Abstract
Thioredoxins (Trxs) are ubiquitous enzymes catalyzing the reduction of disulfide bonds, thanks to a CXXC active site. Among their substrates, 2-Cys methionine sulfoxide reductases B (2-Cys MSRBs) reduce the R diastereoisomer of methionine sulfoxide (MetSO) and possess two redox-active Cys as follows: a catalytic Cys reducing MetSO and a resolving one, involved in disulfide bridge formation. The other MSRB type, 1-Cys MSRBs, possesses only the catalytic Cys, and their regeneration mechanisms by Trxs remain unclear. The plant plastidial Trx CDSP32 is able to provide 1-Cys MSRB with electrons. CDSP32 includes two Trx modules with one potential active site 219CGPC222 and three extra Cys. Here, we investigated the redox properties of recombinant Arabidopsis CDSP32 and delineated the biochemical mechanisms of MSRB regeneration by CDSP32. Free thiol titration and 4-acetamido-4′-maleimidyldistilbene-2,2′-disulfonic acid alkylation assays indicated that the Trx possesses only two redox-active Cys, very likely the Cys219 and Cys222. Protein electrophoresis analyses coupled to mass spectrometry revealed that CDSP32 forms a heterodimeric complex with MSRB1 via reduction of the sulfenic acid formed on MSRB1 catalytic Cys after MetSO reduction. MSR activity assays using variable CDSP32 amounts revealed that MSRB1 reduction proceeds with a 1:1 stoichiometry, and redox titrations indicated that CDSP32 and MSRB1 possess midpoints potentials of −337 and −328 mV at pH 7.9, respectively, indicating that regeneration of MSRB1 activity by the Trx through sulfenic acid reduction is thermodynamically feasible in physiological conditions.
Keywords: Methionine, Oxidation-Reduction, Oxidative Stress, Plant, Reductase, Thiol, Methionine Sulfoxide Reductase, Sulfenic Acid, Thioredoxin
Introduction
Cysteines are key residues in the CXX(C/S) active site of thioredoxins (Trxs)2 and glutaredoxins (Grxs). Trxs are small and ubiquitous protein-disulfide reductases that supply the reducing power needed to break disulfide bonds in physiological partners (1, 2). Grxs, generally reduced by glutathione, are able to reduce protein disulfides and also to carry out the reduction of glutathione-mixed disulfides, a reaction termed deglutathionylation, for which Trxs are not efficient catalysts (3–5). Since the discovery of ribonucleotide reductase as a substrate for Trxs (6) and Grxs (7), both thiol oxidoreductases have been shown to regulate enzymes involved in numerous metabolic pathways, such as apoptosis (8), protein folding (9), or carbon assimilation in photosynthetic organisms (10, 11). Another important function of Trxs and Grxs is their role in the protection against oxidative stress through the regeneration of the activity of peroxiredoxins (Prxs) (12, 13) and methionine sulfoxide reductases (MSRs) (14–16). Prxs and MSRs possess a variable number of redox-active Cys for catalyzing the reduction of their substrates, peroxides and methionine sulfoxide (MetSO), respectively. For both enzymes, the mechanism starts by the oxidation of the “catalytic” Cys to the sulfenic acid form (Cys-SOH) (15, 17–19). In the case of 2-Cys Prxs, MSRAs, and 2-Cys MSRBs, the second step, reduction of Cys-SOH, is achieved by an internal “resolving” Cys, through the formation of an intra- or intermolecular disulfide bond, subsequently reduced by thiol oxidoreductases (15, 17–21). In contrast, 1-Cys Prxs and 1-Cys MSRBs do not possess a potential resolving Cys (15, 19–21). The intervention of glutathione as the Cys-SOH reductant followed by a deglutathionylation step achieved by glutathione S-transferase has been shown for human 1-Cys Prx (22, 23). Similarly, we previously showed that regeneration of Arabidopsis 1-Cys MSRB activity by the GSH/Grx system involves first glutathionylation of the Cys-SOH formed on the catalytic Cys after MetSO reduction and then deglutathionylation by Grxs (21). Interestingly, several lines of evidence indicate that Trxs could also act as reductants for 1-Cys MSRBs, but the underlying biochemical mechanisms remain unclear. For instance, mammalian Trxs are able to covalently interact with substrate-oxidized 1-Cys MSRBs and to regenerate their activity (24). In the case of plant 1-Cys MSRBs, there is generally only one isoform, termed MSRB1, which is located in plastids (20, 25). Arabidopsis MSRB1 is efficiently reduced, as mentioned above, by Grxs (16, 21) but also by a specific plant Trx termed CDSP32 (Chloroplastic Drought-induced Stress Protein of 32 kDa) (16). Plants display a remarkable diversity of Trxs (1), but among the 10 plastidial Trxs assayed, only the unusual CDSP32 is able to regenerate the activity of 1-Cys MSRB1 (16). CDSP32, induced under severe abiotic stress conditions (26, 27), is composed of two Trx modules, with only one potential active redox disulfide center in the C-terminal domain (26). Affinity chromatography and co-immunoprecipitation assays identified plastidial 2-Cys Prxs and 1-Cys MSRB1 as potential targets of CDSP32 (28, 29). Very interestingly, this Trx is able to regenerate the activity of plant and mammalian 1-Cys MSRBs without addition of GSH or any other thiol compound (30), raising the hypothesis that regeneration might proceed through an alternative mechanism, distinct from the glutathionylation/deglutathionylation process used by Grxs (21).
In this work, we performed a biochemical analysis to delineate the reduction mechanism used by CDSP32 in the reduction of 1-Cys MSRB1. Our data show that the Trx, unlike Grxs, is able to directly reduce the sulfenic acid formed on the catalytic Cys of MSRB1 after MetSO reduction.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Nucleotide substitutions at specific positions were performed using the QuikChange mutagenesis method (Stratagene, La Jolla, CA) as reported previously (21) and the pQE30-CDSP32 and pQE30-CDSP32-Cter plasmids (28) as mutagenic PCR templates. Primers for site-directed mutagenesis, containing a modified restriction enzyme site allowing screening, are presented in supplemental Table 1.
Expression and Purification of Recombinant Proteins
Wild-type (WT) and mutated forms of MSRB1 and CDSP32 were purified as described previously (21, 28). Recombinant C-terminal CDSP32 starts from Gly-181 and has a theoretical molecular mass of 14,956.1 Da. CDSP32 proteins were reduced using 20 mm DTTred for 30 min at room temperature and then desalted using HiTrapTM desalting column or IllustraTM NAP-5 Sephadex G-25 (GE Healthcare) in 50 mm K2HPO4/KH2PO4, 100 mm KCl, pH 7.0. Protein concentration was determined using a bicinchoninic acid assay (BC Assay Reagent, Interchim) and by measuring the absorbance at 280 nm using specific extinction coefficients. Protein purity was verified using SDS-polyacrylamide gels stained with ImperialTM protein stain (Pierce).
Determination of CDSP32 Redox Status and AMS Labeling
10 μm WT or C222S CDSP32 was incubated with 1 mm DTTred or DTTox or with 30 μm MSRB1 in the presence or absence of 200 μm N-acetyl-MetSO. After 30 min, proteins were precipitated in 10% trichloroacetic acid. The pellet obtained after centrifugation was washed with 2% trichloroacetic acid and solubilized in 0.1 m Tris-HCl, pH 8.0, 1% (w/v) SDS, 2 mm AMS (Invitrogen). Proteins were separated using 17% bisacrylamide-SDS-polyacrylamide gels and revealed using ImperialTM protein stain.
