Abstract
How most apoptotic stimuli trigger mitochondrial dysfunction remains to be resolved. We screened the entire Bcl-2 network for its involvement in DNA damage-induced apoptosis in HeLa cells. Although the anti-apoptotic member Bcl-xL served as a major suppressor, apoptosis initiated only when both Mcl-1 and Bcl-xL were eliminated. The pro-apoptotic members Bak, Bad, Bim, and Noxa were required for apoptosis induced by DNA damaging agents camptothecin and UV. We, therefore, used a His-tagged Bcl-xL expression system to capture the relevant BH3-only proteins that bind to Bcl-xL in response to DNA damage. Surprisingly, unlike Bad and Bim, which bound Bcl-xL constitutively, Noxa became “Mcl-1-free” and interacted with Bcl-xL after DNA damage but not after death receptor engagement. Similar observations were also made in A431 cells. Importantly, this induced interaction caused cytochrome c release and apoptosis and was directly inhibited by Mcl-1, a protein eliminated or inactivated after DNA damage. These results suggest that the loss/inactivation of Mcl-1 in conjunction with an induced Noxa/Bcl-xL interaction may serve as a trigger for mitochondrial dysfunction during DNA damage-induced apoptosis.
Keywords: Apoptosis, DNA Damage, Mitochondrial Apoptosis, Signal Transduction, siRNA, BH3-only Proteins, Bcl-2 Network
Introduction
The central roles of the Bcl-2 family proteins as major signal transducers, regulators, and effectors of apoptosis through the mitochondria have been well established (1–4). The Bcl-2 family, characterized by the presence of one or more of four Bcl-2 homology (BH)5 domains, consists of the anti-apoptotic and the pro-apoptotic group, which is further divided into the multi-BH domain and the BH3-only proteins (5, 6). The anti-apoptotic members, including Bcl-2, Bcl-xL, Mcl-1, Bcl-w, and A1, suppress apoptosis presumably by sequestering and inhibiting the pro-apoptotic family proteins. The multi-BH domain members Bax, Bak, and Bok, containing BH1–3, are essential effectors for mitochondrial damage (7, 8). Upon apoptotic stimulation, Bax and Bak become activated, form homo-oligomers, and permeabilize the mitochondrial outer membrane. This latter event leads to the release of apoptogenic factors from mitochondria, most notably cytochrome c, which triggers the activation of the effector caspases and the demise of the cell (9).
The BH3-only proteins, including Bad, Bid, Bik, Bim, Bmf, Bnip3, Hrk, Nix, Noxa, and PUMA, are sentinels for diverse stress signals (10, 11). During apoptosis, one or several of the BH3-only proteins undergo transcriptional or post-translational activation and directly or indirectly activate Bax/Bak. Currently, two major competing models have been suggested to account for the activation of Bax/Bak during apoptosis. The “direct model” proposes that some BH3-only proteins, including Bid (tBid), Bim, and PUMA, directly bind to and activate Bax or Bak (12–19). The “indirect model” suggests that Bax and Bak are active by default but are normally sequestered by the anti-apoptotic family proteins, which are differentially bound and inactivated by different BH3-only proteins during apoptosis (20–23).
Although ligands for cell surface death receptors and endoplasmic reticulum stress initiate mitochondria-dependent apoptosis through the BH3-only proteins Bid and Bim, respectively (24–26), the triggering events leading to mitochondrial dysfunction after most other apoptotic stimuli are less clearly understood. DNA damage, a well known inducer of apoptosis, is known to up-regulate two of the pro-apoptotic BH3-only proteins, PUMA and Noxa through the tumor suppressor p53 (27–29). However, recent studies suggest that Noxa targets only Mcl-1 and A1 (22), and the simultaneous loss of PUMA, Bim, and Bid in mouse embryonic fibroblasts did not block apoptosis induced by DNA damage (30). Furthermore, although elimination of Mcl-1 has been shown to be a prerequisite, it is not sufficient for the initiation of apoptosis after DNA damage (23, 31). Thus, how DNA damage causes Bax/Bak activation and mitochondrial dysfunction remains obscure.
To identify the events triggering mitochondrial dysfunction induced by diverse apoptotic stimuli, it is necessary to interrogate the entire Bcl-2 network in the cell (32). In this study, by combining a systematic loss-of-function approach with a series of biochemical assays, we identified the Bcl-2 family proteins that are critically involved in DNA damage-induced apoptosis in HeLa cells. Importantly, we report an induced interaction capable of triggering mitochondrial dysfunction after DNA damage.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies used were Bcl-xL (Cell Signaling, 2762), Mcl-1 (Santa Cruz Biotechnology, Inc., Sc-819), Bcl-2 (Santa Cruz, Sc-509), Bcl-w (Stressgen, AAP-050), Bax (Santa Cruz, Sc-493), Bad (Cell Signaling, 9292), Bcl-2 (Santa Cruz, Sc-509), Bax (Santa Cruz, Sc-493), Bak (Upstate Biotechnology, Inc., 04-433), Bad (Cell Signaling, 9292), PUMA (Sigma, P4743), Bnip3 (Sigma, B7931), BIM (Chemicon, AB17003), Noxa (Imgenex, IMG-349A), Bid (Luo, 1998), β-actin (Sigma, A5441), FLAG (Sigma, F7415), Bak Ab-1 (Calbiochem, AM03T), cytochrome c (BD Pharmingen, 556433), PARP (Cell Signaling, 9542).
Cell Lines and Cell Culture
HeLa cells and the stable pools of HeLa cells overexpressing different proteins were maintained in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal calf serum. HeLa cells were infected with retrovirus containing pRetroX-Tet-On Advanced plasmid (Clontech) followed by neomycin selection. These activator cells were then infected with pRetroX-Tight Puro retroviral vector expressing the gene of interest and selected by puromycin. Tetracycline-inducible cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% antibiotic-free fetal calf serum.
Plasmid Construction
The cDNAs of human Bcl-xL, human Bcl-2, human Bcl-w, and human Mcl-1 were PCR-amplified using Pfu polymerase with 25 cycles of 1 min at 55 °C, 1 min at 94 °C, and 1 min at 72 °C. The forward and reverse primers contain an XhoI site and an EcoRI site, respectively. The PCR products were digested with XhoI and EcoRI then cloned into the Xho1-EcoR1-digested pPURO MaRX II retroviral vector (a gift from Dr. Jing Wang) modified to include a His9 tag. Human Noxa, NoxaL29E, and GFP were PCR-amplified and cloned into the BamHI and EcoRI sites of the pRetroX-Tight Puro retroviral vector following the protocol described above. Fragments of FLAG-GFP-Noxa, FLAG-GFP-NoxaL29E, and FLAG-GFP were obtained by PCR amplification of GFP-Noxa fusion protein or the mutant, and a FLAG tag was introduced in the N-terminal primer. To construct the GST fusion proteins of Noxa and Bcl-xL, the full-length coding regions of wild-type Noxa, Noxa L29E, and Bcl-xL were cloned into the EcoRI and XhoI sites of pGEX-4T1 vector (GE Healthcare). GST-Mcl-1 was generated by ligating the EcoRI/BamHI-digested human Mcl-1 cDNA into EcoRI and BamHI sites of the pGEX-4T1 vector.
siRNA Transfection
siRNA transfections were performed in 35-mm culture dishes using 2.0 × 105 cells per plate and DharmaFECT1 transfection reagent (Dharmacon) in antibiotic-free Dulbecco's modified Eagle's medium supplemented with 10% serum. All siRNA oligos used were ON-TARGET plus siRNA pools of four oligos purchased from Dharmacon. The final siRNA concentration used was 20 nm unless otherwise specified. Forty-eight hours after transfection cells were either harvested for mRNA and protein analysis or treated by apoptotic stimuli.
