Abstract
Cheirosporium gen. nov. is characterised by the production of sporodochial conidiomata, semi-macronematous to macronematous conidiophores that possess several distinct sterile branches, and cheiroid, smooth-walled conidia with rhexolytic secession. The 28S rDNA and ITS rDNA operon of this taxon were amplified and sequenced. A BLAST search revealed low homology between Cheirosporium triseriale and existing sequences in public databases, supporting the hypothesis that the species is new to science. Phylogenetic analysis showed that C. triseriale groups with Dictyosporium and allied species, and nests within the Pleosporales (Dothideomycetes, Ascomycota). Cheirosporium is morphologically distinct from the cheirosporous genera Cheiromyces, Cheiromycina, Dictyosporium, Digitomyces, Digitodesmium and Pseudodictyosporium and these differences are discussed.
Keywords: ascomycetes, Pleosporales, systematics, taxonomy
INTRODUCTION
Freshwater fungi are taxonomically diverse, with more than 1 000 documented species, which include representatives from almost all major fungal classes (Tsui & Hyde 2003, Vijaykrishna et al. 2006). These fungi colonise many different substrates such as submerged plant litter, and aquatic organisms (Vijaykrishna & Hyde 2006). Because most plant litter in freshwater is of terrestrial origin, the aquatic origin of most freshwater fungi is debatable. Many of the known freshwater anamorphic species have also been recorded from terrestrial habitats, although this is not generally the case with teleomorphs (Tsui & Hyde 2003, Vijaykrishna et al. 2006).
Cheirosporous hyphomycetes were studied recently by Tsui et al. (2006) and Kodsueb et al. (2007), while Ho et al. (2000) reviewed cheirosporous genera and provided a key based on morphological characters. Tsui et al. (2006) investigated the phylogenetic relationships among several cheirosporous genera and found that Dictyosporium and allied genera are closely related and form a strong monophyletic grouping in Pleosporales. New species of cheirosporous fungi have also recently been described, such as Aquaticheirospora, including a single species with synnematous conidiomata (Kodsueb et al. 2007). We are studying the freshwater fungi of China (e.g. Cai et al. 2006a, Cai & Hyde 2007), and collected an interesting cheirosporous taxon from Lijiang, Yunnan. Critical morphological examination and phylogenetic analysis based on 28S and ITS rDNA sequences showed that this fungus represents a new ascomycetous anamorph that could not be linked to a teleomorphic genus. It is, therefore, described here as Cheirosporium triseriale gen. & sp. nov.
MATERIALS AND METHODS
Submerged woody substrata were collected by Cai from a small stream in Yunnan, China. Samples were processed and examined following the methods described in Cai et al. (2003). Observations and photomicrographs were made from material mounted in water. Conidia were measured at their widest point. The range between minimum and maximum values for microscopic measurements is given. Mean values are in brackets with ‘n’ being the number of measurements. The holotype is deposited at the Mycological Herbarium of the Chinese Academy of Science (HMAS) in Beijing, China.
DNA extraction, PCR and sequencing
DNA was extracted from the herbarium specimen HMAS 180703 using E.Z.N.A. Forensic DNA Extraction Kit (Omega product No: D3529-01/02). A sporodochial mass was removed from the specimen for extraction using fine forceps, following the manufacturer’s protocols. Partial 28S rDNA and ITS rDNA were amplified using fungal specific primers LROR and LR5 (Vilgalys & Hester 1990) and ITS1 and ITS4 (White et al. 1990), respectively. The polymerase chain reaction (PCR) was carried out in a 50 μL reaction volume and the PCR thermal cycles based on the profile detailed in Cai et al. (2006b). PCR amplification was confirmed on 1 % agarose electrophoresis gels stained with ethidium bromide. Amplicons were then purified using minicolumns, purification resin and buffer, following the manufacturer’s protocols (Amersham product code: 27-9602-01). DNA sequencing was performed using the primers mentioned above in an Applied Biosystem 3730 DNA Analyser at the Genome Research Centre, The University of Hong Kong.