Determination of Thiol and Cysteine Sulfenic Acid Contents
The protein thiol content was determined using the 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) procedure. After 30 min of incubation in the presence of 20 mm DTTred or 20 mm DTTox, proteins (50 μm) were desalted in 30 mm Tris-HCl, pH 8.0, using NAP-5 columns. Thiol measurement was performed after incubation for 30 min in the dark in the presence of 200 μm DTNB, 1% (w/v) SDS. The absorbance was read at 412 nm, and the thiol content was estimated using a molar extinction coefficient of 13,600 m1·cm−1 for 5-thio-2-nitrobenzoic acid (TNB). Cys-SOH content was determined using the specific TNB reagent prepared from reduction of DTNB (31). Pre-reduced MSRs (8–50 μm) were incubated with 70 μm TNB in a final volume of 250 μl of 30 mm Tris-HCl, pH 8.0. Cys-SOH formation was then monitored spectrophotometrically following the decrease in TNB concentration at 412 nm after addition of N-acetyl-MetSO (0.4–1.6 mm) using a molar extinction coefficient of 14,150 m1·cm−1. Absorbance and spectra were recorded using a Cary-50 UV-visible spectrophotometer (Varian, Inc.).
Assays for Trx and MSR Activities
The insulin reduction assay was used to analyze Trx activity as described previously (26) using 5 μm WT and mutated C-terminal CDSP32. Activity of recombinant MSRB proteins using reduced and desalted CDSP32 as an electron donor was determined at 25 °C by monitoring the reduction of the synthetic substrate, dabsyl-MetSO, as reported previously (21).
In Vitro Formation of Heterodimers
After purification, recombinant MSRB1 was pre-oxidized in the presence of 2 mm N-acetyl-MetSO for 15 min at 25 °C and then desalted in 30 mm Tris-HCl, pH 8.0, using NAP-5 columns. Pre-reduced or pre-oxidized MSRB1 (5 or 20 μm) was incubated with 20 μm pre-reduced C-terminal C222S CDSP32, C-terminal WT CDSP32, or C-terminal C219S CDSP32 for 30 min at 25 °C in the presence or absence of 1 mm diamide. Proteins were separated by SDS-PAGE, using NuPAGETM loading buffer, and NuPAGETM 4–12% BisTris gels and MES buffer (Invitrogen). After Coomassie Blue staining, the identity of proteins was determined using peptide mass fingerprints.
Mass Spectrometry Analyses
Bands were excised, destained, and subjected to in gel digestion with trypsin as described previously (32). Peptide mass fingerprints were performed using matrix-assisted laser desorption ionization time-of-flight analysis on a Voyager DE-STR mass spectrometer (PerSeptive Biosystems, Framingham, MA) as described previously (33).
Redox Titrations
Redox titration of CDSP32 was performed using mBBr as reported previously (34). CDSP32 (50 μm) was incubated at defined Eh values in 500-μl aliquots prepared by modifying the proportions of DTTred and DTTox (final DTT concentration, 2 mm). All Em value calculations were based on values of −327 and −385 mV for the Eh of DTT at pH 7.0 and pH 7.9, respectively (35, 36). Titrations were carried out in 0.1 m HEPES-HCl, pH 7.0, or in 0.1 m Tricine-NaOH, pH 7.9. After equilibrating CDSP32 samples in DTT redox buffers for 2 h, 5 μl of a saturated mBBr solution in acetonitrile was added. After 20 min in the dark, CDSP32 was precipitated using the trichloroacetic acid procedure described above, and the protein pellet was dissolved in 0.1 m Tris-HCl buffer, pH 8.0, 1% (w/v) SDS. Fluorescence was measured at 450 nm, after excitation at 380 nm, using an LS50B spectrofluorimeter (PerkinElmer Life Sciences). For redox titration of MSRB1, the enzyme was pre-oxidized using 2 mm N-acetyl-MetSO in 30 mm Tris-HCl, pH 8.0, for 15 min at 25 °C and then desalted in 0.1 m Tricine-NaOH, pH 7.9, using a NAP-5 column. 10 μm pre-oxidized MSRB1 was incubated for 1 h at 25 °C in 500-μl aliquots at defined Eh based on DTTred and DTTox proportions at a final DTT concentration of 10 mm. After desalting on NAP-5 column, protein concentration was determined at 280 nm using an extinction coefficient of 26,930 m−1·cm−1. Aliquots (95 μl) were incubated with 5 μl of 5 mm dabsyl-MetSO for 30 min at 37 °C. The amount of dabsyl-Met formed was determined as described previously (21). Titration data were fitted to the Nernst equation using nonlinear regression. The n value was set to 2 as the disulfide-dithiol exchange, and the oxidation and reduction of Cys sulfenic acid are expected to be two-electron transfer processes (35–38). The redox potential is reported as the mean value ± S.D. of three replicates.
RESULTS
Redox Characterization of WT and Mutated CDSP32
The mature CDSP32 protein is composed of two Trx domains, including only one potential active site, Cys219–Gly220–Pro221–Cys222, in the C-terminal domain (from Gly181 to Tyr302). The N-terminal domain (from Ala59 to Tyr180) contains two Cys at positions 122 and 142, and the C-terminal domain contains another one at position 253. To determine the redox status of CDSP32 cysteines, the free thiol content of WT and mutated CDSP32 was assayed using a DTNB procedure after incubation with DTTred or DTTox and subsequent desalting (Table 1). Reduced WT CDSP32 was found to contain 5.1 thiols, consistent with the reduction of five Cys. After incubation with DTTox, the observed number of free thiols was 2.7, corresponding to a decrease of ∼2 thiols compared with the number in the reduced protein. These results very likely indicate the formation of one disulfide bond in oxidized CDSP32. Consistently, in the case of the C-terminal domain, 3.1 thiols were titrated when the protein was reduced and only 1.0 after incubation with DTTox. These data lead us to conclude that the redox-active CDSP32 Cys are located in the C-terminal part of the protein. Titration experiments with C222S CDSP32 revealed no significant change in the number of free thiols. Indeed, after reductive and oxidative treatments, the numbers of free thiols in the protein lacking the second Cys in the potential active site were 3.9 and 3.5, respectively, indicating that Cys222 is redox-active and very likely able to form a disulfide bond with the other Cys of the potential active site, Cys219.
TABLE 1.
Free thiol content in WT and mutated CDSP32 proteins after treatment with DTTred and DTTox
The free thiol content in CDSP32 was titrated using a standard DTNB assay. Data are expressed in moles of SH·mol of enzyme−1. Data presented are the means ± S.D. (n = 3).
