Abstract
Pertussis toxin (PTX) is recognized as a specific tool that uncouples receptors from Gi and Go through ADP-ribosylation. During the study analyzing the effects of PTX on Ang II type 1 receptor (AT1R) function in cardiac fibroblasts, we found that PTX increases the number of AT1Rs and enhances AT1R-mediated response. Microarray analysis revealed that PTX increases the induction of interleukin (IL)-1β among cytokines. Inhibition of IL-1β suppressed the enhancement of AT1R-mediated response by PTX. PTX increased the expression of IL-1β and AT1R through NF-κB, and a small GTP-binding protein, Rac, mediated PTX-induced NF-κB activation through NADPH oxidase-dependent production of reactive oxygen species. PTX induced biphasic increases in Rac activity, and the Rac activation in a late but not an early phase was suppressed by IL-1β siRNA, suggesting that IL-1β-induced Rac activation contributes to the amplification of Rac-dependent signaling induced by PTX. Furthermore, inhibition of TLR4 (Toll-like receptor 4) abolished PTX-induced Rac activation and enhancement of AT1R function. However, ADP-ribosylation of Gi/Go by PTX was not affected by inhibition of TLR4. Thus, PTX binds to two receptors; one is TLR4, which activates Rac, and another is the binding site that is required for ADP-ribosylation of Gi/Go.
Keywords: Signal Transduction, Calcium, Cardiac Hypertrophy, G Protein-coupled Receptors (GPCR), Phosphodiesterases, Protein Kinase G (PKG)
Introduction
PTX,2 a major virulence factor of Gram-negative bacillus Bordetella pertussis, which causes whooping cough, is well established as a pharmacological tool for a specific inhibitor of Gi signaling. PTX is composed of A-protomer and B-oligomer, and A-protomer exerts ADP-ribosyltransferase activity on the α subunit of heterotrimeric Gi proteins (Gαi), leading to inhibition of receptor-G protein coupling (1, 2), whereas B-oligomer of PTX recognizes and binds carbohydrate-containing receptors that deliver A-protomer into the cytosol (3). However, several reports have demonstrated that PTX has additional effects, such as enhancement of immune responses (4–6), increase in adenosine A1 receptor density (7), and activation of tyrosine kinase, mitogen-activated protein kinase, and NF-κB (8–10). These effects of PTX are reported to be independent of Gi modification.
Angiotensin (Ang) II plays an important role in the regulation of hypertrophy and/or hyperplasia of cardiovascular cells (11–13). In cardiac fibroblasts, Ang II has been demonstrated to stimulate the processes related to extracellular matrix remodeling (14). The biological function of Ang II is mediated by Ang II receptors located on the plasma membrane. Two isoforms (type 1 (AT1) and type 2 (AT2)) of Ang II receptor have been identified, but most of the cardiovascular effects of Ang II are attributed to AT1R (15). AT1R belongs to the Gq-coupled receptor family. Stimulation of AT1R activates phospholipase C and increases [Ca2+]i through the production of inositol 1,4,5-trisphosphate, leading to the modulation of fibroblast activities, such as cell proliferation and extracellular matrix protein synthesis (16).
An increase in AT1R density is one of the features to enhance fibrogenic responses of the heart. For example, an increase in AT1R density has been reported in the heart after myocardial infarction (17, 18) and in hearts from biopsies from patients with spontaneous intracerebral hemorrhage (19). Several cytokines, such as tumor necrosis factor (TNF)-α and interleukin (IL)-1β, have been reported to up-regulate AT1R (17, 20). However, the molecular mechanism responsible for the increase in AT1R density is still unknown.
Many studies suggest that low concentration of ROS acts as a second messenger in the cardiovascular system (21, 22). Stimulation of IL-1β and TNF-α induces ROS production through NADPH oxidase activation (23). A small GTP-binding protein, Rac, regulates the activity of NADPH oxidase (24) and mediates IL-1β- or TNF-α-induced ROS production and NF-κB activation (25). We have previously reported that Rac mediates Ang II-stimulated ROS production through NADPH oxidase activation in cardiac myocytes and cardiac fibroblasts (26, 27). Overexpression of constitutively active Rac1 induces hypertrophic responses in isolated cardiomyocyte and dilated cardiomyopathy in vivo (28, 29). Although a high concentration of hydrogen peroxide (H2O2) is reported to decrease AT1R density (30), it is unknown whether production of low concentration of ROS via Rac-mediated NADPH oxidase activation participates in the receptor-stimulated increase in AT1R density of cardiac cells.
Toll-like receptors (TLRs) play a critical role in both innate and adaptive immunity (31). There are at least 10 TLRs identified so far in humans, which specifically recognize and bind to a variety of pathogenic factors, including lipopolysaccharide. The mouse heart expresses at least six receptors (TLR2, -3, -4, -5, -7, and -9), and the stimulation of these receptors induces activation of NF-κB. TLR2 and TLR4 have been extensively studied in the heart, and both receptors are in part responsible for cardiac dysfunction in certain pathological conditions (32). Recent studies have elucidated that PTX functions as a superior ligand for TLR4 (6, 10). Although stimulation of TLR4 results in production of proinflammatory cytokines, it has not been reported that PTX exerts some pharmacological action(s) through TLR4 in cardiovascular cells, and it is unknown whether PTX-induced ADP-ribosylation of Gi/Go requires TLR4-mediated entry into cells.
During the study of the role of Gi proteins in AT1R-mediated fibrotic responses using rat neonatal cardiac fibroblasts, we found that PTX enhances Ang II-induced increase in [Ca2+]i. Because we previously reported that the treatment with PTX increases Rac activity in rat neonatal cardiac myocytes (26), we hypothesized that Rac is implicated in PTX-induced enhancement of Ang II signaling in cardiac fibroblasts. In this study, we demonstrate that PTX B-oligomer induces Rac activation through a pathway independent of ADP-ribosylation of Gi/Go. PTX increases IL-1β induction through sequential activation of TLR4, Rac, NADPH oxidase, and NF-κB, which leads to AT1R up-regulation through amplification of Rac-dependent signaling in rat cardiac fibroblasts.
EXPERIMENTAL PROCEDURES
Materials, Recombinant Adenoviruses, and Culture of Cardiac Fibroblasts
PTX, simvastatin, and anti-Gαq/11 antibody were purchased from Calbiochem. Ang II was from Peptide Institute. Mastparan-7, ATP, wortmannin, and diphenyleneiodonium (DPI) were purchased from Sigma. Ro-106-9920 was from Tocris. Rat IL-1β and PTX B-oligomer were from Wako. Rabbit anti-rat IL-1β antibody and the rat IL-1β ELISA kit were from Endogen. Anti-Gα11, anti-PLCβ3, anti-IκBα, anti-p65, anti-RhoA, anti-rabbit IgG, and anti-mouse IgG antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). [125I]Ang II, [32P]NAD, and glutathione-Sepharose beads were from Amersham Biosciences. Anti-Rac1 and anti-Rap1 antibodies were from Transduction Laboratories. Anti-Ras antibody was from Upstate Biotechnology. Anti-phospho-Akt and anti-Akt antibodies were from Cell Signaling. Fura2/AM was from Dojindo. 2,7-dichlorofluorescein diacetate and Alexa Fluor 488 goat anti-rabbit antibody were from Molecular Probes. Collagenase and Fugene 6 were from Roche Applied Science. Dual luciferase reagents were from Promega. pNF-κB-Luc and pRL-SV40 were from Stratagene. The sequences coding the Rap1-binding domain of Ral-GDS, Rac-binding domain of p21-activated kinase, Rho-binding domain of rhotekin, or Ras-binding domain of Raf were cloned, sequenced, and ligated into pGEX-4T-1 to make glutathione S-transferase fusion protein constructs. Glutathione S-transferase fusion proteins were expressed at room temperature and purified using gluthathione-Sepharose as described (33). The cDNA encoding GRP1-PH was provided by Dr. Alexander Gray (University of Dundee, Scotland). Recombinant adenoviruses of GRK2 (G protein-coupled receptor kinase 2)-ct, RGS4 (regulator of G protein signaling 4), WT Gαi, Gαi-ct, IκBαm, GFP-fused WT Rac, GFP-fused constitutively active Rac (G12V), DN-Rac (T17N), DN-p47phox, and p115-RGS were produced as described previously (26, 34). Stealth siRNAs oligonucleotides for rat IL-1β, TLR4, and Rac1 were from Invitrogen. Sequences of stealth siRNA used were described in supplemental Table 1. Cardiac fibroblasts were prepared from ventricles of 1–2-day-old Sprague-Dawley rats, as described previously (27).