Retrovirus Production
All retroviruses were produced by transfecting the plasmid DNA of interest into the 293GP packaging cell line. Retroviral infection of HeLa cells was carried out in the presence of Polybrene (10 μg/ml). Infected cells were selected in puromycin (1.2 μg/ml). After 2 days of selection, cultures were expanded and propagated in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal calf serum.
RT-PCR
Total RNA was isolated from HeLa cells using the RNeasy extraction kit (Qiagen) according to the manufacturer's recommendations. Standard PCR protocol was used to amplify DNA from cDNA samples. Primers used are listed in the supplemental Experimental Procedures.
Apoptosis Quantification
Apoptosis was quantified according to nuclear morphology as previously described (33). Briefly, HeLa cells were stained with Hoechst 33342 at 1 μg/ml (Molecular Probes) for 10 min at 37 °C. Two different viewing areas under the microscope were randomly chosen for a picture, with each area containing between 300 and 600 cells. The percentage of cells undergoing nuclear condensation was calculated for each viewing area. At least three independent experiments were carried out for each condition.
Cytochrome c Release Assay
Cytochrome release assay using mitochondria from HeLa cells or Bcl-xL overexpressing-HeLa cells were described in a previous study (25).
Nickel Bead Pulldown Assay
HeLa cells with or without overexpression of various genes were pelleted by centrifugation at 800 × g for 5 min and were resuspended in EBC buffer (50 mm Tris-HCl, pH 7.5, 120 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) with protease inhibitors and phosphatase inhibitors. After a rotation for 45 min at 4 °C, the lysates were centrifuged at 22,000 × g for 20 min, and the supernatant was incubated with 20 μl of nickel-nitrilotriacetic acid-agarose beads (Qiagen). After a rotation of 3 h at 4 °C, the beads were washed twice by EBC buffer containing 10 mm imidazole. 1×SDS loading dye with 250 mm imidazole was used to elute proteins from nickel-nitrilotriacetic acid beads.
Production of Recombinant Proteins
The expression plasmids were transformed into bacteria BL21 (pLys) cells (Novagen). Two hours after induction, Escherichia coli cells were pelleted by centrifugation and resuspended in phosphate-buffered saline. After sonication, the mixture was centrifuged at 20,000 × g, and the supernatant was loaded onto a 1-ml GST affinity column. The bound protein was eluted with GST elution buffer containing 20 mm glutathione, pH 8.0. The eluted recombinant GST fusion proteins were purified in two additional steps: Mono Q ion exchange and Superdex 200 size exclusion chromatography.
RESULTS
Screening Anti-apoptotic Bcl-2 Proteins for Critical Suppressors of DNA Damage-induced Apoptosis in HeLa Cells
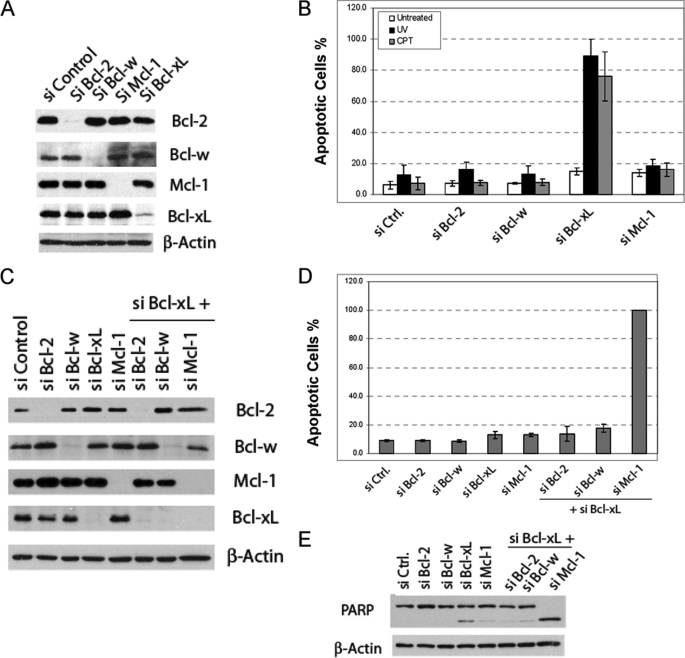
The rate of mitochondria-dependent apoptosis is regulated by both the anti-apoptotic and pro-apoptotic members of the Bcl-2 family (3). To delineate the apoptotic pathway induced by DNA damage, we first sought to identify the inhibitors of this pathway among the anti-apoptotic Bcl-2 family members. Using RNAi, we focused on the identification of proteins that, when eliminated, accelerate DNA damage-induced apoptosis. HeLa cells were transfected with siRNA duplexes against each of the four anti-apoptotic Bcl-2 proteins, Bcl-2, Bcl-w, Bcl-xL, and Mcl-1, and the elimination of these target proteins was monitored by Western blot (Fig. 1A). After transfection, cells were treated with either camptothecin (CPT) or ultraviolet radiation (UV), two known DNA-damaging agents. Apoptosis was quantified 3 h after CPT or UV treatment. Although siRNA against Bcl-2, Bcl-w, Mcl-1, or the control siRNA failed to show any significant effects, siRNA against Bcl-xL dramatically accelerated both CPT- and UV-induced apoptosis, indicating that Bcl-xL is a major endogenous inhibitor for apoptosis induced by DNA damage (Fig. 1B). The 3-h time point allowed us to examine the acceleration of apoptosis by the knockdowns. Similar results were obtained from a 16-h time course experiment after the siRNA knockdown was also carried out (supplemental Fig. S1). Of note, we were unable to detect A1 expression in HeLa cells by either Western blot analysis or RT-PCR (data not shown).
FIGURE 1.
