Abstract
Svp26 is a polytopic integral membrane protein found in the ER and early Golgi compartment. In the Δsvp26 cell, the Golgi mannosyltransferase Ktr3 remains in the ER. Here, we report that two other Golgi mannosyltransferases, Mnn2 and Mnn5 are also mislocalized and found in the ER in the absence of Svp26 and that localization of other mannosyltransferases including Mnn1 are not affected. Mnn2 and Mnn5 bind to Svp26 in vivo as Ktr3 does. Using an in vitro budding assay, the incorporation of Ktr3 and Mnn2 in the COPII vesicles is greatly stimulated by the presence of Svp26. As Svp26 itself is an efficient cargo, Svp26 is likely to support selective incorporation of a set of mannosyltransferases into COPII vesicles by working as their adaptor protein. The domain switching between Svp26-dependent Mnn2 or Ktr3 and Svp26-independent Mnn1 suggests that the lumenal domain of mannosyltransferases, but not the cytoplasmic or transmembrane domain, is responsible for recognition by Svp26.
Keywords: Endoplasmic Reticulum (ER), Golgi, Membrane Proteins, Membrane Trafficking, Yeast, COPII Coat, Saccharomyces Cerevisiae, Adaptor, Localization, Mannosyltransferase
Introduction
The Golgi apparatus is comprised of multiple compartments with different components and functions. These compartments are successively formed by the maturation of the earliest compartment derived from the endoplasmic reticulum (ER)2 through homotypic fusion of the COPII vesicles (1). The maturation of compartments is believed to occur by removing the components of the earlier compartments and acquiring those of the later compartments in a COPI vesicle-dependent manner (2, 3). The newly synthesized Golgi proteins are first translocated in the ER and then selectively incorporated in the COPII vesicles and transported to the earliest Golgi compartment or the cis-Golgi network. When the small G-protein Sar1 is activated by Sec12 on the ER, Sec23/Sec24 subunits are recruited to form the prebudding complex containing the selected cargo proteins (4). Successive binding of Sec13/Sec31 subunits induces ER membrane deformation, budding, and release of the COPII vesicles (5). The retrieval of the Golgi proteins from the compartment at a certain maturation stage is carried out by the COPI vesicles after activation of the G-protein Arf1 and recruitment of coatomer proteins, and the selected cargos are transported to the earlier compartments or to the ER (6, 7).
The first carbohydrates of glycoproteins are transferred from the dolichol intermediates to certain Asn or Ser/Thr residues of the precursor polypeptides in the ER, and then further glycosyl residues are added or modified after moving from the ER to the Golgi (8). The enzymatic specificity of the glycosyltransferases and their localization in the Golgi compartments correlate with the successive addition of sugar residues. Each Golgi enzyme is attributed to a certain specific compartment through the dynamic maturation process by the exit and entry of selected components in the COPI vesicles. Therefore, the selective incorporation of the cargo proteins in the COPII and COPI vesicles should be very important for maintaining the specific protein composition of Golgi compartments. The motif sequences such as LXX(L/M)E for COPII selection (9, 10) or K(X)KXX for COPI selection (11, 12) are found to interact directly with the coat subunits when the motif-containing proteins are incorporated into the vesicles. In addition to the direct selective interaction with the coat subunits (13), incorporation in the vesicles via interaction with adaptor components has been reported (14). A peripheral membrane protein, Vps74, was found to bind to the N-terminal cytoplasmic domain of a subset of mannosyltransferases and is required to maintain their Golgi localization (15, 16). However, we have only limited knowledge about the selective transport mechanism of the Golgi glycosyltransferases.
Svp26 was discovered as a novel function-unknown protein in our global inspection of membrane proteins in the early Golgi compartment (17). Svp26 is an integral membrane protein with four predicted transmembrane segments. Although Svp26 is dispensable for viability of the yeast, we found abnormal hypermannosylation of N-glycosyl chains and mislocalization of Golgi mannosyltransferase Ktr3 to the ER in the Δsvp26 disruptant cell. Immunoprecipitation from the cleared lysate using 1% Triton X-100 indicated specific binding between Ktr3 and Svp26. These results suggest that Svp26 functions to ensure the correct localization of Ktr3 in the Golgi compartments.
We examined whether other mannosyltransferases depend on Svp26 to localize in the Golgi compartments. In this paper, we report that several mannosyltransferases behave as Ktr3 does, and Svp26 helps the selective incorporation of these mannosyltransferases into the COPII vesicles. The lumenal domains of the cargo are responsible for Svp26 dependence.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, Media, and Reagents
Saccharomyces cerevisiae strains used in this study are listed in Table 1. All strains, except for YNY872, which is a derivative of RSY255 (MATα ura3-52 leu2-3, -112, a gift from Dr. Randy Schekman), are based on KA31a, with genes disrupted or epitope tagged by homologous recombination.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype and plasmid | Origin |
|---|---|---|
| KA31a | MATa Δhis3 Δleu2 Δtrp1 Δura3 | Laboratory stock |
| HIY22 | MATa Δhis3 Δleu2 Δtrp1 Δura3 Δsvp26::kanMX4 | Laboratory stock |
| YNY639 | MATa OCH1-3HA::HIS3 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY646 | MATa OCH1-3HA::HIS3 Δsvp26::kanMX4 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY657 | MATa MNN1-3HA::LEU2 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY667 | MATa MNN1-3HA::LEU2 Δsvp26::kanMX4 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY703 | MATa MNN5-3HA::HIS3 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY705 | MATa MNN2-3HA::LEU2 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY707 | MATa MNN5-3HA::HIS3 Δsvp26::kanMX4 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY708 | MATa MNN5-3HA::HIS3 Δsvp26::kanMX4 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY729 | MATa MNN2-3HA::LEU2 Δsvp26::kanMX4 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY741 | As YNY667, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY745 | As YNY729, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY760 | MATa KTR3-3HA::LEU2 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY762 | MATa KTR3-3HA::LEU2 Δsvp26::kanMX4 Δhis3 Δleu2 Δtrp1 Δura3 | This study |
| YNY791 | As HIY22, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY793 | As YNY762, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY872 | MATa 3HA-SVP26::URA3 Δsvp26::kanMX4 leu2-3, -112 ura3-52 | This study |
| YNY924 | MATa MNN2-3HA::LEU2 (multiple copies at LEU2 locus) MNN2-3HA::LEU2 (single copy at MNN2 locus) Δhis3 Δtrp1 Δura3 | This study |
| YNY933 | As YNY646, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY934 | As YNY648, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY935 | As YNY707, pHI130 (SVP26-FLAG TRP1 CEN) | This study |
| YNY976 | MATa KTR3 (N+TM)-KTR3 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
| YNY977 | MATa MNN1 (N+TM)-MNN1 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
| YNY980 | MATa KTR3 (N+TM)-MNN1 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
| YNY983 | MATa MNN1 (N+TM)-KTR3 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
| YNY985 | MATa MNN2 (N+TM)-MNN1 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
| YNY986 | MATa MNN2 (N+TM)-MNN2 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
| YNY987 | MATa MNN1 (N+TM)-MNN2 (lumen)-3HA::URA3 Δsvp26::LEU2 Δhis3 Δleu2 Δtrp1 ura3-52::kanMX4 | This study |
For tagging mannosyltransferases with three copies of the HA epitope at their C termini, an appropriate DNA fragment of the 3′-region of each mannosyltransferase was amplified by PCR and cloned in pYN497 (HIS3 marker) or pYN503 (LEU2 marker). These plasmids carry a coding sequence for triple HA followed by a TDH3 terminator. The sequence of primers used is available upon request. The resulting plasmids were linearized by cutting at a unique restriction site located within a cloned region of each open reading frame and used for the transformation of yeast to obtain desired strains with chromosomally tagged genes by homologous recombination. The N-terminal triple HA-tagged SVP26 construct was inserted under the YPT1 promoter on the integration plasmid pYN570 to create pYN572. This plasmid was used to generate the strain YNY872 that was used in the experiment (see Fig. 5A).
