by transcardiac perfusion of 4% paraformaldehyde in phosphate buffer. The fixed brain was equilibrated in 30% sucrose for cryoprotection. The brain was sectioned into 40-μm thick sagittal brain sections using a cryomicrotome. Tissue sections were blocked and permeabilized for 1 h in 10% normal goat serum in phosphate buffer containing 0.3% Triton X-100. Sections were then incubated with the following primary antibodies overnight at 4 °C: mouse monoclonal antibody K89 against Kv2.1 (NeuroMab, Davis, CA) (21), rabbit anti-Kv2.2long polyclonal antibody (APC-120, Alomone Labs, Jerusalem, Israel) (22), or mouse monoclonal antibody K37 against the N terminus of Kv2.2 (NeuroMab) (23). For K37 staining, sections were preincubated in 0.1 m citric acid, pH 8.8, for 30 min at 80 °C to facilitate the antibody binding. Sections were washed and probed with Alexa-conjugated secondary reagents available for each species and IgG isotype (Invitrogen). After washing, sections were mounted on glass slides for fluorescence imaging. HEK293 cells were cultured on coverslips coated with poly-l-lysine and transfected with Kv2.1 and/or Kv2.2long cDNA using Lipofectamine 2000 (Invitrogen). Two days after transfection, HEK293 cells were fixed in 3% paraformaldehyde with 0.1% Triton X-100 in phosphate-buffered saline. Cells were blocked for 1 h in Tris-buffered saline containing 0.15 m NaCl, 4% nonfat dry milk, 0.05% Tween 20, and 20 mm Tris-HCl, pH 7.4. The cells were then incubated with primary antibodies overnight at 4 °C. Cells were washed and probed with Alexa-conjugated secondary reagents. Fluorescent images were taken with a charge-coupled device camera installed on a Zeiss Axiovert 200M microscope with 10×, 20×, and 63× lenses and a structured illumination imaging system (ApoTome) using Axiovision software (Carl Zeiss). We obtained optically sectioned images using this system. The monoclonal antibodies were obtained from the University of California Davis/National Institutes of Health NeuroMab Facility of the Department of Neurobiology, Physiology, and Behavior, College of Biological Sciences, University of California (Davis, CA). Colocalization analyses were performed using ImageJ. Pearson's coefficient takes into consideration similarity between shapes in two channels while ignoring the intensities of signals. The intensity correlation quotient was obtained as described (24). Line scan analysis was also performed using ImageJ. Fluorescence intensity was measured in 10-pixel (∼1 μm) wide lines and is shown as a percentage of the maximum values in each plot.
Rat Brain Membrane (RBM)3 Fraction
Animals were killed by rapid decapitation (25). The brains were removed and homogenized in ice-cold buffer (320 mm sucrose, 5 mm sodium phosphate, 100 mm NaF, and 1 mm phenylmethylsulfonyl fluoride, pH 7.4) containing protein inhibitor mixture (2 μg/ml aprotinin, 1 μg/ml leupeptin, 2 μg/ml antipain, and 10 μg/ml benzamidine). The homogenates were centrifuged at 800 × g for 10 min, and the resultant supernatants were centrifuged at 39,000 × g for 90 min. The pellets (membrane fraction) were resuspended in the same buffer.
Western Blotting
Samples were prepared in SDS sample buffer (2% SDS, 5% 2-mercaptoethanol, 10% glycerol, and 62.5 mm Tris-HCl, pH 6.8). Proteins were separated on 6 or 7.5% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were washed; blocked with 4% nonfat dry milk in 150 mm NaCl and 20 mm Tris-HCl, pH 8.0, for 30 min; and then incubated with primary antibodies overnight in the blocking buffer at 4 °C. We used mouse monoclonal antibody K89 and the anti-Kv2.2long antibody to detect Kv2 proteins. The membranes were washed and probed with horseradish peroxidase-conjugated secondary antibodies (KPL, Gaithersburg, MD) according to the manufacturer's instructions. Immunoreactive bands were detected with enhanced chemiluminescence (PerkinElmer Life Sciences) and visualized by exposing the membranes to x-ray films.