Identification of anti-apoptotic Bcl-2 proteins critical for inhibiting apoptosis induced by DNA damage in HeLa cells. A, knockdown of the anti-apoptotic Bcl-2 proteins by siRNA is shown. HeLa cells were transfected with the indicated pools of siRNA duplexes. After siRNA transfection, cell lysates were generated and subjected to Western blot analysis. B, effects of siRNA knockdown on DNA damage-induced apoptosis in HeLa cells are shown. After siRNA transfection, HeLa cells were treated with either CPT (75 μm) or UV (20 J/m2). Three hours later, apoptosis was quantified by Hoechst staining. The results are the mean ± S.D. of at least three independent siRNA transfections. These same doses were used in all the subsequent experiments involving CPT and UV. C, combinatorial siRNA knockdown of Bcl-xL and other anti-apoptotic Bcl-2 proteins detected by Western blot is shown. D, effects of the combinatorial siRNA knockdown on the apoptosis of HeLa cells are shown. Forty-eight hours after siRNA transfection, apoptosis was quantified by Hoechst staining. The results are the mean ± S.D. from at least three independent siRNA transfections. The apoptosis after knockdown of Mcl-1 and Bcl-xL was 100% in each experiment. E, PARP cleavage induced by Mcl-1 and Bcl-xL double knockdown was assayed by Western blot analysis of the whole cell lysates using PARP antibody. Ctrl, control.
The above results also suggested that elimination or inactivation of Bcl-xL may represent a rate-limiting step during apoptosis. However, because Bcl-xL knockdown did not cause apoptosis spontaneously, it is possible that inactivation or loss of other anti-apoptotic family proteins might be also necessary to trigger apoptosis. We, therefore, carried out combinatorial siRNA transfections pairing siRNA against Bcl-xL with that of Bcl-2, Bcl-w, or Mcl-1 (Fig. 1C). The pairing between Mcl-1 siRNA and that of Bcl-xL, but not any other pairing, caused robust apoptosis without CPT or UV treatment, indicating that the simultaneous inactivation or elimination of Bcl-xL and Mcl-1 is sufficient for apoptosis induction (Fig. 1, D and E). Together, these results suggest that although Bcl-xL is a primary suppressor for DNA damage-induced apoptosis, both Bcl-xL and Mcl-1 need to be eliminated or inactivated to initiate apoptosis.
Involvement of Multidomain Pro-apoptotic Bcl-2 Family Proteins in DNA Damage-induced Apoptosis
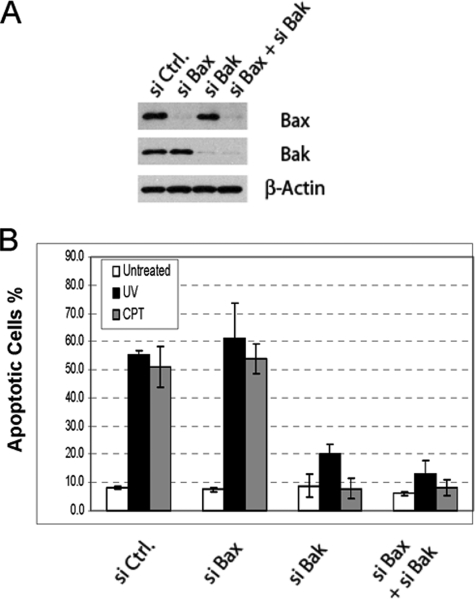
To further delineate the apoptotic pathway of DNA damage-induced apoptosis, we set out to identify the critical pro-apoptotic Bcl-2 family members in this pathway. We first examined the involvement of the two pro-apoptotic multi-BH domain Bcl-2 proteins, Bax and Bak. After siRNA knockdown of Bax and Bak, either separately or in combination, HeLa cells were treated with either UV or CPT. Sixteen hours later, apoptosis was examined by Hoechst staining. Although the knockdown of Bax had no effect, knockdown of either Bak alone or Bax and Bak together abolished apoptosis induced by either UV or CPT (Fig. 2B). Thus, in HeLa cells, Bak, but not Bax, is the major effector for mitochondrial damage induced by DNA damage.
FIGURE 2.
Involvement of multidomain pro-apoptotic Bcl-2 proteins in DNA damage-induced apoptosis. A, shown is siRNA knockdown of Bax, Bak, individually or both in HeLa cells detected by Western blot analysis. Ctrl, control. B, effects of the siRNA knockdown on DNA damage-induced apoptosis in HeLa cells. After siRNA transfection, cells were treated with CPT (75 μm) or UV (20 J/m2). Sixteen hours later apoptosis was quantified by Hoechst staining. The results are the mean ± S.D. from at least three independent siRNA transfections.
Involvement of BH3-only Proteins in CPT-induced Apoptosis
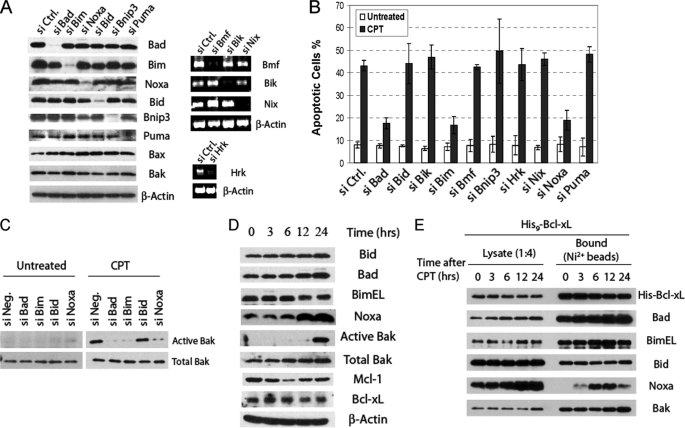
Next, siRNA knockdown was carried out to screen the 10 known BH3-only proteins for their involvement in CPT-induced apoptosis (Fig. 3). Significant knockdown of Bid, Bad, PUMA, Noxa, Bnip3L, and BimEL was verified by Western blot analysis. Whereas antibodies from multiple sources failed to detect Bik, Hrk, Nip3, and Bmf in HeLa cells, we were able to detect the mRNAs as well as the specific knockdown of these genes with RT-PCR (Fig. 3A). After siRNA transfection, cells were treated with CPT for 16 h before apoptosis was quantified. Although siRNA pools against most BH3-only proteins showed no significant effects, siRNAs against Bad, Bim, or Noxa showed strong protection against CPT (Fig. 3B). Using an epitope-specific antibody against active Bak, we found that siRNAs against Bad, Bim, or Noxa significantly inhibited Bak activation (Fig. 3C). Thus, Bad, Bim, and Noxa are critically involved in CPT-induced apoptosis.
FIGURE 3.