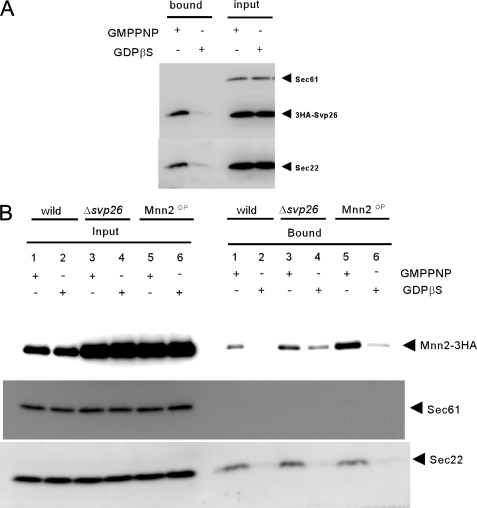
FIGURE 5.
Isolation of the prebudding complex. The ER-enriched membrane fractions prepared from the cells expressing 3HA-SVP26 were incubated with GST-Sar1 and the Sec23-Sec24 complex in the presence of either nonhydrolyzable GTP analog GMPPNP or GDP analog GDPβS as described (4). Membranes were solubilized with digitonin, and proteins bound to GST-Sar1 were collected on the glutathione-Sepharose beads. The input lanes contain 1.25% of the solubilized membranes, and the bound lanes contain the whole materials eluted from the glutathione beads. Sec22 and Sec61 were monitored as a positive and negative control of the experiment, respectively.
YNY924 used in the experiments (see Fig. 4), in which MNN2-3HA is overexpressed from the chromosome, was constructed as described below. A DNA fragment containing a promoter region and the open reading frame of MNN2 was amplified by PCR and then cloned in pYN503 to create pYN586 (MNN2-3HA). This plasmid was linearized by cutting at a unique restriction site within the URA3 region of the vector and used for the homologous insertion at the chromosomal URA3 gene of the yeast strains in which the chromosomal MNN2 is replaced by MNN2-3HA construct. Ten to twelve colonies that grew on an SD-Ura plate were picked, and one that accumulated an appropriate level of Mnn2-3HA in the ER-enriched membrane fraction was selected by Western blotting for further study.
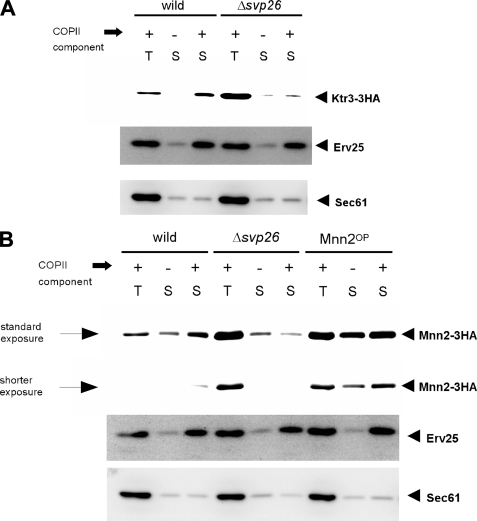
FIGURE 4.
In vitro COPII budding assay using either wild-type or Δsvp26 membranes. A, the ER-enriched membrane fractions prepared from the indicated strains were incubated either in the presence (+) or absence (-) of purified COPII coat components and the incorporation of Ktr3-3HA into COPII vesicles was analyzed by immunoblotting. T contains 2.5% of the total reaction mixture, and S contains 75% of the total COPII vesicle fraction, both of which were collected after the reaction. Erv25 and Sec61 were monitored as a positive and negative control of the experiment, respectively. B, incorporation of Mnn2-3HA into COPII vesicles was analyzed as in A. An assay using microsomal membranes prepared from the Mnn2OP cells were also performed. For detection of Mnn2-3HA, in addition to the standard exposure, a shorter exposure of the same blot is also shown to indicate the differences in intensity more clearly.
To create strains expressing chimeras that were used in Figs. 6 and 7, a BglII site was generated at the junction between the transmembrane and lumenal domains by PCR and used to exchange the DNA fragments to create plasmids that express the HA-tagged chimeric mannosyltransferases. The BglII site introduces additional amino acids Arg-Ser at the junction in chimeras, MNN1-3HA and MNN2-3HA. KTR3-3HA originally has Arg-Ser at this position. In all cases, changes were confirmed by sequencing. The resulting plasmids were linearized and integrated at the URA3 locus as described above, and the colonies with the expression level of chimera constructs comparable to the wild-type genes were selected by Western blotting.