Immunoprecipitation
For the large-scale Kv2.1 affinity purification, RBM (50 mg of protein) was solubilized in lysis buffer containing 20 mm Tris-HCl, 0.15 m NaCl, 1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, and protease inhibitor mixture (2 μg/ml aprotinin, 1 μg/ml leupeptin, 2 μg/ml antipain, and 10 μg/ml benzamidine). A K89 affinity column was prepared using a Seize primary immunoprecipitation kit (Thermo Scientific, Rockford, IL). The lysates were incubated with the column resin for 24 h to remove nonspecific binders. The cleared lysates were then applied to either the K89 affinity column or a control column (with normal mouse IgG), followed by incubation for 4 days at 4 °C. After extensive washes, bound proteins were eluted in boiling SDS sample buffer for 5 min. The eluents were subjected to SDS-PAGE, and the gels were stained with either silver or Coomassie Brilliant Blue G-255. Mass spectrometry analysis was done at the core facility of the University of Maryland (Baltimore). Briefly, excised bands from Coomassie Blue-stained gels were destained and dried in a speed vacuum concentrator. Dried gel pieces were then trypsinized at 37 °C for 18 h. Peptides were extracted into 50% acetonitrile in 5% formic acid and dried in a speed vacuum concentrator. Peptides were desalted and concentrated using a reversed-phase trap column. They were separated in a reversed-phase C18 column during a 90-min linear gradient of 5–90% acetonitrile/water containing 0.1% formic acid at a flow rate of 300 nl/ml. The eluted peptides were directly sprayed into a Finnigan LCQ ion trap mass spectrometer. Tandem mass spectra were acquired for the most intense peptide ion from the previous mass spectra. The acquired mass spectrometry scans were searched against the IPI Protein Sequence Database using the Sorcerer/Sequest search algorithm. The best hit were selected based on probability/percent of coverage and number of peptides or based on Xcore. For immunoprecipitation, HEK293 cells transiently expressing Kv2 or RBM were solubilized at 4 °C for 30 min in lysis buffer. Insoluble materials were removed by centrifugation at 15,000 × g for 15 min at 4 °C. The supernatants were incubated with either mouse monoclonal antibody K89 or the anti-Kv2.2long antibody at 1 μg/ml for 2 h at 4 °C and then incubated with protein G or A immobilized on agarose beads for 2 h at 4 °C with agitation. After extensive washes, bound proteins were eluted and subjected to Western blotting.
Construction of Non-conducting Pore Mutants of Kv2.1 and Kv2.2
Rat Kv2.1 and Kv2.2 in the RBG4 vector were obtained from James Trimmer (University of California, Davis). We found that the clone encodes the genomic sequence of Kv2.2. Two pore mutations (W365C/Y380T) were introduced to Kv2.1 by PCR. Codons TGG (Trp365) and TAC (Tyr380) were mutated to TGC (Cys) and ACC (Thr), respectively. The mutant was cloned into the pIRES-mCherry vector (Clontech). The Kv2.1 W365C/Y380T construct was sequenced in its entirety to ensure that no additional mutations were introduced. Wild-type Kv2 sequences were also cloned into the pIRES-AcGFP vector for patch-clamp experiments.
Electrophysiology and Data Analysis
HEK293 cells were transiently transfected with either wild-type Kv2.1 or wild-type Kv2.2long (in the pIRES-AcGFP vector) with or without the pore mutants. Outward Kv2 channel currents were recorded using the whole-cell voltage-clamp technique. We chose cells that express comparable levels of green fluorescent protein. Currents were recorded with an MultiClamp 700A amplifier (Molecular Devices, Sunnyvale, CA), sampled at 20 kHz, and filtered at 2 kHz with a series resistance of <5 megohms. Patch pipettes were pulled from borosilicate glass tubing to give a resistance of 1–2 megohms when filled with the pipette solution (135 mm KCl, 2 mm MgCl2, 10 mm EGTA, 10 mm glucose, and 10 mm HEPES, pH 7.3). The extracellular buffer contained 140 mm NaCl, 4 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm glucose, and 10 mm HEPES, pH 7.4. All recordings were performed at room temperature (∼25 °C). The membrane potential was held at −80 mV and depolarized (or hyperpolarized) from the holding potential of −80 mV to voltages between −90 and +70 mV in 10-mV increments for 200 ms. Linear leak components were identified from the hyperpolarizing pulse and subtracted. Peak outward sustained K+ currents were measured 100 ms into the pulse at each depolarizing potential.
RESULTS
Two Different Sequences of rat Kv2.2
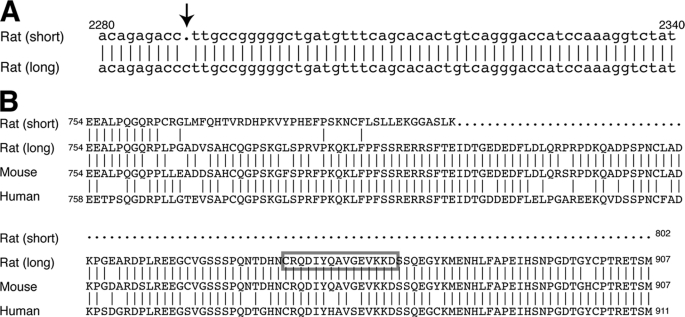
Kv2.2 mRNA was first cloned from a rat brain cDNA library (9) and identified to encode an 802-amino acid long protein. However, we recently found that there are two mRNA sequences of rat Kv2.2 registered in the NCBI Database. One originated from the cloned cDNA sequence (accession number M77482), and the other from the rat genomic sequence (accession number NM_054000). They differ after nucleotide 2288 (number based on the coding region) because of a single-nucleotide deletion (Fig. 1A). The resultant two protein sequences have either 802 (from the original cDNA) or 907 (from the genomic sequence) amino acid residues. In the previous analyses of Kv2.2 localization in the brain, antibodies raised against the cytoplasmic C terminus of the shorter form (Kv2.2short) were used because this region differs substantially from that of Kv2.1 (data not shown). Studies reported that Kv2.2short exhibits a subcellular localization distinct from that of Kv2.1 (14, 26). However, the epitope sequence is present only in Kv2.2short but not in the novel longer form (Kv2.2long) (Fig. 1B). Therefore, we evaluated the subcellular localization of Kv2.2long in the cerebral cortex of rat using a rabbit antibody whose epitope is specific in Kv2.2long (Fig. 1B) (22).