Selective involvement of BH3-only proteins in CPT-induced apoptosis. A, siRNA knockdown of the BH3-only proteins is shown. After siRNA transfection, the six proteins on the left were examined by Western blot with their respective antibodies. On the right, mRNAs of Bmf, Bik, Nix, and Hrk were amplified by RT-PCR with gene-specific primers to give rise to DNA fragments of expected sizes (supplemental Experimental Procedures). Ctrl, control. B, effects of the siRNA knockdown on CPT-induced apoptosis in HeLa cells are shown. The siRNA transfection, CPT treatment, and apoptosis detection were as described in Fig. 2B. The results are the mean ± S.D. from at least three independent siRNA transfections. C, shown are the effects of siRNA knockdown on Bak activation after CPT treatment. After siRNA transfection, cells were either treated with CPT (75 μm) or vehicle for 16 h. Bak activation was detected by immunoprecipitation with the epitope-specific antibody Ab-1 followed by Western blot analysis as described under “Experimental Procedures.” D, time course of the relevant Bcl-2 family proteins after CPT treatment is shown. HeLa cell lysates were generated at different time points after CPT treatment and analyzed by Western blot. E, shown is a nickel bead pulldown analysis of His9-Bcl-xL cells after CPT treatment. HeLa cells overexpressing Bcl-xL with a His9 tag (His9-Bcl-xL) were treated with CPT for the indicated times and harvested in EBC buffer containing 0.5% Nonidet P-40. Cell lysates were subjected to nickel bead pulldown. The bound proteins were eluted with imidazole and analyzed by Western blot.
To explore the potential mechanisms of how Bad, Bim, and Noxa are involved in the activation of Bak after CPT treatment, we examined these proteins as well as Bcl-xL and Mcl-1 in whole cell lysates by Western blot in a detailed time course analysis. At parallel time points, Bak activation was monitored (Fig. 3D). Bak activation reached the highest level between 12 and 24 h. Importantly, although levels of Bad and Bim showed little changes, the Noxa level was significantly elevated after 12 h of CPT treatment. Bcl-xL levels remained constant throughout the time course, whereas Mcl-1 level decreased initially but gradually recovered after 12 h.
As Bad, Bim, and Noxa are the only three BH3-only proteins critically involved in CPT-induced apoptosis, what are their targets? Because Bcl-xL is the major inhibitor in this process and the elimination of both Bcl-xL and Mcl-1 suffices to induce apoptosis, we initially attempted to examine the interaction between endogenous Bcl-xL and these three BH3-ony proteins after CPT treatment. Although Bad and BimEL constitutively interacted with endogenous Bcl-xL, no interaction was detected between Noxa and endogenous Bcl-xL after CPT treatment. To enhance the sensitivity of this assay, we resorted to HeLa cells engineered to overexpress a polyhistidine-tagged Bcl-xL (His9-BclxL). We reasoned that the initial triggering protein should bind to the His9-Bcl-xL in a CPT-inducible fashion. At different time points after CPT treatment, whole cell lysates were generated and were subjected to pulldown by nickel beads. As shown in Fig. 3E, the association of Bad, Bid, Bim, and Bak with His9-Bcl-xL remained constant throughout the time course. Surprisingly, although no association was detected before CPT treatment, Noxa became associated with Bcl-xL 3 h after treatment, reaching maximum levels between 6 and 12 h. Of importance, these interactions were strictly dependent on His9-Bcl-xL, as no interactions were detected by the nickel bead pulldown in control HeLa cells (supplemental Fig. S2). In addition, we verified this CPT-induced interaction in CHAPS buffer, which is known to preserve the conformation of the Bcl-2 family proteins (supplemental Fig. S3). Together, these results suggest that although Bad and Bim may contribute to apoptosis triggering by sequestering Bcl-xL constantly, Noxa may trigger apoptosis by interacting with and inactivating Bcl-xL after CPT treatment.
Involvement of Bad, Bim, and Noxa in UV-induced Apoptosis
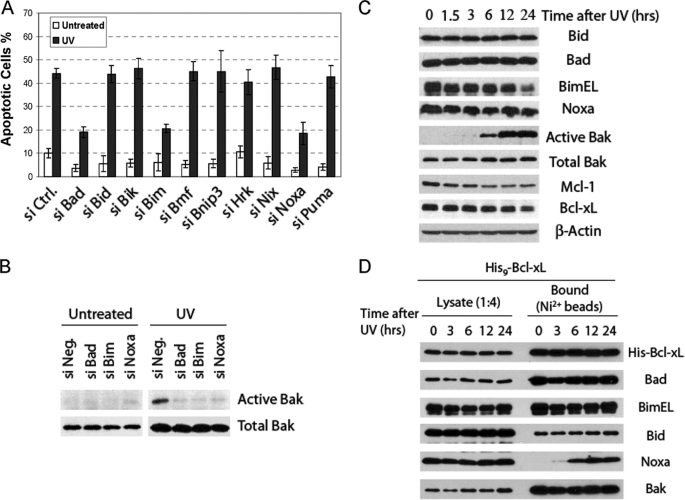
Next, we screened the BH3-only proteins for their involvement in apoptosis induced by UV, another known DNA damaging agent. Similar to the finding in CPT-induced apoptosis, of the 10 known BH3-only proteins, loss of Bad, Bim, or Noxa, strongly suppressed apoptosis induced by UV (Fig. 4A). Consistent with this observation, Bak activation after UV treatment was compromised in cells transfected with siRNAs against Bim, Noxa, or Bad (Fig. 4B). To explore the potential mechanisms of how Bad, Bim, and Noxa are involved in the activation of Bak after UV, we monitored these proteins as well as Bcl-xL, Mcl-1, and Bak in a detailed time course analysis. As shown in Fig. 4C, Bak activation started at 6 h and was maximal at 12 h. In contrast to results with CPT-treated cells (Fig. 3D), the level of Noxa remained relatively constant throughout the time course. Also, Mcl-1 levels decreased over time, consistent with a proteasome-mediated degradation of Mcl-1 after UV treatment (31).
FIGURE 4.
Involvement of Bim, Bad, and Noxa in UV-induced apoptosis. A, effects of siRNA knockdown on UV-induced apoptosis in HeLa cells are shown. After siRNA transfection, cells were treated with UV (20 J/m2). Twelve hours later, Hoechst dye was added to the cells to quantify for apoptosis. The results are the mean ± S.D. from at least three independent siRNA transfections. Ctrl, control. B, effects of siRNA knockdown on Bak activation after UV treatment are shown. Immunoprecipitation and detection of active Bak was carried out similarly to Fig. 3C. C, time course of the relevant Bcl-2 family proteins after UV treatment is shown. HeLa cell lysates were generated at different time points after UV treatment and analyzed by Western blot. D, shown is binding of Bak, Bad, Bid, BimEL, and Noxa to Bcl-xL in a HeLa cell line overexpressing His-tagged Bcl-xL. His9-Bcl-xL HeLa cells were treated with UV and harvested in EBC buffer containing 0.5% Nonidet P-40 at the indicated time points. Cell lysates were analyzed by a nickel bead pulldown assay as described in Fig. 3E.