| No. of Cys | No. of free thiols measured |
|||
|---|---|---|---|---|
| DTTred | DTTox | Difference (DTTred − DTTox) | ||
| CDSP32 | 5 | 5.1 ± 0.1 | 2.7 ± 0.1 | 2.4 ± 0.1 |
| CDSP32 C terminus | 3 | 3.1 ± 0.2 | 1.0 ± 0.2 | 2.1 ± 0.3 |
| C222S CDSP32 | 4 | 3.9 ± 0.4 | 3.5 ± 0.4 | 0.4 ± 0.6 |
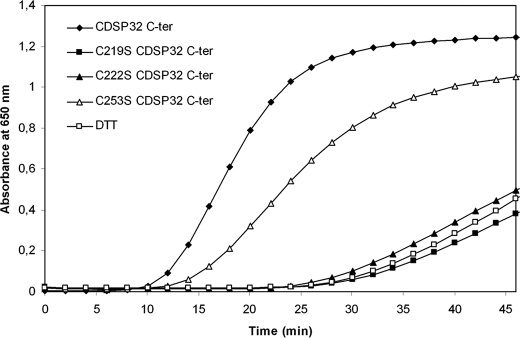
The thioredoxin activity of WT and mutated C-terminal CDSP32 was assayed using insulin as a substrate to investigate the role of the three Cys present in this domain (Fig. 1). Note that we previously reported the inability of whole CDSP32 to reduce the disulfide bonds in insulin (26). In the presence of C-terminal WT CDSP32, insulin precipitation occurred after 10 min, and reduction proceeded at a rate of 0.114 A650 unit·min−1 as reported previously (26). In contrast, C-terminal C219S CDSP32 and C222S CDSP32 were found unable to reduce insulin underlining the critical role of Cys219 and Cys222 in the reaction. When using C-terminal C253S CDSP32, insulin precipitation took place after 12 min with a rate lower than that of the WT form (0.063 A650 unit·min−1), revealing that Cys253 is not required for insulin reduction. Taking into consideration the thiol titration data and the conservation of the Trx-active site, these results indicate that Cys219 and Cys222 are the catalytic and resolving redox-active Cys, respectively, in CDSP32. Furthermore, they indicate that Cys253 is not required for CDSP32 disulfide reductase activity.
FIGURE 1.
Thioredoxin activity of C-terminal WT and mutated CDSP32. Reduction of insulin (1 g·liter−1) by 5 μm C-terminal CDSP32 was monitored as the increase in turbidity at 650 nm due to precipitation, in 0.1 m K2HPO4, 2 mm EDTA, pH 7.0, in a final volume of 700 μl. The reaction was started by adding 0.7 mm DTT.
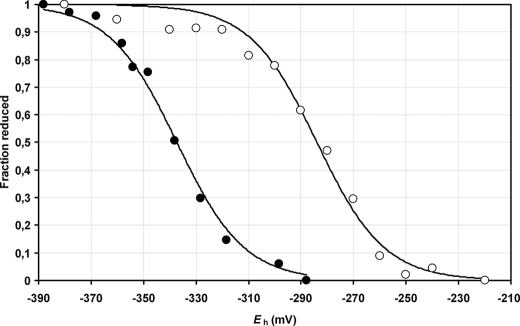
We then characterized the CDSP32 redox properties by determining the redox midpoint potential (Em) of C-terminal CDSP32 using mBBr to label free thiols at 25 °C at two pH values, 7.0 and 7.9 (Fig. 2). Titration data best fitted to the Nernst equation for a single two-electron redox couple (n settled to 2). The calculated redox midpoint potentials (Em) were −284 ± 3 and −337 ± 2 mV at pH 7.0 and 7.9, respectively. The difference of −53 ± 4 mV is consistent with the −59 mV/pH unit variation expected for the process of disulfide bond reduction, which involves the uptake of two protons per two electrons (35, 36). These potential values are in the range of those measured for canonical Trxs of plants and other organisms (43–45).
FIGURE 2.
Redox titration of C-terminal CDSP32 domain. Titration was performed in HEPES, pH 7.0 (○), and Tricine, pH 7.9 (●). The percentages of reduced fraction as a function of Eh were fitted to the Nernst equation (n = 2) using nonlinear regression. Data are represented as mean values ± S.D. (n = 3).
Redox-dependent Interaction between MSRB1 and CDSP32
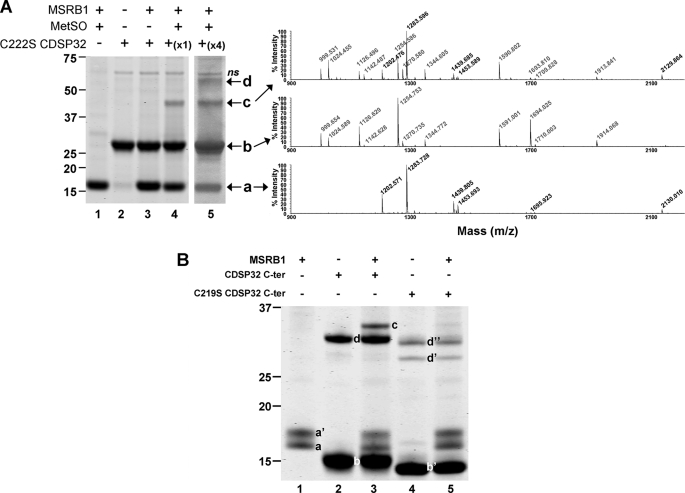
The plastidial 1-Cys MSRB1 was first identified as able to interact with CDSP32 via a redox-based mechanism in affinity chromatography experiments (29). Moreover, activity assays showed that CDSP32 provides MSRB1 with electrons (16, 30). To delineate the regeneration mechanism of MSRB1 by CDSP32, we first investigated whether both proteins could form redox-dependent heterodimeric complexes. For this purpose, MSRB1, either reduced or substrate-oxidized, was incubated with reduced C222S CDSP32 in ratios of 1:1 and 1:4. A mutated form of the Trx lacking the resolving Cys was used to stabilize the complex formed through an intermolecular disulfide bridge as shown for many targets of Trxs (24, 39). Then proteins were analyzed using nonreducing SDS-PAGE and Coomassie Blue staining (Fig. 3A) and by Western analysis (data not shown). When incubated alone, oxidized MSRB1 (Fig. 3A, band a, lane 1) or reduced C222S CDSP32 (Fig. 3A, band b, lane 2) migrated as monomers of ∼17 and ∼29 kDa, respectively. When reduced MSRB1 was incubated with C222S CDSP32, only the two monomers were visible after staining (Fig. 3A, bands a and b, lane 3). Treatment of MSRB1 with N-acetyl-MetSO before incubation with C222S CDSP32 resulted in the appearance of a third band at ∼45 kDa, corresponding to the size of an MSRB1-CDSP32 heterodimer (Fig. 3A, band c, lanes 4 and 5). Note also a fourth band at ∼60 kDa corresponding to a CDSP32 dimer (Fig. 3A, band d, lane 5). Peptide mass fingerprints confirmed that the 45-kDa band contains peptides from both proteins, whereas only MSRB1 peptides are present in band a and CDSP32 peptides in bands b and d (Fig. 3A). The complex formation was also assayed using WT and C219S C-terminal domains of CDSP32 in the presence of diamide, a compound triggering the formation of disulfide bridges (Fig. 3B). Oxidized MSRB1 migrated as two monomers (Fig. 3B, bands a and a′, lanes 1, 3, and 5), for which peptide mass fingerprints are indistinguishable (data not shown). Reduced C-terminal WT CDSP32 migrated as monomers and dimers of ∼15 and 30 kDa, respectively (Fig. 3B, lane 2, bands b and d). A band of 32 kDa was observed when MSRB1 was incubated with C-terminal WT CDSP32 (Fig. 3B, lane 3, band c), and peptide mass fingerprints revealed the presence of both proteins in this band. The mutated C219S form of C-terminal CDSP32 migrated as a monomer with an apparent size slightly lower than that of the WT form (Fig. 2B, lane 4, band b′) and as two dimers (Fig. 3B, bands d′ and d″, lane 5), but incubation with MSRB1 did not lead to the apparition of a band corresponding to a heterodimer (Fig. 3B, lane 5). Altogether, these data clearly show that CDSP32 forms a complex with oxidized MSRB1 through its catalytic cysteine, Cys219, very likely via a disulfide bond as indicated by the experiments performed using diamide.
FIGURE 3.