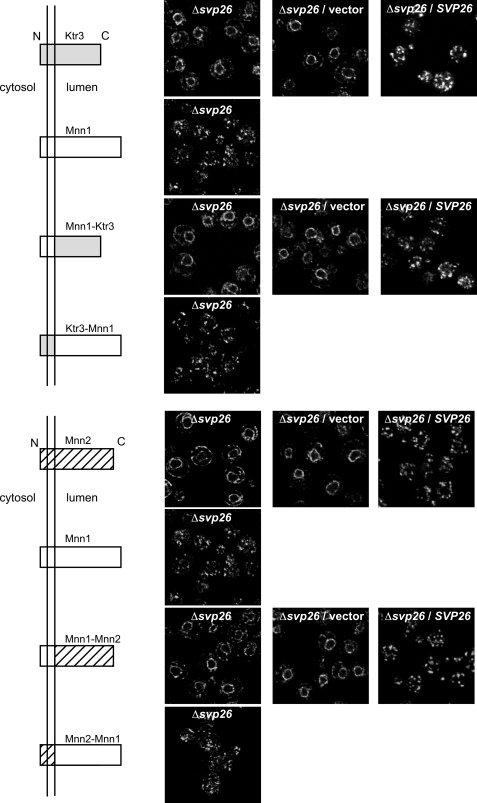
FIGURE 6.
Localization of the chimera mannosyltransferases in the presence or absence of Svp26. Top, localization of the chimera proteins composed of Ktr3 and Mnn1. In the ΔSVP26 strain, the chimera with the lumenal domain of Ktr3, but not the chimera with the lumenal domain of Mnn1, showed mislocalization to the ER. Introduction of the SVP26 gene on a CEN plasmid recovered the normal Golgi distribution of the Mnn1-Ktr3 chimera. Bottom, localization of the chimera proteins composed of Mnn2 and Mnn1. Similarly to the results at the top, the chimera with the lumenal domain of Mnn2, but not the chimera with the lumenal domain of Mnn1, showed mislocalization to the ER. N and C indicate the N terminus and C terminus of the chimeras, respectively.
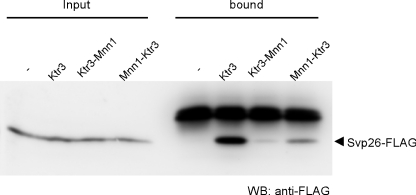
FIGURE 7.
Interaction of Svp26 with the chimeras between Ktr3 and Mnn1. Co-immunoprecipitation of Svp26 with wild-type Ktr3, Ktr3-Mnn1, and Mnn1-Ktr3 was examined as in Fig. 3. HA-tagged chimeras were immunoprecipitated from digitonin-solubilized cell lysate with anti-HA mAb, and co-precipitated Svp26-FLAG was detected by immunoblotting with anti-FLAG mAb. WB, Western blotting.
Yeast cells were grown in YPD (1% Bacto yeast extract (BD Biosciences, Franklin Lakes, NJ), 2% Bacto peptone (BD Biosciences), and 2% glucose) or SD (0.17% yeast nitrogen base without amino acids (BD Biosciences), 0.5% ammonium sulfate, 2% glucose, and appropriate supplements) medium at 30 °C unless other temperatures were indicated. YPD was used as a growth medium. Solid media were made with 1.5% agar. Escherichia coli DH5a (F−, supE44 ΔlacU169 ϕ80lacZDM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) was used in plasmid propagation. E. coli was grown in an LB (1% Bacto tryptone (BD Biosciences), 0.5% Bacto yeast extract (BD Biosciences) and 0.5% NaCl) medium. Digitonin was purchased from Wako Pure Chemical Industries (Japan).
Antibodies and Immunoblotting
Antisera against Sec22, Sec61, and Erv25 were kindly provided by Dr. Dieter Gallwitz (Max Planck Institute, Göttingen, Germany), Dr. Randy Schekman (The University of California, Berkeley) and Dr. Charles Barlowe (Dartmouth Medical School), respectively. Anti-myc (9E10, Berkeley antibody), anti-HA (12CA5, Roche Diagnostics) and anti-FLAG (M2, Sigma-Aldrich) monoclonal antibodies were also purchased. Rabbit polyclonal antibody to Mnn9 was described previously (18). For Western blotting, these antisera were used at a dilution of 1:1000. After incubation with peroxidase-conjugated secondary antibodies (KPL), detection was performed using enhanced chemiluminescence (Thermo Scientific).
Indirect Immunofluorescence
The localizations of HA-tagged mannosyltransferases and Mnn9 were observed by indirect immunofluorescence as described previously (19). Anti-HA mouse monoclonal antibody or rabbit anti-Mnn9 antibodies diluted to 1:50 were used as primary antibodies. Alexa 488-conjugated goat antibody to mouse immunoglobulin G (Molecular Probes) or Alexa 488-conjugated goat antibody to rabbit immunoglobulin G (Molecular Probes) were both diluted to 1:250 and used as secondary antibodies.
Sucrose Density Gradient Fractionation
Cells were converted to spheroplasts, suspended in an ice-cold 1× JR lysis buffer (20 mm HEPES, 50 mm potassium acetate, 0.2 m sorbitol, 2 mm EDTA, pH 7.4) (20) containing protease inhibitors (1 μg/ml each of chymostatin, aprotinin, leupeptin, pepstatin A, antipain, 1 mm benzamidine, and 1 mm phenylmethylsulfonyl fluoride), and lysed by 10 strokes in a Dounce homogenizer. The lysate was centrifuged at 400 × g for 3 min to remove unlysed cells. 0.2 ml of the supernatant was loaded onto a sucrose step gradient, which was generated using the following steps (all sucrose solutions were made (w/v, %) in 20 mm HEPES-KOH, pH 7.4, 50 mm potassium acetate, 2 mm EDTA): 0.25 ml 60%, 0.5 ml 50%, 1 ml 46%, and 0.25 ml 18% sucrose. After 6 h of centrifugation in a Beckman TLS55 rotor at 100,000 × g, six fractions of 0.35 ml were sequentially collected from the top of the gradient. Aliquots of each fraction were mixed with SDS sample buffer, and proteins were resolved by SDS-PAGE and detected by immunoblotting using anti-Mnn9, anti-Sec61, and anti-HA antibodies. Enhanced chemiluminescence signals were captured by an image analyzer equipped with a cooled charge-coupled device camera (LAS-1000plus; Fuji Film, Tokyo, Japan), and digital images were quantified using ImageJ software and graphed in Microsoft Excel.