FIGURE 1.
Two sequences of rat Kv2.2. A, sequence alignment of rat Kv2.2 derived from the original cDNA (short) and the genomic DNA (long) sequences. The original cDNA lacks the cytosine indicated by the arrow. B, sequence alignment of rat Kv2.2 proteins derived from the original cDNA (short) and the genomic DNA (long) sequences, as well as those from the mouse and human genomic sequences. Vertical lines indicate amino acid residues identical to the longer form of Kv2.2. The box indicates the epitope of the antibody used in this study.
Colocalization of Kv2.1 and Kv2.2long in Rat Brain Neurons
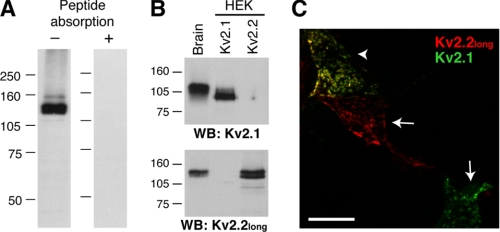
First, the specificity of the anti-Kv2.2long polyclonal antibody was verified as described previously (27, 28). RBM fractions were prepared and subjected to Western blotting using the anti-Kv2.2long antibody. As shown in Fig. 2A, a major band was observed at ∼140 kDa, which is larger than the size expected from the primary amino acid sequence (102 kDa), indicating post-translational modifications of Kv2.2. Indeed, alkaline phosphatase treatment of RBM resulted in a downward shift of the immunoreactive band from 140 to 110 kDa (data not shown), indicating constitutive phosphorylation of Kv2.2 in the brain, like Kv2.1 (29, 30). Preincubation with the antigen peptide completely blocked the binding of the antibody, verifying the specificity of the antibody (Fig. 2A). To examine whether the anti-Kv2.2long antibody cross-reacts with Kv2.1, lysates of HEK293 cells solely expressing Kv2.1 or Kv2.2long were probed with either the mouse anti-Kv2.1 (K89) or anti-Kv2.2long antibody by Western blotting (Fig. 2B). Rat Kv2.1 and Kv2.2long exhibit 48% identity in the C termini, but the epitope sequence of the anti-Kv2.2long antibody is missing in Kv2.1 (data not shown). Likewise, the K89 and anti-Kv2.2long antibodies detected Kv2.1 and Kv2.2, respectively, expressed in HEK293 cells, but there was no detectable cross-reactivity of the antibodies. This was also verified in the immunostaining of HEK293 cells coexpressing Kv2.1 and Kv2.2long (Fig. 2C).
FIGURE 2.
Specificity of the anti-Kv2.2long antibody. A, RBM fractions were solubilized in lysis buffer (see “Experimental Procedures”) and subjected to Western blotting with (right lane) or without (left lane) preincubation of the antibody solution with the antigen peptide. B, HEK293 (HEK) cell lysates expressing either Kv2.1 or Kv2.2long were subjected to Western blotting (WB) using the K89 (upper panel) or anti-Kv2.2long (lower panel) antibody. Molecular mass markers (in kilodaltons) are indicated. C, HEK293 cells doubly transfected with Kv2.1 and Kv2.2long were immunostained. The arrows indicate cells stained solely by either the K89 or anti-Kv2.2long antibody, and the arrowhead indicates a doubly labeled cell. Non-transfected cells in the field were not labeled (not shown). The specificity was also verified in singly transfected cells as well (not shown).
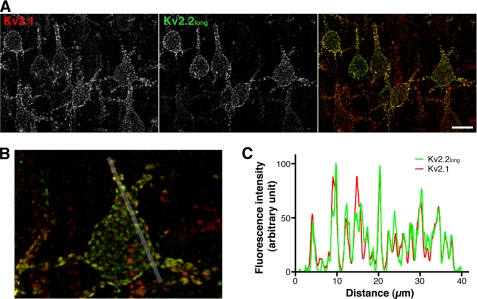
With the specificity of the antibodies verified, subcellular localizations of Kv2.1 and Kv2.2long in cortical neurons were investigated. Rat brain sagittal sections were doubly immunostained with the anti-Kv2.1 (K89) and anti-Kv2.2long antibodies. Kv2.1 was relatively homogeneously expressed throughout the cerebral cortex as reported previously (17), whereas Kv2.2 expression was detected in a subset of cortical pyramidal neurons (∼50% of Kv2.1-positive neurons). The expression of Kv2.2 was especially apparent in the layer V pyramidal neurons. This is consistent with previous findings by others, using single-cell reverse transcription-PCR, that ∼60% of cortical neurons express Kv2.2 (16). In the pyramidal neurons, Kv2.1 formed large clusters in the somata and proximal dendrites (Fig. 3A) as reported previously (18). Our immunostaining results showed that Kv2.2 is also localized in large clusters in the somata and proximal dendrites, in contrast to the previously reported localization of Kv2.2short. In neurons positive for both Kv2.1 and Kv2.2long, we found a substantial overlap of Kv2.1 and Kv2.2long in discrete clusters (Fig. 3B). Quantitative image analyses revealed that Kv2.1 and Kv2.2long exhibit a high degree of colocalization with the following colocalization coefficients: a Pearson's coefficient (the correlation of the intensity distributions between two channels) of 0.66 ± 0.03 and an intensity correlation quotient (the degree of synchronous changes in signal intensity between two channels) (24) of 0.29 ± 0.01 in a colocalization analysis (p < 0.01, when the intensity correlation quotient was compared with that obtained from random noise images; n = 5). Line scanning analysis also clearly indicated their colocalization in clusters (Fig. 3C). These results indicate that Kv2.1 and Kv2.2long coexist in these microstructures in the same membrane domains of cortical neurons.