Sequence alignment and phylogenetic analysis
Sequences were aligned using BioEdit (Hall 1999). Two data sets were analysed; the 28S rDNA data set and ITS/5.8S rDNA data set. Novel sequences (EU413953, EU413954), together with reference sequences obtained from GenBank, were aligned using Clustal X (Thomson et al. 1997). Reference sequences used in this study are mainly from Lumbsch et al. (2005) and Tsui et al. (2006), both studies providing 5 or more reference sequences. Alignment was manually adjusted to allow maximum alignment and minimise gaps.
Phylogenetic analyses were performed using maximum parsimony as implemented in PAUP v. 4.0b10 (Swofford 2002). Ambiguously aligned regions, 26 and 49 characters in 28S and ITS data sets respectively, were excluded from all analyses. Characters were equally weighted and gaps were treated as missing data. Trees were derived using the heuristic search option with TBR branch swapping and 1 000 random sequence additions. Maxtrees were unlimited, branches of zero length were collapsed and all parsimonious trees were saved. Clade stability was assessed using a bootstrap (BT) analysis with 1 000 replicates, each with 10 replicates of random stepwise addition of taxa. Kishino-Hasegawa tests (KH Test) (Kishino & Hasegawa 1989) were performed in order to determine whether trees were significantly different. Trees were figured using Treeview (Page 1996).
The model of evolution was estimated by using Mrmodeltest 2.2 (Nylander 2004). Posterior probabilities (PP) (Rannala & Yang 1996, Zhaxybayeva & Gogarten 2002) were determined by Markov Chain Monte Carlo sampling (BMCMC) in MrBayes v. 3.0b4 (Huelsenbeck & Ronquist 2001). Six simultaneous Markov chains were run for 1 000 000 generations and trees were sampled every 100th generation (resulting 10 000 total trees). The first 2 000 trees, which represented the burn-in phase of the analyses, were discarded and the remaining 8 000 trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree.
RESULTS
The LSU sequence obtained from the herbarium specimen consisted of 724 nucleotides. A BLAST search in NCBI (www.ncbi.nih.gov/blast) showed that this sequence is most similar to Aquaticheirospora lignicola (AY736378, 93 % identity), Munkovalsaria appendiculata (AY772016, 89 %), Leptosphaeria calvescens (AY849944, 89 %) and Montagnula opulenta (AY678086, 89 %). The 28S rDNA data set consisted of 40 sequences. The final data set included 537 characters after alignment. Only part of the Cheirosporium triseriale 28S rDNA sequence was used in the analysis, as most of the included reference sequences from NCBI are shorter than ours. Parsimony analysis resulted in 28 equally parsimonious trees. The KH test showed that these trees were not significantly different. One of these trees is shown in Fig. 1.
Fig. 1.
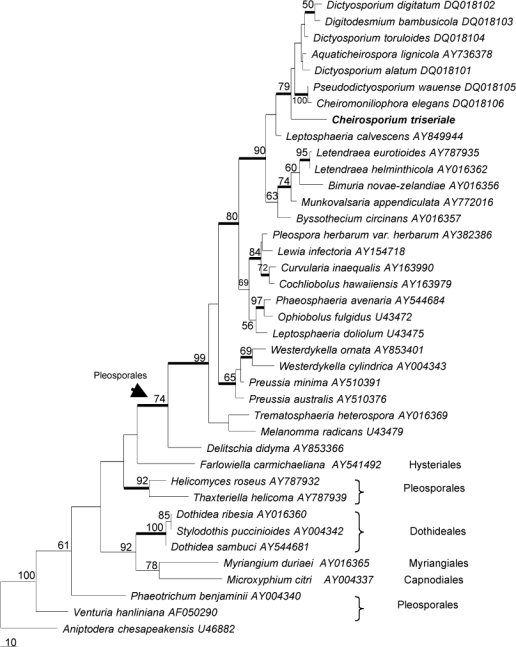
Phylogenetic tree generated from parsimony analysis based on 28S rDNA sequences. Data were analysed with random addition sequence, and treating gaps as missing data. Bootstrap values ≥ 50 % are shown above or below branches. Thickened branches indicate Bayesian posterior probabilities ≥ 95 %. The tree is rooted with Aniptodera chesapeakensis.