Quantification of Intracellular Ca2+ and ROS Concentration
[Ca2+]i was measured by the method described previously (35). Briefly, cells (5 × 104) were plated on a 3 × 10-mm microcoverglass (MATSUNAMI) and loaded with 1 μm fura-2/AM in the cultured medium at 37 °C for 30 min. Cells were washed with HEPES-buffered salt solution containing 107 mm NaCl, 6 mm KCl, 1.2 mm MgSO4, 0.5 mm EGTA, 20 mm HEPES (pH 7.4), and 11.5 mm glucose. Measurement of intracellular ROS concentration was performed in 2 mm Ca2+-containing HEPES-buffered salt solution with a fluorescent dye, 2,7-dichlorofluorescein diacetate, as described previously (27). Fluorescence images were recorded and analyzed with a video image analysis system (Aquacosmos, Hamamatsu Photonics). The peak changes (ΔF/F0) of dichlorofluorescein fluorescence intensity were identified as values obtained by subtracting the basal fluorescence intensity (F0) from the maximal intensity during a 15-min PTX treatment.
Measurement of IL-1β mRNA and Protein Expression
Expression of IL-1β mRNA and protein was measured by real time reverse transcription-PCR and ELISA, as described previously (36). For the preparation of real time reverse transcription-PCR analysis, cells (3 × 105) plated on 6-well dishes were treated with PTX for 24 h and lysed with 400 μl of RLT buffer (Qiagen). For ELISA, cells (1 × 105) on 12-well dishes were treated with PTX (100 ng/ml) in 500 μl of medium, and cells were then collected together with medium. After cells were homogenized with a 26-gauge syringe, 100 μl of supernatants were used. Assays were performed according to the manufacturer's instructions.
Microarray Analysis
Cells (1 × 106) plated on 35-mm dishes were treated with PTX for 24 h and lysed with 400 μl of RLT buffer. Total RNA was extracted with the RNeasy minikit (Qiagen) and RNase-free DNase set (Qiagen). Total RNA was converted to biotin-labeled cRNA, which was hybridized to the rat genome U34A GeneChip (Affymetrix) for 16–24 h at 45 °C. The hybridization signals on the microarray were scanned and computed at a target intensity of 500 by a GeneChip Scanner 3000 and GeneChip Operating Software (Affymetrix), respectively. The data analysis was performed as follows. At the first step, probe sets without expression in the fibroblasts, which were indicated as absent by absolute analysis in more than half of the replicates in both the control and PTX-treated groups, were eliminated from the data set. Then, if the difference in the mean signal intensity of a given probe set was equal to the cut-off (1.25-fold) or more between the control and PTX-treated groups and if its p value calculated by Student's t test was less than 0.05, that probe set was employed. At the last step, probe sets with an annotation “signal transduction” (GO:0007165) in the AmiGO data base (available on the World Wide Web) were extracted, using the NetAffx Gene Ontology Mining Tool (available on the World Wide Web).
Ang II Binding Assay
Measurement of Ang receptor binding was performed according to the previous report (15) with a slight modification. After various treatments for 24 h, cardiac fibroblasts were rinsed with 10 ml of ice-cold phosphate-buffered saline and mechanically detached in 1 ml of ice-cold lysis buffer containing 10 mm Tris, pH 7.4, 5 mm EDTA, 5 mm EGTA, 1 μg/ml benzamidine, 10 μg/ml soybean trypsin inhibitor (type II-S), and 5 μg/ml leupeptin. The cell lysate was centrifuged at 45,000 × g for 10 min at 4 °C. The pellet containing crude membrane fraction was resuspended in 1 ml of ice-cold lysis buffer with a Potter type homogenizer, frozen, and stored at −80 °C until use. After the concentration of membrane protein was determined, membrane protein (20 μg) was used for the binding studies. The membrane was incubated with 0.1 nm 125I-Ang II in 75 mm Tris, pH 7.4, 12.5 mm MgCl2, 2 mm EDTA, and increasing concentrations of unlabeled Ang II (0–14 nm) for 1 h at 25 °C. Nonspecific binding was determined in the presence of 1 μm unlabeled Ang II. The reaction mixture was filtered over Whatman GF/C filters. The filters were washed with ice-cold buffer containing 25 mm Tris, pH 7.4, and 1 mm MgCl2. The bound 125I-Ang II on the filters was measured with a γ-counter. The values of Kd and Bmax were calculated by Prism software (GraphPad Software, San Diego, CA).
Measurement of NF-κB Activity
After adenovirus was infected at 100 MOI for 2 h in serum-free medium, fibroblasts (3 × 105 cells) in a 24-well plate were transiently co-transfected with 0.45 μg of pNF-κB-Luc and 0.05 μg of pRL-SV40 control plasmid, using Fugene 6 (27). Luciferase activity was measured 48 h after transfection with dual luciferase reagents.
Measurement of Small GTPase Activities
Activation of small G proteins was determined as described previously (26). Activated Rac, Rho, Ras, and Rap1 were pulled down with 5 μg of glutathione S-transferase-fused Rac-interacting domain of p21-activated kinase (PAK-CRIB), Rho-binding domain of rhotekin (34), Ras-binding domain of Raf-1 (37), and Rap1-binding domain of Ral-GDS (38), respectively. Pulled-down small G proteins were detected with anti-Rac1, anti-RhoA, anti-Ras and anti-Rap1 antibodies. For knockdown of Rac1, cells were transfected with a mixture of Rac1 siRNAs (50 nm each) for 72 h.
Confocal Visualization of GFP-fused Proteins and NF-κB p65 Subunit
Cells (1 × 105) plated on glass bottom 35-mm dishes were infected for 24 h with GFP, GFP-Rac, GFP-constitutively active Rac, GFP fusion protein with PX domain of p40phox (p40phox-PX), and p40phox-PX (R105K). After the treatment with PTX (100 ng/ml) for 24 h, cells were fixed by 10% formaldehyde neutral buffer solution. For localization of NF-κB, cells were stained with anti-p65 antibody. Fluorescence images were measured at an excitation wavelength of 488 nm with a laser-scanning confocal imaging system (Carl Zeiss LSM510).