We also used the His9-Bcl-xL pulldown assay to examine the relevant BH3-only proteins for their involvement in the inactivation of Bcl-xL after UV. His9-Bcl-xL cells were treated with UV and harvested at different time points. Among the three BH3-only proteins involved, both Bad and Bim bound Bcl-xL constitutively (Fig. 4D). In contrast, although no interaction was detected before treatment, a dramatic increase in the binding of Noxa to Bcl-xL preceding Bak activation was observed after UV treatment. Similar to the observation in CPT-treated cells, this UV-induced interaction was verified in CHAPS buffer (supplemental Fig. S3). Taken together, these results identify Bad, Bim, and Noxa as BH3-only proteins critical for the apoptotic pathway in response to UV. Importantly, the UV-dependent binding of Noxa to Bcl-xL may serve as a triggering event leading to mitochondrial dysfunction.
BH3-dependent Bcl-xL Binding, Cytochrome c Release, and Apoptosis Induction by Noxa
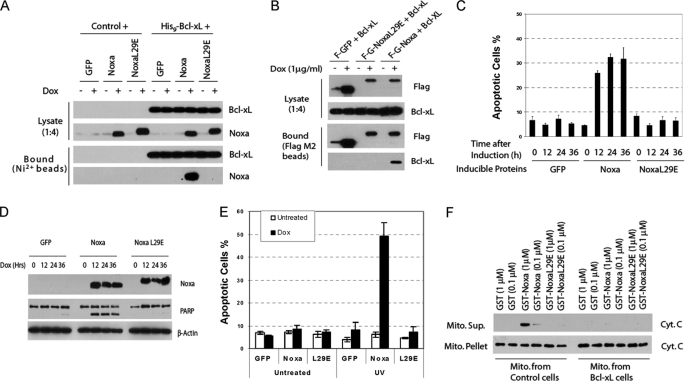
To investigate the specificity and function of the DNA damage-induced interaction between Noxa and Bcl-xL (Figs. 3 and 4), we established a stable pool of cells expressing Noxa or its BH3 mutant (L29E) under the control of a doxycycline (Dox)-inducible promoter (“Tet-on”). We first examined the interaction between Noxa and Bcl-xL by retroviral expression of His9-Bcl-xL in these cells. Noxa bound to His9-Bcl-xL after Dox induction (Fig. 5A). In contrast, no binding was detected when the BH3 domain mutant (L29E) Noxa was used. We also tested this interaction in a reciprocal fashion using an untagged Bcl-xL and a Tet-on controlled FLAG-tagged GFP-Noxa fusion protein or its BH3 mutant (Fig. 5B). We were able to pull down the expressed Bcl-xL by the FLAG-tagged GFP-Noxa but not by its BH3 mutant (L29E).
FIGURE 5.
BH3-dependent Bcl-xL binding, cytochrome release, and apoptosis induction by Noxa. A, pulldown of Noxa by Bcl-xL is BH3-dependent. HeLa cells expressing the indicated proteins under the control of tetracycline (Tet-on) were infected with a retrovirus expressing either vector or His9-Bcl-xL. After induction with Dox (1 μg/ml) for 8 h, cell lysates were subjected to nickel bead pulldown as described in Fig. 3E. B, pulldown of Bcl-xL by Noxa is BH3-dependent. HeLa cells expressing FLAG-GFP, FLAG-GFP-Noxa, or FLAG-GFP-NoxaL29E under the control tetracycline were infected with a retrovirus expressing either vector or Bcl-xL. After Dox induction, cell lysates were subjected to nickel bead pulldown as described in Fig. 3E. C, BH3-dependent apoptotic activity of Noxa is shown. The Tet-on HeLa cells expressing GFP, Noxa, or NoxaL29E were induced by Dox for the indicated times. Apoptosis was quantified by Hoechst staining. The results are the mean ± S.D. from at least three independent experiments. D, BH3-dependent PARP cleavage activity of Noxa is shown. Cell lysates from C were analyzed by Western blot. E, sensitization to UV-induced apoptosis by Noxa is shown. The Tet-on HeLa cells expressing GFP, Noxa, or NoxaL29E were induced by Dox for 3 h before being subjected to UV (20 J/m2) treatment. Six hours later, apoptosis was quantified by Hoechst staining. The results are the mean ± S.D. from two independent experiments. F, BH3-dependent in vitro cytochrome c release activity of Noxa is shown. Mitochondria were isolated from regular or Bcl-xL-overexpressing HeLa cells. Recombinant GST or GST-Noxa fusion protein (supplemental Fig. S4) was incubated with mitochondria at 30 °C for 1 h. The supernatant (Sup.) and the mitochondrial (Mito.) pellets were analyzed by Western blot using a cytochrome c antibody.
Next, HeLa cells expressing GFP, Noxa, or NoxaL29E under Tet-on control were used to examine the apoptotic activity of Noxa. Noxa, but not GFP or Noxa L29E, displayed a significant apoptotic activity as well as PARP cleavage (Fig. 5, C and D). To investigate the involvement of Noxa in UV-induced apoptosis, we induced the Noxa expression for 3 h before treating the cells with UV. The expression of Noxa greatly sensitized the cells to UV-induced apoptosis (Fig. 5E). Combined with the effects of the siRNA knockdowns of Bcl-xL or Mcl-1 (Fig. 1), these results strongly suggested that Noxa targets not only Mcl-1 but also Bcl-xL in vivo.
To further examine the biochemical activity of Noxa, we generated recombinant GST-Noxa and GST-NoxaL29E (supplemental Fig. S4) and tested their effects on mitochondria in vitro. GST-Noxa, but not GST or GST-Noxa (L29E), induced a significant cytochrome c release from HeLa cell mitochondria. This release was completely inhibited using mitochondria from Bcl-xL-overexpressing HeLa cells (Fig. 5F). These results indicate that Noxa is capable of interacting with Bcl-xL, causing cytochrome c release and inducing apoptosis in a BH3-dependent fashion.
