Abstract
Three species of Mycosphaerella, namely M. eumusae, M. fijiensis, and M. musicola are involved in the Sigatoka disease complex of bananas. Besides these three primary pathogens, several additional species of Mycosphaerella or their anamorphs have been described from Musa. However, very little is known about these taxa, and for the majority of these species no culture or DNA is available for study. In the present study, we collected a global set of Mycosphaerella strains from banana, and compared them by means of morphology and a multi-gene nucleotide sequence data set. The phylogeny inferred from the ITS region and the combined data set containing partial gene sequences of the actin gene, the small subunit mitochondrial ribosomal DNA and the histone H3 gene revealed a rich diversity of Mycosphaerella species on Musa. Integration of morphological and molecular data sets confirmed more than 20 species of Mycosphaerella (incl. anamorphs) to occur on banana. This study reconfirmed the previously described presence of Cercospora apii, M. citri and M. thailandica, and also identified Mycosphaerella communis, M. lateralis and Passalora loranthi on this host. Moreover, eight new species identified from Musa are described, namely Dissoconium musae, Mycosphaerella mozambica, Pseudocercospora assamensis, P. indonesiana, P. longispora, Stenella musae, S. musicola, and S. queenslandica.
Keywords: Mycosphaerella, phylogeny, Sigatoka disease complex, taxonomy
INTRODUCTION
The genus Mycosphaerella is phylogenetically heterogeneous (Crous et al. 2007a), contains more than 3000 names (Aptroot 2006), and has been linked to more than 30 well-known anamorphic genera (Crous et al. 2006a, b, 2007a, b, Arzanlou et al. 2007a). Species of Mycosphaerella inhabit different ecological niches as saprobes, plant pathogens or endophytes (Farr et al. 1995, Verkley & Starink-Willemse 2004, Crous et al. 2004b, 2006a, 2007a, b), and have a worldwide distribution from tropical and subtropical to warm and cool regions (Crous 1998, Crous et al. 2000, 2001). Plant-pathogenic species of Mycosphaerella are among the most common and destructive plant pathogens occurring on a wide range of hosts including trees, herbaceous plants, and plantation crops. The invasion of leaf and stem tissue and concomitant distortion of the host plant physiology cause considerable economic losses (Park et al. 2000, Goodwin et al. 2001, Maxwell et al. 2005, Cortinas et al. 2006, Crous et al. 2006a, b, Hunter et al. 2006).
The Sigatoka disease complex, which is the most serious and economically important leaf spot disease of banana, is attributed to species of Mycosphaerella. Mycosphaerella musicola (anamorph Pseudocercospora musae) which causes (yellow) Sigatoka disease, M. fijiensis (anamorph P. fijiensis) which causes the black Sigatoka disease, and M. eumusae (anamorph P. eumusae), which causes eumusae leaf spot disease (reviewed in Jones 2000, 2003, Crous & Mourichon 2002) are the major constituents of the Sigatoka disease complex. The disease reduces the photosynthetic capacity of the plant as a consequence of necrotic leaf lesions, and induces physiological alterations of the plant, resulting in reduced crop yield and fruit quality. All three species emerged on bananas during the last century and became major constraints to commercial production worldwide. The chronology of disease records around the world and genetic structure of pathogen population suggests that South-East Asia, where the host genus Musa is indigenous, is the centre of origin for all three fungal species (Mourichon & Fullerton 1990, Carlier et al. 1996, Hayden et al. 2003, Rivas et al. 2004).
Yellow Sigatoka disease was first reported on banana in Java in 1902. The disease spread rapidly to all banana-growing regions during the following 20 years, and has since reached the limits of its distribution worldwide (reviewed in Jones 2000, 2003). The fungus responsible for the disease was described as Cercospora musae. In 1941 Leach established the connection between C. musae and its teleomorph, Mycosphaerella musicola. Mulder (in Mulder & Stover 1976) validated the species descriptions, while the anamorph was transferred to Pseudocercospora as P. musae (Deighton 1976). In the early 1960s, another, even more severe leaf spot disease on banana appeared in the Fiji Islands, which Rhodes (1964) described as black leaf streak disease and later became known as the black Sigatoka disease. Morelet (1969) validated the species name as M. fijiensis, while Deighton (1976) placed its anamorph in Pseudocercospora as P. fijiensis. In 1974 a new variety of M. fijiensis was described from Honduras and named as M. fijiensis var. difformis. Deighton (1979) placed both varieties in the genus Paracercospora, based on the slight thickening observed on the rims of scars and conidial hila. However, this feature was not supported by DNA phylogeny, and as there were many intermediate morphological forms, the genus Paracercospora was again reduced to synonymy under Pseudocercospora by Crous et al. (2001). Mycosphaerella eumusae (Pseudocercospora eumusae), was recognised as a new constituent of the Sigatoka complex of banana in the mid-1990s (Carlier et al. 2000, Crous & Mourichon 2002, Jones 2003). Presently, M. eumusae is known from parts of South-East Asia, Indian Ocean Islands and Nigeria, where it could co-exist with the other two species. Besides the three primary agents of the Sigatoka disease complex, several additional species of Mycosphaerella (or their anamorphs) have been described from Musa, but for the majority of these species no culture or herbarium specimen is available, and the pathological relevance of those species remains unclear (reviewed in Jones 2000, Crous et al. 2003, Aptroot 2006).
The identity and distribution of the various Mycosphaerella species associated with leaf spots of banana are not yet fully understood, which is mainly due to the difficulties experienced by scientists who have to identify them by conventional methods and without specialist taxonomic support. Furthermore, because these species are morphologically highly similar and frequently co-occur on the same lesion, pathogen recognition and subsequent disease management have proven to be rather difficult. To enable the development of specific molecular-based diagnostic tools for pathogen recognition, all related species present on the same host have to be considered. Recently, Arzanlou et al. (2007a) developed a highly sensitive set of Taqman probes to distinguish M. fijiensis from M. musicola and M. eumusae in leaf material. Little attention has been given to date, however, to other species of Mycosphaerella that occur on Musa spp. Because several Mycosphaerella species can co-occur in the same lesion (Crous 1998), it is quite possible that there may be other species of Mycosphaerella associated with the Sigatoka disease complex. The aim of the present study was, therefore, to employ a multi-gene DNA sequence typing approach on a global set of Mycosphaerella isolates to distinguish the various species occurring on banana. To this end morphological and cultural growth data were integrated with DNA sequence data from the internal transcribed spacer region of the rDNA operon, and partial actin, histone H3, and small subunit mitochondrial ribosomal DNA gene sequences.
MATERIALS AND METHODS
Isolates
Isolates (Table 1) were obtained by isolation from infected symptomatic banana leaves, or supplied as pure cultures by the following departments and institutes: The Horticulture and Food Research Institute of New Zealand, Auckland, New Zealand; Centre de coopération internationale en recherché agronomique pour le développement (CIRAD, Montpellier, France); University of Florida, Tropical Research & Education Centre (USA); Forestry and Agricultural Biotechnology Institute (FABI, Pretoria, South Africa). Isolates were recovered from infected banana leaves as single ascospores or conidia. Germinating spores were examined 24 h after germination on 2 % malt extract agar (MEA; Sigma-Aldrich Chemie, Zwijndrecht, The Netherlands) plates under a stereomicroscope, and single-spore cultures were established on fresh MEA plates following the protocol of Crous (1998).
Table 1.
Isolates of Mycosphaerella or its anamorphs used for DNA analysis and morphological studies.