Formation of heterodimers between MSRB1 and CDSP32. A, formation of heterodimers between MSRB1 and C222S CDSP32. Pre-reduced or pre-oxidized MSRB1 was incubated with pre-reduced C222S CDSP32 in a 1:1 molar ratio. Coomassie Blue-stained nonreducing 4–12% polyacrylamide gel (left panel) and peptide mass fingerprints for identification of proteins present in each band (right panel) are shown. a, MSRB1 monomer; b, C222S CDSP32 monomer; c, MSRB1-C222S CDSP32 heterodimer; d, CDSP32 dimer; ns, nonspecific. B, formation of heterodimer between MSRB1 and C-terminal WT and C219S CDSP32 in the presence of diamide. Assays were performed as in A except that incubation was in a 1:4 molar ratio (MSRB1:CDSP32 C terminus). a and a′, MSRB1 monomers; b, C-terminal WT CDSP32 monomer; b′, C-terminal C219S CDSP32 monomers; c, C-terminal MSRB1-CDSP32 heterodimer; d, C-terminal WT CDSP32 dimer. Molecular masses are indicated on the left (kDa).
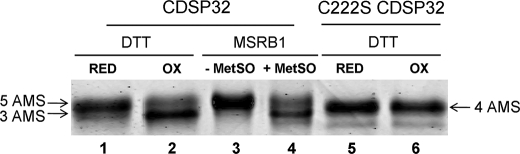
To determine the redox status of CDSP32 cysteines in the process of MSRB1 reduction, we used the thiol-specific reagent AMS. We compared the migration profiles of WT and C222S CDSP32 after DTT treatment or after incubation with MSRB1 in the presence or absence of N-acetyl-MetSO. Proteins were solubilized in a buffer containing AMS and separated in high resolution nonreducing SDS-polyacrylamide gels (Fig. 4). AMS alkylation led to an increase of nearly 0.5 kDa per Cys-SH. Accordingly, we observed a faster migration of DTTox-treated WT CDSP32 (Fig. 4, lane 2) compared with the reduced protein (Fig. 4, lane 1), revealing a decreased number of accessible Cys. Based on thiol titration data (Table 1), the size decrease corresponds to the absence of two free thiols in the DTTox-treated protein. When using C222S CDSP32, no migration variation was observed either in the presence of DTTred or DTTox (Fig. 4, lanes 5 and 6), indicating that Cys222 is very likely involved in a disulfide bridge with Cys219, consistently with data collected from thiol titration and insulin reduction assays. This was further confirmed in similar experiments using a C-terminal CDSP32 form lacking Cys219, which showed no change in migration after incubation with DTTox and AMS treatment (supplemental Fig. 1). In other respects, in the presence of MSRB1 when N-acetyl-MetSO was omitted, the migration of WT CDSP32 was identical to that observed after DTTred treatment (Fig. 4, lane 3), revealing complete reduction of the protein. The addition of N-acetyl-MetSO led to a faster migration of CDSP32 corresponding to that of CDSP32 treated with DTTox. Note that N-acetyl-MetSO has no effect on CDSP32 when incubated without MSRB1 (data not shown) and that MSRB1 monomer was excluded from the gel to obtain a correct separation of AMS-alkylated CDSP32 monomers. These results show that oxidized MSRB1 modifies the redox status of CDSP32 in a way similar to that of DTTox. On the basis of thiol and redox titration data, we conclude that the reduction of MSRB1 by CDSP32 results in the oxidation of the two Cys in the Trx-active site, Cys219 and Cys222, very likely through the formation of a disulfide bond between the two cysteines.
FIGURE 4.
Migration patterns of AMS-labeled CDSP32 after incubation with DTTred, DTTox, or MSRB1. WT and C222S CDSP32 were incubated with either DTTred or DTTox (lanes 1, 2 and 5, 6, respectively). WT CDSP32 was incubated with MSRB1 in the presence or absence of N-acetyl-MetSO. After trichloroacetic acid precipitation and AMS labeling, 0.25 μg of CDSP32 was loaded on a nonreducing 17% gel, and staining was achieved using Coomassie Blue.
Catalytic Parameters of MSRB1 Reduction by CDSP32
To further characterize the roles of CDSP32 redox-active Cys participating in MSRB1 reduction, we compared the ability of pre-reduced WT and mutated Trx forms to supply MSRB1 with electrons in the absence of any other reducer. These assays were monitored by following MetSO reduction using dabsyl-MetSO as a substrate mimicking peptide-bound MetSO. When varying the concentration of WT or of mutated C222S CDSP32, Met production was found to follow hyperbolic saturation curves (data not shown). In the presence of only pre-reduced MSRB1, we measured a stoichiometry of ∼1 mol of Met formed per mol of enzyme (Table 2). Saturation with WT CDSP32 led to the reduction of 10.3 mol of Met per mol of MSRB1 (Table 2). We calculated an apparent constant of semi-saturation of MSRB1 by CDSP32 (S0.5) of 54.1 μm. The C-terminal part of CDSP32 alone was also able to supply MSRB1 with electrons, and saturation led to the formation of 6 mol of Met per mol of MSRB1, with an S0.5 of 15.7 μm. The full-length CDSP32, in which Cys219 was replaced by Ser, was no more able to reduce MSRB1 in the presence or absence of a chemical reducer (data not shown). Very interestingly, the two mutated forms of the full-length protein, in which Cys222 or Cys222 and Cys253 were changed to Ser, were still able to supply MSRB1 with electrons. At saturating concentrations, the amounts of Met formed were equal to 95 and 79% of those recorded with WT CDSP32, for C222S CDSP32, and C222S/C253S CDSP32, respectively. The S0.5 values for both mutated forms were nearly twice higher than that of the WT protein (Table 2). Altogether, these results show that CDSP32 directly reduces MSRB1 and that among the five Cys present in CDSP32, only the catalytic one, Cys219, is required for the activity of MSRB1 reduction.
TABLE 2.
Stoichiometry of MSRB1 reduction by CDSP32
The stoichiometry values were calculated using hyperbolic saturation curves. The amount of Met formed was obtained at saturating concentrations of CDSP32 (“stoichiometry per mol of MSRB1”) or for concentration values lower than S0.5 (“stoichiometry per mol of CDSP32”). Data presented are mean values ± S.D. (n = 3).
| Reductant | Stoichiometry per mol of MSRB1 | S0.5 | Stoichiometry per mol of CDSP32 |
|---|---|---|---|
| moles of Met formed·mol of MSRB1−1 | μm | moles of Met formed·mol of MSRB1−1·mol of CDSP32−1 | |
| No reductant | 1.1 ± 0.1 | ||
| CDSP32 C terminus | 6.0 ± 1.2 | 15.7 ± 3.0 | 1.15 ± 0.05 |
| CDSP32 | 10.3 ± 0.9 | 54.1 ± 3.1 | 0.97 ± 0.14 |
| C222S CDSP32 | 9.8 ± 0.1 | 108.1 ± 20.2 | 0.43 ± 0.07 |
| C222S/C253S CDSP32 | 8.2 ± 0.1 | 93.7 ± 11.9 | 0.48 ± 0.02 |
Then we calculated the stoichiometry of MSRB1 reduction by CDSP32 by plotting the number of Met moles formed per mol of MSRB1 as a function of the CDSP32/MSRB1 molar ratio for CDSP32 concentration values lower than the calculated S0.5 (Table 2). The obtained data follow a linear regression model, the slope of which corresponds to the number of Met moles formed for a CDSP32/MSRB1 ratio of 1 (Table 2). For C-terminal WT CDSP32 and CDSP32, the slope value, very close to 1, indicates that 1 mol of CDSP32 is sufficient for 1 mol of MSRB1 to reduce 1 MetSO mol. In the case of C222S CDSP32 and C222S/C253S CDSP32, the slope values are equal to ∼0.5, showing that 2 mol of Trx are necessary for 1 MSRB1 mol to reduce 1 mol of substrate (Table 2). These results indicate that WT CDSP32 regenerates MSRB1 activity through an equimolar process and that Trx forms lacking Cys222 or Cys222 and Cys253 use an alternative mechanism involving two Trx molecules to reduce MSRB1.