In Vitro COPII Vesicle Budding Assay and Prebudding Complex Isolation
Purification of COPII coat components Sar1, Sec23/24, and Sec13/31, and the vesicle budding assay were performed as described (20). Microsomal membranes were prepared from the SVP26 or Δsvp26 cells expressing either KTR3-3HA or MNN2-3HA. Microsomal membranes were washed once with 0.5 m NaCl in B88 and then two times with B88 containing protease inhibitors and incubated for 30 min at 25 °C in the presence or absence of the purified coat components. After the incubation, vesicles were separated from the donor membranes by centrifugation at 10,000 × g for 5 min. The supernatant was further centrifuged at 100,000 × g for 1 h to collect the COPII vesicles. The pellets were analyzed by SDS-PAGE and immunoblotting.
Isolation of the prebudding complex was performed as described (4). Microsomal membranes (100 μg protein) prepared from cells expressing 3HA-SVP26 were washed with 0.5 m NaCl as above and were then incubated for 20 min at 25 °C with purified GST-Sar1 (3 μg) and Sec23/24 (5 μg) in the presence of 0.1 mm guanine nucleotide analog (GMPPNP or GDPβS) and an ATP regeneration system in a total reaction volume of 200 μl. The membranes were sedimented (12,000 × g, 20 min) through 0.1 m sucrose/B88-8 (B88, pH 8.0), and the pellet was solubilized with 1% digitonin in B88-8 for 10 min at 25 °C. Insoluble material was removed by centrifugation (12,000 × g, 10 min), and the supernatant was incubated with glutathione-Sepharose (GE Healthcare) for 40 min at 4 °C under gentle rotation. The beads were washed with 1% digitonin in B88-8, and the bound material was eluted by boiling for 1 min in SDS sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting.
Immunoprecipitation
Cells were grown at 30 °C in the YPD medium to an A600 nm = 1.0, and 9 optical density units of cells were collected and washed once with water. Cells were then suspended in B88 (20 mm HEPES, 150 mm potassium acetate, 5 mm magnesium acetate, 250 mm sorbitol, pH 6.8) containing protease inhibitors (as above) and disrupted by rigorously agitating with glass beads using a Multi-beads shocker (Yasui Kikai, Japan) three times for 1 min with 1-min intervals at 4 °C between each burst. Unbroken cells were removed by centrifugation at 400 × g for 5 min. The cleared lysate was mixed with one-ninth the volume of 10% (w/w) digitonin (final 1%) and kept on ice for 10 min. After centrifugation at 10,000 × g for 5 min, the supernatant was mixed with anti-HA mAb and incubated at 4 °C for 60 min with gentle rotation. Protein A-Sepharose beads (GE Healthcare) washed three times with B88 containing 0.2% digitonin were added, and the incubation was continued at 4 °C overnight. The beads were washed five times with B88 containing 0.5% digitonin and then boiled in SDS sample buffer for 1 min to obtain the sample. The sample was subjected to SDS-PAGE, and the indicated proteins were detected by Western blotting.
RESULTS
Golgi Localization of Mannosyltransferases Mnn2 and Mnn5 Depends on Svp26
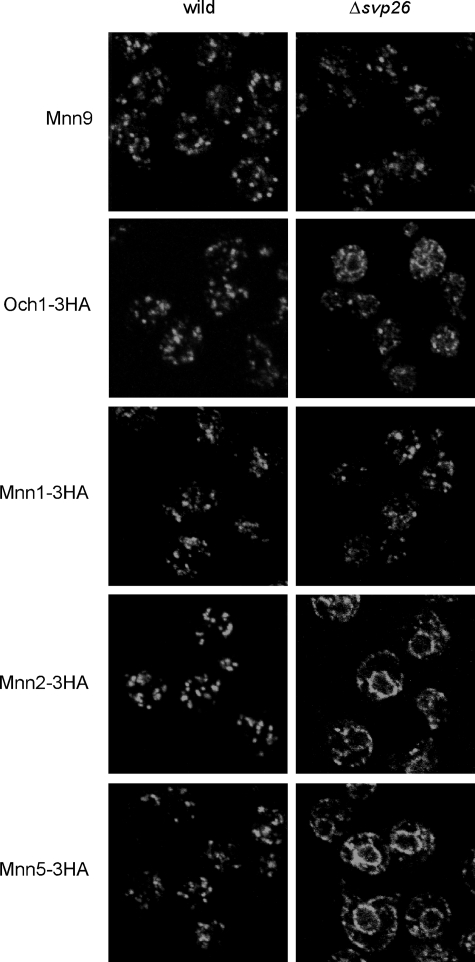
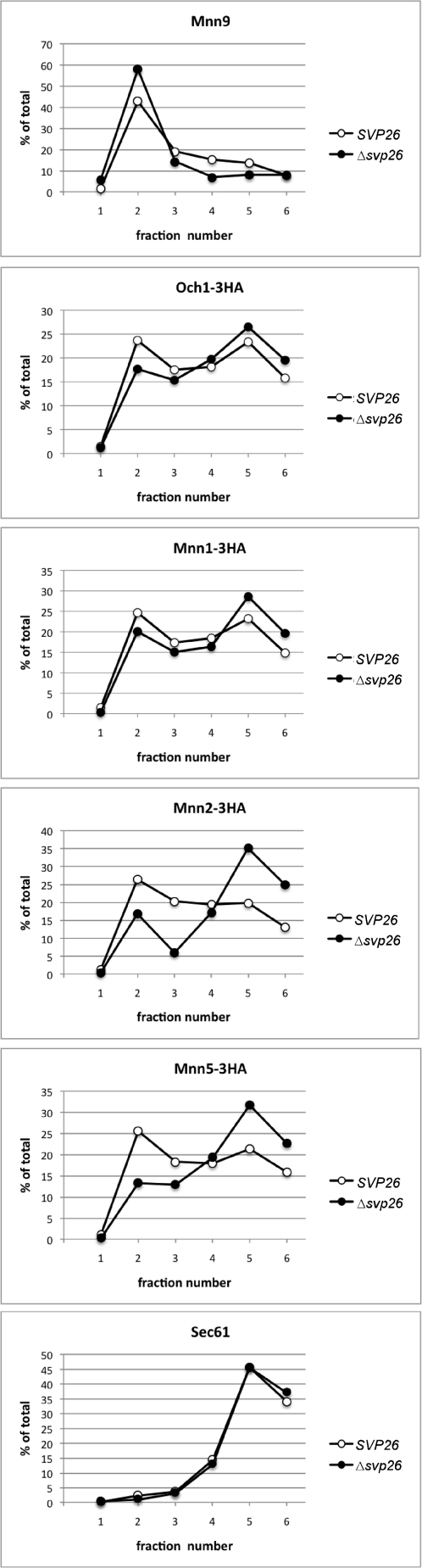
The most significant phenotypes of Δsvp26 cells found previously were hypermannosylation of N-glycosyl chains on several glycoproteins including invertase and mislocalization of Ktr3 from the Golgi to ER (17). As Ktr3 is an O-mannosyltransferase (21), we examined whether the Golgi localization of N-mannosyltransferases depends on the presence of Svp26, which may cause excess N-mannosylation. All Golgi N-mannosyltransferases are type II membrane proteins (8), but their molecular masses are divergent, and amino acid sequences have low homology. We observed the localization of tagged proteins encoded by the modified structural genes that were integrated in the yeast chromosome of the wild-type or Δsvp26 cells by immunofluorescence microscopy (Fig. 1). In both the Δsvp26 cells and wild-type cells, the localization of Mnn1 and Mnn9 were punctate, as reported previously (18, 22, 23), which is a typical pattern of Golgi-localized proteins. The N-mannosyltransferases Mnn2 and Mnn5 had double ring signals in most of the Δsvp26 cells, as we reported previously for Ktr3, which indicates mislocalization from the Golgi to ER (17). The signal of the early Golgi mannosyltransferase Och1, which transfers the first α-1,6-mannosyl residue to the core N-glycosyl chain (24), was found predominantly in Golgi punctae, but was also observed weakly in ER