| Species | Accession number1 | Source | Origin | GenBank numbers (ITS, ACT, HIS, mtSSU) |
|---|---|---|---|---|
| Cercospora apii | CPC 12682; CBS 119395 | Musa cv. Cavendish | Western Bangladesh | EU514222, —, —, — |
| CPC 12683 | Musa cv. Cavendish | Western Bangladesh | EU514223, —, —, — | |
| CPC 12684 | Musa cv. Cavendish | Western Bangladesh | EU514224, —, —, — | |
| Dissoconium musae | X1021; CBS 122453 | Musa cv. Nendran (Plantain) AAB | India | EU514225, EU514296, EU514349, EU514402 |
| X1022; CBS 122454 | Musa cv. Nendran (Plantain) AAB | India | EU514226, EU514297, EU514350, EU514403 | |
| Mycosphaerella citri | X126; CBS 122455 | Citrus sp. | USA, Florida | EU514227, —, —, — |
| X742; CBS 122456 | Musa cv. SH 3436 AAAA | Tonga | EU514228, —, —, — | |
| X743 | Musa cv. SH 3362 AA | Tonga | EU514229, —, —, — | |
| Mycosphaerella colombiensis | X24 | Musa cultivar | Mozambique | EU514230, —, —, — |
| X215 | — | South Africa | EU514231, EU514298, EU514351, EU514404 | |
| Mycosphaerella communis | X1023 | Musa cv. Valery: Cavaendish | Trinidad | EU514232, EU514299, EU514352, EU514405 |
| Mycosphaerella eumusae | S1030B | Musa cultivar | Mauritius | EU514233, EU514300, EU514353, EU514406 |
| S1037B | Musa cultivar | Mauritius | EU514234, EU514301, EU514354, EU514407 | |
| S1037C | Musa cultivar | Mauritius | EU514235, EU514302, EU514355, EU514408 | |
| S1037G | Musa cultivar | Mauritius | EU514236, —, —, — | |
| S1037H | Musa cultivar | Mauritius | EU514237, —, —, — | |
| X208; CIRAD 1156; CPC 4579 | Musa cultivar | Mauritius | EU514238, —, —, — | |
| X209; CBS 114825; CIRAD 1157; CPC 4580 | Musa cultivar | Mauritius | EU514239, —, —, — | |
| X865; CIRAD 535 | Musa cultivar | India | EU514240, EU514303, EU514356, EU514409 | |
| X866; CBS 121377; CIRAD 458 | Musa cultivar | Malaysia | EU514241, EU514304, EU514357, EU514410 | |
| X867; CBS 121378; CIRAD 459 | Musa cultivar | Malaysia | EU514242, EU514305, EU514358, EU514411 | |
| X869; CBS 121380; CIRAD 563 | Musa cultivar | Sri Lanka | EU514243, EU514306, EU514359, EU514412 | |
| X870; CBS 121381; CIRAD 485 | Musa cultivar | Thailand | EU514244, EU514307, EU514360, EU514413 | |
| X873; CBS 121382; CIRAD 487 | Musa cultivar | Thailand | EU514245, EU514308, EU514361, EU514414 | |
| X875; CIRAD 671 | Musa cultivar | Vietnam | EU514246, EU514309, EU514362, EU514415 | |
| X876; CBS 121383; CIRAD 744 | Musa cultivar | Mauritius | EU514247, EU514310, EU514363, EU514416 | |
| Mycosphaerella fijiensis | CIRAD 86; CBS 120258; C86a | Musa cultivar | Cameroon | EU514248, —, —, — |
| X84 | Musa cultivar | Colombia | EU514249, EU514311, EU514364, EU514417 | |
| X92 | Musa cultivar | Colombia | EU514250, EU514312, EU514365, EU514418 | |
| X104 | Musa cultivar | Colombia | EU514251, EU514313, EU514366, EU514419 | |
| X110 | Musa cultivar | Colombia | EU514252, EU514314, EU514367, EU514420 | |
| X843; CIRAD 11 | Musa cultivar | Honduras | EU514253, EU514315, EU514368, EU514421 | |
| X847; CBS 121362; CIRAD 364 | Musa cultivar | Taiwan | EU514254, EU514316, EU514369, EU514422 | |
| X850; CIRAD 355 | Musa cultivar | Ivory Coast | EU514255, EU514317, EU514370, EU514423 | |
| Mycosphaerella lateralis | S1024 | Musa cultivar | Mauritius | EU514256, —, —, — |
| Mycosphaerella mozambica | X34; CBS 122464 | Musa cultivar | Mozambique | EU514257, EU514318, EU514371, EU514424 |
| X884; CBS 121391; UQ438 | Musa cultivar | Australia | EU514258, EU514319, EU514372, EU514425 | |
| Mycosphaerella musae | X398; CBS 122458 | Musa cv. Cavendish AAA | Tonga | EU514259, EU514320, EU514373, EU514426 |
| X813 | Musa cultivar | Malawi | EU514260, EU514321, EU514374, EU514427 | |
| X814; CBS 122459 | Musa cultivar | Malawi | EU514261, EU514322, EU514375, EU514428 | |
| X818; CBS 122460 | Musa cultivar | Malawi | EU514262, —, —, — | |
| X819; CBS 122461 | Musa cultivar | Malawi | EU514263, —, —, — | |
| X879; CBS 121386; CIRAD 64 | Musa cultivar | Malawi | EU514264, EU514323, EU514376, EU514429 | |
| Mycosphaerella musicola | X42; CBS 116634; IMI 123823 | Musa cultivar | Cuba | EU514265, —, —, — |
| X63 | Musa cultivar | Windward Islands | EU514266, EU514324, EU514377, EU514430 | |
| X67 | Musa cultivar | Windward Islands | EU514267, EU514325, EU514378, EU514431 | |
| X588 | Musa cv. Williams | Australia | EU514268, EU514326, EU514379, EU514432 | |
| X589 | Musa cv. Williams | Australia | EU514269, EU514327, EU514380, EU514433 | |
| X596 | Musa cv. SH-3362 AA | Australia | EU514270, EU514328, EU514381, EU514434 | |
| X602 | Musa cv. Lakatan | Australia | EU514271, EU514329, EU514382, EU514435 | |
| X857; CBS 121371; UQ430 | Musa cultivar | Australia | EU514272, EU514330, EU514383, EU514436 | |
| X858; CBS 121372; UQ433 | Musa cultivar | Australia | EU514273, EU514331, EU514384, EU514437 | |
| X860; CBS 121374; UQ2003 | Musa cultivar | Australia | EU514274, EU514332, EU514385, EU514438 | |
| Mycosphaerella thailandica | X22 | Musa cultivar | Windward Islands | EU514275, EU514333, EU514386, EU514439 |
| X53 | Musa cultivar | Australia | EU514276, EU514334, EU514387, EU514440 | |
| X882; CBS 121389; CIRAD 81 | Musa cultivar | Mozambique | EU514277, EU514335, EU514388, EU514441 | |
| X883; CBS 121390; CIRAD 1165 | Musa cultivar | Brazil | EU514278, EU514336, EU514389, EU514442 | |
| Passalora loranthi | X28; CBS 122465 | Musa cultivar | Camaroon | EU514279, EU514337, EU514390, EU514443 |
| X138; CBS 122466 | Citrus sp. | Mozambique | EU514280, EU514338, EU514391, EU514444 | |
| Pseudocercospora assamensis | X988; #9; CBS 122467 | Musa cultivar | India, Assam | EU514281, EU514339, EU514392, EU514445 |
| Pseudocercospora indonesiana | X991; #11-5; CBS 122473 | Musa cultivar | Indonesia, Western Sumatra | EU514282, EU514340, EU514393, EU514446 |
| X992; #11-6; CBS 122474 | Musa cultivar | Indonesia, Western Sumatra | EU514283, —, —, — | |
| Pseudocercospora longispora | X474; CBS 122469 | Musa cv. Pisang Mas AA | Malaysia | EU514284, EU514341, EU514394, EU514447 |
| X475; CBS 122470 | Musa cv. Pisang Mas AA | Malaysia | EU514285, EU514342, EU514395, EU514448 | |
| Pseudocercospora sp. | X1083; CBS 122468 | Ravenala madagascariensis | India | EU514286, —, —, — |
| Stenella musae | X45 | Musa cultivar | Windward Islands | EU514287, EU514343, EU514396, EU514449 |
| X47; CBS 122476 | Musa cultivar | Windward Islands | EU514288, EU514344, EU514397, EU514450 | |
| X55 | Musa cultivar | Windward Islands | EU514289, —, —, — | |
| X70; CBS 122478 | Musa cultivar | Windward Islands | EU514290, EU514345, EU514398, EU514451 | |
| X745; CBS 122477 | Musa cv. TU8 AAAA | Tonga | EU514291, EU514346, EU514399, EU514452 | |
| X877; CBS 121384; CIRAD 41 | Musa cultivar | Martinique | EU514292, EU514347, EU514400, EU514453 | |
| X878; CBS 121385; CIRAD 56 | Musa cultivar | Martinique | EU514293, EU514348, EU514401, EU514454 | |
| Stenella musicola | X1019; CBS 122479 | Musa cv. Grand Nain AAA (Cav.) | India | EU514294, —, —, — |
| Stenella queenslandica | X1084; CBS 122475 | Musa banksii | Australia | EU514295, —, —, — |
1 CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CIRAD: Centre de coopération internationale en recherche agronomique pour le développement, Montpellier, France; CPC: Culture collection of Pedro Crous, housed at CBS; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, UK; UQ: University of Queensland, Australia; X, S: Culture collection of Mahdi Arzanlou, housed at CBS.
DNA phylogeny
Genomic DNA was isolated from fungal mycelia grown on MEA, using the FastDNA kit (BIO101, Carlsbad, CA, USA) according to the manufacturer’s protocol. The primers ITS1 and ITS4 (White et al. 1990) were used to amplify part of the internal transcribed spacer region (ITS) of the nuclear ribosomal RNA operon, including the 3′ end of the 18S rRNA gene, the first ITS region, the 5.8S rRNA gene, the second ITS region, and the 5′ end of the 28S rRNA gene. A part of the actin gene (ACT) was amplified with primers ACT-512F and ACT-783R (Carbone & Kohn 1999), a part of the small subunit mitochondrial ribosomal DNA (mtSSU) with primers MNS1 and MNS2 (Li et al. 1994), and a part of the histone H3 (HIS) gene with primers CYLH3F and CYLH3R (Crous et al. 2004b). Amplification reactions were performed with each primer set in a total reaction volume of 25 μl, which was composed of 1 × PCR Buffer (Applied Biosystems, Foster City, USA), variable MgCl2 concentrations, 60 μM dNTPs, 0.2 μM of each forward and reverse primer, 1.5 U of Taq DNA polymerase (Roche Diagnostics, Indianapolis, USA) and 1–10 ng of genomic DNA. PCR cycle conditions were 5 min of 95 °C, followed by 36 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 60 s, and a final elongation at 72 °C for 7 min. Amplicons were sequenced using both PCR primers with a DYEnamic ET Terminator Cycle Sequencing kit (Amersham Biosciences, Roosendaal, the Netherlands) according to the manufacturer’s recommendations, and sequences were analysed on an ABI Prism 3700 DNA Sequencer (Perkin-Elmer, Norwalk, Foster City, CA).
The resulting nucleotide sequences were analysed and automatically aligned using BioNumerics v. 4.5 (Applied Maths, Kortrijk, Belgium) followed by manual improvement by eye where necessary. Phylogenetic analyses were performed with PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2003), using the neighbour-joining algorithm with the uncorrected (“p”), the Kimura 2-parameter and the HKY85 substitution models. Alignment gaps longer than 10 bases were coded as single events for the phylogenetic analyses; the remaining gaps were treated as missing data. Any encountered ties were randomly broken. Phylogenetic relationships were also inferred with the parsimony algorithm using the heuristic search option with simple (ITS alignment) or 100 random taxa additions (combined alignment) and tree bisection and reconstruction (TBR) as the branch-swapping algorithm; alignment gaps were treated as missing (combined alignment) or as a fifth character state (ITS alignment) and all characters were unordered and of equal weight. Branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. Other measures calculated included tree length, consistency index, retention index and rescaled consistency index (TL, CI, RI and RC, respectively). The robustness of the obtained trees was evaluated by 10 000 000 fast stepwise (ITS alignment) or 1000 bootstrap heuristic bootstrap replications (combined alignment). Sequences were deposited in GenBank (Table 1) and the alignments in TreeBASE (www.treebase.org).