In Vitro PTX-catalyzed ADP-ribosylation Assay
In vitro ADP-ribosylation of Gαi proteins by PTX was performed as described previously (39) with a slight modification. Briefly, cardiac fibroblasts pretreated with or without 100 ng/ml PTX for 24 h were harvested with ice-cold lysis buffer containing 50 mm Tris (pH 7.5), 5 mm EDTA, 5 mm EGTA, 10 μg/ml benzamidine, 5 μg/ml aprotinin, and 5 μg/ml leupeptin. After centrifugation at 15,000 rpm for 10 min at 4 °C, the pellet was resuspended in lysis buffer. PTX was preactivated by incubation in the solution containing 50 mm Tris (pH 7.5), 5 mm ATP, 20 mm dithiothreitol, and 1 mg/ml bovine serum albumin for 30 min at 30 °C. Then activated PTX was added to the assay mixture, including 100 μg of the membrane, and incubated for 60 min at 30 °C. The final concentrations of all reagents in the assay mixture were as follows: 50 mm Tris (pH 7.5), 50 μm GDP, 10 mm thymidine, 5 μm NAD, 0.5 μm [32P]NAD, 20 μg/ml PTX, 0.2 mg/ml bovine serum albumin, 1 mm ATP, and 4 mm dithiothreitol. The reaction was stopped by the addition of an excessive amount of ice-cold 50 mm Tris (pH 7.5), and the samples were centrifuged at 15,000 rpm for 10 min at 4 °C. The pellet was solubilized in SDS sample buffer, boiled, and subjected to 12% SDS-PAGE. Radioactive bands were detected by filmless autoradiographic analysis (BAS2000 system, Fujifilm).
Statistical Analysis
The results are presented as mean ± S.E. from at least three independent experiments. The representative data of time course experiments were plotted from one of three similar experiments that were performed with more than 20 cells. The mean values were compared with control by one-way analysis followed by Dunnet's t test (for three or more groups) or Student's t test (for two groups).
RESULTS
PTX Enhances Ang II-induced Ca2+ Release through AT1R Up-regulation
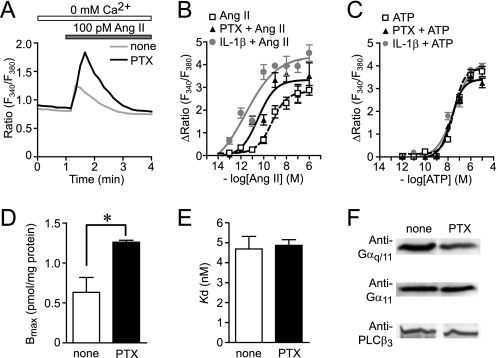
During the study of AT1R function in cardiac fibroblasts, we found that treatment with PTX enhances transient increase in [Ca2+]i induced by Ang II at low concentration in the absence of extracellular Ca2+ (Fig. 1A). The EC50 value of Ang II for the changes in [Ca2+]i increases was 464 ± 44 pm in control cells, whereas the EC50 value was decreased to 91 ± 33 pm in PTX-pretreated cells (Fig. 1B). However, the ATP-induced Ca2+ release was not affected by PTX (Fig. 1C). These results suggest that PTX selectively enhances Ca2+ response induced by AT1R stimulation. We also found that treatment with PTX for 24 h resulted in a 2-fold increase in maximal 125I-Ang II binding activity (Bmax) in comparison with PTX-untreated membrane (Fig. 1D). PTX increased AT1R density in a time-dependent manner, and more than 18 h was required for a 2-fold increase in AT1R density (supplemental Fig. 1). The PTX-induced increase in Bmax was completely suppressed by CV11974 (1 μm, AT1R-selective blocker) but not by PD123319 (1 μm, AT2R-selective blocker) (data not shown). The Kd value was not affected by PTX (Fig. 1E), indicating that the PTX-induced enhancement of AT1R function is not explained by structural changes in AT1R. It has been reported that the increased expression of Gαq/11 and PLCβ3 is involved in the enhancement of Ang II-induced Ca2+ responses in the ischemic heart (40, 41). However, PTX did not affect the expression levels of Gαq, Gα11, and PLCβ3 (Fig. 1F). These results suggest that the enhancement of Ang II-induced Ca2+ release in PTX-treated cells is due to AT1R up-regulation but not up-regulation of components of the Gαq-PLCβ pathway.
FIGURE 1.
PTX enhances Ca2+ responses by Ang II through AT1R up-regulation. A, average time courses of Ca2+ response induced by Ang receptor stimulation with Ang II (100 pm) in control and PTX-treated cells. B and C, peak increases in [Ca2+]i (ΔRatio) plotted against various concentrations of Ang II (B) and ATP (C) in control, PTX-treated, and IL-1β-treated cells. Cells were treated with PTX (100 ng/ml) or IL-1β (10 ng/ml) for 24 h before agonist stimulation. D and E, increases in AT1R density induced by PTX (100 ng/ml) for 24 h. The Bmax (D) and Kd (E) values for Ang II binding were calculated with GraphPad Prism software. F, effects of PTX on expression of Gαq/11 and PLCβ3. *, p < 0.05 versus PTX-untreated cells. Error bars, S.E.
Because commercially available PTX contaminates with other endotoxins, including lipopolysaccharide, it is possible that other endotoxins contribute to enhancement of AT1R function. Thus, we examined the effects of denatured PTX or PTX purchased from another manufacturer (Sigma) on AT1R functions. Pretreatment of PTX with heat significantly reduced the enhancement of Ang II-induced Ca2+ release induced by PTX (supplemental Fig. 1). In contrast, the Ang II-induced Ca2+ release was also enhanced by PTX purchased from Sigma as well as that induced by PTX from Calbiochem. These results suggest that PTX proteins per se induce AT1R up-regulation in cardiac fibroblasts.
IL-1β Production Induced by PTX Treatment
To examine whether PTX treatment induces production of a factor(s) that participates in up-regulation of AT1R, we performed microarray analysis of mRNAs from PTX-treated fibroblasts. For each gene, we calculated the average intensity in expression for both control and PTX-treated cells and plotted the ratio of these two induction values. Genes were chosen whose expression was at least 1.25-fold increased or decreased as compared with control cells. The probe sets of 405 genes showed significant changes by PTX treatment. Genes were then assigned to several groups according to their function, and we picked out 70 genes in the gene cluster that is termed “signal transduction” in the AmiGO data base (supplemental Table 2). PTX treatment selectively increased AT1R mRNA (Fig. 2A) but not other G protein-coupled receptors. Among genes increased by PTX treatment, IL-1α and IL-1β mRNAs showed a marked increase in expression (Fig. 2A). Real-time PCR confirmed the strong induction of IL-1β mRNA by PTX treatment (Fig. 2B). Although PTX is reported to increase IL-12 expression by inhibition of Gi signaling in T lymphocytes (42), PTX did not significantly increase mRNA expression of other cytokines (supplemental Table 1). Treatment with mastoparan-7 or the expression of WT Gαi or inhibitory polypeptides of Gi signaling (Gαi-ct, a polypeptide that specifically inhibits receptor-Gi protein coupling (39); RGS4, a GTPase-activating protein that specifically binds the GTP-bound form of Gαi and Gαq (43); and GRK2-ct, a Gβγ (βγ subunit of heterotrimeric G protein)-sequestering polypeptide (44)) did not increase IL-1β mRNA expression (Fig. 2B). We also confirmed that the expression of Gαi-ct did not enhance Ang II-induced Ca2+ release (data not shown), and the treatment with B-oligomer of PTX enhanced Ang II-induced Ca2+ release (Fig. 2C). Furthermore, ELISA revealed that the treatment with PTX actually increased the expression of IL-1β protein levels, whereas the expression of IL-1α protein was below the detection level in PTX-treated cardiac fibroblasts (Fig. 2D). These results suggest that PTX selectively induces IL-1β production, and Gi modification is not required for PTX-induced IL-1β production.
FIGURE 2.