FIGURE 3.
Colocalization of Kv2.1 and Kv2.2long in discrete clusters. A, rat brain sections doubly immunostained with the anti-Kv2.1 (K89) and anti-Kv2.2long antibodies. The images show pyramidal neurons in cortical layer V. Scale bar, 20 μm. B, high power image of a cortical pyramidal neuron stained with the K89 (red) and anti-Kv2.2long (green) antibodies. The white line indicates the 40-μm segment used for the line scan analysis in C. C, line scan analysis of the distribution of Kv2.1 and Kv2.2long. Fluorescence intensity is shown as a percentage of the maximum signal.
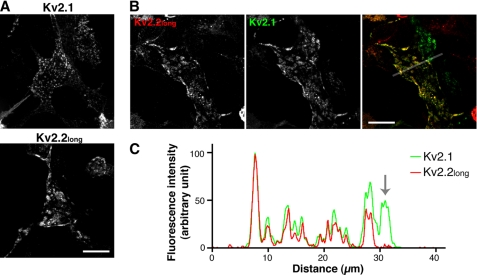
It has been previously reported that Kv2.1 forms large neuron-like clusters in HEK293 cells (31, 32). Using this model system, we further confirmed the clustered localization of Kv2.2long and its colocalization with Kv2.1. As shown in Fig. 4A, Kv2.2long formed large clusters reminiscent of those formed by Kv2.1. Furthermore, Kv2.1 and Kv2.2long were nearly completely colocalized in clusters in cells expressing both of the subunits (Fig. 4, B and C). Together, these results strongly indicate that Kv2.2long exhibits a subcellular localization similar to that of Kv2.1 and that they may form heteromeric delayed rectifier channels, in contrast to the current view of Kv2 channels.
FIGURE 4.
Clustering of Kv2.2long in HEK293 cells. A, clustered localization of Kv2.1 and Kv2.2long in HEK293 cells expressing individual subunits. Cells were fixed and immunostained with either the K89 or anti-Kv2.2long antibody. B, co-clustering of Kv2.1 and Kv2.2long in doubly transfected HEK293 cells stained with the K89 (green) and anti-Kv2.2long (red) antibodies. The line indicates the 40-μm segment used in C. Scale bars = 20 μm. C, line scanning analysis of the distribution of Kv2.1 and Kv2.2long in the cell shown in B. The arrow indicates the segment that covers the cell expressing only Kv2.1. The anti-Kv2.2long antibody showed only background level immunoreactivity within this segment.
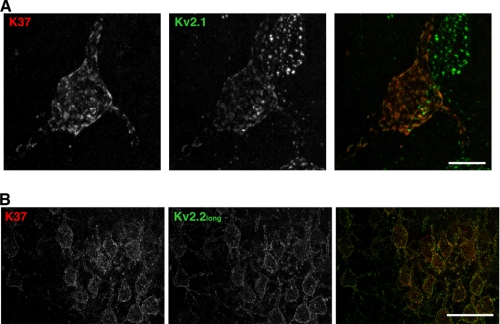
Finally, we examined the colocalization of Kv2.1 and Kv2.2long in neurons using mouse monoclonal antibody K37 raised against the N-terminal 61 residues of Kv2.2, which are identical between Kv2.2short and Kv2.2long. Therefore, this antibody was expected to label both Kv2.2short and Kv2.2long. As shown in Fig. 5A, antibody K37 labeling showed clustered localization of Kv2.2 in the somata and proximal dendrites of cortical pyramidal neurons, similar to that observed with the rabbit anti-Kv2.2long antibody. We did not detect Kv2.2 labeling in distal dendrites, which was observed in previous studies using different antibodies. Double labeling with the K37 and anti-Kv2.2long antibodies showed virtually identical staining patterns in cortical neurons (Fig. 5B). These results indicate that Kv2.2long is the predominant form of Kv2.2 expressed in rat cortical pyramidal neurons.
FIGURE 5.