Characterization of Cysteine Sulfenic Acid of MSRB1
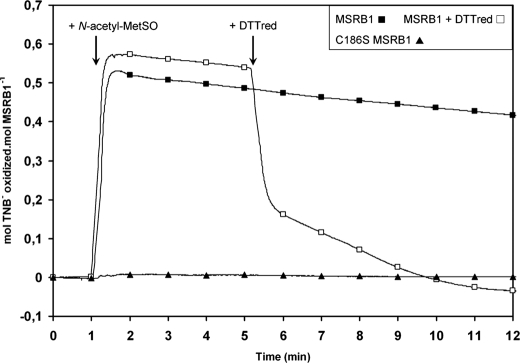
The regeneration mechanism of MSRB1 activity was then investigated using the Cys-SOH-specific reagent, TNB (17, 31), to delineate the redox changes in MSRB1 induced by N-acetyl-MetSO. When MSRB1, first incubated with TNB, was treated with a saturating concentration of N-acetyl-MetSO, TNB oxidation occurred and remained relatively stable (Fig. 5). The level of oxidation recorded was 0.57 ± 0.04 mol of TNB oxidized per mol of MSRB1. These data reveal the formation of a stable sulfenic acid, followed by the formation of MSRB1-TNB adduct and the concomitant release of one H2O molecule. When subsequently adding DTTred, we observed a decrease in the number of moles of TNB oxidized per mol of MSRB1 down to the basal level, corresponding to reduction and release of the TNB moiety. When performing similar experiments using C186S MSRB1, no change in the oxidized TNB/MSRB1 mole ratio was noticed, indicating no formation of sulfenic acid due to the absence of the catalytic Cys. In the case of WT enzyme, the calculated ratio of moles of TNB oxidized per mol of MSRB1 is nearly twice lower than expected regarding the theoretical 1-Cys MSRB mechanism for reducing MetSO, which involves 1 mol of enzyme per mol of substrate. To get a deeper insight regarding TNB titration, we used an alternative method consisting in quantifying the TNB moiety linked to MSRB1. The enzyme was incubated with TNB and N-acetyl-MetSO, and then excess reagents were removed using trichloroacetic acid precipitation. After solubilization, a residual absorbance at 412 nm of 0.66 ± 0.02 was detected (supplemental Fig. 2), indicating a residual absorbance of the TNB moiety linked on MSRB1. When adding DTT to the protein solution, the calculated ratio was 0.94 ± 0.05, showing that ∼1 mol of TNB was linked to 1 mol of MSRB1, and thus that the catalytic Cys186 is converted to a sulfenic acid form after reduction of MetSO.
FIGURE 5.
Characterization of Cys-SOH of MSRBs. The formation of cysteine sulfenic acid in MSRB1 was determined using TNB after addition of N-acetyl-MetSO as a substrate and monitored as the number of moles of oxidized TNB per mol of MSRB1. Typical traces were acquired using WT MSRB1 (■), WT MSRB1 and subsequently 1 mm DTTred (□), and C186S MSRB1 (▲).
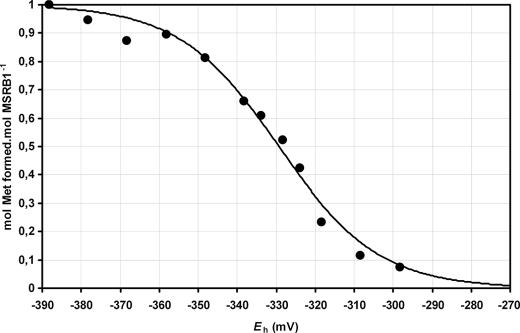
Finally, we determined the Em of MSRB1 at pH 7.9. For this purpose, MSRB1 was first pre-oxidized using dabsyl-MetSO as a substrate to form Cys-SOH, desalted to remove the excess of MetSO, then incubated with DTT in various dithiol-disulfide ratios, and desalted a second time before determination of the stoichiometry of dabsyl-Met production (Fig. 6). The calculated Em was −328 ± 2 mV. To our knowledge, this is the first study reporting a redox midpoint potential for a MSRB protein and a Cys-SH/Cys-SOH couple. Considering the redox potential measured for the CDSP32 Trx (−337 ± 2 mV), the value obtained here for MSRB Cys-SH/Cys-SOH is compatible with an efficient reduction of oxidized MSRB1 by CDSP32.
FIGURE 6.
Redox titration of MSRB1 at pH 7.9. The stoichiometry data (number of Met moles formed per mol of MSRB1) were fitted as a function of Eh to the Nernst equation using nonlinear regression. Data are represented as mean values ± S.D. (n = 3).
DISCUSSION
The aim of this study was to delineate the mechanism used by the CDSP32 Trx in the regeneration of Arabidopsis thaliana plastidial 1-Cys MSRB1 activity. The results presented here show that CDSP32 regenerates MSRB1 through the direct reduction of the sulfenic acid formed on the catalytic Cys, without the help of any other thiol compound. Notably, we recently showed that MSRB1 activity is also regenerated by the GSH/Grx system but via a glutathionylation step of the sulfenic acid and then by Grx reduction of the GSH-MSR adduct (16, 21). Thus, MetSO reductases displaying only one redox-active cysteine would be regenerated in planta by at least two types of electron donors and through distinct regeneration processes. Taking into consideration the concentration of reduced glutathione compared with that of CDSP32, the GSH/Grx system could be the main MSRB1 electron provider upon optimal growth conditions. CDSP32 could act more specifically during stress responses when the pool of reduced GSH decreases and/or when GSH is needed for other functions, such as the protection of carbon metabolic enzymes such as glyceraldehyde-3-phosphate dehydrogenase (40). Indeed, accumulation of CDSP32 has been shown to occur during environmental constraints, such as severe water deficit or photooxidative treatments, which result in decreases in the pool of reduced glutathione (26, 27, 41). Thus, the Trx could provide MSRB1 with electrons and ensure the MSRB activity level required for protection during environmental constraints (42).