Involvement of IL-1β production in PTX-induced enhancement of Ca2+ response by Ang II stimulation. A, effects of PTX on the expression of AT1R, IL-1α, and IL-1β mRNAs. After cells were treated with PTX (100 ng/ml) for 24 h, total RNA was extracted. The expression of mRNAs was determined with microarray analysis. ID numbers of primer probe sets are shown in parenthesis. B, effects of respective reagents on IL-1β mRNA expression in cardiac fibroblasts. Cells were treated with PTX (100 ng/ml) for 24 h, treated with mastoparan-7 (10 μm) for 12 h, or infected with LacZ, WT Gαi, Gαi-ct, RGS4, and GRK2-ct at 300 MOI for 48 h. The -fold increases were calculated by the values of untreated cells (none) set as 1. C, effects of B-oligomer of PTX on Ang II-induced Ca2+ releases. Cells were treated with B-oligomer (1 or 10 nm) for 24 h before Ca2+ measurement. D, time course of PTX-induced expression of IL-1β protein. E, effects of IL-1β siRNAs, DN-Rac, and wortmannin (WTM) on PTX-induced IL-1β production. Two different siRNAs were used. F and G, effects of IL-1β neutral antibody (F) or IL-1β siRNAs (G) on Ang II-induced Ca2+ responses in control, PTX-treated, or IL-1β-treated cells. Cells were treated with PTX (100 ng/ml) or IL-1β (1 ng/ml) for 24 h before Ang II (100 pm) stimulation with or without anti-IL-1β antibody (500 μg/ml). Cells were transfected with IL-1β siRNAs (100 nm) 48 h before PTX treatment. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus PTX-untreated, B-oligomer-untreated, control siRNA-treated, PTX-treated, or IL-1β-treated cells. Error bars, S.E.
IL-1β Mediates PTX-induced Enhancement of Ang II-induced Ca2+ Response
Because it has been reported that IL-1β increases AT1R density in cardiac fibroblasts (17, 45), the cells were treated with IL-1β. Treatment with IL-1β (10 ng/ml) enhanced Ang II-induced Ca2+ release (EC50 = 31 ± 26 pm) but not ATP-induced Ca2+ release, in rat cardiac fibroblasts (Fig. 1, B and C). These effects of IL-1β are similar to the effects of PTX treatment, and the enhancement by IL-1β seems to be consistent with the findings that PTX treatment increased the induction of IL-1β mRNA and protein. Thus, we examined whether PTX-induced IL-1β production participates in the enhancement of AT1R function. The PTX-induced IL-1β production was suppressed by the treatment with IL-1β siRNAs (Fig. 2E). The enhancement of Ang II-induced Ca2+ release by IL-1β treatment was almost completely suppressed by anti-IL-1β neutral antibody (Fig. 2F), indicating that the antibody sufficiently inhibits IL-1β-mediated responses. The enhancement of Ang II-induced Ca2+ release by PTX was also suppressed by anti-IL-1β antibody and IL-1β siRNAs (Fig. 2G), indicating that PTX-induced IL-1β secretion mediates the enhancement of Ang II-induced Ca2+ release.
Involvement of NF-κB in PTX-induced IL-1β Expression
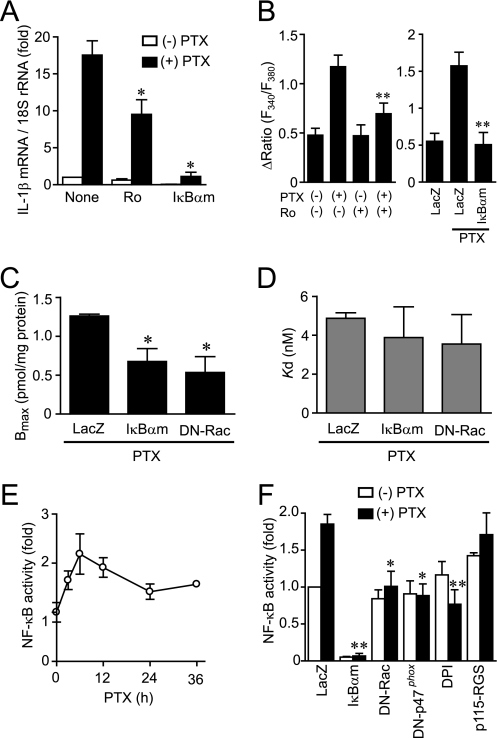
As the promoter regions of IL-1β and AT1R contain a putative NF-κB binding site (46–48), we next examined the involvement of NF-κB in PTX-induced IL-1β production. As shown in Fig. 3A, PTX-induced increase in IL-1β mRNA expression was suppressed by the treatment with Ro-106-9920, a selective inhibitor of IκB phosphorylation, and by the expression of a dominant negative IκB, IκBαm. Because Ro-106-9920 showed cytotoxic effects at higher concentration, we could not increase the concentration to observe complete inhibition of the IL-1β induction. The enhancement of AT1R function by PTX was suppressed by Ro-106-9920 and IκBαm (Fig. 3B), and the PTX-induced increase in AT1R density was suppressed by IκBαm (Fig. 3, C and D). Because an inhibition of NADPH oxidase activity suppresses NF-κB activation and IL-1β production induced by Gα13 activation (36), we next examined the involvement of NADPH oxidase. Treatment with PTX increased NF-κB-dependent luciferase activity (Fig. 3, E and F). This NF-κB activation was suppressed by the treatment with DPI or by the expression of dominant negative (DN)-Rac and DN-p47phox, both of which are essential for NADPH oxidase activation (24), but not by p115-RGS, a Gα13-inhibitory polypeptide (26). These results suggest that PTX induces NF-κB activation through Rac-NADPH oxidase pathway and that NF-κB mediates PTX-induced IL-1β production and AT1R up-regulation.
FIGURE 3.
Requirement of NF-κB for PTX-induced enhancement of Ca2+ response by Ang II stimulation. A, effects of Ro106-9920 and IκBαm on PTX-induced IL-1β mRNA expression. Cells were pretreated for 20 min with Ro106-9920 (1 μm) or infected with IκBαm (100 MOI) for 48 h before the treatment of PTX (100 ng/ml) for 24 h. B, effects of NF-κB inhibitors on Ang II-induced Ca2+ responses in PTX-treated cells. C and D, effects of IkBαm and DN-Rac on PTX-induced increase in AT1R density. Cells were infected with adenoviruses expressing IkBαm or DN-Rac 24 h before PTX treatment. AT1R density was determined with receptor binding assay. E, time course of PTX-induced changes in NF-κB-dependent luciferase activity. F, effects of IκBαm, DN-Rac, DN-p47phox, DPI, and p115-RGS on PTX-induced NF-κB activation. Cells were infected with LacZ, IκBαm, DN-Rac, DN-p47phox, or p115-RGS at 100 MOI for 48 h or pretreated with DPI (5 μm) for 20 min before the addition of PTX (100 ng/ml) for 6 h. *, p < 0.05; **, p < 0.01 versus PTX-untreated or LacZ-expressing cells. Error bars, S.E.