rings in approximately half of the Δsvp26 cells. The distribution of fluorescent signals in the intracellular compartments replicated the distribution of the enzymes in the sucrose density gradient fractionation (Fig. 2). Mnn9 was most abundant in fraction 2 and showed very similar distribution in both the wild-type and Δsvp26 cells. This suggests that proteins in the early Golgi compartments are recovered in fraction 2 of the wild-type and Δsvp26 cells. Och1, Mnn1, Mnn2, and Mnn5 are widely distributed with two peaks at fractions 2 and 5, which likely corresponds to their presence in different Golgi compartments. The distribution of Och1 and Mnn1 had similar overall distributions in the wild-type and Δsvp26 cells, which suggests that Och1 and Mnn1 are also in other Golgi compartments than Mnn9-containing compartments independently of Svp26. As shown by the distribution of Sec61, proteins in the ER are recovered mainly in the fraction 5. Considerable amounts of Mnn2 and Mnn5 in fraction 2 in the wild type were observed in fraction 5 in Δsvp26, which, based on the distribution of Mnn9 and Sec61, correlates well with the microscopy images of the ER ring localization of these proteins in Δsvp26 cells. These results indicate a clear difference in the dependence of N-mannosyltransferases on Svp26 for their Golgi localization.
FIGURE 1.
Localization of mannosyltransferases in the wild-type or Δsvp26 cells. Mannosyltransferases Mnn2, Mnn5, Mnn1, and Och1 were tagged with three tandem copies of HA at the C terminus by homologous recombination using integration plasmids either in the wild-type or Δsvp26 strains. HA-tagged mannosyltransferases were visualized by immunofluorescence straining with mouse anti-HA mAb. Mnn9 was visualized with rabbit anti-Mnn9 antiserum.
FIGURE 2.
Subcellular fractionation of mannosyltransferases in the wild-type or Δsvp26 strain. The cell lysate of strains used in Fig. 1 were fractionated on a sucrose density gradient composed of 0.25 ml 60%, 0.5 ml 50%, 1 ml 46%, and 0.25 ml 18% sucrose. After 6 h of centrifugation in a Beckman TLS55 rotor at 100,000 × g, six fractions of 0.35 ml were sequentially collected from the top of the gradient. Aliquots of each fraction were analyzed by SDS-PAGE followed by immunoblotting using anti-HA, anti-Mnn9, and anti-Sec61 antibodies. Mnn9 and Sec61 were detected on the same immunoblot used for Mnn5-3HA. The signal intensity of indicated proteins was quantified with ImageJ software and graphed using Microsoft Excel. The top of the gradient corresponds to fraction 1.
Svp26-dependent Golgi Proteins Mnn2 and Mnn5 Bind to Svp26
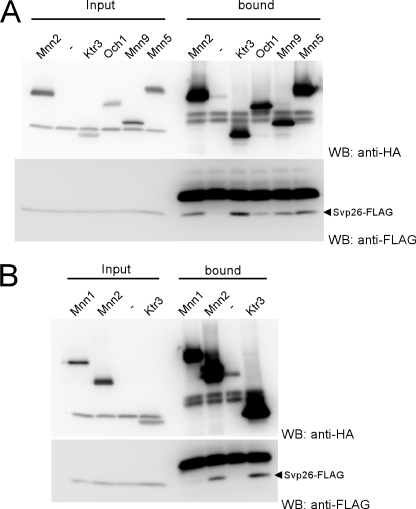
Mnn2 and Mnn5 are mislocalized and found in the ER of the Δsvp26 cells (Fig. 1). As we reported previously by co-immunoprecipitation experiments from the cleared lysate containing 1% Triton X-100, Ktr3 binds to Svp26 (17). We examined the interaction between Mnn2 or Mnn5 and Svp26 by co-immunoprecipitation in lysate containing 1% digitonin to observe any weak or transient interactions. A significant amount of Svp26 co-immunoprecipitated with Mnn2, although less than the amount that co-immunoprecipitated with Ktr3 (Fig. 3A). No Svp26 was detected in the immunoprecipitate of Mnn1, which does not depend on Svp26 for its Golgi localization (Fig. 3B). As shown in Figs. 1 and 2, Mnn5 is mislocalized to the ER in the Δsvp26 cells. The amount of Svp26 that was found in the Mnn2 immunoprecipitate was comparable to the amount seen in the immunoprecipitate of Mnn5 (Fig. 3A). Compared with the negative control sample, a small amount of Svp26 was detected in the immunoprecipitate of Och1 and Mnn9, which show little or no apparent mislocalization in the Δsvp26 cells (Fig. 3A).
FIGURE 3.
Physical interaction of Svp26 with mannosyltransferases. Co-precipitation of Svp26 with Mnn5, Mnn2, Mnn9, Ktr3, and Och1 (A) and lack of co-precipitation with Mnn1 (B). Mannosyltransferases were tagged at the C termini with three copies of the HA epitope on the chromosome of the Δsvp26 strain, which were then transformed with a CEN plasmid expressing the SVP26-FLAG construct from its own promoter. The total cell lysate prepared by agitation with glass beads was solubilized with 1% digitonin, and HA-tagged mannosyltransferases were precipitated with anti-HA mAb and co-precipitated Svp26-FLAG was detected with an anti-FLAG mAb. Each input lane contains 1% of the total material used for the precipitation. WB, Western blotting.