Morphology
Growth rates and colony morphology were recorded from colonies grown on MEA plates after 30 d incubation in darkness at 24 °C. Colony colours (surface and reverse) were assessed after growth on MEA and oatmeal agar (OA, Gams et al. 2007) using the colour charts of Rayner (1970). Microscopic observations were made from colonies cultivated on MEA and OA. Preparations were mounted in lactic acid and studied under a light microscope (× 1000 magnification). The 95 % confidence intervals were derived from 30 observations of spores formed on MEA or OA, with extremes given in parentheses. All cultures obtained in this study are maintained in the culture collection of the Centraalbureau voor Schimmelcultures (CBS) in Utrecht, the Netherlands or the working collections of Pedro Crous (CPC) or Mahdi Arzanlou (X, S numbers) at CBS (Table 1). Nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org) (Crous et al. 2004a).
RESULTS
DNA phylogeny
Two alignments of DNA sequences were subjected to phylogenetic analyses. The first alignment consisted of ITS sequences generated in this study as well as sequences obtained from the NCBI GenBank nucleotide sequence database. The ITS alignment consisted of a total number of 113 sequences (including one outgroup); 508 characters including alignment gaps were subjected to the analyses. Of these characters, 224 were parsimony-informative, 42 variable and parsimony-uninformative, and 242 were constant. Trees supporting the same clades were obtained irrespective of the analysis method used. The parsimony analysis yielded 11 780 equally most parsimonious trees that mainly differed in the order of taxa at the terminal nodes; one of the trees is presented in Fig. 1 (TL = 861 steps; CI = 0.569; RI = 0.934; RC = 0.532).
Fig. 1.
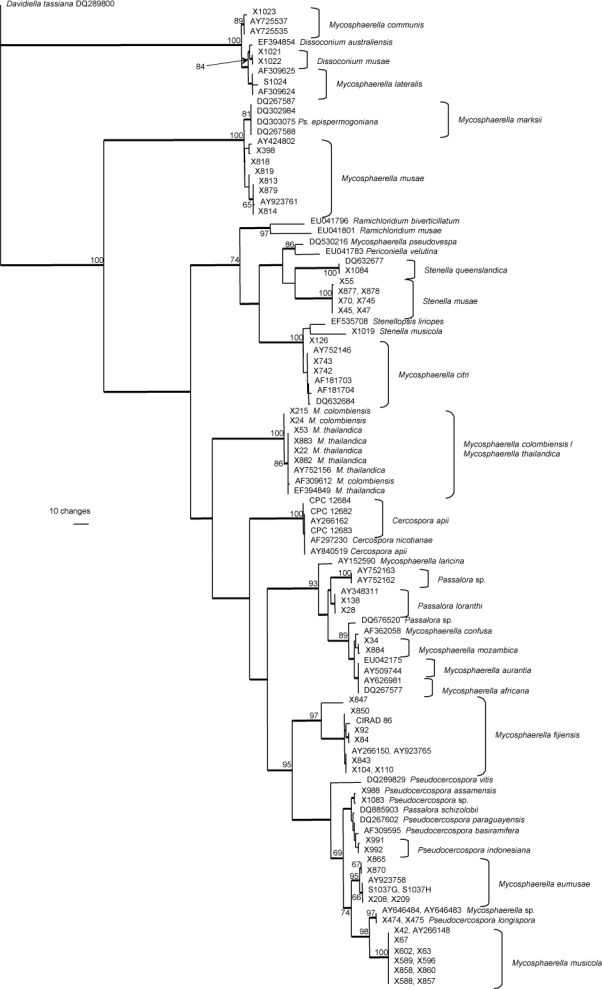
One of 11 780 equally most parsimonious trees obtained from a heuristic search with simple taxon additions of the ITS sequence alignment. The scale bar shows 10 changes, and bootstrap support values (65 % and higher) from 10 000 000 fast stepwise replicates are shown at the nodes. Thickened lines indicate the strict consensus branches. The tree was rooted to sequences of Davidiella tassiana strain CPC 11600 (GenBank accession number DQ289800). M. = Mycosphaerella and Ps. = Pseudocercospora.
The sequence data in the second alignment were analysed as one combined set consisting of 1648 characters (incl. alignment gaps) (number of included characters: ITS: 509, ACT: 188, HIS: 375, mtSSU: 576). This second alignment included 54 sequences (including the outgroup) and of the 1648 characters, 517 were parsimony-informative, 93 were variable and parsimony-uninformative, and 1038 were constant. Trees supporting the same clades were obtained irrespective of the analysis method used. The parsimony analysis yielded eight equally most parsimonious trees that mainly differed in the order of taxa at the terminal nodes; one of the trees is presented in Fig. 2 (TL = 1513 steps; CI = 0.654; RI = 0.901; RC = 0.589). Similar to the results obtained for the ITS alignment, the same lineages were found with the combined alignment. The ACT and HIS data were found to be more variable within species than the ITS and mtSSU data (data not shown for individual loci, variation within clades in Fig. 2). The phylogenetic results obtained are discussed where applicable in the descriptive notes below.
Fig. 2.
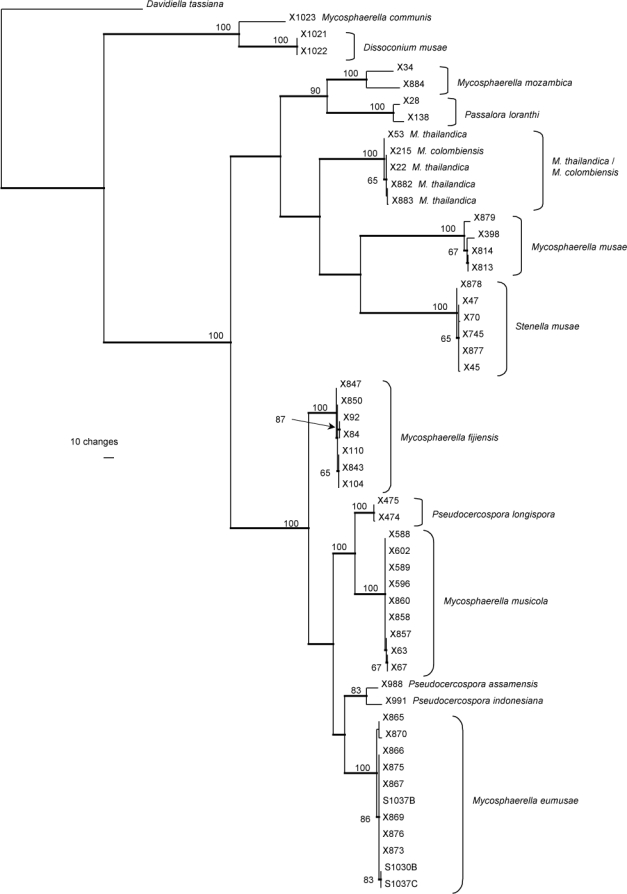
One of eight equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the combined (ITS, ACT, HIS, mtSSU) sequence alignment. The scale bar shows 10 changes, and bootstrap support values (65 % and higher) from 1000 replicates are shown at the nodes. Thickened lines indicate the strict consensus branches. The tree was rooted to sequences of Davidiella tassiana strain CPC 11600 (GenBank accession number DQ289800, DQ289867, EF679665, EU514455, respectively). M. = Mycosphaerella.
Taxonomy
The results of this study showed a rich diversity of Mycosphaerella spp. on Musa. Phylogenetic analyses revealed that more than 20 species of Mycosphaerella or its anamorphs occur on banana, including species known from hosts other than banana, namely Cercospora apii, Mycosphaerella citri, M. communis, M. lateralis, M. thailandica, and Passalora loranthi (Fig. 1). Furthermore, eight species proved to be morphologically and phylogenetically distinct from the species presently known from banana. These new species are described below.
Cercospora apii Fresen., Beitr. Mykol. 3: 91. 1863
= Cercospora hayi Calp., Studies on the Sigatoka disease of bananas and its fungus pathogen, Atkins Garden and Research Laboratory, Cuba: 63. 1955.
Specimens examined. Cuba, Musa paradisiaca var. sapientum, 1955, L. Calpouzos, holotype FH, ex-type culture ATCC 12234. – India, Bangladesh, Musa cv. Cavendish, Oct. 2005, I. Buddenhagen, CBS H-20035, culture CBS 119395.
Notes — In their treatment of the genus Cercospora, Crous & Braun (2003) considered C. hayi to be a synonym of the older name, Cercospora apii, which is known to have a wide host range. Based on a comparison of DNA sequence data with the ex-type strain of C. apii (GenBank AY840519; Groenewald et al. 2006), this synonymy appears to be correct.