Immunolocalization of Kv2.2 using an antibody against the N-terminal domain. A, sagittal brain sections were stained with the anti-Kv2.1 antibody (K89) and antibody K37 raised against the N terminus of Kv2.2. The epitope of antibody K37 is conserved in Kv2.2short and Kv2.2long. Scale bar = 10 μm. B, brain sections were stained with the rabbit anti-Kv2.2long antibody and K37 antibodies. The images were taken in the cerebral cortex. Scale bar = 50 μm.
Molecular Interaction of Kv2.1 and Kv2.2long
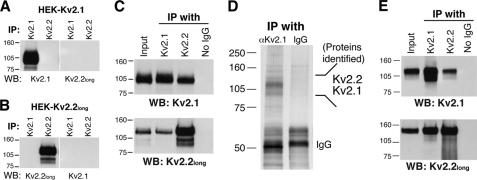
Considering the high degree of colocalization, we hypothesized that Kv2.1 and Kv2.2long form functional heteromeric channels. To test this, we examined whether they can physically interact in HEK293 cells using co-immunoprecipitation analysis. We first confirmed the specificity of the antibodies in the immunoprecipitation application. Lysates from HEK293 cells expressing either Kv2.1 or Kv2.2long were subjected to immunoprecipitation with either the anti-Kv2.1 (K89) or anti-Kv2.2long antibody. There was no detectable cross-reactivity in this type of application (Fig. 6, A and B). We then performed co-immunoprecipitation assays using cells coexpressing Kv2.1 and Kv2.2long. As shown in Fig. 6C, the anti-Kv2.1 antibody (K89) co-immunoprecipitated Kv2.2long along with Kv2.1. Furthermore, the anti-Kv2.2long antibody also co-immunoprecipitated Kv2.1 together with Kv2.2long. These results were highly reproducible in three independent experiments (data not shown). To exclude the possibility that the interaction occurs following cell lysis but not in situ, we separately prepared lysates from cells expressing either Kv2.1 or Kv2.2long, mixed these lysates together, and performed the co-immunoprecipitation assay. There was no detectable co-immunoprecipitation (supplemental Fig. 1), indicating that the binding of Kv2.1 and Kv2.2long occurs in situ, but not after cell lysis.
FIGURE 6.
Co-immunoprecipitation of Kv2.1 and Kv2.2long from brain tissues and HEK293 cells. A and B, specificity of the antibodies in the immunoprecipitation application. Lysates from HEK293 cells expressing either Kv2.1 (A) or Kv2.2long (B) were subjected to immunoprecipitation (IP) with either the anti-Kv2.1 (K89) or anti-Kv2.2long antibody. Bound proteins were detected by Western blotting (WB) using the subtype-specific antibodies as indicated. C, co-immunoprecipitation of Kv2.1 and Kv2.2long from HEK293 cells. Cells expressing both Kv2.1 and Kv2.2long subunits were solubilized in 0.1% Triton X-100 lysis buffer. Either the K89 or anti-Kv2.2long antibody was added to the lysates and incubate for 2 h at 4 °C. Protein A or G beads were added, and the lysates were further incubated for 2 h at 4 °C. Proteins precipitated with beads were eluted and detected by Western blotting using the K89 and anti-Kv2.2long antibodies. D, co-purification of Kv2.2 with Kv2.1 from rat brain tissues. A silver-stained gel image of affinity-purified samples for mass spectrometry analysis is presented. For the mass spectrometry analysis, a Coomassie Blue-stained gel was used. See “Experimental Procedures” for details. E, reciprocal co-immunoprecipitation of Kv2.1 and Kv2.2long from rat brain lysates. Control experiments in C and D were performed without the addition of the antibodies during immunoprecipitation.
We then examined whether they are associated in brain neurons. Kv2.1 was purified from rat brain lysates using an affinity column conjugated with antibody K89. Bound proteins were eluted and separated by SDS-PAGE, and proteins were then stained with Coomassie Brilliant Blue. Multiple bands appeared at ∼100 kDa but not in the eluent from the control column (Fig. 6D). These bands were excised, trypsinized, and subjected to mass spectrometry analysis for protein identification (Table 1). The analysis revealed that the 120-kDa major band contained Kv2.1, showing the high efficiency of Kv2.1 affinity purification. Intriguingly, peptides of the Kv2.2 subunit were recovered from the band above the Kv2.1 band (∼140 kDa). This was consistent in two independent affinity-purified samples. We also confirmed their interaction using co-immunoprecipitation assays. The K89 and anti-Kv2.2long antibodies reciprocally immunoprecipitated Kv2.1 and Kv2.2long from rat brain lysates as shown in Fig. 6E. These results demonstrate that Kv2.1 and Kv2.2long interact in brain neurons.
TABLE 1.
Peptide identification by mass spectrometry
| Size | Protein | Identification probability | No. of unique peptides | No. of total spectra | Sequence coverage |
|---|---|---|---|---|---|
| % | % | ||||
| ∼140 kDa | Kv2.2 | 99.80a | 2 | 9 | 2.37 |
| ∼120 kDa | Kv2.1 | 100.00 | 6 | 30 | 9.38 |
a Kv2.2 protein identification probabilities were computed using the shorter Kv2.2 sequence available in the data base and are thus potentially incomplete.