Rac Mediates PTX-induced IL-1β Production and AT1R Up-regulation
We have previously reported in rat neonatal cardiomyocytes that PTX increases basal Rac activity (26). Because the PTX-induced NF-κB activation and IL-1β production was suppressed by DN-Rac (Figs. 2 and 3), we next examined whether PTX increases Rac activity in cardiac fibroblasts. Rac was activated from 10 min after PTX treatment and still activated at 24 h after the treatment (Fig. 4A). We also found that PTX did not affect the activities of other small G proteins, Ras, Rap1, and RhoA (supplemental Fig. 2). It has been reported that phosphatidylinositol (PI) 3-kinase participates in PTX B-oligomer-induced antiapoptotic action against HIV-Tat infection in NK cells (49). We confirmed that PTX B-oligomer increased Rac activity in cardiac fibroblasts (supplemental Fig. 3). Pretreatment with wortmannin completely suppressed PTX-induced Rac activation (Fig. 4B). The activated Rac has been reported to translocate from cytosol to the plasma membrane through recognition of membrane phospholipids, such as PI 3-phosphate (PI-3-P), PI 4-phosphate, PI 5-phosphate, and PI 3,4,5-trisphosphate, through the carboxyl-terminal polybasic region of Rac (50–52). Confocal imaging revealed that PTX actually translocated GFP-fused WT Rac from cytosol to the plasma membrane, as observed with constitutively active Rac (Fig. 4C). Pretreatment with wortmannin inhibited PTX-induced membrane localization of Rac. To demonstrate the involvement of PI-3-P, p40phox-PX, a specific marker for PI-3-P, was expressed (53). Under the basal condition, p40phox-PX was predominantly localized in the PI-3-P-enriched early endosome and nucleus (Fig. 4D). Treatment with PTX for 10 min promoted the translocation of p40phox-PX from early endosome to the plasma membrane. However, PTX did not affect the localization of p40phox-PX (R105K), a mutant that cannot recognize PI-3-P. The localization of the PH domain of GRP1 (54), a marker for PI 3,4,5-trisphosphate, and that of the PH domain of PLCδ1, a marker for PI 4,5-bisphosphate, were not changed by PTX (data not shown). These results suggest that PTX-induced PI-3-P production through PI 3-kinase activation is required for translocation and activation of Rac.
FIGURE 4.
PTX induces Rac activation. A, time course of Rac activation induced by PTX (100 ng/ml). B, effects of wortmannin on PTX-induced Rac activation. Cells were pretreated with wortmannin (WTM; 100 nm) for 10 min before PTX stimulation. C, localization of GFP-fused wild type Rac and constitutively active Rac (CA-Rac) (G12V) with or without PTX stimulation. D, localization of GFP-fused PX domain of p40phox (p40phox-PX), and PI-3-P interaction-deficient mutant (p40phox-PX (R105K)) with or without PTX treatment. *, p < 0.05 versus PTX-untreated cells. Error bars, S.E.
Involvement of Rac in PTX-induced ROS Production
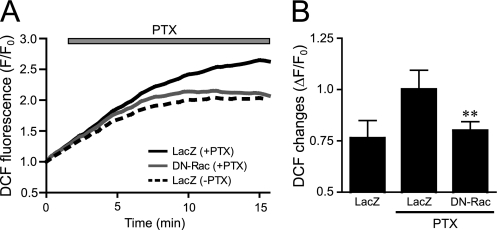
One of the targets for Rac is NADPH oxidase. Because the PTX-induced NF-κB activation was suppressed by DPI, DN-Rac, and DN-p47phox (Fig. 3F), Rac-mediated activation of NADPH oxidase may participate in PTX-induced NF-κB activation. We found that PTX gradually increased dichlorofluorescein fluorescence intensity, indicating ROS production in cardiac fibroblasts (Fig. 5). The expression of DN-Rac completely suppressed this ROS production, suggesting that PTX activates Rac and turns on a signaling cascade downstream of Rac.
FIGURE 5.
Effects of DN-Rac on PTX-induced ROS production. A and B, average changes (A) and peak increases (B) in PTX-induced F/F0 of dichlorofluorescein from time course experiments. The increases in PTX-induced fluorescence of dichlorofluorescein were calculated by the value of maximal fluorescence intensity (F) during 20 min of stimulation and initial value of fluorescence, F0. *, p < 0.05; **, p < 0.01 versus PTX-untreated or LacZ-expressing cells. Error bars, S.E.
Essential Role of Rac in PTX-induced AT1R Up-regulation
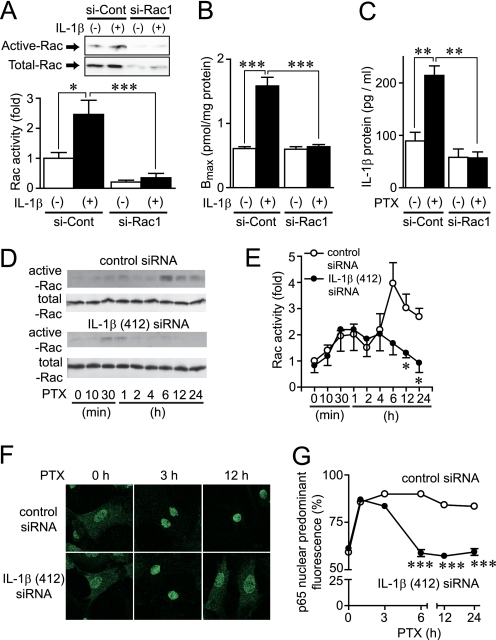
Because PTX-induced ROS production, NF-κB activation, and increase in AT1R density were inhibited by DN-Rac, Rac may play a central role in regulation of AT1R density. Inhibitors of 3-hydroxy-3-methylglutaryl-CoA reductase (statins) are known to suppress the activity of Rho family G proteins by inhibition of isoprenylation (55). It has been reported that simvastatin inhibits Rac activity in the H9c2 cell line and rat neonatal cardiomyocytes (56, 57). Simvastatin is also reported to reduce AT1R density in vascular smooth muscle cells (58). Therefore, we examined whether simvastatin inhibits IL-1β-induced up-regulation of AT1R by inhibition of Rac. Treatment with simvastatin completely suppressed the IL-1β-induced Rac activation (supplemental Fig. 4). Consistent with this result, the IL-1β-induced up-regulation of AT1R was also suppressed by simvastatin and DN-Rac. The Ang II-induced Ca2+ release was also enhanced in IL-1β-treated cells, and this enhancement was completely suppressed by simvastatin. These results suggest that simvastatin suppresses IL-1β-induced up-regulation of AT1R by inhibition of Rac activity. To prove the requirement of Rac in AT1R up-regulation more directly, we used Rac1 siRNAs. Knockdown of Rac1 almost completely suppressed IL-1β-induced Rac activation (Fig. 6A), increase in AT1R density (Fig. 6B), and enhancement of AT1R-stimulated Ca2+ responses (supplemental Fig. 4). Thus, Rac1 may predominantly regulate AT1R up-regulation by agonist stimulation. Because IL-1β induces Rac activation and PTX-induced IL-1β production was completely suppressed by knockdown of Rac1 (Fig. 6C), we hypothesize that PTX-induced IL-1β production plays a role in amplification of Rac activation. Treatment with IL-1β siRNA suppressed the PTX-induced Rac activation at a late phase of activation (from 6 h after the treatment) but did not suppress Rac activation at an early phase of activation (Fig. 6, D and E). Furthermore, IL-1β siRNA also suppressed PTX-induced nuclear localization of NF-κB in a late phase but not an early phase (Fig. 6, F and G). These results suggest that PTX-induced IL-1β production participates in the sustained activation of Rac and NF-κB, which is essential for AT1R up-regulation.
FIGURE 6.
Amplification of Rac-mediated signaling by PTX-induced IL-1β production. A, cells were transfected with siRNAs for Rac1 (si-Rac1) or their randomized controls (si-Cont) for 72 h before 5-min stimulation with IL-1β (10 ng/ml). B, effects of si-Rac1 on the maximal increases in AT1R density by IL-1β stimulation. Cells were treated with IL-1β for 24 h before membrane preparation. C, effects of si-Rac1 on PTX-induced production of IL-1β proteins. Cells were treated with PTX (100 ng/ml) for 90 min. D and E, effects of IL-1β siRNA on PTX-induced Rac activation. Cells were transfected with IL-1β (412) siRNA (100 nm) for 48 h before treatment with PTX (100 ng/ml). F and G, effects of IL-1β (412) siRNA on PTX-induced nuclear localization of the NF-κB p65 subunit. More than 100 cells were scanned and quantified the subcellular localization of p65 using Photoshop (13, 27). *, p < 0.05; ***, p < 0.001 versus IL-1β-treated or control siRNA-treated cells. Error bars, S.E.