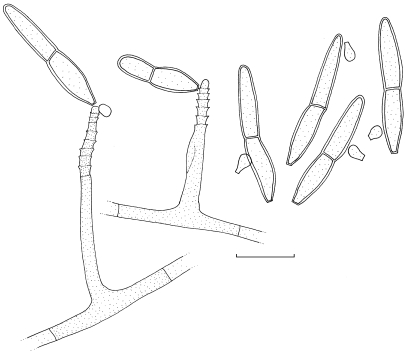
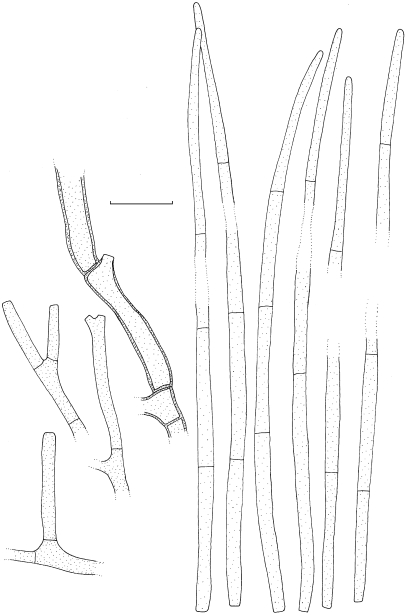
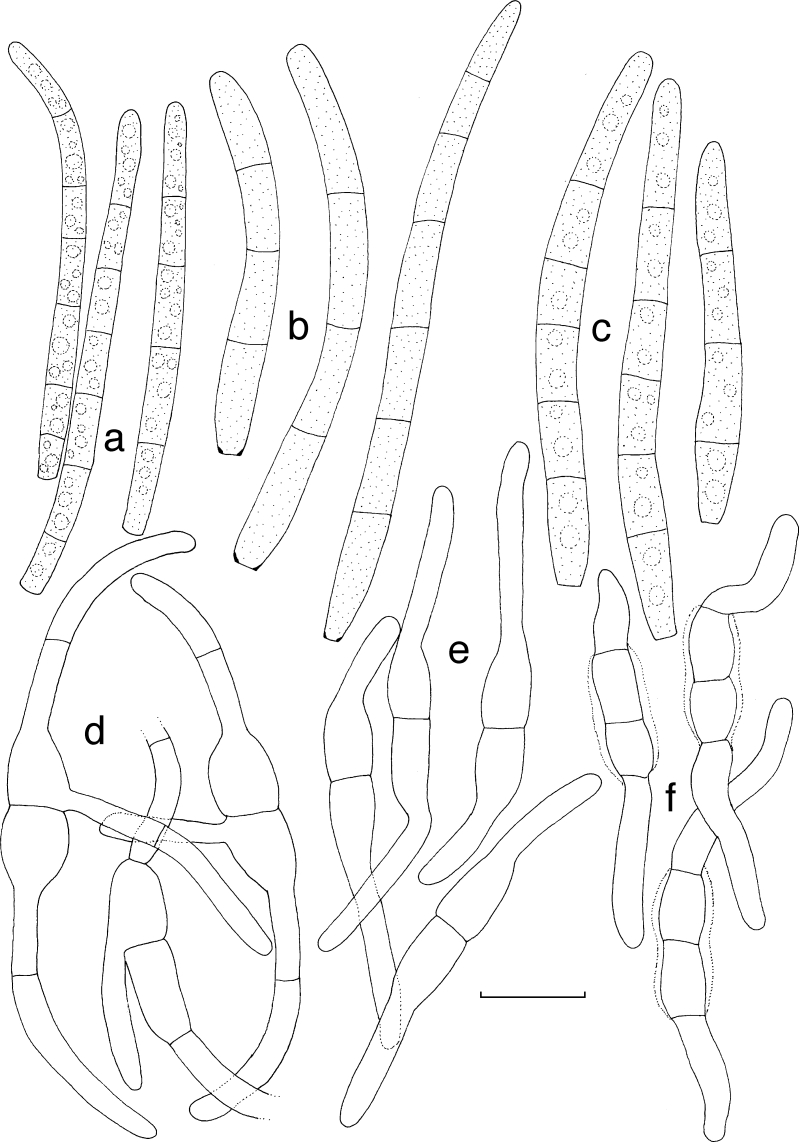
Dissoconium musae Arzanlou & Crous, sp. nov. — MycoBank MB505972; Fig. 3, 4
Fig. 3.
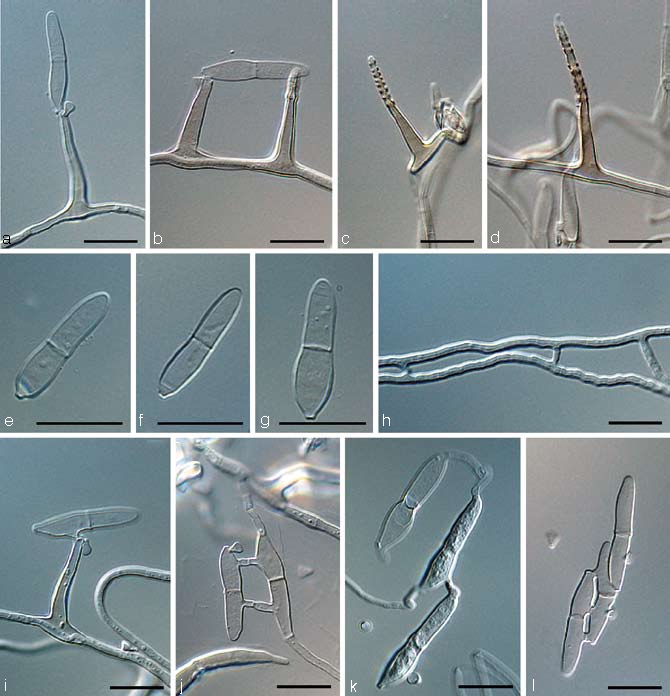
Dissoconium musae (CBS 122453). a–d. Conidiophores with sympodially proliferating conidiogenous cells, which produce primary and secondary conidia in pairs; e–g. primary conidia with truncate base; h–l. anastomoses between hyphae, primary and secondary conidia and primary conidia. — Scale bar = 10 μm.
Fig. 4.
Dissoconium musae (CBS 122453). — Scale bar = 10 μm.
Dissoconio communi simile, sed coloniis in vitro tarde crescentibus (usque ad 10 mm diam post 30 dies ad 24 °C in agaro maltoso).
Etymology. Named after its host plant, Musa.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae hyaline to subhyaline, thin-walled, smooth, forming a dense network with numerous anastomoses, 2–3 μm wide; aerial hyphae subhyaline, smooth, 2–3 μm wide. Conidiophores arising orthotropically from vegetative hyphae, often reduced to conidiogenous cells and continuous with supporting hyphae, thin-walled, smooth, pale brown, unbranched, straight, subulate to lageniform, tapering towards the apex, (10–)19–25(–53) × (2.5–)3–5 μm. Conidiogenous cells terminal, proliferating sympodially (but appearing as annellides under the light microscope), giving rise to a short conidium-bearing rachis, loci somewhat darkened and thickened. Conidia forming in sympodial order in pairs on a conidiogenous cell; the primary conidium is 2-celled, while the secondary conidium is aseptate; primary conidia pale olivaceous-brown, thin-walled, smooth, ellipsoidal to obclavate, 1-septate, apex obtuse, base obconically-truncate, (11–)22–26(–35) × (3–)4–5 μm, hilum unthickened; about 1 μm diam. Secondary conidia 1-celled, pale olivaceous-brown, pyriform to turbinate, 4–5 × 3–4 μm, base truncate, flat, unthickened, about 0.5 μm diam. Both conidial types are discharged forcibly in pairs and then anastomose on the agar surface. Anastomosis between primary conidia occurs as well and primary conidia may show multiple anastomoses. Primary conidia germinate from both ends and produce several conidiogenous cells and conidia (microcyclic conidiation). Germination of secondary conidia was not observed.
Cultural characteristics — Colonies on MEA slow-growing, reaching 10 mm diam after 30 d at 24 °C, erumpent, unevenly folded, with sparse aerial mycelium, colonies with granulate margin; surface hazel to isabelline in centre, and vinaceous-buff in outer region; brown-vinaceous in reverse. Colonies on OA reaching 25 mm diam after 30 d at 24 °C, effuse, with moderate aerial mycelium, later become powdery in centre, surface hazel; olivaceous in reverse.
Specimen examined. India, Tamil Nadu, Tiruchirapally, Musa cv. Nendran (Plantain) AAB, 2005, I. Buddenhagen, holotype CBS H-20036, culture ex-type X1021 = CBS 122453.
Notes — The genus Dissoconium is characterised by producing pairs of forcibly discharged primary and secondary conidia on sympodially proliferating conidiogeneous cells. Sympodial proliferation of the conidiogenous cells gives rise to a conidium-bearing rachis, which resembles that encountered in the genus Ramichloridium. The recent revision of the genus Ramichloridium and allied genera (Arzanlou et al. 2007b) revealed that R. apiculatum, the type species of the genus, is phylogenetically close to the species in the genus Dissoconium. However, Dissoconium is morphologically distinct from Ramichloridium by producing two types of forcibly discharged conidia. So far, seven species of Dissoconium have been described from different substrates (de Hoog et al. 1991, Jackson et al. 2004). Dissoconium musae is phylogenetically distinct from the other species of this genus, but morphologically similar to D. commune and D. dekkeri (teleomorph: Mycosphaerella lateralis), from which it differs based on its slower growth rate in culture.
Mycosphaerella eumusae Crous & Mour., Sydowia 54: 36. 2002
Anamorph. Pseudocercospora eumusae Crous & Mour., Sydowia 54: 36. 2002.
Specimen examined. Reunion, on leaves of Musa sp., 2001, J. Carlier, PREM 57314 (holotype of teleomorph), PREM 57315 (holotype of anamorph), cultures ex-type (CIRAD 1156, 1157 = CPC 4579, 4580 = CBS 114824, CBS 114825).
Notes — Based on the DNA sequence data obtained in this study (Fig. 2), it appears that M. eumusae is heterogeneous as presently circumscribed. Further studies would be required to determine if the phylogenetic variation also correlates with differences in morphology.
Mycosphaerella fijiensis M. Morelet, Ann. Soc. Sci. Nat. Archéol. Toulon Var 21: 105. 1969
= Mycosphaerella fijiensis var. difformis J.L. Mulder & R.H. Stover, Trans. Brit. Mycol. Soc. 67: 82. 1976.
Anamorph. Pseudocercospora fijiensis (M. Morelet) Deighton, Mycol. Pap. 140: 144. 1976.
Basionym. Cercospora fijiensis M. Morelet, Ann. Soc. Sci. Nat. Archéol. Toulon Var 21: 105. 1969.
≡ Paracercospora fijiensis (M. Morelet) Deighton, Mycol. Pap. 144: 51. 1979.
= Cercospora fijiensis var. difformis J.L. Mulder & R.H. Stover, Trans. Brit. Mycol. Soc. 67: 82. 1976.
≡ Paracercospora fijiensis var. difformis (J.L. Mulder & R.H. Stover) Deighton, Mycol. Pap. 144: 52. 1979.
Specimens examined. Hawaii, on leaves of Musa sp., D.S. Meredith & J.S. Lawrence, holotype IMI 136696. – Cameroon, date and collector unknown, epitype designated here CBS H-20037, culture ex-epitype CIRAD 86 = CBS 120258.
Note — The specimen and associated strain designated here as epitype, represent the strain that was selected by the Mycosphaerella consortium to obtain the full genome sequence of M. fijiensis (www.jgi.doe.gov/sequencing/why/CSP2006/mycosphaerella.html).