CDSP32 is composed of two Trx domains with only one potential Trx-active site in the C-terminal domain, including Cys219 and Cys222. The N-terminal domain contains two Cys, and the C-terminal domain possesses one supplementary Cys at position 253. To determine the role of each Cys in the activity and redox status of CDSP32, we characterized WT and mutated forms by thiol titration and insulin reduction assays. The thiol titration results showed that only two Cys are involved in the protein redox status and that they are located in the C-terminal domain (Table 1). When using C222S CDSP32, no noticeable difference in thiol content was observed after oxidation (Table 1), revealing that Cys222 is involved in the redox status of CDSP32, very likely through the formation of a disulfide bridge with Cys219 upon oxidation, as reported for the resolving Cys in many Trxs (9). This hypothesis is supported by insulin reduction assays, in which we observed that only C-terminal WT and C253S CDSP32 are able to reduce the hormone (Fig. 1), indicating that Cys219 and Cys222 are absolutely required. Moreover, in regeneration assays of MSR activity (Table 2), the C-terminal CDSP32 domain and a mutated CDSP32 form, in which the Cys253 is changed to a Ser, are able to provide MSRB1 with electrons, showing that the two N-terminal Cys and the most C-terminal one are dispensable. These data lead us to conclude that these three Cys do not participate in the CDSP32 redox status and in the reduction of the substrates tested in this study. However, we cannot exclude potential roles for these Cys in the interaction with other partners. To further characterize the redox properties of the Trx, we determined the oxidation-reduction midpoint potentials (Em) of C-terminal CDSP32 at pH 7.0 and 7.9, the physiological pH values in the stroma, using an mBBr procedure (Fig. 2). Consistent with a two electrons process, Em values were −284 ± 3 and −337 ± 2 mV at pH 7.0 and 7.9, respectively. These values are significantly less negative than those of the canonical plastidial Trxs f and m (approximately −360 mV at pH 7.9) (43, 44) and lower than the redox midpoint potential of human Trx1 (approximately −290 mV) (45) but in the same range of those recorded for Escherichia coli Trx (46) or plastidial Trxs x and y (43, 44). Interestingly, Trxs x and y are efficient reducers of the plastidial 2-Cys Prx (43) and Prx Q (44, 47), respectively, two previously identified targets of CDSP32 (28, 29). At pH 7.9, the redox midpoint potentials of 2-Cys Prx and Prx Q are around −370 mV (47, 48), a value ∼35 mV more negative than those of CDSP32, Trx x and Trx y, showing that efficient reduction of the disulfide bond formed in oxidized Prxs can occur only when the pool of Trxs is highly reduced. On the contrary, the redox midpoint potential of MSRB1 is −328 ± 2 mV at pH 7.9 (Fig. 6), a value slightly less electronegative than that of CDSP32 (−337 ± 2 mV). This indicates that MSRB1 reduction by CDSP32 is thermodynamically feasible at pH 7.9, the physiological value observed in illuminated chloroplasts (49). The analysis of C-terminal CDSP32 redox titration curves revealed that the Trx could sustain 50% of MSRB1 activity even if only 30% of the total pool of protein is reduced (Fig. 2). Taking into consideration the redox midpoint potentials of plastidial Prxs, the reduction of MSRB1 by CDSP32 appears far more favorable than that of 2-Cys Prx and Prx Q, which in comparison exhibit lower Em values.
Contrary to other CDSP32 targets, MSRB1 possess only one redox-active Cys at position 186. We previously reported that incubation of MSRB1 with N-acetyl-MetSO leads to an increase of 16 Da in the protein mass, compared with that of the reduced form. Moreover, thiol titration showed that only one Cys is oxidized in MSRB1 after MetSO reduction (21). These data argue for the formation of a stable sulfenic acid on Cys186 after substrate reduction. In this study, we used the Cys-SOH-specific reagent, TNB, to further characterize the action of N-acetyl-MetSO on MSRB1 catalytic Cys (Fig. 5). The results acquired with WT and C186S MSRB1 are consistent with the formation of a stable sulfenic acid on Cys186 (Fig. 5). For WT MSRB1, the calculations revealed that only ∼0.5 Cys-SOH was titrated per mol of oxidized MSRB (Fig. 5), a value twice lower than that expected regarding the catalytic mechanism used by MSRBs (21, 50). Similarly, ∼0.5 Cys-SOH, instead of a value of 1, was recorded for the mutated poplar MSRA possessing only the catalytic Cys. Nevertheless, the thiol titration assays performed with this protein were consistent with the oxidation of only one Cys after substrate reduction (51). In other respects, the quantification of TNB moiety linked on MSRB1 indicated that the reagent conserves partial absorbance when linked to the protein (supplemental Fig. 2), thus explaining the low ratio titrated with MSRB1 and poplar MSRA. These TNB assays, in full agreement with free thiol titration data and mass spectrometry experiments, clearly demonstrate the presence of a stable sulfenic acid on Cys186 in MSRB1 (21).
MSRB1 was initially identified by affinity chromatography as a potential CDSP32 target using a C222S CDSP32 bait and DTTred elution (29), suggesting that the interaction proceeds through the formation of a transient MSRB1-CDSP32 heterodimer linked through a disulfide bond. However, we could not exclude that the interaction between the two proteins might be due to a noncovalent linkage. But several lines of evidence reported in this study give high credence for the formation of a disulfide bridge. Indeed, in agreement with affinity chromatography data (29), incubation of C222S CDSP32 with MSRB1 after N-acetyl-MetSO treatment leads to the formation of a heterodimer, as shown by protein electrophoresis and peptide mass fingerprints (Fig. 3A). In similar experiments performed in the presence of diamide, a heterodimer was observed when using C-terminal WT CDSP32, whereas no complex was formed with the mutated form of C-terminal CDSP32 devoid of catalytic Cys219 (Fig. 3B). These results strongly argue for the formation of a transient disulfide bond between the catalytic Cys of both partners after MetSO reduction and subsequent formation of sulfenic acid on Cys186 of MSRB1.
We then characterized the mechanism used by CDSP32 in the regeneration of MSRB1 activity. The use of AMS, as an alkylating agent in nonreducing SDS-PAGE, revealed that incubation of CDSP32 with either DTTox or oxidized MSRB1 promotes a similar change in the Trx redox status (Fig. 4), which, taking into consideration the thiol titration data, corresponds to the formation of a disulfide bridge involving Cys219 and Cys222. When performing activity assays with pre-reduced Trx forms (Table 2), 10- and 6-fold increases of MSRB1 activity were measured in the presence of WT whole CDSP32 and the C-terminal domain, respectively. These results clearly show that both N-terminal Cys122 and Cys142 are not necessary for the regeneration of MSRB1 activity. The efficiency difference could originate from an altered folding of the C-terminal domain. Unexpectedly, full-length CDSP32 proteins, in which the resolving Cys222 alone or together with Cys253, was changed to Ser, are able to regenerate MSRB1 activity nearly as efficiently as WT CDSP32 (Table 2). These data reveal that Cys222 is not necessary to regenerate MSRB1 activity in vitro. The calculation of the stoichiometry showed that 1 mol of WT CDSP32 is sufficient to allow reduction of 1 mol of MetSO by 1 mol of MSRB1, whereas 2 mol of C222S or C222S/C253S CDSP32 are required (Table 2). The main information arising from these data is that WT CDSP32 reduces MSRB1 very likely thanks to the two redox-active Cys219 and Cys222 in an equimolar process involving the formation of an intramolecular disulfide bond. In the case of mutated forms lacking the resolving Cys, the regeneration could involve two Trx molecules. Consistent with this hypothesis, dimers of the Trx have been observed when incubating oxidized MSRB1 with WT or mutated CDSP32 (Fig. 3 and data not shown). The formation of Trx dimers linked through a disulfide bond formed between both catalytic Cys has been also noticed in vitro, when incubating human 1-Cys MSRB3 with a mutated Trx lacking the resolving Cys (24).