As we have shown for Ktr3, both Mnn2 and Mnn5 (which localize to the Golgi in an Svp26-dependent manner) show physical interaction with Svp26. The lower amount of Svp26 that was co-immunoprecipitated with Och1 compared with Mnn2 or Mnn5 may correspond to a weak mislocalization of Och1 to the ER in Δsvp26 cells compared with the stronger ER-mislocalization observed for Mnn2 or Mnn5. As will be noted in the “Discussion,” Mnn9 forms multiple-subunit glycosyltransferase complexes (18, 23), which may be related to the Svp26-independent Golgi localization of Mnn9.
Svp26 Stimulates in Vitro Incorporation of Ktr3 and Mnn2 Proteins in COPII Vesicles
There are two potential mechanisms to maintain the localization of a Golgi protein, i.e. to facilitate the arrival of the protein to, or to prevent its exit from, the Golgi. In our previous report, we favored the model that Svp26 interferes with the exit of Ktr3 from the Golgi, because our sec12 secretion block experiment indicated that Svp26 exclusively localizes in the Golgi (17). However, Svp26 was found in two peak fractions when the wild-type cell lysate was fractionated by sucrose density gradient centrifugation. Compared with the fractionation of marker proteins, Svp26 is found in the ER and early Golgi fractions (data not shown) (25). Although the signal of Svp26 in the unit area is not enough to be detected by our fluorescence microscopy, we conclude that a considerable amount of Svp26 is present in the ER. Therefore, we investigated whether Svp26 may help Ktr3 exit from the ER to the Golgi.
First, we examined whether the efficiency to incorporate the mannosyltransferases in COPII vesicles is affected by the presence or absence of Svp26. To test this, we used the in vitro COPII vesicle formation system with purified coat subunits and an ER-enriched membrane preparation (26). When the wild-type ER-enriched membrane fraction was used, Ktr3 and Mnn2 were released from the ER efficiently by the addition of purified coat subunits (Figs. 4, A and B). Without the addition of COPII coat components, much less Ktr3 and Mnn2 was released from the ER, compared with the positive control Erv25. The negative control Sec61 was not released in either condition. Therefore, we conclude that Ktr3 and Mnn2 were efficiently incorporated in the COPII vesicles. On the contrary, for unknown reasons, no stimulation in the release of Mnn1 and Mnn5 was found by the addition of COPII coat subunits, and we could not apply this test for these proteins (data not shown). It may be possible that other components that are lost in the purified in vitro budding system are necessary for their incorporation in vesicles.
The ER-enriched membrane fraction from the Δsvp26 cells contains significantly higher amounts of Ktr3 than the wild-type fraction. The ratio of Ktr3 signals in the Δsvp26 to wild-type cells in Fig. 4A is calculated to be 2.8. This is consistent with the microscopic observation that most of the Ktr3 signal is found not in the Golgi but in the ER, in Δsvp26 cells. Very little Ktr3 was released from this Δsvp26 ER fraction by the addition of purified COPII subunits (Fig. 4A). As Erv25 (27) is released in both the Δsvp26 and wild-type fractions, the formation and release of COPII vesicles occur normally. These results suggest that the presence of Svp26 strongly stimulates Ktr3 incorporation into COPII vesicles.
Similarly, incorporation of Mnn2 into the COPII vesicles was analyzed (Fig. 4B). Mnn2 was efficiently incorporated into the COPII vesicles when the wild-type ER-enriched membranes were used as donor membranes. In contrast, much less Mnn2 was incorporated into the COPII vesicles when ER-enriched membranes of the Δsvp26 strain were used. When the efficiency of incorporation (a ratio of the signal intensity in lane S to the signal intensity in lane T) in the presence of the COPII components using the wild-type membranes was set to 1, the efficiency using the Δsvp26 membrane was 0.20. To exclude the possibility that accumulation of Mnn2 in the Δsvp26 ER membranes and not the absence of Svp26 may indirectly interfere with the incorporation of Mnn2 into the COPII vesicles, we created an MNN2-3HA-overexpressing strain (Mnn2OP) by incorporating multiple copies of the MNN2-3HA construct into the genome of the wild-type strain. A ratio of the amount of Mnn2 in the ER-enriched membrane fraction from the Mnn2OP cells to that present in the Δsvp26 membrane fraction was 0.77. The efficiencies of incorporation of Mnn2 into COPII vesicles using membranes from Δsvp26 and Mnn2OP cells were calculated to be 0.20 and 0.82, respectively, when the efficiency of the wild type was set to 1. Similar results were obtained by independent experiments. We conclude that the incorporation of Mnn2 into COPII vesicles depends on the presence of Svp26 and is not strongly inhibited by accumulation of Mnn2 in Δsvp26 cells.
In the COPII vesicle formation system without adding the Sec13-Sec31 complex, a prebudding complex containing activated Sar1, the Sec23-Sec24 complex, and selected cargo proteins can be formed in the ER membrane. This complex can be isolated by the pull down of GST-Sar1 in the presence of nonhydrolyzable GTP analog GMPPNP but not in the presence of GDP analog GDPβS (4). We found that efficiency of the incorporation (bound/input signal ratio) of Svp26 in the prebudding complex was 1.44 when that of the positive control Sec22 was set to 1. No detectable amount of the negative control Sec61 was incorporated (Fig. 5A). Therefore, Svp26 is a protein that is more efficiently incorporated in the COPII prebudding complex than Sec22.
We sought to examine whether Mnn2 is also incorporated into this prebudding complex in an Svp26-dependent manner, and so we performed the assay using the microsomal membranes from the same strains used in Fig. 4B. The membranes of the wild type, Δsvp26 and Mnn2OP contain 6.2, 19.3, and 19.3 arbitrary units of Mnn2 signals, respectively (Fig. 5B). The ratios of Mnn2 signals that are recovered on the activated Sar1-bound beads are 1, 0.49, and 0.91, respectively, when signal of the wild type was set to 1. We conclude that Mnn2 is incorporated in the prebudding complex in an Svp26-dependent manner. Svp26 is likely to function as an adaptor that links Mnn2 to the coat components of the COPII vesicles; however, we cannot rule out the possibility of indirect interactions.