Mcl-1 Regulates the Interaction between Noxa and Bcl-xL
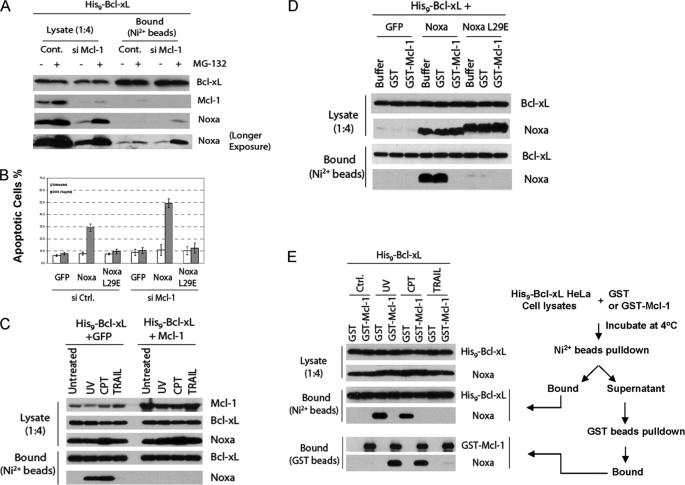
Given the strong affinity between Noxa and Mcl-1 (22), we hypothesized that Mcl-1 might be a negative regulator for this interaction. We, therefore, lowered the expression of Mcl-1 by siRNA in His9-Bcl-xL cells and tested the Noxa/Bcl-xL interaction. Interestingly, knockdown of Mcl-1 in HeLa cells caused a strong decrease of endogenous Noxa, suggesting that Mcl-1 might regulate the stability of Noxa (Fig. 6A). Therefore, MG-132 was added to elevate the expression of Noxa. Knockdown of Mcl-1 in combination with MG-132 significantly enhanced the binding of Noxa to His9-Bcl-xL (Fig. 6A). Furthermore, we found that eliminating Mcl-1 significantly enhanced Noxa induced apoptosis, supporting the role of Mcl-1 as an endogenous inhibitor for Noxa/Bcl-xL interaction (Fig. 6B). We next tested the effect of an elevated level of Mcl-1 on the induced interaction between Noxa and Bcl-xL. HeLa cells co-expressing His9-Bcl-xL with either GFP or Mcl-1 were generated and treated with UV or CPT. As a control, these cells were treated with TRAIL, which induces apoptosis in HeLa cells through the cell surface death receptors. Although TRAIL failed to induce any interaction, both UV and CPT induced a robust Noxa/Bcl-xL interaction, which is completely abolished by the expression of Mcl-1 (Fig. 6C).
FIGURE 6.
Mcl-1 inhibits the interaction between Noxa and Bcl-xL through sequestration. A, effects of Mcl-1 knockdown on Noxa/Bcl-xL interaction are shown. After siRNA transfection, MG-132 (10 μm) or vehicle was added to the His9-Bcl-xL cells. Two hours later cell lysates were subjected to nickel bead pulldown as described in Fig. 3E. Cont, control. B, effects of Mcl-1 knockdown on the apoptotic activity of Noxa are shown. After siRNA transfection, the Tet-on HeLa cells expressing the indicated proteins were induced by Dox for 24 h. Apoptosis was quantified by Hoechst staining. The results are the mean ± S.D. from at least three independent experiments. Ctrl, control. C, inhibition of DNA damage-induced Noxa/Bcl-xL interaction by Mcl-1 in HeLa cells. His9-Bcl-xL cells co-expressing either GFP or Mcl-1 were treated with UV (20 J/m2), CPT (75 μm), or TRAIL (10 ng/ml). Twelve hours later, cell lysates were subjected to nickel bead pulldown as described in Fig. 3E. D, shown is inhibition of Noxa/Bcl-xL interaction by Mcl-1 in vitro. HeLa cells co-expressing His9-Bcl-xL with GFP, Noxa, or NoxaL29E were generated by retroviral infection. Cell lysates were incubated with buffer, GST, or GST-Mcl-1 at 30 °C for 1 h before being subjected to nickel bead pulldown and Western blot analysis. E, inhibition of the DNA damage induced Noxa/Bcl-xL interaction by Mcl-1 through sequestration. His9-Bcl-xL cells were treated with UV, CPT, or TRAIL as described in C. Cell lysates were incubated with either GST or GST-Mcl-1. The mixtures were subject to nickel bead pulldown. The unbound fractions were subjected to GST bead pulldown. The lysates and bound proteins were analyzed by Western blot.
To understand the mechanism of this inhibition, we resorted to the in vitro system using extracts from cells coexpressing His9-Bcl-xL with GFP, Noxa, or NoxaL29E under the tetracycline control. After Dox induction, whole cell lysates were harvested for the nickel bead pulldown assay. Although a robust Noxa/Bcl-xL interaction was detected in the presence of GST or buffer, the addition of GST-Mcl-1 completely suppressed the interaction (Fig. 6D). We also tested the effect of Mcl-1 on the induced Noxa/Bcl-xL interaction using whole cell lysates from the His9-Bcl-xL cells treated with CPT, UV, or TRAIL. His9-Bcl-xL was able to pull down a significant amount of Noxa after CPT and UV but not TRAIL treatment. However, the addition of GST-Mcl-1 completely abolished this induced interaction (Fig. 6E). After the nickel bead pulldown, the supernatant was subjected to GST pulldown. Endogenous Noxa was found associated with GST-Mcl-1 but only after UV or CPT (Fig. 6E), indicating that GST-Mcl-1 inhibited the induced interaction by sequestering Noxa away from His9-Bcl-xL. Meanwhile, the dramatic increase of Noxa captured by GST-Mcl-1 in vitro after UV or CPT treatment suggests the generation of a large excess of Noxa over endogenous Mcl-1 in the lysates. Nevertheless, these results suggest that Mcl-1 is a direct inhibitor of the UV- or CPT-induced Noxa/Bcl-xL interaction through sequestration of Noxa.
Generation of Mcl-1-free Noxa and Formation of Bcl-xL/Noxa Complex after DNA Damage
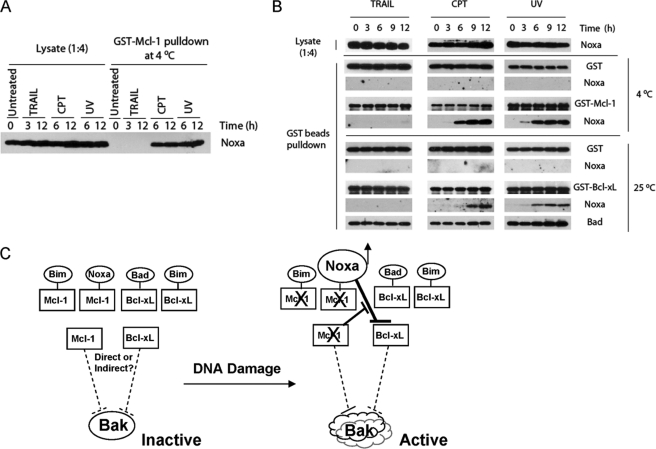
Considering the strong affinity between endogenous Noxa and Mcl-1 (supplemental Fig. S5) and the potent inhibitory effect of Mcl-1 on Noxa/Bcl-xL interaction, it is critical to know if Mcl-1-free Noxa can be generated after DNA damage. We, therefore, incubated GST-Mcl-1 with extracts from HeLa cells that were either untreated or treated by CPT, UV, or TRAIL. Using extracts from untreated cells, no Noxa was detected in the GST-Mcl-1 pulldown. However, 6 and 12 h after UV and CPT treatment, respectively, large amounts of endogenous Noxa were captured in the cell extracts by GST-Mcl-1, indicating the generation of Mcl-1-free Noxa after DNA damage. In contrast, TRAIL treatment did not produce any Mcl-1-free Noxa.