Kv2.1 and Kv2.2long Form Heteromeric Delayed Rectifier K+ Channels
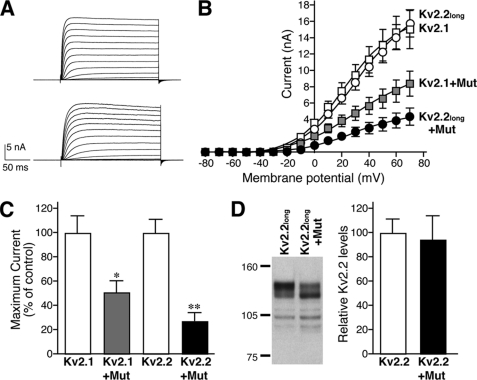
The immunoprecipitation experiments suggested the interaction of the two Kv2 subunits. However, it was not conclusive whether or not the Kv2.1 and Kv2.2 subunits form heteromeric channels. To directly address this, we took advantage of the non-conducting pore mutant of Kv2.1 (W365C/Y380T) (19, 33). If they truly form heteromeric channels, the non-conducting mutant of Kv2.1 should be able to suppress the conductance of the wild-type Kv2.2long channels. We therefore expressed either wild-type Kv2.1 or Kv2.2long with Kv2.1 W365C/Y380T in HEK293 cells and recorded ionic currents in voltage-clamp recordings. Kv2.2long expressed in HEK293 cells exhibited delayed rectifier currents (Fig. 7A), which were similar to those of homomeric Kv2.1 channels in the current-voltage relationship (Fig. 7B). Kv2.1 W365C/Y380T itself did not conduct detectable currents (data not shown), as reported previously. Coexpression of Kv2.1 W365C/Y380T significantly inhibited the current amplitude of wild-type Kv2.1 channels (p < 0.01; n = 6) (Fig. 7B), demonstrating the inhibitory efficacy of the mutant. This non-conducting mutant also significantly and dramatically suppressed the conductance of the wild-type Kv2.2long channels (Fig. 7, B and C) by nearly 70% (p < 0.001; n = 6). Western blot analysis confirmed that this inhibition was not due to a reduction in the expression level of wild-type Kv2.2long. The level of wild-type Kv2.2long was not affected by the coexpression of the Kv2.1 pore mutant (Fig. 7D). These results strongly suggest that Kv2.1 and Kv2.2long, which are colocalized in large surface clusters in neurons, form heteromeric delayed rectifier channels.
FIGURE 7.
Formation of heteromeric Kv2 channels in HEK293 cells. A, delayed rectifier currents encoded by either Kv2.1 (upper panel) or Kv2.2long (lower panel) homomeric channels in HEK293 cells. B, inhibition of the wild-type Kv2 channels by the Kv2.1 non-conducting pore mutant. HEK293 cells were transfected with either wild-type Kv2.1 or Kv2.2long together with either the non-conducting mutant of Kv2.1 (Mut) or an empty vector at a DNA ratio of 1:10. Steady-state current amplitudes were recorded as described under “Experimental Procedures” at different membrane potentials. C, inhibition of Kv2 channel currents at +60 mV by the non-conducting mutant of Kv2.1. Values are mean ± S.E. (n = 6). *, p = 0.0022 (versus Kv2.1); **, p = 0.0002 (versus Kv2.2) and p = 0.033 (versus Kv2.1 + mutant). D, the levels of Kv2.2long are unchanged by the coexpression of the Kv2.1 mutant. Cells were transfected with Kv2.2long and either an empty vector or the Kv2.1 mutant. Kv2.2long was detected by Western blotting using the anti-Kv2.2long antibody. The levels of Kv2.2long were measured and are expressed as a percentage of the vector control.
DISCUSSION
Previous studies have shown that Kv2.2 is uniformly localized in the dendrites of mammalian neurons (14, 16, 26), whereas Kv2.1 is highly concentrated in the somata and proximal dendrites in discrete surface clusters (17). Together with the fact that the non-conducting mutant of rat Kv2.1 does not inhibit rat Kv2.2 channels expressed in HEK293 cells (19), it has been thought that Kv2.1 and Kv2.2 do not form heteromeric channels. It was puzzling, however, because Kv2 subunits of Xenopus do form functional heteromeric channels (33). We believe that our results have solved this conundrum. In this work, we report that there are two different mRNA sequences of rat Kv2.2 in the data base: the original cDNA sequence (9) and one derived from the genomic sequence. We found that the original sequence has a single-nucleotide deletion compared with the genomic sequence. The cDNA sequence with the deletion results in a frameshift in the protein sequence after amino acid residue 764, providing a 105-amino acid shorter Kv2.2 polypeptide (Kv2.2short) than that deduced from the genomic sequence (Kv2.2long). We currently do not know whether the deletion is due to a cloning artifact or is the result of endogenous RNA editing. However, immunolabeling using antibody K37 indicated that Kv2.2long is the predominant form expressed in cortical neurons, although we cannot exclude the possibility that K37 preferentially reacts with Kv2.2long.