PTX Stimulates TLR4, Leading to Rac Activation
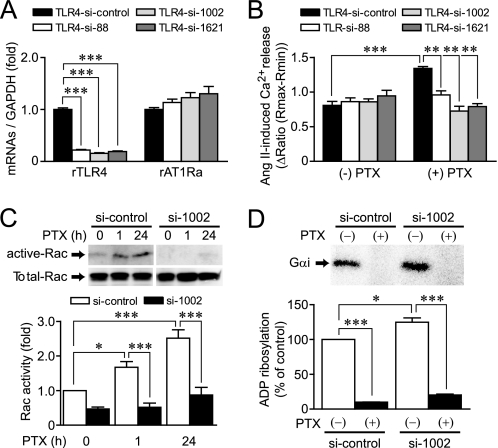
We next examined which receptor(s) functions as a target of PTX in cardiac fibroblasts. Because TLR4 is reported to work as a putative candidate receptor of B-oligomer (10), we examined whether stimulation of TLR4 is required for PTX-induced AT1R up-regulation in cardiac fibroblasts. Treatment with TLR4 siRNAs (si-88, si-1002, and si-1621) significantly decreased TLR4 mRNA levels but did not decrease AT1R mRNA levels (Fig. 7A). The PTX-induced enhancement of Ang II-induced Ca2+ release and increase in Rac activity were completely abolished by TLR4 siRNA treatment (Fig. 7, B and C). In contrast, PTX-induced ADP-ribosylation of Gαi proteins was not suppressed but preferably enhanced by TLR4 knockdown (Fig. 7D). These results suggest that TLR4 mediates PTX-induced Rac activation and AT1R up-regulation, but TLR4 does not mediate PTX-induced ADP-ribosylation of Gαi proteins.
FIGURE 7.
Roles of TLR4 in PTX-induced Rac activation and ADP-ribosylation of Gi/Go. A, effects of TLR4 siRNAs on the expression of TLR4 and AT1R mRNAs. B, effects of TLR4 siRNAs on PTX-induced enhancement of Ang II-induced Ca2+ responses. Cells were treated with PTX for 24 h after siRNA treatment for 48 h. C, effects of TLR4 siRNA (si-1002) on PTX-induced Rac activation. D, effects of TLR4 siRNA on PTX-induced ADP-ribosylation of Gαi proteins. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E.
DISCUSSION
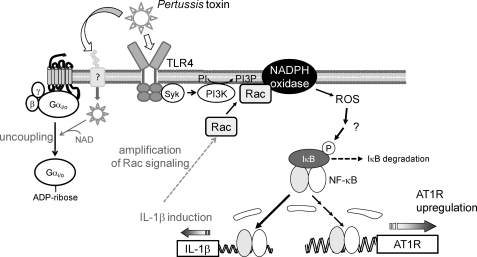
In this study, we demonstrated a novel action of PTX that enhances AT1R-stimulated Ca2+ response through AT1R up-regulation independently of ADP-ribosylation in rat cardiac fibroblasts. Using PTX as a powerful tool for analyzing the mechanism of AT1R up-regulation, we demonstrated that stimulation of TLR4 by PTX B-oligomer enhances AT1R function. Previous reports have suggested that Syk (spleen tyrosine kinase) and PI 3-kinase participate in TLR4-mediated responses (49, 59). We found that PTX-induced Rac activation was completely suppressed by inhibition of Syk (supplemental Fig. 3) and PI 3-kinase (Fig. 5), suggesting that Syk and PI 3-kinase mediate PTX-induced Rac activation. We also found that Rac-mediated NF-κB activation through ROS production plays a central role in the regulation of AT1R density. The PTX-induced NF-κB activation was suppressed by DPI and the dominant negative mutants of Rac and p47phox (Fig. 3). Because DPI is an inhibitor of NADPH oxidase and Rac and p47phox are essential components of NADPH oxidase activation, the origin of PTX-induced ROS production may be NADPH oxidase. In addition, PTX induced degradation of IκBα proteins in a time-dependent manner, which was abolished by Rac inhibition (supplemental Fig. 3). Although molecular mechanism underlying ROS-mediated NF-κB activation is still unknown, this result implies that ROS-mediated inhibition of mitogen-activated protein kinase phosphatases may be involved (60). Because the promoter regions of IL-1β and AT1R contain a putative NF-κB binding site, AT1R up-regulation may be induced by direct interaction of AT1R promoter with NF-κB. However, PTX-induced enhancement of Ca2+ response by AT1R stimulation was almost completely suppressed by anti-IL-1β antibody and IL-1β siRNAs (Fig. 2). Thus, IL-1β released from fibroblasts by PTX treatment may be the main mechanism of PTX-induced AT1R up-regulation. Furthermore, Rac1 inhibition suppressed PTX-induced IL-1β production, and IL-1β inhibition suppressed PTX-induced Rac activation at a late but not an early phase (Fig. 6). Thus, Rac-mediated IL-1β production may amplify Rac-dependent signaling through IL-1β-mediated Rac activation. These results suggest that PTX induces AT1R up-regulation through a TLR4 → PI 3-kinase → Rac → NADPH oxidase → ROS → NF-κB → IL-1β-dependent signal pathway (Fig. 8).
FIGURE 8.
Schema of TLR4-mediated AT1R up-regulation induced by PTX. PTX induces ROS production through sequential activation of TLR4, Syk, PI 3-kinase (PI3K), Rac, and NADPH oxidase. Although the mechanism of NF-κB activation induced by ROS is still unknown, ROS mediate NF-κB-dependent expression of IL-1β. Induction of IL-1β also induces Rac activation through IL-1 receptor stimulation, leading to amplification of Rac-dependent signaling. Sustained activation of Rac may be required for PTX-induced AT1R up-regulation in rat cardiac fibroblasts. A-protomer of PTX enters the cells through unidentified binding site, and ADP-ribosylates Gi/Go proteins.
We revealed that stimulation of TLR4 mediates PTX-induced AT1R up-regulation. It is thought that MyD88 and Trif-related adaptor molecule mediate TLR4-mediated NF-κB activation (32). However, it has recently been reported that oxidized LDL induces NADPH oxidase-dependent ROS production through TLR4 stimulation in macrophages (59). These authors have also demonstrated that Syk but not MyD88 is responsible for TLR4-mediated ROS production. In addition, another study has shown that stimulation of TLR4 by PTX B-oligomer induces activation of MyD88-independent signaling pathways (10). Thus, PTX induces stimulation of TLR4 that preferentially activates the Syk-dependent Rac signaling pathway.
PTX is frequently used as a specific tool to examine the involvement of Gi in cellular signaling. Abolishment of TLR4 by siRNA did not affect PTX-mediated ADP-ribosylation of Gi and Go (Fig. 7D). Thus, PTX binds to two receptors; one is TLR4 that activates Rac and another is the binding site that liberates the A-protomer into cells. So far, the Gi/Go-independent signaling pathway is not usually considered when PTX is used in vitro and in vivo. Because PTX activates Rac in addition to ADP-ribosylation of Gi and Go, it is no longer thought that PTX is a specific inhibitor of receptor-Gi signaling.