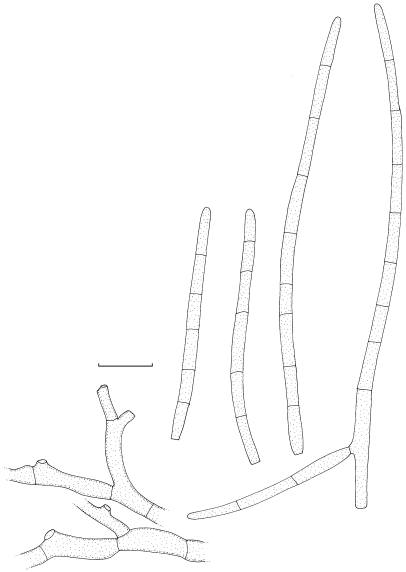
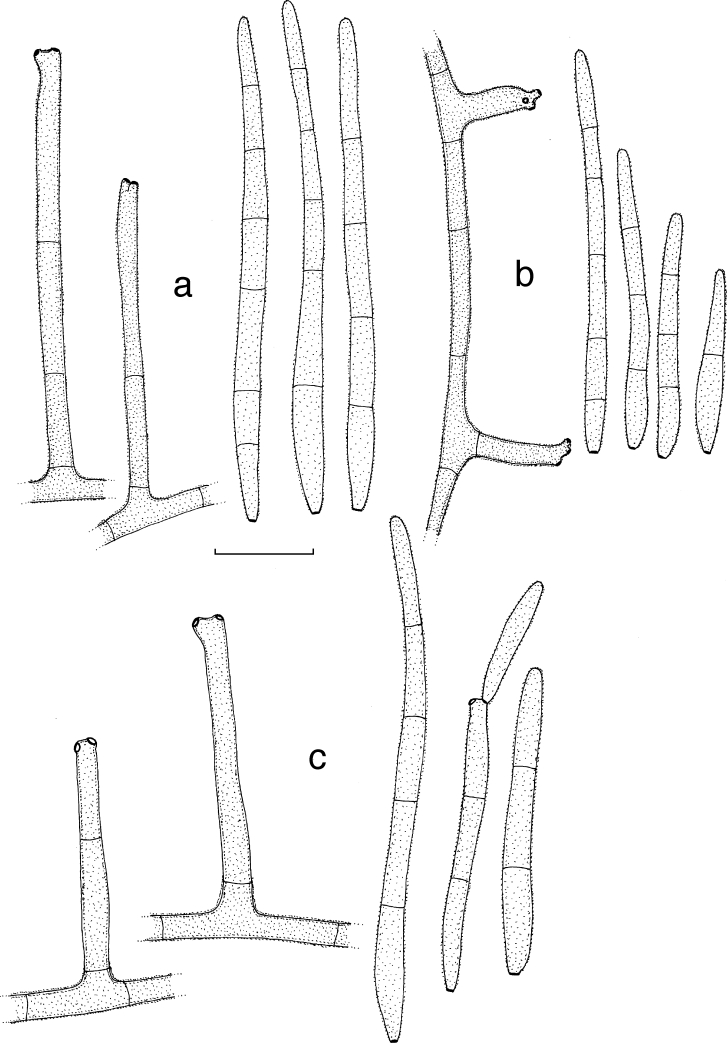
Mycosphaerella mozambica Arzanlou & Crous, sp. nov. — MycoBank MB505973; Fig. 5, 6
Fig. 5.
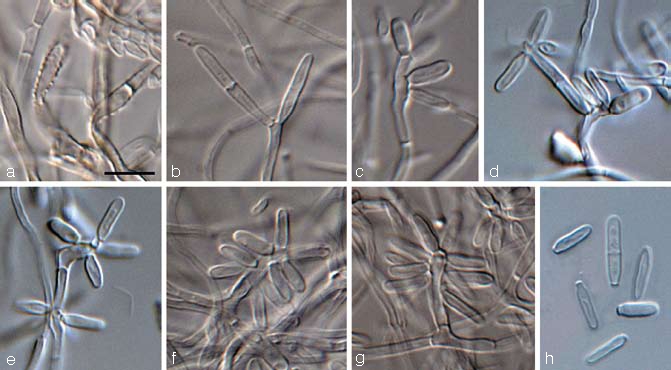
Mycosphaerella mozambica (CBS 122464). a. Verruculose hyphae; b–e. unbranched or loosely branched conidiophores with sympodially proliferating conidiogenous cells; f–g. sympodially proliferating conidiogenous cells give rise to short conidium-bearing rachis; h. conidia with truncate base. — Scale bars = 10 μm.
Fig. 6.
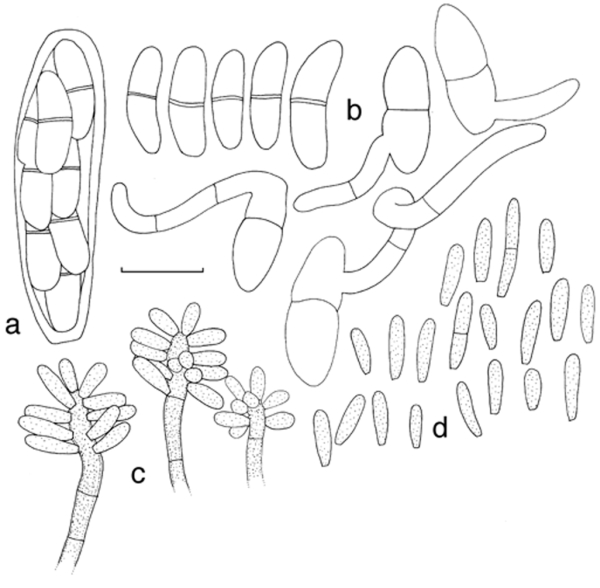
Mycosphaerella mozambica (CBS 122464). a. Ascus with biseriate ascospores; b. ascospore germination pattern; c. conidiophores with sympodially proliferating conidiogenous cells, which give rise to short conidium-bearing rachis; d. conidia. — Scale bar = 10 μm.
Anamorph. Ramichloridium-like.
Ascosporae rectae vel curvatae, fusoideo-ellipsoideae utrinque obtusae, ad septum medianum vix constrictae, (9–)10–11(–12) × 3–3.5(–4) μm.
Etymology. Named after the country of origin, Mozambique.
In vivo: Leaf spots amphigenous, irregular to subcircular,1–7 mm diam, grey to pale brown on adaxial surface, grey on abaxial surface, with dark brown margins. Ascomata amphigenous, intermingled among those of M. musicola, dark brown, subepidermal, becoming erumpent, globose, 70–90 μm diam; wall consisting of 2–3 layers of medium brown textura angularis. Asci aparaphysate, fasciculate, bitunicate, subsessile, obovoid to broadly ellipsoid, straight to slightly curved, 8-spored, 28–35 × 7–9 μm. Ascospores bi- to tri-seriate, overlapping, hyaline, non-guttulate, thin-walled, straight to curved, fusoid-ellipsoidal with obtuse ends, widest in middle of apical cell, medianly 1-septate, not to slightly constricted at the septum, tapering towards both ends, but more prominently towards the lower end, (9–)10–11(–12) × 3–3.5(–4) μm; ascospores becoming distorted upon germination after 24 h on MEA, becoming constricted at the septum, 6–7 μm wide with irregular, wavy germ tubes, growing 90 ° to the long axis, and not arising from the polar ends of the spore.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae hyaline to subhyaline, thin-walled, smooth or slightly rough, 2–4 μm wide; aerial hyphae pale olivaceous, smooth or finely verruculose. Conidiophores arising from unbranched or loosely branched hyphae, occasionally reduced to conidiogenous cells or integrated, hyaline, subcylindrical, 2–2.5 μm wide and up to 35 μm long. Conidiogenous cells integrated, terminal, polyblastic, sympodial, loci aggregated, flat, not protuberant (not denticle-like), unthickened, but somewhat darkened. Conidia solitary, obovoid, ellipsoidal, obclavate 0(–1)-septate, hyaline, thin-walled, smooth, (5–)9–12(–22) × 2–2.5(–3) μm; hilum truncate, flat, broad, unthickened, slightly darkened, about 1 μm diam. Although rarely observed, older conidia can become elongated, obclavate, and up to 4-septate.
Cultural characteristics — Colonies on MEA reaching 45 mm diam after 30 d at 24 °C; erumpent, folded, with moderate velvety to hairy aerial mycelium, with smooth, entire margins; surface pale vinaceous to mouse-grey; brown-vinaceous in reverse. Colonies on OA reaching 51 mm diam after 30 d at 24 °C; effuse, with sparse aerial mycelium and entire edge; surface vinaceous-buff to vinaceous, and pale vinaceous in reverse.
Specimens examined. Mozambique, Chimoio, Bairro, on leaf of Musa cv. 2003, A. Viljoen, holotype CBS H-20039, culture ex-type X34 = CBS 122464; CBS H-20040, CBS H-20041, CBS H-20042.
Notes — Sympodially proliferating conidiogenous cells are somewhat confusing with other morphologically similar genera such as Ramichloridium and Veronaea. The type species and most of the taxa referred to these genera are dematiaceous. The scars in Ramichloridium are subhyaline and slightly prominent. Veronaea has pigmented, truncate, flat loci and conidia with truncate bases. A recent revision of Ramichloridium and allied genera (Arzanlou et al. 2007b) revealed the type species of Ramichloridium, R. apiculatum, to be allied to the Dissoconium clade in Capnodiales, while the type species of Veronaea, V. botryosa, resides in Chaetothyiales. Mycosphaerella mozambica appeared to occur quite commonly on the banana samples investigated from Mozambique. Based on DNA sequence data, the ex-type strain appears similar to an isolate collected in Australia (CBS 121391 = X884). Unfortunately, however, the latter strain was sterile, so this could not be confirmed based on morphology.
Mycosphaerella musae (Speg.) Syd. & P. Syd., Philipp. J. Sci., C 8: 482. 1913
Basionym. Sphaerella musae Speg., Anales Mus. Nac. Hist. Nat. Buenos Aires 19: 354. 1909.
= Sphaerella musae Sacc., Atti Accad. Sci. Veneto-Trentino-Istriana, Ser. 3, 10: 67. 1917, homonym.
Specimen examined. Argentina, Jujuy, Orán, on leaves of Musa sapientum, Mar. 1905, holotype LPS, slide ex-type IMI 91165.