Using specific immunoreagents, we found that Kv2.2long is highly colocalized in discrete surface clusters with Kv2.1 in the somata and proximal dendrites of rat cortical neurons. Clustering might be an intrinsic characteristic of Kv2.2long, like Kv2.1, because it can form clusters in HEK293 cells in the absence of Kv2.1. It has been previously shown that the clustering of Kv2.1 requires the internal sequence in the cytoplasmic C-terminal tail, the proximal restriction and clustering signal (15). The critical residues in the proximal restriction and clustering signal are well conserved in the C-terminal tail of Kv2.2 (data not shown) in both the shorter and longer forms. However, it has been shown that Kv2.2short does not form clusters in HEK293 cells (32). We speculate that the altered protein sequence in the C terminus of Kv2.2short results in a large structural rearrangement, which prevents it from forming clusters.
The substantial colocalization of Kv2.1 and Kv2.2long brought about the possibility that they are indeed expressed as heteromeric channels. Our co-immunoprecipitation assays and the experiments using the non-conducting mutant clearly demonstrated that Kv2.1 and Kv2.2long are physically associated in brain neurons in situ and form heteromeric channels when coexpressed. This is highly consistent with the findings of Blaine and Ribera (33) that the channel current of Xenopus Kv2.2 is significantly inhibited by a Xenopus Kv2.1 non-conducting mutant. The tetramerization (T1) domain of Kv1 channel subunits has been located in the cytoplasmic N-terminal tail (34). It has been recently reported that the N-terminal domain of Kv2.1 is also critical in forming a heterotetramer with Kv6.3, a modulatory subunit (35). In addition to the difference in the C-terminal tail, two amino acid residues at positions 161 and 162 in the N-terminal domain also differ between Kv2.2short (Asp and Gly) and Kv2.2long (Gln and Arg). This may account for the lack of heteromerization of Kv2.1 and Kv2.2short, although these residues are not close to the conserved histidine residue (at amino acid 109 in Kv2.2) that is critical for the binding of Kv6.3 to Kv2.1. However, it should be noted that Schnitzler and co-workers (35) also reported that the N-terminal tail of Kv2.2short can in fact interact with that of Kv2.1 in the yeast two-hybrid assay. There is also accumulating evidence that the N- and C-terminal tails of Kv2.1 subunits interact to change the biophysical and biochemical properties of the channel (36–38). Therefore, it is possible that the C termini of Kv2 subunits facilitate the formation of channel tetramers and that this may be disrupted in Kv2.2short.
One interesting aspect of the Kv2.1-Kv2.2 interaction is that the non-conducting mutant of Kv2.1 showed a greater inhibition on wild-type Kv2.2long than on Kv2.1 (p < 0.05; n = 6) (Fig. 5C). Because it has been elegantly shown that Kv2.1 and Kv9.3, another modulatory subunit, have a fixed stoichiometry of 3:1 (39), we speculate that Kv2.1 and Kv2.2long also have a similar stoichiometry, in which Kv2.1 is dominant in a tetramer. With wild-type Kv2.1 and its non-conducting mutant, their stoichiometry follows a binomial distribution, and thus, the degree of inhibition would be variable. However, if Kv2.1 and Kv2.2long preferably have a fixed stoichiometry of 3:1, the Kv2.1 non-conducting mutant becomes dominant in Kv2.2long/Kv2.1 mutant tetramers. Therefore, the mutant would exhibit a much greater inhibitory efficacy on the Kv2.2long channel current than on wild-type Kv2.1 (Fig. 7B). This is still a speculation and requires further research.
Acknowledgments
We thank Drs. Jeanne Nerbonne and James Trimmer for providing the cDNA constructs and for the inspiration to start this work. We also thank the members of the Misono laboratory, Scott Thompson's laboratory, Thomas Blanpied's laboratory, Matthew Trudeau's laboratory, Mankyo Chung's laboratory, and Andrea Meredith's laboratory for helpful discussions and other technical assistance.
This work was supported by the American Heart Association. The University of California Davis/National Institutes of Health NeuroMab Facility is supported by National Institutes of Health Grant U24NS050606.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- RBM
- rat brain membrane.