Svp26 Recognizes the Lumenal Domain of Mannosyltransferase
All Golgi mannosyltransferases identified so far in S. cerevisiae are type II membrane proteins (8). Their polypeptides can be divided into three parts, i.e. N-terminal cytoplasmic, short transmembrane, and large lumenal domains. The lumenal domain is believed to contribute to their enzyme activity (28). We sought to determine which domain is responsible for the differences in Ktr3 or Mnn2 (that depend on Svp26 for Golgi localization) andMnn1 (that does not need Svp26 to localize in the Golgi) by exchanging their domains (Fig. 6). Ktr3 has an Arg-Ser sequence that can be encoded by a BglII-cleavable nucleotide sequence AGATCT at the boundary of the transmembrane and lumenal domains. We introduced a BglII site at the corresponding region of the coding sequences of Mnn1 and Mnn2 and constructed chimera genes between MNN1 and either of KTR3 or MNN2. The genes were integrated at the URA3 locus on the chromosome, and chimera proteins were produced similarly in ample amounts to be detectable by immunoblotting and immunofluorescent staining. The chimera here will be referred to by combining the name of the mannosyltransferase that provides the N-terminal and lumenal domains. In the wild-type cell, all chimera proteins were observed in cytoplasmic punctae by immunofluorescent staining. This suggests that they are all localized to Golgi compartments. In the Δsvp26 cells, Mnn1-Ktr3 and Mnn1-Mnn2 chimeras are clearly mislocalized to the ER, whereas Ktr3-Mnn1 and Mnn2-Mnn1 are localized in the Golgi. The localization was rescued by the introduction of the wild-type SVP26 by a CEN plasmid. As the ER mislocalization of chimera proteins corresponds to the origin of the lumenal domain, this domain is responsible for the Svp26-dependent localization of these proteins.
By immunoprecipitation from the cleared lysate, Svp26 binds to Mnn1-Ktr3 but not to Ktr3-Mnn1. This fits the proposed model that the luminal domain of the Svp26-dependent protein (Ktr3) binds to Svp26, but the Svp26-independent luminal domain (Mnn1) does not (Fig. 7).
DISCUSSION
The Golgi O-mannosyltransferase Ktr3 is found in the ER in the Δsvp26 cells. Here, we examined whether the Golgi localization of the five N-mannosyltransferases, Och1, Mnn1, Mnn2, Mnn5, and Mnn9 are also Svp26-dependent. Mnn2 and Mnn5, like Ktr3, are found in the ER in the absence of Svp26. The Golgi localization of Mnn1 and Mnn9 is not affected. A small amount of Och1 was observed in the ER in half of the cells. We tested whether any of the five N-mannosyltransferases bind to Svp26 by immunoprecipitation from the cleared lysate containing 1% digitonin. Although co-immunoprecipitation from such lysate could not exclude indirect interaction because digitonin leaves many membrane protein complexes intact, Svp26 was not detected in the immunoprecipitate of Mnn1, the Golgi localization of which is independent of Svp26. On the other hand, a significant amount of Svp26 was co-immunoprecipitated with Mnn2 and Mnn5, which suggests that their Golgi localization and ability to interact with Svp26 are somehow related. These interactions are likely to be weaker than the Ktr3-Svp26 interaction, as no Svp26 interaction was detected when using Triton X-100 solubilized membranes. A small amount of Svp26 was detected in the immunoprecipitate of Och1 that shows weak ER mislocalization in the Δsvp26 disruptant. A small amount of Svp26 was also found in the immunoprecipitate of Mnn9, although the Golgi localization of Mnn9 is not affected by the loss of Svp26. Mnn9 exists in the stable complex with Van1 (M-pol I, V-complex) or with four proteins including Anp1 (M-pol II, A-complex) (18, 23). This complex formation may be more important for the Golgi localization of Mnn9 than its binding to Svp26, or there may be other proteins that bind to and assist the exit of other subunits of the complex, which function redundantly with Svp26 in facilitating the entry of the Mnn9-containing complex into COPII vesicles.
Our in vitro budding experiment suggested that the selective incorporation of Ktr3 and Mnn2 in the COPII vesicles is clearly stimulated by Svp26. At the very least, the incorporation of cargo into the COPII vesicle is a stage of Svp26 dependence for Ktr3 and Mnn2. The ER membrane fraction of the Δsvp26 disruptant contains a larger amount of these mannosyltransferases than the wild-type ER fraction (Fig. 5). It is likely that the efficiency of the exit from the ER is retarded in the Δsvp26 disruptant, and Ktr3 and Mnn2 accumulate in the ER. Svp26 is a good cargo for COPII vesicles and efficiently exits from the ER in the in vitro budding assay (data not shown) (25). Ktr3, Mnn2, and Mnn5 can bind to Svp26. It is most probable that the interaction with Svp26 physically stimulates their incorporation into the COPII vesicles and exit from the ER.
It is reported that certain sequence motifs of the cargo molecules are recognized by the Sec24 subunit of the COPII coat (29). These motifs are, for example, found in the cytoplasmic domain of soluble NSF attachment protein receptor (SNARE) proteins (9, 10). Also, because Svp26 is efficiently incorporated in the prebudding complex, it may have some sequence motif or structural characteristics that will be recognized by the coat subunits. The Svp26-dependent Golgi mannosyltransferases may therefore indirectly contact with the coat proteins through the direct interaction with Svp26. In other words, Svp26 mediates the interaction between the mannosyltransferase cargo and coat proteins. Therefore, we propose Svp26 acts as a sorting adaptor for COPII coats. Recently, it has been reported that the exit of the vacuolar alkaline phosphatase precursor proPho8 from the ER via COPII vesicle depends on Svp26/Erv26 (30–32). Investigation of this process revealed that Svp26 acts as the adaptor or receptor for the COPII coat. Also, another type II membrane protein, Gda1, which is a guanosine diphosphatase located in the Golgi, is thought to be dependent on Svp26 for its exit from the ER (31). It is possible that some other proteins with little or no affinity to the coat subunits utilize Svp26 as the adaptor for the COPII coat to efficiently exit the ER. The presence of adaptors for coat proteins is important for accepting a wide variety of proteins into the COPII coat.
Svp26 binds to proteins with different molecular masses and with little sequence homology and stimulates the exit of the interacting proteins from the ER. The nature of the interaction between Svp26 and its interacting proteins is largely unknown. However, by the domain exchanging of Svp26-dependent Ktr3 or Mnn2 and Svp26-independent Mnn1, we found the lumenal domains are responsible for the dependence on Svp26 for ER exit. Recently, it has been reported that the lumenal domain of proPho8 interacts with Svp26 (32). It may be a common feature that Svp26 recognizes the lumenal domains of its interacting proteins.