Notes — Mycosphaerella musae is reported to be the causal organism of Mycosphaerella speckle disease. However, as shown in the present study (Fig. 1), several distinct species appear to be able to induce these symptoms. Further collections would thus be required to recollect this species. All cultures examined in the present study were sterile.
Mycosphaerella musicola R. Leach ex J.L. Mulder, Trans. Brit. Mycol. Soc. 67: 77. 1976
Basionym. Mycosphaerella musicola R. Leach, Trop. Agric. (Trinidad) 18: 92. 1941 (nom. nud.).
Anamorph. Pseudocercospora musae (Zimm.) Deighton, Mycol. Pap. 140: 148. 1976.
Basionym. Cercospora musae Zimm., Centralbl. Bakteriol. Parasitenk. 2. Abt. 8: 219. 1902.
= Cercospora musae Massee, Bull. Misc. Inform. Kew 28: 159. 1914.
Specimens examined. Jamaica, on leaves of Musa sapientum, Jan. 1959, R. Leach, holotype IMI 75804a. – Cuba, on leaves of Musa sp., epitype designated here CBS H-20038, culture ex-epitype IMI 123823 = CBS 116634.
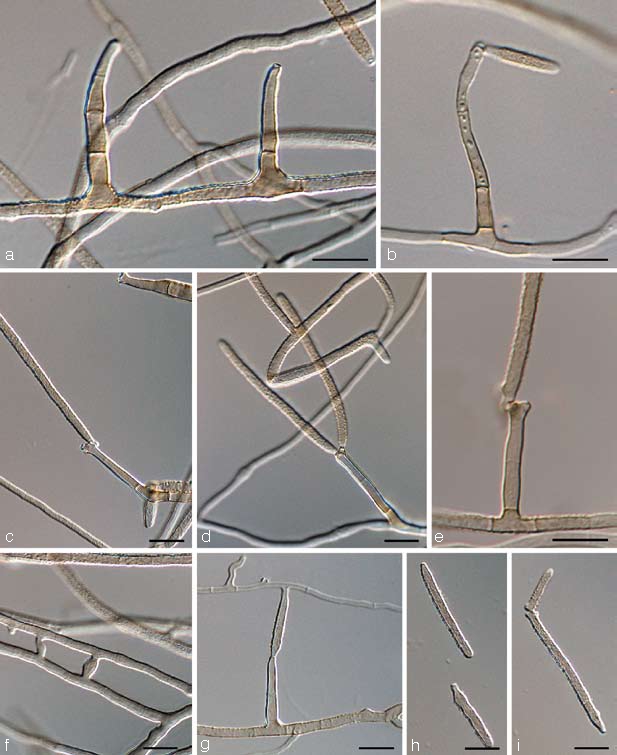
Pseudocercospora assamensis Arzanlou & Crous, sp. nov. — MycoBank MB505974; Fig. 7, 8
Fig. 7.
Pseudocercospora assamensis (CBS 122467). a. Conidiophore with sympodial and percurrent growth of conidiogenous cell; b–c. conidia. — Scale bar = 10 μm.
Fig. 8.
Pseudocercospora assamensis (CBS 122467). — Scale bar = 10 μm.
Pseudocercosporae musae similis, sed conidiis longioribus et angustioribus, (30–)59–70(–83) × 2–3 μm.
Etymology. Named after the locality of origin, India, Assam.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae smooth, branched, septate, medium brown, 2.5–4 μm wide; aerial hyphae thin-walled, smooth, medium brown. Conidiophores solitary, arising from superficial hyphae, medium brown, thin-walled, smooth, unbranched or branched above, 0–1-septate, subcylindrical, straight, up to 20 μm long, 2–3 μm wide. Conidiogenous cells integrated, terminal, or conidiophores reduced to conidiogenous cells, subcylindrical, tapering to truncate or bluntly rounded apices, medium brown, smooth, proliferating sympodially; conidial scars inconspicuous. Conidia solitary, pale brown, smooth, subcylindrical, with truncate bases and bluntly rounded apices, thin-walled with irregular swellings in older conidia, straight or curved, pluriseptate, (30–)59–70(–83) × 2–3 μm; hila about 1 μm wide, neither thickened nor darkened-refractive; microcyclic conidiation observed.
Cultural characteristics — Colonies on MEA reaching 47 mm diam after 30 d at 24 °C. Colonies elevated at the centre, with abundant aerial mycelium, and entire, smooth margin; surface pale mouse-grey to mouse-grey, olivaceous in reverse. Colonies on OA reaching 35 mm diam after 30 d at 24 °C; effuse, with moderate, velvety aerial mycelium, and entire, smooth margins; surface pale mouse-grey, and iron-grey in reverse.
Specimen examined. India, Assam, Naojan, on leaf of Musa cv. Nanderan (Plantain), 2005, I. Buddenhagen, holotype CBS H-20044, culture ex-type X988 = CBS 122467.
Notes — Based on its characteristic conidial shape and dimensions, P. assamensis appears distinct from those species presently known from this host. Pseudocercospora musae conidia are shorter and above all wider (10–80 × 2–6 μm; Carlier et al. 2000) than in P. longispora. Pseudocercospora longispora has much longer and somewhat wider conidia.
Pseudocercospora indonesiana Arzanlou & Crous, sp. nov. — MycoBank MB505975; Fig. 9, 10
Fig. 9.
Pseudocercospora indonesiana (CBS 122473). a–d. Conidia; e. intercalary conidiogenous cell. — Scale bar = 10 μm.
Fig. 10.
Pseudocercospora indonesiana (CBS 122473). — Scale bar = 10 μm.
Pseudocercosporae longisporae similis, sed conidiis modice brunneis, hyphis tenuitunicatis, modice brunneis, non inflatis et non monilioidibus-muriformibus, coloniis in vitro celeriter crescentibus (usque ad 27 mm diam post 30 dies ad 24 °C in agaro maltoso).
Etymology. Named after its country of origin, Indonesia.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae thin-walled, smooth, branched, septate, medium brown, 2.5–4 μm wide; aerial hyphae, thin-walled, smooth, medium brown. Conidiophores solitary, arising from superficial hyphae, medium brown, smooth, unbranched, 0–2-septate, subcylindrical, straight, up to 30 μm long, 2–2.5 μm wide. Conidiogenous cells integrated, terminal, subcylindrical, tapering to truncate or bluntly rounded apices, medium brown, smooth, proliferating sympodially, frequently reduced to conidiogenous loci; conidial scars inconspicuous. Conidia solitary, pale brown, smooth, subcylindrical, bases truncate, apices bluntly rounded, thin-walled, straight or curved, guttulate, 3–7-septate, (40–)78–95(–120) × 2–3 μm; hila unthickened, neither darkened nor refractive.
Cultural characteristics — Colonies on MEA reaching 27 mm diam after 30 d at 24 °C. Colonies low convex, with abundant aerial mycelium, and entire, smooth margin; surface pale mouse-grey to mouse-grey; in reverse dark mouse-grey. Colonies on OA reaching 35 mm diam after 47 d at 24 °C; effuse, with moderate aerial mycelium, and entire, smooth margins; surface pale mouse-grey; in reverse olivaceous-black.
Specimen examined. Indonesia, Western Sumatra, Kumango, on leaf of Musa cv. Buai, 2004, I. Buddenhagen, holotype CBS H-20045, culture ex-type X992 = CBS 122473.
Notes — Pseudocercospora indonesiana is phylogenetically distinct from the other species of Pseudocercospora occurring on Musa. Morphologically it has longer conidia than P. musae (teleomorph M. musicola) and P. assamensis, though they are very similar to those of P. longispora; it can, however, be distinguished from the latter by having medium brown conidia (those of P. longispora being pale brown), and its faster growth rate on MEA and OA.
Pseudocercospora longispora Arzanlou & Crous, sp. nov. — MycoBank MB505976; Fig. 11, 12
Fig. 11.
Pseudocercospora longispora (CBS 122469). a–e. Conidia. — Scale bar = 10 μm.
Fig. 12.
Pseudocercospora longispora (CBS 122469). — Scale bar = 10 μm.
Pseudocercosporae musae similis, sed conidiis longioribus, 82–120 × 2.5–4 μm.
Etymology. Named after its characteristically long conidia.
In vitro on OA: Mycelium submerged and superficial; submerged hyphae smooth, branched, septate, medium brown, thin-walled, 2–3 μm wide; aerial hyphae smooth, medium brown; hyphal cells become thick-walled, swollen, forming dark-brown monilioid, muriform cells, 5–17 × 7–12 μm. Conidiophores solitary, arising from superficial hyphae; conidiophores medium brown, smooth, unbranched or branched above, 0–2-septate, subcylindrical, straight, up to 30 μm long, 2–3 μm wide. Conidiogenous cells integrated, terminal, subcylindrical, tapering to truncate or bluntly rounded apices, medium brown, smooth, forming conidia by sympodial proliferation, rarely by means of percurrent proliferation; conidial scars inconspicuous. Conidia solitary, pale brown, thin-walled, smooth, cylindrical to subcylindrical, widest in the middle of conidium, tapering towards the apex, bases truncate, straight, multi-septate, 82–120 × 2.5–4 μm; hila about 1 μm diam, neither thickened nor darkened-refractive.
Cultural characteristics — Colonies reaching 15 mm diam after 30 d at 24 °C. Colonies erumpent, with moderate aerial mycelium, and entire, smooth edges; surface buff to rosy-buff, mouse-grey to dark grey; in reverse dark mouse-grey. Colonies on OA reaching 15 mm diam after 30 d at 24 °C, effuse, with abundant aerial mycelium, and entire, smooth margins; surface pale mouse-grey; in reverse dark mouse-grey.
Specimen examined. Malaysia, Felcra Plantation, Melaka, Musa cv. Pisang Byok AAA/AAB, July 1988, D.R. Jones, holotype CBS H-20043, culture ex-type X475 = CBS 122470.
Notes — Pseudocercospora longispora resembles P. musae (teleomorph Mycosphaerella musicola) in its colony morphology on MEA and OA. However, in P. musae conidia are much shorter (10–80 × 2–6 μm; Carlier et al. 2000) than in P. longispora.