REFERENCES
- 1.Hille B. (2001) Ionic Channels of Excitable Membranes, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 2.Trimmer J. S., Rhodes K. J. (2001) in Potassium Channels in Cardiovascular Biology (Archer S. L., Rusch N. J. eds) pp. 163–175, Plenum Press, New York [Google Scholar]
- 3.Yu F. H., Yarov-Yarovoy V., Gutman G. A., Catterall W. A. (2005) Pharmacol. Rev. 57, 387–395 [DOI] [PubMed] [Google Scholar]
- 4.Isacoff E. Y., Jan Y. N., Jan L. Y. (1990) Nature 345, 530–534 [DOI] [PubMed] [Google Scholar]
- 5.Ruppersberg J. P., Schroter K. H., Sakmann B., Stocker M., Sewing S., Pongs O. (1990) Nature 345, 535–537 [DOI] [PubMed] [Google Scholar]
- 6.Christie M. J., North R. A., Osborne P. B., Douglass J., Adelman J. P. (1990) Neuron 4, 405–411 [DOI] [PubMed] [Google Scholar]
- 7.Sheng M., Liao Y. J., Jan Y. N., Jan L. Y. (1993) Nature 365, 72–75 [DOI] [PubMed] [Google Scholar]
- 8.Frech G., VanDongen A. M., Schuster G., Brown A. M., Joho R. H. (1989) Nature 340, 642–645 [DOI] [PubMed] [Google Scholar]
- 9.Hwang P. M., Glatt C. E., Bredt D. S., Yellen G., Snyder S. H. (1992) Neuron 8, 473–481 [DOI] [PubMed] [Google Scholar]
- 10.Stocker M., Kerschensteiner D. (1998) Biochem. Biophys. Res. Commun. 248, 927–934 [DOI] [PubMed] [Google Scholar]
- 11.Post M. A., Kirsch G. E., Brown A. M. (1996) FEBS Lett. 399, 177–182 [DOI] [PubMed] [Google Scholar]
- 12.Kramer J. W., Post M. A., Brown A. M., Kirsch G. E. (1998) Am. J. Physiol. 274, C1501–C1510 [DOI] [PubMed] [Google Scholar]
- 13.Salinas M., de Weille J., Guillemare E., Lazdunski M., Hugnot J. P. (1997) J. Biol. Chem. 272, 8774–8780 [DOI] [PubMed] [Google Scholar]
- 14.Hwang P. M., Fotuhi M., Bredt D. S., Cunningham A. M., Snyder S. H. (1993) J. Neurosci. 13, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim S. T., Antonucci D. E., Scannevin R. H., Trimmer J. S. (2000) Neuron 25, 385–397 [DOI] [PubMed] [Google Scholar]
- 16.Guan D., Tkatch T., Surmeier D. J., Armstrong W. E., Foehring R. C. (2007) J. Physiol. 581, 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimmer J. S. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10764–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakoshi H., Trimmer J. S. (1999) J. Neurosci. 19, 1728–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malin S. A., Nerbonne J. M. (2002) J. Neurosci. 22, 10094–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misonou H., Menegola M., Buchwalder L., Park E. W., Meredith A., Rhodes K. J., Aldrich R. W., Trimmer J. S. (2006) J. Comp. Neurol. 496, 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misonou H., Mohapatra D. P., Park E. W., Leung V., Zhen D., Misonou K., Anderson A. E., Trimmer J. S. (2004) Nat. Neurosci. 7, 711–718 [DOI] [PubMed] [Google Scholar]
- 22.Johnston J., Griffin S. J., Baker C., Skrzypiec A., Chernova T., Forsythe I. D. (2008) J. Physiol. 586, 3493–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maletic-Savatic M., Lenn N. J., Trimmer J. S. (1995) J. Neurosci. 15, 3840–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., Lau A., Morris T. J., Guo L., Fordyce C. B., Stanley E. F. (2004) J. Neurosci. 24, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misonou H., Mohapatra D. P., Menegola M., Trimmer J. S. (2005) J. Neurosci. 25, 11184–11193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang P. M., Cunningham A. M., Peng Y. W., Snyder S. H. (1993) Neuroscience 55, 613–620 [DOI] [PubMed] [Google Scholar]
- 27.Rhodes K. J., Trimmer J. S. (2006) J. Neurosci. 26, 8017–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saper C. B. (2005) J. Comp. Neurol. 493, 477–478 [DOI] [PubMed] [Google Scholar]
- 29.Misonou H., Menegola M., Mohapatra D. P., Guy L. K., Park K. S., Trimmer J. S. (2006) J. Neurosci. 26, 13505–13514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park K. S., Mohapatra D. P., Misonou H., Trimmer J. S. (2006) Science 313, 976–979 [DOI] [PubMed] [Google Scholar]
- 31.O'Connell K. M., Tamkun M. M. (2005) J. Cell Sci. 118, 2155–2166 [DOI] [PubMed] [Google Scholar]
- 32.Mohapatra D. P., Trimmer J. S. (2006) J. Neurosci. 26, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaine J. T., Ribera A. B. (1998) J. Neurosci. 18, 9585–9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bixby K. A., Nanao M. H., Shen N. V., Kreusch A., Bellamy H., Pfaffinger P. J., Choe S. (1999) Nat. Struct. Biol. 6, 38–43 [DOI] [PubMed] [Google Scholar]
- 35.Mederos Y., Schnitzler M., Rinné S., Skrobek L., Renigunta V., Schlichthörl G., Derst C., Gudermann T., Daut J., Preisig-Müller R. (2009) J. Biol. Chem. 284, 4695–4704 [DOI] [PubMed] [Google Scholar]
- 36.Ju M., Stevens L., Leadbitter E., Wray D. (2003) J. Biol. Chem. 278, 12769–12778 [DOI] [PubMed] [Google Scholar]
- 37.Wray D. (2004) Eur. Biophys. J. 33, 194–200 [DOI] [PubMed] [Google Scholar]
- 38.Mohapatra D. P., Siino D. F., Trimmer J. S. (2008) J. Neurosci. 28, 4982–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerschensteiner D., Soto F., Stocker M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6160–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]