As the lumenal domain of mannosyltransferase is believed to function as the catalytic domain of the enzyme (28), it is very important when and how the interaction with Svp26 ends after the mannosyltransferase arrives in the Golgi compartment and what effect the interaction with Svp26 may have on the enzyme activity. The N-glycosyl chains of invertase and the modified hen egg lysozyme accept hypermannosylation in the Δsvp26 disruptant (17), the mechanism of which is completely unknown. If Svp26 acts not only as the sorting adaptor for COPII coats but also as a negative regulator of the enzyme activity to control the degree of mannosylation in the early Golgi, the mannosyltransferases that arrive in the Golgi by a default transport pathway may cause the addition of more mannose in the Δsvp26 disruptant.
Even in the absence of Svp26, a small amount of Svp26-dependent mannosyltransferase is delivered in the Golgi by the “default” traffic pathway. This may mean that COPII vesicles nonselectively incorporate components in the ER. Alternatively, there may be a COPII-independent delivery of proteins from the ER to Golgi. A small amount of Mnn2 is released from the ER-enriched fraction even without the addition of purified COPII subunits for an unknown reason (Fig. 4B). This phenomenon is more significant in the Mnn2OP membrane. Interestingly, this release without the addition of the COPII component was significantly decreased in the Δsvp26 ER fraction that has similarly accumulated Mnn2. Therefore, the uncharacterized delivery mechanism, if it exists, seems to also be Svp26-dependent in the case of Mnn2.
Acknowledgments
We thank Drs. Dieter Gallwitz, Randy Schekman, and Charles Barlowe for antibodies; Drs. Hironori Inadome and Hiroyuki Adachi for helpful discussions; and Christopher J. Noakes and Dr. Keisuke Sato for critical reading of the manuscript.
This work was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (to Y. N. and K. Y.), a grant from the RIKEN Institute of Japan (to K. Y.), and a grant from the Noda Institute of Scientific Research (to Y. N. and K. Y.).
- ER
- endoplasmic reticulum
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- mAb
- monoclonal antibody
- GMPPNP
- guanosine 5′-(β,γ-imido)triphosphate
- GDPβS
- guanosine 5′-(β-thio)diphosphate.
REFERENCES
- 1.Pelham H. R. (1998) Trends Cell Biol. 8, 45–49 [DOI] [PubMed] [Google Scholar]
- 2.Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., Glick B. S. (2006) Nature 441, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 3.Matsuura-Tokita K., Takeuchi M., Ichihara A., Mikuriya K., Nakano A. (2006) Nature 441, 1007–1010 [DOI] [PubMed] [Google Scholar]
- 4.Kuehn M. J., Herrmann J. M., Schekman R. (1998) Nature 391, 187–190 [DOI] [PubMed] [Google Scholar]
- 5.Salama N. R., Yeung T., Schekman R. W. (1993) EMBO J. 12, 4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynor E. C., Graham T. R., Emr S. D. (1998) Biochim. Biophys. Acta. 1404, 33–51 [DOI] [PubMed] [Google Scholar]
- 7.Todorow Z., Spang A., Carmack E., Yates J., Schekman R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13643–13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean N. (1999) Biochim. Biophys. Acta. 1426, 309–322 [DOI] [PubMed] [Google Scholar]
- 9.Mossessova E., Bickford L. C., Goldberg J. (2003) Cell 114, 483–495 [DOI] [PubMed] [Google Scholar]
- 10.Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., Schekman R. (2003) Cell 114, 497–509 [DOI] [PubMed] [Google Scholar]
- 11.Cosson P., Letourneur F. (1994) Science 263, 1629–1631 [DOI] [PubMed] [Google Scholar]
- 12.Letourneur F., Gaynor E. C., Hennecke S., Démollière C., Duden R., Emr S. D., Riezman H., Cosson P. (1994) Cell 79, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 13.Springer S., Schekman R. (1998) Science 281, 698–700 [DOI] [PubMed] [Google Scholar]
- 14.Sato K., Nakano A. (2007) FEBS Lett. 581, 2076–2082 [DOI] [PubMed] [Google Scholar]
- 15.Schmitz K. R., Liu J., Li S., Setty T. G., Wood C. S., Burd C. G., Ferguson K. M. (2008) Dev. Cell 14, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu L., Tai W. C., Chen L., Banfield D. K. (2008) Science 321, 404–407 [DOI] [PubMed] [Google Scholar]
- 17.Inadome H., Noda Y., Adachi H., Yoda K. (2005) Mol. Cell Biol. 25, 7696–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto H., Yoda K. (1997) Biochem. Biophys. Res. Commun. 241, 682–686 [DOI] [PubMed] [Google Scholar]
- 19.Sato K., Noda Y., Yoda K. (2009) Mol. Biol. Cell 20, 4444–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimoni Y., Schekman R. (2002) Methods Enzymol. 351, 258–278 [DOI] [PubMed] [Google Scholar]
- 21.Lussier M., Sdicu A. M., Bussereau F., Jacquet M., Bussey H. (1997) J. Biol. Chem. 272, 15527–15531 [DOI] [PubMed] [Google Scholar]
- 22.Graham T. R., Seeger M., Payne G. S., MacKay V. L., Emr S. D. (1994) J. Cell Biol. 127, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jungmann J., Munro S. (1998) EMBO J. 17, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama K., Nagasu T., Shimma Y., Kuromitsu J., Jigami Y. (1992) EMBO J. 11, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bue C. A., Bentivoglio C. M., Barlowe C. (2006) Mol. Biol. Cell 17, 4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M. F., Ravazzola M., Amherdt M., Schekman R. (1994) Cell 77, 895–907 [DOI] [PubMed] [Google Scholar]
- 27.Belden W. J., Barlowe C. (1996) J. Biol. Chem. 271, 26939–26946 [DOI] [PubMed] [Google Scholar]
- 28.Wiggins C. A., Munro S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Votsmeier C., Gallwitz D. (2001) EMBO J. 20, 6742–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bue C. A., Barlowe C. (2009) J. Biol. Chem. 284, 24049–24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anand V. C., Daboussi L., Lorenz T. C., Payne G. S. (2009) Mol. Biol. Cell 20, 1592–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dancourt J., Barlowe C. (2009) Traffic 10, 1006–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]