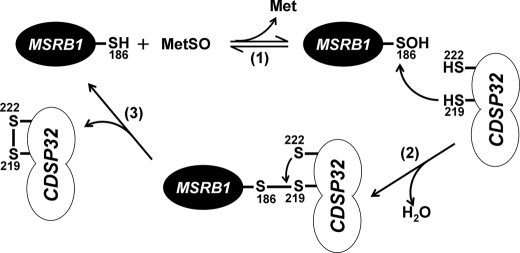
Based on our results, we propose a model for the reduction of MSRB1 by CDSP32 (Fig. 7) which involves the following: (i) the formation of a stable sulfenic acid on Cys186 of MSRB1 after reduction of MetSO; (ii) the direct reduction of Cys-SOH by the catalytic Cys219 of CDSP32 and the formation of a transient heterodimer; and then (iii) the release of reduced MSRB1 after the formation of an intramolecular disulfide bond between Cys219 and Cys222 in CDSP32. Then, although there is no direct evidence, oxidized CDSP32 would be regenerated by the ferredoxin-dependent Trx reductase, like most other plastidial Trxs (52). The CDSP32 protein is present only in photosynthetic eukaryotes, and except for Chlamydomonas reinhardtii, the two Cys of the C-terminal active site are conserved (supplemental Fig. 3), indicating that the proposed mechanism is very likely similar in other organisms. It is worth mentioning that this mechanism is highly different from that used by the GSH/Grx system, which proceeds first through the reduction of the cysteine-sulfenic acid in MSRB1 by glutathionylation and does not involve the formation of a heterodimeric Grx-MSR complex (21).
FIGURE 7.
Model for the regeneration of MSRB1 by the CDSP32 Trx. The first step consists of MetSO reduction with the concomitant release of 1 mol of Met and the formation of a stable sulfenic acid intermediate on catalytic Cys186 of MSRB1 (1). The sulfenic acid is then attacked by the catalytic Cys219 of CDSP32 leading to the liberation of one molecule of water and to the formation of a transient intermolecular disulfide bond (2). Then the resolving Cys222 of CDSP32 attacks the Cys219 to form an intramolecular disulfide bond and release the reduced MSRB1 (3).
Our data show the direct reduction of a Cys-SOH form by a Trx and raise the questions whether it is a general process conserved for most Trx types or whether the example described here, involving an unusual Trx and one specific target, is an exception. In the case of plant Trxs, many plastidial isoforms, like Trxs f, m, and y, appear as unable to reduce Cys-SOH in MSRB1 (16). However, the formation of a stable cysteine-sulfenic acid in proteins and its subsequent reduction by Trxs have been proposed for several proteins such as E. coli 1-Cys Prx bacterioferritin co-migratory protein (53), human 1-Cys MSRBs (24), protein-tyrosine phosphatase 1B (54), or the E. coli periplasmic ld-transpeptidase YbiS (55), indicating that this biochemical feature is very likely conserved for some Trx types in other organisms. In other respects, oxidation of Cys into Cys-SOH is implicated in several fundamental metabolic pathways (56). For instance, E. coli OxyR transcription factor, responsible for the oxidative stress response, is regulated through oxidation of an important Cys into a stable Cys-SOH form (57), and mitotic protein-tyrosine phosphatases are inactivated after Cys-SOH formation (58). We could thus presume that Trxs, besides their activity of disulfide reductases, fulfill other important functions through the reduction of cysteine-sulfenic acid forms.
Acknowledgments
We are very grateful to Prof. David D. Knaff and team (Texas Tech University, Lubbock) for their help in redox midpoint potential determination, Patricia Henri (Commissariat à l'Energie Atomique et aux Energies Alternatives, Institut de Biologie Environnementale et Biotechnologie, Service de Biologie Végétale et Microbiologie Environnmenetales, and Laboratoire d'Ecophysiologie Moléculaire des Plantes) for technical assistance, Noëlle Bécuwe (CEA, IBEB, SBVME, LEMP) for mutagenesis, and Rémy Puppo (CEA, IBEB, SBVME, LEMP) for helpful advice in high pressure liquid chromatography experiments. We are also grateful to Dr. Nicolas Rouhier (Nancy University) for critical reading of the manuscript.
This work was supported by Agence Nationale de la Recherche (Génoplante) Grants GNP05010G (to E. L. and P. R.) and ANR-08-BLAN-0153 (to M. Z., C. H. M., P. L. M., and S. D. L.) and by Région Provence-Alpes-Côte d'Azur (to L. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- Trx
- thioredoxin
- AMS
- 4-acetamido-4′-maleimidyldistilbene-2,2′disulfonic acid
- DTNB
- 5,5-dithiobis(2-nitrobenzoic acid)
- DTTred
- reduced dithiothreitol
- DTTox
- oxidized dithiothreitol (trans-4,5-dihydroxy-1,2-dithiane)
- GSH
- reduced glutathione
- Grx
- glutaredoxin
- MetSO
- methionine sulfoxide
- MSR
- methionine sulfoxide reductases
- Prx
- peroxiredoxins
- TNB
- 5-thio-2-nitrobenzoic acid
- WT
- wild type
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MES
- 4-morpholineethanesulfonic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- mBBr
- monobromobimane.
REFERENCES
- 1.Meyer Y., Siala W., Bashandy T., Riondet C., Vignols F., Reichheld J. P. (2008) Biochim. Biophys. Acta 1783, 589–600 [DOI] [PubMed] [Google Scholar]
- 2.Vieira Dos Santos C., Rey P. (2006) Trends Plant Sci. 11, 329–334 [DOI] [PubMed] [Google Scholar]
- 3.Jung C. H., Thomas J. A. (1996) Arch. Biochem. Biophys. 335, 61–72 [DOI] [PubMed] [Google Scholar]
- 4.Rouhier N., Lemaire S. D., Jacquot J. P. (2008) Annu. Rev. Plant Biol. 59, 143–166 [DOI] [PubMed] [Google Scholar]
- 5.Zaffagnini M., Michelet L., Massot V., Trost P., Lemaire S. D. (2008) J. Biol. Chem. 283, 8868–8876 [DOI] [PubMed] [Google Scholar]
- 6.Laurent T. C., Moore E. C., Reichard P. (1964) J. Biol. Chem. 239, 3436–3444 [PubMed] [Google Scholar]
- 7.Holmgren A. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 2275–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J. J., Lee Y. J. (2003) Biochem. J. 373, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt C., Lillig C. H., Holmgren A. (2008) Biochim. Biophys. Acta 1783, 641–650 [DOI] [PubMed] [Google Scholar]
- 10.Couturier J., Koh C. S., Zaffagnini M., Winger A. M., Gualberto J. M., Corbier C., Decottignies P., Jacquot J. P., Lemaire S. D., Didierjean C., Rouhier N. (2009) J. Biol. Chem. 284, 9299–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemaire S. D., Michelet L., Zaffagnini M., Massot V., Issakidis-Bourguet E. (2007) Curr. Genet. 51, 343–365 [DOI] [PubMed] [Google Scholar]
- 12.Chae H. Z., Chung S. J., Rhee S. G. (1994) J. Biol. Chem. 269, 27670–27678 [PubMed] [Google Scholar]