To directly test the interaction between Noxa and Bcl-xL in vitro, we added GST or GST-Bcl-xL into HeLa cell lysates harvested at different time points after TRAIL, UV, and CPT (Fig. 7B). Because the Mcl-1-free Noxa is likely to form a complex with endogenous Bcl-xL, we carried out the incubation at room temperature to allow for exchange of Noxa between endogenous Bcl-xL and the GST-Bcl-xL. GST-Bcl-xL was able to bind endogenous Noxa 6 h after UV or CPT treatment but not after TRAIL, roughly coinciding with the appearance of the Mcl-1-free Noxa as detected by GST-Mcl-1 pulldown at 4 °C. Taken together, these results indicate that DNA damage caused a dramatic increase of Mcl-1-free Noxa, which can subsequently interact with Bcl-xL.
FIGURE 7.
Generation of Mcl-1-free Noxa and its binding to Bcl-xL after DNA damage. A, the appearance of Mcl-1-free Noxa after DNA damage is shown. HeLa cells were treated with TRAIL (10 ng/ml), CPT (75 μm), or UV (20 J/m2) and harvested at the indicated time points. Cell lysates (1.0 mg) were incubated with GST-Mcl-1 at 4 °C for 1 h. The mixtures were subjected to GST pulldown. The bound proteins were analyzed with Western blot using Noxa antibody. B, the association of Noxa with Bcl-xL in vitro after DNA damage is shown. HeLa cells were treated with TRAIL, CPT, or UV and harvested at the indicated time points. The whole cell lysates (1.6 mg) were incubated with either GST-Mcl-1 (100 nm) at 4 °C or with GST-Bcl-xL (120 nm) at 25 °C for 1 h before GST pulldown. In both cases GST was used as a negative control. The bound proteins were eluted with glutathione and analyzed with Western blot analysis. C, shown is a model for DNA damage-induced apoptosis through the Bcl-2 network in HeLa cells. Bcl-xL and Mcl-1 are the major inhibitors of DNA damage-induced apoptosis in HeLa cells. They exist in the cell as inactive and active forms. Although the inactive forms are constitutively bound by BH3-only proteins, the active forms inhibit Bak activation either directly by sequestering Bak or indirectly by sequestering the activator BH3-only proteins. During DNA damage, up-regulation of Noxa combined with the elimination or inactivation of Mcl-1 generated Mcl-1-free Noxa, which in turn binds to and inactivates Bcl-xL. The inactivation of both Mcl-1 and Bcl-xL leads to Bak activation.
DISCUSSION
How various stress signals are translated into mitochondrial damage remains one of the core questions in the study of apoptosis. In this work we used a combination of loss-of-function and biochemical approaches to systematically analyze the entire Bcl-2 network for its involvement in DNA damage-induced apoptosis. Our results identify and define the actions of selective Bcl-2 family proteins responsible for triggering mitochondrial dysfunction after DNA damage (Fig. 7C).
Selective Involvement of the Bcl-2 Family Proteins in DNA Damage-induced Apoptosis
Through siRNA screening, we were able to identify the critical Bcl-2 family proteins involved in DNA damage-induced apoptosis. Among the anti-apoptotic family proteins, Bcl-xL, whose knockdown greatly sensitized cells, appears to be the primary suppressor of DNA damage-induced apoptosis in HeLa cells. However, because the elimination of both Bcl-xL and Mcl-1 was sufficient to initiate apoptosis, Mcl-1 qualified as the other gateway protein guarding against apoptosis. This conclusion is supported by the strong synergy in apoptosis induction between Noxa and Bad (21, 22).
The identification of Bad as a critical player in DNA-damage-induced apoptosis is surprising because Bad−/− mice displayed only a weak apoptosis phenotype in lymphoid cells (34). However, the clear blockade of DNA damage-induced apoptosis by the knockdown of Bad in HeLa cells suggests that the involvement of Bad may be cell type-specific. It is possible that in some cancer cells, a fraction of Bcl-xL is bound by Bad constitutively (supplemental Fig. S5 and Fig. 7C). Therefore, the elimination of Bad is expected to increase the amount of active Bcl-xL in the cell. Similarly, because endogenous Bim and Bcl-xL formed a complex before treatment (supplemental Fig. S5), the knockdown of Bim should also lead to a similar increase of active Bcl-xL, which in turn inhibits apoptosis. Our finding is consistent with the phenotype of the Bim−/− mice, which displayed mild but significant resistance to DNA damage-induced apoptosis in multiple cell types (35). Although the induced interaction between Noxa and Bcl-xL was surprising, the blockade of DNA damage-induced apoptosis after Noxa knockdown is consistent with previous studies in multiple cell types (36–38). In A431 cells, we also found that Bim, Bad, and Noxa are necessary for UV-induced apoptosis (supplemental Fig. S6). In addition, we found that Noxa is critically involved in BxPC3 cells (supplemental Fig. S7). Because the cells used in this study do not contain functional p53, these results do not exclude the possibility that p53 may play an important role in DNA damage-induced apoptosis. It has been demonstrated that wild-type p53 perturbs the Bcl-2 network by directly interacting with Bax or releasing PUMA from Bcl-xL after DNA damage (39).
Induced Interaction between Noxa and Bcl-xL after DNA Damage
The dramatic induction of association between Noxa and Bcl-xL appears to be tightly regulated. First, Mcl-1 negatively regulates this interaction. The in vivo and in vitro studies (Fig. 6) strongly suggest that Mcl-1 is a major endogenous inhibitor of the Noxa/Bcl-xL interaction. Combined with the observation that Mcl-1 elimination or inactivation occurs at an early stage after DNA damage, these results place Mcl-1 as an apical anti-apoptotic Bcl-2 protein (31), protecting Bcl-xL from Noxa.
Second, the induced interaction is positively regulated by additional events that elevate or maintain the Noxa level in response to DNA damage. It was apparent that the elimination of Mcl-1 alone is not sufficient for the induction of Noxa/Bcl-xL interaction or apoptosis (Fig. 6, A and B). After CPT or UV treatment, whereas Mcl-1 decreased at least in the early time points, Noxa was either up-regulated or kept at the same level. Interestingly, in HeLa cells overexpressing Mcl-1, we observed a strong up-regulation of Noxa after UV treatment, suggesting that a transcriptional up-regulation of Noxa may be a general phenomenon for DNA damage (supplemental Fig. S8). However, this up-regulation does not appear to be mediated by p53, as knockdown of p53 in HeLa cells did not affect apoptosis induced by DNA damage (data not shown). Nonetheless, the differential regulation of Noxa and Mcl-1 seems sufficient for the induced Noxa/Bcl-xL interaction as well as the induction of apoptosis after DNA damage (Figs. 5 and 6). It is also worth mentioning that this induced interaction is not restricted to HeLa cells. Using A431 cells, we were also able to observe a UV-induced Noxa/Bcl-xL interaction (supplemental Fig. S9).