The ITS sequence consisted of 533 nucleotides. The BLAST search in NCBI showed that this sequence is most comparable to Dictyosporium digitatum (DQ018089, 83 %), Dictyosporium toruloides (DQ018093, 82 %), Dictyosporium heptasporum (DQ018090, 82 %) and Dictyosporium giganticum (DQ018095, 82 %) (Tsui et al. 2006). The ITS rDNA data set consisted of 28 sequences. The final data set comprised 718 characters after alignment. Parsimony analysis resulted in three equally parsimonious trees. The KH test showed that these trees were not significantly different. One of the trees is shown in Fig. 2.
Fig. 2.
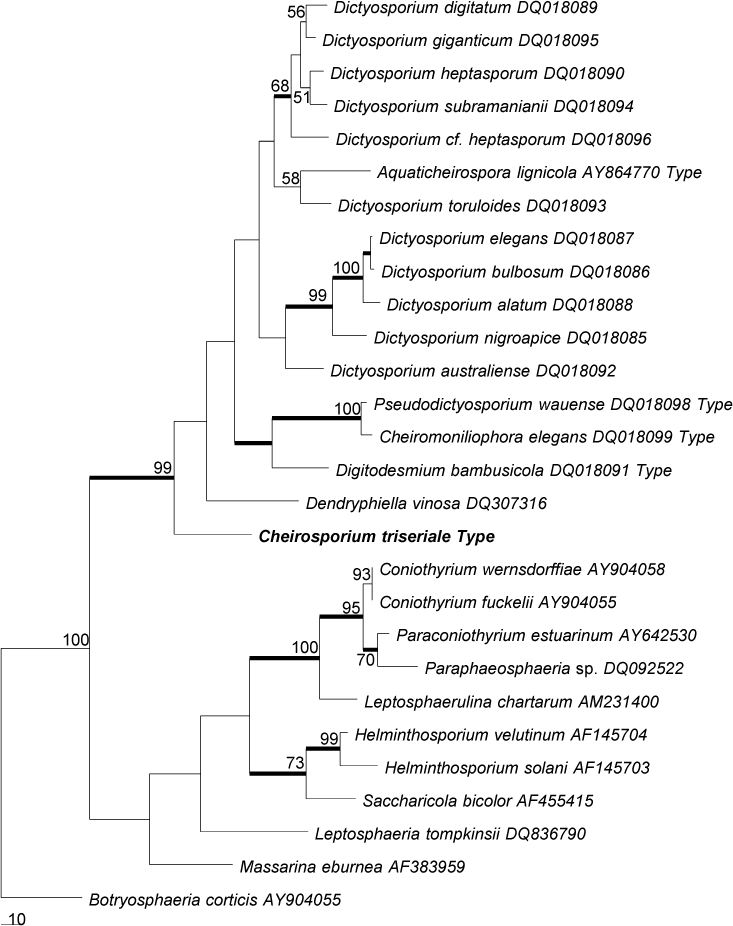
Phylogenetic tree generated from parsimony analysis based on ITS rDNA sequences. Data were analysed with random addition sequence and treating gaps as missing data. Bootstrap values ≥ 50 % are shown above or below branches. Thickened branches indicate Bayesian posterior probabilities ≥ 95 %. The tree is rooted with Botryosphaeria corticis.
The topologies generated from Bayesian analyses are essentially similar to that of the parsimony analyses. The Bayesian trees are therefore not shown, but the statistically supported clades are marked in the parsimony trees (Fig. 1, 2).
Taxonomy
Cheirosporium triseriale gen. & sp. nov.
Cheirosporium L. Cai & K.D. Hyde, gen. nov. — MycoBank MB506570.
Coloniae in substrato naturali sporodochiales, dispersae, punctiformes, brunneae, atro-brunneae vel nigrae. Conidiophora semi-macronematosa vel macronematosa, mononematosa, septata, flexuousa, irregulariter ramosa, ramis steriles vel fertiles; pars sterilis elongatis, obclavata. Cellulae conidiogenae monoblasticae, terminales, determinatae, non-prolificantes. Conidia acrogena, holoblastica, solitaria, sicca, olivacea vel brunnea, cheiroidea, levia. Conidiorum secessio rhexolytica.
Etymology. Referring to the cheiroid conidia in this genus.