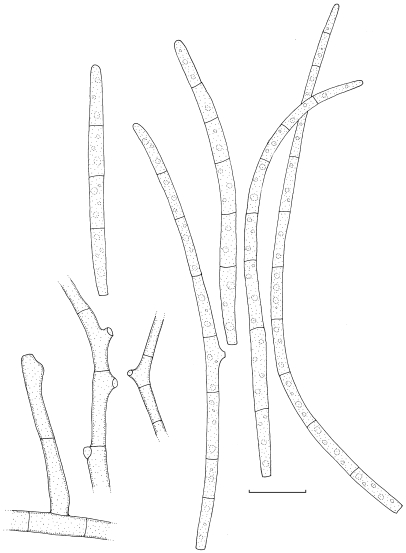
Stenella musae Arzanlou & Crous, sp. nov. — MycoBank MB505977; Fig. 13, 14a
Fig. 13.
Stenella musae (CBS 122477). a–d. Conidiophores with sympodially proliferating conidiogenous cells; e–f. conidia. — Scale bar = 10 μm.
Fig. 14.
a. Stenella musae (CBS 122477); b. Stenella queenslandica (CBS 122475); c. Stenella musicola (CBS 122479). — Scale bar = 10 μm.
Conidiophora ex hyphis superficialibus oriunda, modice brunnea, tenuitunicata, verruculosa vel verrucosa, 0–3-septata, subcylindrica, recta vel geniculata-sinuosa, non ramosa, ad 30 μm longa et 2–2.5 μm lata. Cellulae conidiogenae integratae, terminales, interdum intercalares, modice brunneae, verruculosae, subcylindricae, apicem versus attenuatae, sympodiales, locis truncatis, subdenticulatis, 1–1.5 μm diam, inspissatis et fuscatis-refringentibus praeditae. Conidia solitaria, dilute brunnea, verruculosa, tenuitunicata, subcylindrica vel obclavata, recta vel curvata, 0–7-septata, (7–)27–40(–70) × 1.5–3 μm, hilo inspissato obscuriore refringente, 1–1.5 μm diam praedita.
Etymology. Named after its host, Musa.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae smooth to verrucose, thin-walled, subhyaline to medium brown, 2–3 μm wide, with thin septa; aerial hyphae coarsely verrucose, olivaceous-brown to medium brown, rather thick-walled, 2–2.5 μm wide, with thin septa. Conidiophores arising from superficial hyphae, medium brown, rather thick-walled, finely verrucose to verruculose, 0–3-septate, subcylindrical, straight to geniculate-sinuous, unbranched, up to 30 μm long, 2–2.5 μm wide. Conidiogenous cells integrated, terminal, sometimes intercalary, unbranched, medium brown, finely verruculose, subcylindrical, tapering towards flat-tipped, subdenticulate apical loci, 1–1.5 μm diam, proliferating sympodially; loci thickened, darkened, refractive. Conidia solitary, thin-walled, pale brown, finely verrucose, subcylindrical to obclavate, with subobtuse apex, and long obconically subtruncate to obconically subtruncate base, straight to curved, 0–7-septate, (7–)27–40(–70) × 1.5–3 μm; hilum thickened, darkened, refractive, 1–1.5 μm diam.
Cultural characteristics — Colonies on MEA reaching 30 mm diam after 30 d at 24 °C. Colonies erumpent, unevenly folded, with moderate aerial mycelium, and entire, smooth margin; surface pale mouse-grey to mouse-grey; in reverse dark mouse-grey. Colonies on OA reaching 48 mm diam after 30 d at 24 °C; effuse, with moderate aerial mycelium, and entire margins; surface pale mouse-grey to mouse-grey, and dark mouse-grey in reverse.
Specimens examined. Tonga, ACIAR Plot, Tongatapu, Musa cv. TU8 AAAA, Mar. 1990, R.A. Fullerton, holotype CBS H-20047, culture ex-type X745 = CBS 122477. – Windward Islands, St Lucia, on Musa cv., 2003, E. Reid, culture X47 = CBS 122476.
Notes — Stover (1994) discussed and illustrated a Stenella sp. from banana, and named it ‘Cercospora non-virulentum’, which was considered as a prevalent co-inhabitant with Black Leaf Streak and Sigatoka. Mycosphaerella musae is the causal agent of Mycosphaerella Speckle disease of banana (Carlier et al. 2000). A comparison made between strains isolated from Mycosphaerella Speckle disease symptoms (presumed M. musae), and ‘Cercospora non-virulentum’ isolates in culture, suggested that the two species are identical, both producing brown, verruculose conidia with thickened scars on agar medium (Stover 1994). An inoculation assay carried out by using a mixture of conidia and mycelium of ‘Cercospora non-virulentum’ on banana ‘Cavendish Valery’ leaves resulted in leaf spot symptoms after 70 d incubation, resembling those obtained using ascospores derived from ‘M. musae’ strains.
Because ‘Cercospora non-virulentum’ was never validly published, it is difficult to make a comparison with Stenella musae. However, based on the description provided by Stover (1994), S. musae has shorter conidia (7–70 × 1.5–3 μm) than ‘Cercospora non-virulentum’ (55–200 × 2.6–3.2 μm).
A further complication lies in the fact that several phylogenetically distinct species of Mycosphaerella have in the past been isolated from Mycosphaerella Speckle disease symptoms of banana. All the ‘M. musae’ isolates examined in this study were sterile, and thus could not be used for morphological comparison. Mycosphaerella musae was originally described from Musa sapientum leaves collected in Argentina. An examination of the type (IMI 91165) shows ascospores to be straight to slightly curved, fusoid-ellipsoidal with narrowly obtuse ends, being widest at the median septum (Fig. 15). Further collections would thus be required to clarify the identity of this species.
Fig. 15.
Ascospores of Mycosphaerella musae (IMI 91165). — Scale bar = 10 μm.
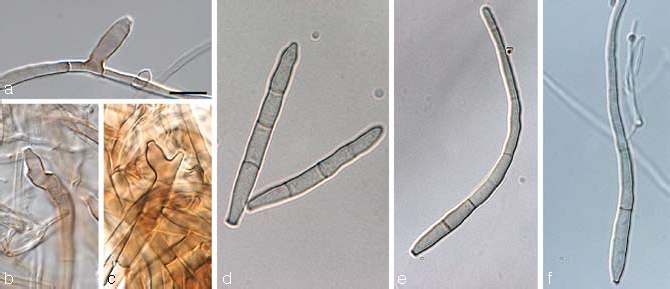
Stenella musicola Arzanlou & Crous, sp. nov. — MycoBank MB505978; Fig. 14c, 16
Fig. 16.
Stenella musicola (CBS 122479). a–e. Conidiophores with sympodially proliferating conidiogenous cells and darkened, thickened loci; f–g. hyphal anastomoses; h–i. conidia. — Scale bar = 10 μm.
Stenellae musae similis, sed conidiophoris leviter longioribus et latioribus, (18–)30–36(–45) × (2–)2.5–3(–4) μm, conidiis saepe longioribus, (7–)37–57(–120) × 2–4 μm. A Stenellae queenslandica conidiophoris 0–2-septatis et conidiis 2–4 μm latis differt.
Etymology. Named after its host, Musa.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae smooth to verrucose, thin-walled, subhyaline to olivaceous brown, 2–3 μm wide, with thin septa; aerial hyphae coarsely verrucose, olivaceous-brown, rather thick-walled, 2–2.5 μm wide, with thin septa. Conidiophores arising from superficial hyphae, pale brown, rather thick-walled, finely verruculous, 0–2-septate, occasionally continuous with supporting hyphae, subcylindrical, straight to geniculate-sinuous, unbranched, (18–)30–36(–45) × (2–)2.5–3(–4) μm. Conidiogenous cells integrated, terminal, sometimes intercalary, un-branched, pale brown, smooth or finely verruculose, cylindrical to subcylindrical, sometimes swollen at the apex, with flat-tipped apical loci, proliferating sympodially; 1–1.5 μm diam, loci thickened, darkened, refractive. Conidia solitary, rarely in unbranched chains, medium brown, thin-walled, finely verruculose subcylindrical to obclavate, with subobtuse apex, and long obconically subtruncate to obconically subtruncate base, straight to curved, 0–pluri-septate, (7–)37–57(–120) × 2–4 μm; hilum thickened, darkened, refractive, 1–1.5 μm wide.
Cultural characteristics — Colonies on MEA reaching 28 mm diam after 30 d at 24 °C; effuse, slightly raised at the centre, with moderate, velvety to hairy aerial mycelium; folded, with entire smooth margin; surface pale mouse-grey to mouse-grey; in reverse dark mouse-grey. Colonies on OA reaching 39 mm diam after 30 d at 24 °C; effuse, with moderate velvety to hairy aerial mycelium, and entire, smooth margins; surface pale mouse-grey to mouse-grey, and olivaceous in reverse.
Specimen examined. India, Tamil Nadu, Tiruchirapally, on leaf of Musa cv. Grand Nain AAA (Cav.), 2005, I. Buddenhagen, holotype CBS H-20046, culture ex-type X1019 = CBS 122479.
Notes — Stenella musicola morphologically also resembles S. citrigrisea (teleomorph Mycosphaerella citri), which is known from Citrus (Pretorius et al. 2003). It differs from the later species, however, based on its conidial dimensions. In S. musicola conidia range from (7–)37–57(–120) × 2–4 μm, while in S. citrigrisea conidia are longer and narrower, namely 25–200 × 1.5–3 μm. The three new Stenella species on Musa spp. are morphologically very similar and only gradually differentiated in the size and septation of the conidiophores and conidia.
Stenella queenslandica Arzanlou & Crous, sp. nov. — MycoBank MB505979; Fig. 14b, 17
Fig. 17.
Stenella queenslandica (CBS 122475). a. Conidiophore with terminal conidiogenous cell; b–d. conidia. — Scale bar = 10 μm.
Stenellae musae similis, sed conidiis longioribus, 51–83 × 2–2.5 μm. A Stenella musicola conidiophoris 1–4-septatis et conidiis saepe longioribus et angustioribus, 51–83 × 2–2.5 μm, differt.
Etymology. Named after Queensland, the state in Australia where this fungus was collected.