Another important finding of this study is that Rac is a physiological mediator of AT1R up-regulation induced by IL-1β stimulation. The inhibition of Rac suppressed the increase in AT1R density and the enhancement of Ang II-induced Ca2+ response by IL-1β stimulation (Fig. 6 and supplemental Fig. 4). Because other agonists that up-regulate AT1R, such as Ang II and TNF-α, also increase Rac activity, Rac-mediated AT1R up-regulation may be a common mechanism among various stimuli. Statins are inhibitors of 3-hydroxy-3-methylglutaryl-CoA reductase and appear to have pleiotropic effects on the cardiovascular system that are independent of their ability to decrease serum cholesterol (14, 55). These include inhibition of cardiac hypertrophy and left ventricular dysfunction, anti-inflammatory effects, and antioxidative effects (55, 61). Recent studies have demonstrated that statins inhibit ROS production and myocardial apoptosis by inhibition of Rac (57). Up-regulation of AT1R is thought to be one of the features involved in cardiac remodeling. Thus, the present results suggest a novel mechanism in which statins inhibit cardiac fibrosis by inhibition of AT1R up-regulation in cardiac fibroblasts. Statins also inhibit Rho activity by inhibition of isoprenylation. However, we could not detect the activation of Rho, Ras, and Rap1 by PTX treatment (supplemental Fig. 2). Thus, inhibition of Rac is essential for the inhibition of AT1R up-regulation by statin.
In conclusion, we demonstrated a novel action of PTX that induces AT1R up-regulation independently of ADP-ribosylation of Gi/Go. This mechanism includes TLR4-mediated Rac activation, ROS production, and NF-κB activation. Activation of NF-κB induces IL-1β production, resulting in amplification of Rac signaling, which leads to increase in AT1R density. The involvement of the TLR4-Rac signaling pathway in the regulation of AT1R density will provide a possible novel target for inhibiting cardiac remodeling. In addition, we have provided pharmacologically important information indicating that PTX per se influences G protein-coupled receptor signaling independently of Gαi inhibition. Activation of the TLR4-Rac signaling pathway by PTX suggests that we should consider pharmacological actions of PTX in addition to a specific inhibitor of Gi/Go-mediated signal transduction.
Acknowledgment
We thank Miyuki Toyotaka for analyzing the localization of the NF-κB p65 subunit.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M. Nishida, M. Nakaya, and H. K.); a grant-in-aid for scientific research on Innovative Areas (to M. Nishida); a grant-in-aid for scientific research on Priority Areas (to H. K.); and grants from the Nakatomi Foundation, Sapporo Bioscience Foundation, and Naito Foundation (to M. Nishida).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–4.
The data discussed in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE5017.
- PTX
- pertussis toxin
- Ang II
- angiotensin II
- AT1R
- Ang II type 1 receptor
- DN-Rac and DN-p47phox
- dominant negative Rac and p47phox, respectively
- DPI
- diphenyleneiodonium
- GFP
- green fluorescent protein
- ct
- carboxyl terminal region
- IκBαm
- non-phosphorylated form of IκBα, which works as a dominant negative mutant
- IL
- interleukin
- MOI
- multiplicity of infection
- NF-κB
- nuclear factor κB
- IκB
- inhibitor of κB
- PH
- pleckstrin homology
- PI
- phosphatidylinositol
- PI-3-P
- PI 3-phosphate
- PLC
- phospholipase C
- PX
- phox homology
- Ro-106-9920
- 6-(phenylsulfinyl)tetrazolo[1,5-b]pyridazine
- ROS
- reactive oxygen species
- TNF-α
- tumor necrosis factor-α
- WT
- wild type
- ELISA
- enzyme-linked immunosorbent assay
- siRNA
- small interfering RNA.
REFERENCES
- 1.Katada T., Ui M. (1982) J. Biol. Chem. 257, 7210–7216 [PubMed] [Google Scholar]
- 2.Kurose H., Katada T., Amano T., Ui M. (1983) J. Biol. Chem. 258, 4870–4875 [PubMed] [Google Scholar]
- 3.Tamura M., Nogimori K., Yajima M., Ase K., Ui M. (1983) J. Biol. Chem. 258, 6756–6761 [PubMed] [Google Scholar]
- 4.Lando Z., Teitelbaum D., Arnon R. (1980) Nature 287, 551–552 [DOI] [PubMed] [Google Scholar]
- 5.Linthicum D. S., Munoz J. J., Blaskett A. (1982) Cell Immunol. 73, 299–310 [DOI] [PubMed] [Google Scholar]
- 6.Racke M. K., Hu W., Lovett-Racke A. E. (2005) Trends Immunol. 26, 289–291 [DOI] [PubMed] [Google Scholar]
- 7.Jajoo S., Mukherjea D., Pingle S., Sekino Y., Ramkumar V. (2006) J. Pharmacol. Exp. Ther. 317, 1–10 [DOI] [PubMed] [Google Scholar]
- 8.Li H., Wong W. S. (2001) Biochem. Biophys. Res. Commun. 283, 1077–1082 [DOI] [PubMed] [Google Scholar]
- 9.Melien O., Sandnes D., Johansen E. J., Christoffersen T. (2000) J. Cell Physiol. 184, 27–36 [DOI] [PubMed] [Google Scholar]
- 10.Wang Z. Y., Yang D., Chen Q., Leifer C. A., Segal D. M., Su S. B., Caspi R. R., Howard Z. O., Oppenheim J. J. (2006) Exp. Hematol. 34, 1115–1124 [DOI] [PubMed] [Google Scholar]
- 11.Timmermans P. B., Wong P. C., Chiu A. T., Herblin W. F., Benfield P., Carini D. J., Lee R. J., Wexler R. R., Saye J. A., Smith R. D. (1993) Pharmacol. Rev. 45, 205–251 [PubMed] [Google Scholar]
- 12.de Gasparo M., Catt K. J., Inagami T., Wright J. W., Unger T. (2000) Pharmacol. Rev. 52, 415–472 [PubMed] [Google Scholar]
- 13.Onohara N., Nishida M., Inoue R., Kobayashi H., Sumimoto H., Sato Y., Mori Y., Nagao T., Kurose H. (2006) EMBO J. 25, 5305–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown R. D., Ambler S. K., Mitchell M. D., Long C. S. (2005) Annu. Rev. Pharmacol. Toxicol. 45, 657–687 [DOI] [PubMed] [Google Scholar]
- 15.Villarreal F. J., Kim N. N., Ungab G. D., Printz M. P., Dillmann W. H. (1993) Circulation 88, 2849–2861 [DOI] [PubMed] [Google Scholar]
- 16.Sakata Y., Hoit B. D., Liggett S. B., Walsh R. A., Dorn G. W., 2nd (1998) Circulation 97, 1488–1495 [DOI] [PubMed] [Google Scholar]
- 17.Gurantz D., Cowling R. T., Varki N., Frikovsky E., Moore C. D., Greenberg B. H. (2005) J. Mol. Cell Cardiol. 38, 505–515 [DOI] [PubMed] [Google Scholar]
- 18.Nio Y., Matsubara H., Murasawa S., Kanasaki M., Inada M. (1995) J. Clin. Invest. 95, 46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamani M. H., Cook D. J., Tuzcu E. M., Abdo A., Paul P., Ratliff N. B., Yu Y., Yousufuddin M., Feng J., Hobbs R., Rincon G., Bott-Silverman C., McCarthy P. M., Young J. B., Starling R. C. (2004) Am. J. Transplant. 4, 1097–1102 [DOI] [PubMed] [Google Scholar]