Conidiomata on the natural substratum sporodochial, scattered, punctiform, brown, dark brown or black. Mycelium immersed or partly superficial, consisting of branched, septate, smooth, thin-walled, subhyaline to light brown hyphae. Stroma none. Setae and hyphopodia absent. Conidiophores semi-macronematous to macronematous, mononematous, septate, flexuous, irregularly branched, branches sterile or fertile; sterile branches with an elongate, relatively enlarged, obclavate cell. Conidiogenous cells monoblastic, terminal, determinate, not proliferating. Conidia acrogenous, holoblastic, solitary, dry, olivaceous to brown, cheiroid, smooth-walled. Conidial secession rhexolytic.
Type species. Cheirosporium triseriale.
Cheirosporium triseriale L. Cai & K.D. Hyde, sp. nov. — MycoBank MB506551; Fig. 3a–j
Fig. 3.
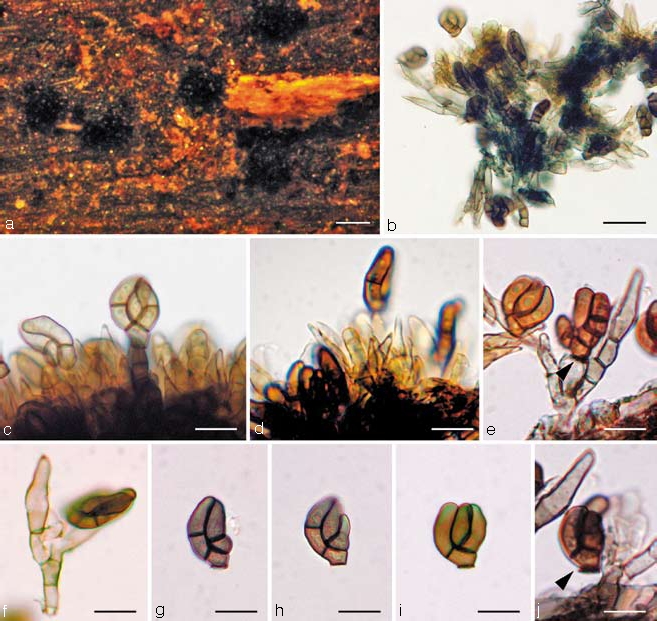
Cheirosporium triseriale. a. Sporodochial conidiomata on the host surface; b. squash mount of the conidiophores and conidia; c, d. conidiophores bearing conidia (note numerous sterile apices of the conidiophores); e, f. individual conidiophores. Note the sterile branches; g–j. conidia. — Scale bars: a = 200 μm, b = 40 μm, c–j = 20 μm.
Sporodochia in substrato naturali 150–300 μm diam, dispersa, punctiformia, brunnea, atro-brunnea vel nigra. Conidiophora 50–120 × 4–6 μm, semi-macronematosa vel macronematosa, mononematosa, hyalina vel subhyalina, levia, septata, flexuousa, irregulariter ramosa, ramis steriles vel fertiles; pars sterilis elongatis, obclavata. Cellulae conidiogenae monoblasticae, terminales, determinatae, hyalinae vel subhyalinae, doliiformes vel late cylindracae, non-prolificantes. Conidia 15–25 × 11–15 μm, acrogena, holoblastica, solitaria, sicca, olivacea vel brunnea, cheiroidea, levia, basi truncata triramifera, complanata, ramis 0–1-septatis, cylindricis. Conidiorum secessio rhexolytica.
Etymology. Referring to the conidia of this species, which produce three arms of cells in series.
Sporodochia on the natural substratum 150–300 μm diam, scattered, punctiform, brown, dark brown or black. Mycelium immersed or partly superficial, consisting of branched, septate, smooth, thin-walled, subhyaline to light brown hyphae. Stroma none. Setae and hyphopodia absent. Conidiophores 50–120 × 4–6 μm, semi-macronematous to macronematous, mononematous, hyaline to subhyaline, smooth-walled, septate, flexuous, irregularly branched, branches sterile or fertile; sterile branches with an elongate, relatively enlarged, obclavate, apical cell, 12–18 μm long, 4.5–6 μm wide. Conidiogenous cells monoblastic, terminal, determinate, hyaline to subhyaline, doliiform or broad-cylindrical, not proliferating. Conidia 15–25 × 11–15 μm (av. = 19 × 12 μm, n = 30), acrogenous, holoblastic, solitary, dry, olivaceous to brown, cheiroid, mostly flattened in one plane, smooth-walled, truncate at the base, with three laterally appressed, 0–1-septate, cylindrical arms, arising from a common basal cell. Conidial secession rhexolytic.