The interaction between Noxa and Bcl-xL appears weak as we were unable to detect Noxa in immunoprecipitation using Bcl-xL antibody after DNA damage. However, because both Noxa and Bcl-xL are found on the mitochondrial outer membrane, their interaction in the microenvironment may be underestimated in the detergent extracts (40). Moreover, Noxa may not need to bind all the Bcl-xL proteins because a fraction of the Bcl-xL molecules are already complexed with the BH3-only proteins in some cancer cells.
It is worth noting that most of our studies were carried out in HeLa cells. It is possible that in some other cell types, especially different cancer cells, a different set of critical proteins might be involved depending on a different composition of the Bcl-2 network. However, it appears that Noxa is critically involved in DNA damage induced apoptosis in most cell types. Based on our results, the induced Noxa/Bcl-xL interaction should at least contribute to the triggering of mitochondrial dysfunction after DNA damage.
Triggering Events Leading to Mitochondrial Dysfunction after DNA Damage
After DNA damage, two major events seem to be necessary and sufficient for the induction of apoptosis. One is the elimination or inactivation of Mcl-1, and the other is the binding of Noxa to Bcl-xL. The latter event is preceded by the generation of the large excess of Mcl-1-free Noxa (Fig. 7C). As a major rate-limiting factor for Bak activation and apoptosis, Bcl-xL appears to be inactivated by Noxa after DNA damage (Fig. 7C). According to the direct activation model, this inactivation may cause Bcl-xL to release Bim, which in turn directly activates Bak. On the other hand, the indirect activation model predicts that Noxa may displace Bak from Bcl-xL, therefore, allowing Bak to undergo spontaneous activation. Whereas the weak interaction between Noxa and Bcl-xL in detergent lysates makes it difficult to detect a displacement of either Bim or Bak from Bcl-xL by Noxa, considering the localization of both Noxa and Bcl-xL on the mitochondrial outer membrane, such a displacement is certainly a possibility (40). Alternatively, it remains possible that Bcl-xL may prevent Bax/Bak activation through other unknown mechanisms (41). Thus, a mere neutralization of Bcl-xL by Noxa in conjunction with a loss or inactivation of Mcl-1should be sufficient to trigger Bak activation and subsequent apoptosis (Fig. 1) (32, 42).
Overall, this study defined the critical components of the DNA damage-induced mitochondrial pathway. Our results also suggest that an inducible Noxa/Bcl-xL contributes to apoptosis triggering. It will be of great interest to examine the generation of Mcl-1-free Noxa and the induced Noxa/Bcl-xL interaction in other apoptotic pathways.
Acknowledgments
We thank Drs. Robert Lewis, Joyce Solheim, and Manabu Furukawa for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM76237 and GM76237-04S1. This work was also supported by a pilot grant from the Nebraska Center for Cellular Signaling (to X. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S9.
- BH
- Bcl-2 homology
- CPT
- camptothecin
- Dox
- doxycycline
- PARP
- poly(ADP-ribose) polymerase
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- RT
- reverse transcription
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand.
REFERENCES
- 1.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 2.Green D. R., Kroemer G. (2004) Science 305, 626–629 [DOI] [PubMed] [Google Scholar]
- 3.Adams J. M., Cory S. (2007) Curr. Opin. Immunol. 19, 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green D. R., Reed J. C. (1998) Science 281, 1309–1312 [DOI] [PubMed] [Google Scholar]
- 5.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 6.Gross A., McDonnell J. M., Korsmeyer S. J. (1999) Genes Dev. 13, 1899–1911 [DOI] [PubMed] [Google Scholar]
- 7.Lindsten T., Ross A. J., King A., Zong W. X., Rathmell J. C., Shiels H. A., Ulrich E., Waymire K. G., Mahar P., Frauwirth K., Chen Y., Wei M., Eng V. M., Adelman D. M., Simon M. C., Ma A., Golden J. A., Evan G., Korsmeyer S. J., MacGregor G. R., Thompson C. B. (2000) Mol. Cell 6, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X., Wang X. (2004) Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 10.Huang D. C., Strasser A. (2000) Cell 103, 839–842 [DOI] [PubMed] [Google Scholar]
- 11.Bouillet P., Strasser A. (2002) J. Cell Sci. 115, 1567–1574 [DOI] [PubMed] [Google Scholar]
- 12.Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 14.Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 15.Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. (2006) Mol. Cell 24, 199–210 [DOI] [PubMed] [Google Scholar]
- 16.Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 17.Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. (2008) Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Certo M., Del Gaizo Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S. A., Letai A. (2006) Cancer Cell 9, 351–365 [DOI] [PubMed] [Google Scholar]
- 19.Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 20.Willis S. N., Adams J. M. (2005) Curr. Opin. Cell Biol. 17, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 23.Cuconati A., Mukherjee C., Perez D., White E. (2003) Genes Dev. 17, 2922–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Zhu H., Xu C. J., Yuan J. (1998) Cell 94, 491–501 [DOI] [PubMed] [Google Scholar]
- 25.Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 26.Puthalakath H., O'Reilly L. A., Gunn P., Lee L., Kelly P. N., Huntington N. D., Hughes P. D., Michalak E. M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., Strasser A. (2007) Cell 129, 1337–1349 [DOI] [PubMed] [Google Scholar]
- 27.Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. (2000) Science 288, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 28.Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 29.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 30.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 31.Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. (2003) Genes Dev. 17, 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chipuk J. E., Green D. R. (2008) Trends Cell Biol. 18, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George N. M., Evans J. J., Luo X. (2007) Genes Dev. 21, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranger A. M., Zha J., Harada H., Datta S. R., Danial N. N., Gilmore A. P., Kutok J. L., Le Beau M. M., Greenberg M. E., Korsmeyer S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9324–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Köntgen F., Adams J. M., Strasser A. (1999) Science 286, 1735–1738 [DOI] [PubMed] [Google Scholar]
- 36.Naik E., Michalak E. M., Villunger A., Adams J. M., Strasser A. (2007) J. Cell Biol. 176, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibue T., Takeda K., Oda E., Tanaka H., Murasawa H., Takaoka A., Morishita Y., Akira S., Taniguchi T., Tanaka N. (2003) Genes Dev. 17, 2233–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villunger A., Michalak E. M., Coultas L., Müllauer F., Böck G., Ausserlechner M. J., Adams J. M., Strasser A. (2003) Science 302, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 39.Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004) Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 40.Leber B., Lin J., Andrews D. W. (2007) Apoptosis 12, 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minn A. J., Kettlun C. S., Liang H., Kelekar A., Vander Heiden M. G., Chang B. S., Fesik S. W., Fill M., Thompson C. B. (1999) EMBO J. 18, 632–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cory S., Adams J. M. (2002) Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]