- 20.Peng J., Gurantz D., Tran V., Cowling R. T., Greenberg B. H. (2002) Circ. Res. 91, 1119–1126 [DOI] [PubMed] [Google Scholar]
- 21.Finkel T. (1999) J. Leukoc. Biol. 65, 337–340 [DOI] [PubMed] [Google Scholar]
- 22.Griendling K. K., Ushio-Fukai M. (2000) Regul. Pept. 91, 21–27 [DOI] [PubMed] [Google Scholar]
- 23.Sundaresan M., Yu Z. X., Ferrans V. J., Sulciner D. J., Gutkind J. S., Irani K., Goldschmidt-Clermont P. J., Finkel T. (1996) Biochem. J. 318, 379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumimoto H. (2008) FEBS J. 275, 3249–3277 [DOI] [PubMed] [Google Scholar]
- 25.Sulciner D. J., Irani K., Yu Z. X., Ferrans V. J., Goldschmidt-Clermont P., Finkel T. (1996) Mol. Cell Biol. 16, 7115–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida M., Tanabe S., Maruyama Y., Mangmool S., Urayama K., Nagamatsu Y., Takagahara S., Turner J. H., Kozasa T., Kobayashi H., Sato Y., Kawanishi T., Inoue R., Nagao T., Kurose H. (2005) J. Biol. Chem. 280, 18434–18441 [DOI] [PubMed] [Google Scholar]
- 27.Fujii T., Onohara N., Maruyama Y., Tanabe S., Kobayashi H., Fukutomi M., Nagamatsu Y., Nishihara N., Inoue R., Sumimoto H., Shibasaki F., Nagao T., Nishida M., Kurose H. (2005) J. Biol. Chem. 280, 23041–23047 [DOI] [PubMed] [Google Scholar]
- 28.Pracyk J. B., Tanaka K., Hegland D. D., Kim K. S., Sethi R., Rovira I. I., Blazina D. R., Lee L., Bruder J. T., Kovesdi I., Goldshmidt-Clermont P. J., Irani K., Finkel T. (1998) J. Clin. Invest. 102, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussman M. A., Welch S., Walker A., Klevitsky R., Hewett T. E., Price R. L., Schaefer E., Yager K. (2000) J. Clin. Invest. 105, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichiki T., Takeda K., Tokunou T., Iino N., Egashira K., Shimokawa H., Hirano K., Kanaide H., Takeshita A. (2001) Arterioscler. Thromb. Vasc. Biol. 21, 1896–1901 [DOI] [PubMed] [Google Scholar]
- 31.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 32.Chao W. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H1–H12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurose H., Arriza J. L., Lefkowitz R. J. (1993) Mol. Pharmacol. 43, 444–450 [PubMed] [Google Scholar]
- 34.Nishida M., Sato Y., Uemura A., Narita Y., Tozaki-Saitoh H., Nakaya M., Ide T., Suzuki K., Inoue K., Nagao T., Kurose H. (2008) EMBO J. 27, 3104–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida M., Sugimoto K., Hara Y., Mori E., Morii T., Kurosaki T., Mori Y. (2003) EMBO J. 22, 4677–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagamatsu Y., Nishida M., Onohara N., Fukutomi M., Maruyama Y., Kobayashi H., Sato Y., Kurose H. (2006) J. Pharmacol. Sci. 101, 144–150 [DOI] [PubMed] [Google Scholar]
- 37.Chiloeches A., Paterson H. F., Marais R., Clerk A., Marshall C. J., Sugden P. H. (1999) J. Biol. Chem. 274, 19762–19770 [DOI] [PubMed] [Google Scholar]
- 38.Franke B., Akkerman J. W., Bos J. L. (1997) EMBO J. 16, 252–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arai K., Maruyama Y., Nishida M., Tanabe S., Takagahara S., Kozasa T., Mori Y., Nagao T., Kurose H. (2003) Mol. Pharmacol. 63, 478–488 [DOI] [PubMed] [Google Scholar]
- 40.Ju H., Zhao S., Tappia P. S., Panagia V., Dixon I. M. (1998) Circulation 97, 892–899 [DOI] [PubMed] [Google Scholar]
- 41.Bai H., Wu L. L., Xing D. Q., Liu J., Zhao Y. L. (2004) Chin. Med. J. 117, 88–93 [PubMed] [Google Scholar]
- 42.He J., Gurunathan S., Iwasaki A., Ash-Shaheed B., Kelsall B. L. (2000) J. Exp. Med. 191, 1605–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruyama Y., Nishida M., Sugimoto Y., Tanabe S., Turner J. H., Kozasa T., Wada T., Nagao T., Kurose H. (2002) Circ. Res. 91, 961–969 [DOI] [PubMed] [Google Scholar]
- 44.Nishida M., Maruyama Y., Tanaka R., Kontani K., Nagao T., Kurose H. (2000) Nature 408, 492–495 [DOI] [PubMed] [Google Scholar]
- 45.Cowling R. T., Gurantz D., Peng J., Dillmann W. H., Greenberg B. H. (2002) J. Biol. Chem. 277, 5719–5724 [DOI] [PubMed] [Google Scholar]
- 46.Monks B. G., Martell B. A., Buras J. A., Fenton M. J. (1994) Mol. Immunol. 31, 139–151 [DOI] [PubMed] [Google Scholar]
- 47.Kaiser P., Rothwell L., Goodchild M., Bumstead N. (2004) Anim. Genet. 35, 169–175 [DOI] [PubMed] [Google Scholar]
- 48.Cowling R. T., Zhang X., Reese V. C., Iwata M., Gurantz D., Dillmann W. H., Greenberg B. H. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H1176–H1183 [DOI] [PubMed] [Google Scholar]
- 49.Zocchi M. R., Contini P., Alfano M., Poggi A. (2005) J. Immunol. 174, 6054–6061 [DOI] [PubMed] [Google Scholar]
- 50.Ueyama T., Eto M., Kami K., Tatsuno T., Kobayashi T., Shirai Y., Lennartz M. R., Takeya R., Sumimoto H., Saito N. (2005) J. Immunol. 175, 2381–2390 [DOI] [PubMed] [Google Scholar]
- 51.van Hennik P. B., ten Klooster J. P., Halstead J. R., Voermans C., Anthony E. C., Divecha N., Hordijk P. L. (2003) J. Biol. Chem. 278, 39166–39175 [DOI] [PubMed] [Google Scholar]
- 52.Brown G. E., Stewart M. Q., Liu H., Ha V. L., Yaffe M. B. (2003) Mol. Cell 11, 35–47 [DOI] [PubMed] [Google Scholar]
- 53.Honbou K., Minakami R., Yuzawa S., Takeya R., Suzuki N. N., Kamakura S., Sumimoto H., Inagaki F. (2007) EMBO J. 26, 1176–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gray A., Van Der Kaay J., Downes C. P. (1999) Biochem. J. 344, 929–936 [PMC free article] [PubMed] [Google Scholar]
- 55.Rikitake Y., Liao J. K. (2005) Circ. Res. 97, 1232–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ichiki T., Takeda K., Tokunou T., Funakoshi Y., Ito K., Iino N., Takeshita A. (2001) Hypertension 37, 535–540 [DOI] [PubMed] [Google Scholar]
- 57.Laufs U., Kilter H., Konkol C., Wassmann S., Böhm M., Nickenig G. (2002) Cardiovasc. Res. 53, 911–920 [DOI] [PubMed] [Google Scholar]
- 58.Ito M., Adachi T., Pimentel D. R., Ido Y., Colucci W. S. (2004) Circulation 110, 412–418 [DOI] [PubMed] [Google Scholar]
- 59.Bae Y. S., Lee J. H., Choi S. H., Kim S., Almazan F., Witztum J. L., Miller Y. I. (2009) Circ. Res. 104, 210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 61.Maack C., Kartes T., Kilter H., Schäfers H. J., Nickenig G., Böhm M., Laufs U. (2003) Circulation 108, 1567–1574 [DOI] [PubMed] [Google Scholar]