Habitat — Saprobic on submerged woody bamboo culms.
Known distribution — China.
Teleomorph — Unknown.
Specimen examined. China, Yunnan, Lijiang, Heilongtan Park (26° 30′ N, 100° 13′ E), on submerged wood in a small stream, 16 July 2005, L. Cai, holotype herb. HMAS 180703, paratype herb. IFRD 100001.
DISCUSSION
Cheirosporium triseriale is unique among cheirosporous hyphomycetes because of its unusual combination of morphological characters. These include:
-
–
The conidiophores of Ch. triseriale branch to form two to several distinct sterile branches, in which there is a distinct, enlarged, hyaline, obclavate apical cell. This character has not been reported from cheirosporous genera. Similar sterile branches occur among conidiophores in species of genera such as Cylindrocladium, Falcocladium, Vesicladiella and Tubercularia, and in the first three, sterile branches have been regarded as a diagnostic generic character (Seifert 1985, Crous & Wingfield 1994, Crous et al. 1994).
-
–
In Ch. triseriale, conidial secession is typically rhexolytic (Fig. 3e, j), which differs from other cheirosporous genera.
-
–
The conidiophores of Ch. triseriale are semi-macronematous to macronematous.
The most similar genus to Cheirosporium is Digitomyces, which includes species that also produce 2–3-armed cheirosporous conidia (Mercado et al. 2003). Digitomyces was established for Dictyosporium verrucosum (Tzean & Chen 1989, Goh et al. 1999, Mercado et al. 2003), characterised by verruculose conidia and micronematous to macronematous conidiophores (Mercado et al. 2003). Digitomyces differs from Cheirosporium in lacking sterile branches, and the conidial secession is schizolytic with a truncate conidial base. The verrucose cell wall of conidia in Digitomyces is also different to that of Cheirosporium, but wall ornamentation has been shown to have less phylogenetic significance in distinguishing genera (e.g. Cai et al. 2006b) Whether Digitomyces merits generic rank awaits further investigation, and it shares many morphological similarities with Dictyosporium.
Cheirosporium triseriale should also be compared with species of Dictyosporium, because of their similar conidial morphology (Sutton 1985, Goh et al. 1999). In addition to the sterile branches and the rhexolytic conidial secession, Cheirosporium is further distinguished by its semi-macronematous to macronematous conidiophores (Goh et al. 1999). Cheiromyces and Cheiromycina can also be differentiated from Cheirosporium by the production of distoseptate conidia (Berkeley 1875, Sutton & Muhr 1986). Species of Pseudodictyosporium have macronematous conidiophores but they produce enteroblastic conidiogenous cells and lack sporodochial conidiomata (Matsushima 1971). Species of Digitodesmium produce sporodochial conidiomata but the conidial arms are discrete and mostly with narrow, incurved apical cells (Kirk 1981, Ho et al. 1999).
The partial sequences of 28S rDNA and ITS rDNA confirmed the relative taxonomic distance of the new taxon from other cheirosporous genera. No close relatives were found. The highest ITS sequence similarity between Cheirosporium triseriale and sequences available in public databases is only 83 % with Dictyosporium digitatum (DQ018089). The morphological distinction and low sequence homology with existing taxa indicates the need to establish a new genus for this taxon. Parsimony analyses provided indications of its phylogenetic position. In both 28S rDNA and ITS rDNA trees, Ch. triseriale clustered with high bootstrap support in Pleosporales. This is consistent with its morphological characters, because most cheirosporous genera have affinities with the Pleosporales (Tsui et al. 2006).