- 13.Rouhier N., Gelhaye E., Sautiere P. E., Brun A., Laurent P., Tagu D., Gerard J., de Faÿ E., Meyer Y., Jacquot J. P. (2001) Plant Physiol. 127, 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- 14.Brot N., Weissbach L., Werth J., Weissbach H. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 2155–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar R. A., Koc A., Cerny R. L., Gladyshev V. N. (2002) J. Biol. Chem. 277, 37527–37535 [DOI] [PubMed] [Google Scholar]
- 16.Vieira Dos Santos C., Laugier E., Tarrago L., Massot V., Issakidis-Bourguet E., Rouhier N., Rey P. (2007) FEBS Lett. 581, 4371–4376 [DOI] [PubMed] [Google Scholar]
- 17.Boschi-Muller S., Azza S., Sanglier-Cianferani S., Talfournier F., Van Dorsselear A., Branlant G. (2000) J. Biol. Chem. 275, 35908–35913 [DOI] [PubMed] [Google Scholar]
- 18.Rouhier N., Jacquot J. P. (2002) Photosynth. Res. 74, 259–268 [DOI] [PubMed] [Google Scholar]
- 19.Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 20.Tarrago L., Laugier E., Rey P. (2009) Mol. Plant. 2, 202–217 [DOI] [PubMed] [Google Scholar]
- 21.Tarrago L., Laugier E., Zaffagnini M., Marchand C., Le Maréchal P., Rouhier N., Lemaire S. D., Rey P. (2009) J. Biol. Chem. 284, 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manevich Y., Feinstein S. I., Fisher A. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3780–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ralat L. A., Manevich Y., Fisher A. B., Colman R. F. (2006) Biochemistry 45, 360–372 [DOI] [PubMed] [Google Scholar]
- 24.Kim H. Y., Kim J. R. (2008) Biochem. Biophys. Res. Commun. 371, 490–494 [DOI] [PubMed] [Google Scholar]
- 25.Vieira Dos Santos C., Cuiné S., Rouhier N., Rey P. (2005) Plant Physiol. 138, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rey P., Pruvot G., Becuwe N., Eymery F., Rumeau D., Peltier G. (1998) Plant J. 13, 97–107 [DOI] [PubMed] [Google Scholar]
- 27.Broin M., Cuiné S., Peltier G., Rey P. (2000) FEBS Lett. 467, 245–248 [DOI] [PubMed] [Google Scholar]
- 28.Broin M., Cuiné S., Eymery F., Rey P. (2002) Plant Cell 14, 1417–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rey P., Cuiné S., Eymery F., Garin J., Court M., Jacquot J. P., Rouhier N., Broin M. (2005) Plant J. 41, 31–42 [DOI] [PubMed] [Google Scholar]
- 30.Ding D., Sagher D., Laugier E., Rey P., Weissbach H., Zhang X. H. (2007) Biochem. Biophys. Res. Commun. 361, 629–633 [DOI] [PubMed] [Google Scholar]
- 31.Turell L., Botti H., Carballal S., Ferrer-Sueta G., Souza J. M., Durán R., Freeman B. A., Radi R., Alvarez B. (2008) Biochemistry 47, 358–367 [DOI] [PubMed] [Google Scholar]
- 32.Preger V., Tango N., Marchand C., Lemaire S. D., Carbonera D., Di Valentin M., Costa A., Pupillo P., Trost P. (2009) Plant Physiol. 150, 606–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Augusto L. A., Decottignies P., Synguelakis M., Nicaise M., Le Maréchal P., Chaby R. (2003) Biochemistry 42, 3929–3938 [DOI] [PubMed] [Google Scholar]
- 34.Krimm I., Lemaire S., Ruelland E., Miginiac-Maslow M., Jaquot J. P., Hirasawa M., Knaff D. B., Lancelin J. M. (1998) Eur. J. Biochem. 255, 185–195 [DOI] [PubMed] [Google Scholar]
- 35.Hirasawa M., Schürmann P., Jacquot J. P., Manieri W., Jacquot P., Keryer E., Hartman F. C., Knaff D. B. (1999) Biochemistry 38, 5200–5205 [DOI] [PubMed] [Google Scholar]
- 36.Hutchison R. S., Ort D. R. (1995) Methods Enzymol. 252, 220–228 [DOI] [PubMed] [Google Scholar]
- 37.Claiborne A., Mallett T. C., Yeh J. I., Luba J., Parsonage D. (2001) Adv. Protein Chem. 58, 215–276 [DOI] [PubMed] [Google Scholar]
- 38.Carballal S., Radi R., Kirk M. C., Barnes S., Freeman B. A., Alvarez B. (2003) Biochemistry 42, 9906–9914 [DOI] [PubMed] [Google Scholar]
- 39.Balmer Y., Schürmann P. (2001) FEBS Lett. 492, 58–61 [DOI] [PubMed] [Google Scholar]
- 40.Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., Miginiac-Maslow M., Noctor G., Trost P., Lemaire S. D. (2007) FEBS J. 274, 212–226 [DOI] [PubMed] [Google Scholar]
- 41.Eymery F., Rey P. (1999) Plant Physiol. Biochem. 37, 305–312 [Google Scholar]
- 42.Laugier E., Tarrago L., Dos Santos C. V., Eymery F., Havaux M., Rey P. (2010) Plant J. 61, 271–282 [DOI] [PubMed] [Google Scholar]
- 43.Collin V., Issakidis-Bourguet E., Marchand C., Hirasawa M., Lancelin J. M., Knaff D. B., Miginiac-Maslow M. (2003) J. Biol. Chem. 278, 23747–23752 [DOI] [PubMed] [Google Scholar]
- 44.Collin V., Lamkemeyer P., Miginiac-Maslow M., Hirasawa M., Knaff D. B., Dietz K. J., Issakidis-Bourguet E. (2004) Plant Physiol. 136, 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson W. H., Pohl J., Montfort W. R., Stuchlik O., Reed M. S., Powis G., Jones D. P. (2003) J. Biol. Chem. 278, 33408–33415 [DOI] [PubMed] [Google Scholar]
- 46.Krause G., Holmgren A. (1991) J. Biol. Chem. 266, 4056–4066 [PubMed] [Google Scholar]
- 47.Rouhier N., Gelhaye E., Gualberto J. M., Jordy M. N., De Fay E., Hirasawa M., Duplessis S., Lemaire S. D., Frey P., Martin F., Manieri W., Knaff D. B., Jacquot J. P. (2004) Plant Physiol. 134, 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.König J., Baier M., Horling F., Kahmann U., Harris G., Schürmann P., Dietz K. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werdan K., Heldt H. W., Milovancev M. (1975) Biochim. Biophys. Acta 396, 276–292 [DOI] [PubMed] [Google Scholar]
- 50.Boschi-Muller S., Gand A., Branlant G. (2008) Arch. Biochem. Biophys. 474, 266–273 [DOI] [PubMed] [Google Scholar]
- 51.Rouhier N., Kauffmann B., Tete-Favier F., Palladino P., Gans P., Branlant G., Jacquot J. P., Boschi-Muller S. (2007) J. Biol. Chem. 282, 3367–3378 [DOI] [PubMed] [Google Scholar]
- 52.Jacquot J. P., Eklund H., Rouhier N., Schürmann P. (2009) Trends Plant Sci. 14, 336–343 [DOI] [PubMed] [Google Scholar]
- 53.Jeong W., Cha M. K., Kim I. H. (2000) J. Biol. Chem. 275, 2924–2930 [DOI] [PubMed] [Google Scholar]
- 54.Lee S. R., Kwon K. S., Kim S. R., Rhee S. G. (1998) J. Biol. Chem. 273, 15366–15372 [DOI] [PubMed] [Google Scholar]
- 55.Depuydt M., Leonard S. E., Vertommen D., Denoncin K., Morsomme P., Wahni K., Messens J., Carroll K. S., Collet J. F. (2009) Science 326, 1109–1111 [DOI] [PubMed] [Google Scholar]
- 56.Poole L. B., Karplus P. A., Claiborne A. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 57.Kim S. O., Merchant K., Nudelman R., Beyer W. F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J. S. (2002) Cell 109, 383–396 [DOI] [PubMed] [Google Scholar]
- 58.Denu J. M., Tanner K. G. (1998) Biochemistry 37, 5633–5642 [DOI] [PubMed] [Google Scholar]