The 28S rDNA phylogeny shows that Ch. triseriale nests in a clade with Cheiromoniliophora elegans, Pseudodictyosporium wauense, Aquaticheirospora lignicola, Digitodesmium bambusicola and several Dictyosporium species (Tsui et al. 2006). This is indicative of the close relationships among these cheirosporous genera. Similar findings were obtained from phylogenies derived from the ITS rDNA data set. In the ITS rDNA tree, Ch. triseriale groups with other cheirosporous genera in the Pleosporales. Cheirosporium triseriale is basal and phylogenetically distant in the clade. In both trees, Ch. triseriale clustered with high bootstrap support in the clade of cheirosporous anamorphs.
Our phylogenetic analyses indicate that the current taxonomy of anamorphic cheirosporous genera needs further evaluation. For example, the generic status of Aquaticheirospora should be reconsidered. Dictyosporium and Aquaticheirospora have similar conidial form and type of conidiogenesis (Kodsueb et al. 2007), but differ because the latter has synnematous conidiomata. In our analyses, Aquaticheirospora lignicola nested amongst Dictyosporium species and lacks sufficient phylogenetic distance to warrant separate generic status (Fig. 2), indicating that the synnematous character may not be of importance in generic delineation. In their phylogenetic analysis, Kodsueb et al. (2007) did not include any cheirosporous anamorphs because these were not available in GenBank at that time. Similarly, our ITS trees also reveal little phylogenetic distance between the monotypic genera Pseudodictyosporium and Cheiromoniliophora. The two species have 98.7 % similarity in the ITS locus. Morphologically, Pseudodictyosporium and Cheiromoniliophora are similar in having macronematous conidiophores, non-sporodochial conidiomata and flattened conidia with three arms. They are distinguished by the nature of the conidiogenous cells, which are holoblastic and proliferate sympodially in Cheiromoniliophora and enteroblastic and proliferate percurrently in Pseudodictyosporium. It has been suggested that the two genera should possibly be synonymised (Tsui et al. 2006).
In the ITS tree, the branch leading into the clade with Ch. triseriale at its base is strongly supported. One may argue that Aquaticheirospora, Pseudodictyosporium, Cheiromoniliophora, Digitodesmium and Cheirosporium might be all congeneric with Dictyosporium. Based on our present understanding of this group of fungi, it is not possible to fully resolve the generic boundaries of these genera. Regardless of the subjectivity in taxonomic practice, the important implication of this study is that these cheirosporous anamorphs merit further study of evolutionary biology. The morphological characters used to distinguish these genera should also be re-evaluated using multiple-gene phylogenies.
Acknowledgments
The University of Hong Kong is thanked for providing LC a postgraduate scholarship. This research was funded by Hong Kong Research Grant Council no. HKU7230/02M. Helen Leung and Heidi Kong at the University of Hong Kong are thanked for technical help.
REFERENCES
- Berkeley MJ. 1875. Notices of North American fungi. Grevillea 3: 97 – 112 . [Google Scholar]
- Cai L, Hyde KD. 2007. Anamorphic fungi from freshwater habitats in China: Dictyosporium tetrasporum and Exserticlava yunnanensis spp. nov., and two new records for Pseudofuscophialis lignicola and Pseudobotrytis terrestris. Mycoscience 48: 290 – 296 . [Google Scholar]
- Cai L, Ji KF, Hyde KD. 2006a. Variation between freshwater and terrestrial fungal communities on decaying bamboo culms. Antonie van Leeuwenhoek International Journal of General and Molecular Microbiology 89: 293 – 301 . [DOI] [PubMed] [Google Scholar]
- Cai L, Jeewon R, Hyde KD. 2006b. Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycological Research 110: 137 – 150 . [DOI] [PubMed] [Google Scholar]
- Cai L, Zhang KQ, McKenzie EHC, Hyde KD. 2003. Freshwater fungi from bamboo and wood submerged in the Liput river in the Philippines. Fungal Diversity 13: 1 – 12 . [Google Scholar]
- Crous PW, Wingfield MJ. 1994. A monograph of Cylindrocladium, including anamorphs of Calonectria. Mycotaxon 51: 341 – 435 . [Google Scholar]
- Crous PW, Wingfield MJ, Alfenas AC, Silveira SF. 1994. Cylindrocladium naviculatum sp. nov. and two new vesiculate hyphomycete genera, Falcocladium and Vesicladiella. Mycotaxon 50: 441 – 458 . [Google Scholar]
- Goh TK, Hyde KD, Ho WH, Yanna 1999. A revision of the genus Dictyosporium with descriptions of three new species. Fungal Diversity 2: 65 – 100 . [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95 – 98 . [Google Scholar]
- Ho WH, Hodgkiss IJ, Hyde KD. 2000. Cheiromyces lignicola, a new cheirosporous anamorphic species from Hong Kong. Mycologia 92: 582 – 588 . [Google Scholar]
- Ho WH, Hyde KD, Hodgkiss IJ. 1999. Digitodesmium recurvum, a new species of chirosporous hyphomycetes from Hong Kong. Mycologia 91: 900 – 904 . [Google Scholar]
- Huelsenbeck JP, Ronquist FR. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Biometrics 17: 754 – 755 . [DOI] [PubMed] [Google Scholar]
- Kirk PM. 1981. New or interesting microfungi. Transactions British Mycological Society 76: 71 – 88 . [Google Scholar]
- Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order of Hominoidea. Journal of Molecular Evolution 29: 170 – 179 . [DOI] [PubMed] [Google Scholar]
- Kodsueb R, Lumyong S, Ho WH, Hyde KD, McKenzie EHC, Jeewon R. 2007. Morphological and molecular characterization of Aquaticheirospora and phylogenetics of Massarinaceae (Pleosporales). Botanical Journal of the Linnean Society 155: 283 – 296 . [Google Scholar]
- Lumbsch HT, Schmitt I, Lindemuth R, Miller A, Mangold A, Fernandez F, Huhndorf S. 2005. Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomyceta). Molecular Phylogenetics and Evolution 34: 512 – 524 . [DOI] [PubMed] [Google Scholar]
- Matsushima T. 1971. Microfungi of the Solomon Islands and Papua-New Guinea Published by the author, Kobe: . [Google Scholar]
- Mercado SA, Calduch M, Gene J, Guarro J, Delgado G. 2003. Digitomyces, a new genus of hyphomycetes with cheiroid conidia. Mycologia 95: 860 – 864 . [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest 2.0. Program distributed by the author Evolutionary Biology Centre, Uppsala University; . [Google Scholar]
- Page RDM. 1996. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357 – 358 . [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304 – 311 . [DOI] [PubMed] [Google Scholar]
- Seifert K. 1985. A monograph of Stilbella, and some allied hyphomycetes. Studies in Mycology 27: 1 – 235 . [Google Scholar]
- Sutton BC. 1985. Notes on some deuteromycete genera with cheiroid or digitate brown conidia. Proceedings of the Indian Academy of Science (Plant Science) 94: 229 – 244 . [Google Scholar]
- Sutton BC, Muhr LE. 1986. Cheiromycina flabelliformis gen. et sp. nov. on Picea from Sweden. Nordic Journal of Botany 6: 831 – 836 . [Google Scholar]
- Swofford DL. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Version 4b10 Sinauer Associates, Sunderland, USA: . [Google Scholar]
- Thomson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876 – 4882 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui CKM, Berbee ML, Jeewon R, Hyde KD. 2006. Molecular phylogeny of Dictyosporium and allied genera inferred from ribosomal DNA. Fungal Diversity 21: 157 – 166 . [Google Scholar]
- Tsui CKM, Hyde KD. 2003. Freshwater mycology. Fungal Diversity Research Series 10: 1 – 350 . [Google Scholar]
- Tzean SS, Chen JL. 1989. Two new species of Dictyosporium from Taiwan. Mycological Research 92: 497 – 502 . [Google Scholar]
- Vijaykrishna D, Hyde KD. 2006. Inter- and intra stream variation of lignicolous freshwater fungi in tropical Australia. Fungal Diversity 21: 203 – 224 . [Google Scholar]
- Vijaykrishna D, Jeewon R, Hyde KD. 2006. Molecular taxonomy, origins and evolution of freshwater ascomycetes. Fungal Diversity 23: 351 – 390 . [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238 – 4246 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, USA: . [Google Scholar]
- Zhaxybayeva O, Gogarten JP. 2002. Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. Genomics 3: 1 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]