In vitro on MEA: Mycelium submerged and superficial; submerged hyphae smooth, thin-walled, subhyaline to olivaceous-brown, 2–3 μm wide, with thin septa; aerial hyphae coarsely verrucose, olivaceous-brown, rather thick-walled, 2–2.5 μm wide, with thin septa. Conidiophores arising from superficial hyphae, pale brown, thin-walled, finely verrucose, 1–4-septate, occasionally reduced to conidiogenous cells, subcylindrical, straight to geniculate-sinuous, unbranched, up to 40 μm long and 2–3 μm wide. Conidiogenous cells integrated, terminal, sometimes intercalary, unbranched, pale brown, smooth or finely verruculose, cylindrical, tapering to a bluntly rounded apex with flat-tipped apical loci that proliferate sympodially; loci thickened, darkened, refractive about 1 μm diam. Conidia solitary, medium brown, thin-walled, verruculose, subcylindrical to obclavate, with subobtuse to obtuse apex and long obconically subtruncate to obconically subtruncate base, straight to curved, 0–multi-septate, 51–83 × 2–2.5 μm; hilum thickened, darkened, refractive, 0.5–1 μm wide.
Cultural characteristics — Colonies on MEA reaching 24 mm diam after 30 d at 24 °C. Colonies effuse, slightly elevated at the centre with abundant aerial mycelium, and entire, smooth margins; surface mouse-grey to dark mouse-grey; dark mouse-grey in reverse. Colonies on OA reaching 41 mm diam after 30 d at 24 °C, colonies effuse, with moderate aerial mycelium, and entire, smooth margin; surface olivaceous-grey; iron-grey in reverse.
Specimen examined. Australia, Queensland, Mount Lewis, Mount Lewis Road, 16° 34′ 47.2″ S, 145° 19′ 7″ E, 538 m alt., on Musa banksii leaf, Aug. 2006, P.W. Crous, W. Gams & B. Summerell, holotype CBS H-20050, culture ex-type CBS 122475.
Notes — The ITS sequence of Stenella queenslandica is identical to that of Mycosphaerella obscuris (Burgess et al. 2007), a pathogen of Eucalyptus known from Vietnam and Indonesia. However, the latter fungus is a species of Teratosphaeria with a Readeriella anamorph (CBS 119973), which appears to be a synonym of T. suttonii (Crous & Wingfield 1997, Crous et al. 2007a, b), and the deposited sequences (DQ632676, DQ632677) belong to another species.
DISCUSSION
The present study is the first multi-gene DNA phylogenetic study of a global set of Mycosphaerella isolates associated with the Sigatoka disease complex of banana. Considering that Sigatoka diseases are the economically most important diseases of banana and the main constraint for banana production worldwide (reviewed in Jones 2000), there was a huge paucity of knowledge relating to the identity of other Mycosphaerella species occurring on banana. Even though several species of Mycosphaerella have in the past been described from Musa, the majority has never been known from culture (Pont 1960, Stover 1963, 1969, 1977, 1980, 1994, Mulder & Stover 1976, Pons 1987, Crous et al. 2003, Aptroot 2006, Arzanlou et al. 2007a). The integration of DNA analyses and morphology in the present study revealed more than 20 species of Mycosphaerella to occur on banana. Five of these species were shown to have wider host ranges than banana only, and we described a further eight new species of Mycosphaerella from various Musa collections.
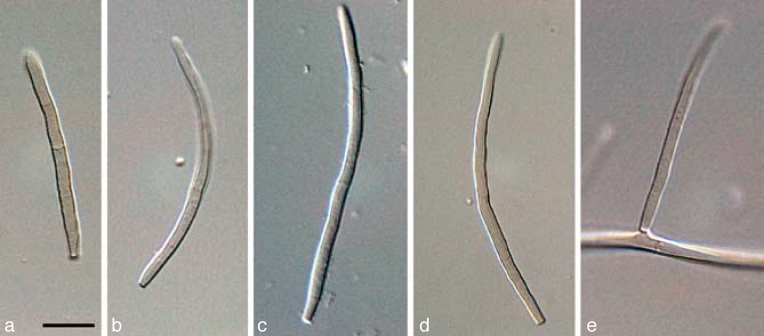
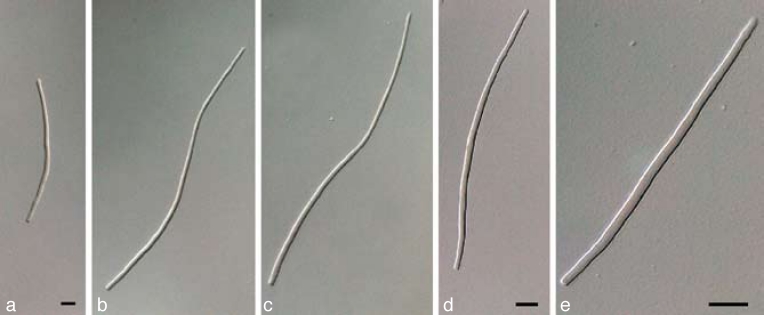
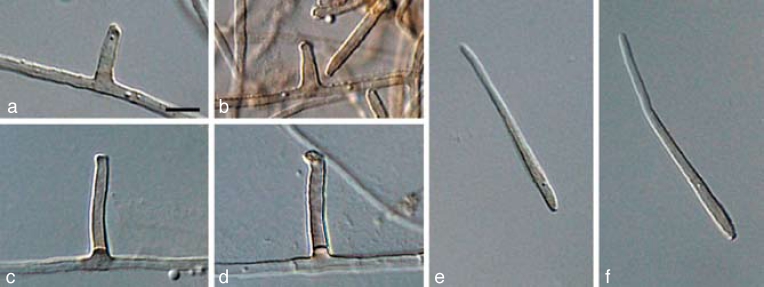
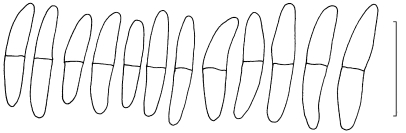
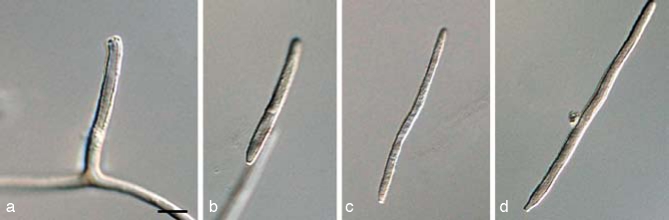
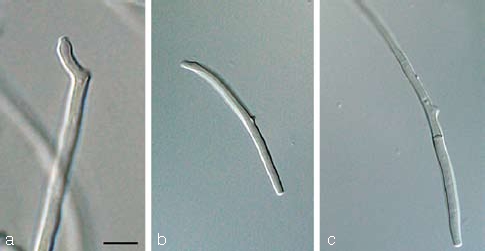
The three primary agents of the Sigatoka disease complex, M. eumusae, M. fijiensis, and M. musicola can be distinguished based on their conidial morphology and ascospore germination patterns (reviewed in Jones 2000, Crous & Mourichon 2002). Conidia of M. fijiensis are medium brown, and have a characteristic thickening along the basal rim of the hilum, which is absent in M. musicola and M. eumusae. These two species have medium and pale brown conidia, respectively. Ascospores of M. fijiensis and M. musicola germinate from both polar ends, do not become distorted (4–5 μm wide), with a germ tube parallel to the long axis of the spore. However, in M. musicola a mucoid sheath surrounds the germinating ascospores, and the germ tubes are more irregular in width than in M. fijiensis. Ascospores of M. eumusae show some distortion upon germination (5–6 μm wide), and frequently germinate by means of 3–4 germ tubes, which grow parallel or lateral to the long axis of the spore (Fig. 18, 19). Thus, all of these species can be identified based on a combination of morphology and cultural characteristics, but proper identification remains problematic to the non-specialist. Hence the DNA barcodes generated in this study, along with the Taqman probes (Arzanlou et al. 2007a) is an alternative method of identification.
Fig. 18.
a–c. Conidia in Pseudocercospora eumusae, P. fijiensis, and P. musae, respectively; d–f. ascospore germination pattern in M. eumusae, M. fijiensis and M. musicola, respectively. — Scale bar = 10 μm.
Fig. 19.
Pseudocercospora fijiensis. a–c. Conidiophores with sympodially proliferating conidiogenous cells; d–f. obclavate conidia with darkened hilum. — Scale bar = 10 μm.
Besides the three primary agents of the Sigatoka complex disease, which have Pseudocercospora anamorphs, three additional Pseudocercospora species were described from Musa in the present study. One of these, Pseudocercospora longispora, has in the past been confused with P. musae (teleomorph M. musicola) and has been isolated from similar Sigatoka disease lesions. Although these species can be distinguished based on differences in conidial size and shape, these characters overlap among the various Pseudocercospora species, making explicit identification solely possible by means of additional markers such as DNA sequence data (Fig. 2).
Much confusion still surrounds the identity of M. musae. According to Stover (1994), M. musae is identical to a Stenella species called ‘Cercospora non-virulentum’. This species was considered as a prevalent co-inhabitant with black Sigatoka and yellow Sigatoka. A comparison made between isolates isolated from Mycosphaerella Speckle disease symptoms, revealed several phylogenetically distinct species to be associated with this disease. In the present study we treated four Stenella species, three of which proved to be new on banana. None of these three new species fit with the description provided for ‘Cercospora non-virulentum’ by Stover (1994), which appears to represent yet another undescribed species of Stenella. Further collections would thus be required to resolve the status of ‘Cercospora non-virulentum’ and M. musae.
Data obtained in the present study revealed three species of Dissoconium on Musa, of which one is described as new. The recent revision of the genus Ramichloridium and allied genera (Arzanlou et al. 2007b) revealed that R. apiculatum, type species of Ramichloridium, has phylogenetic affinity with the genus Dissoconium. However, the latter genus is morphologically distinct from Ramichloridium by producing forcibly discharged pairs of primary and secondary conidia. Thus far seven species of Dissoconium have been described from different substrates, and as in the Pseudocercospora species occurring on Musa, identification is best achieved by means of molecular sequence data.
It is interesting to note that up to six species have been reported during the course of the present study as occurring on hosts other than Musa. Although our present data suggest the causal agents of Sigatoka to be highly specific to banana, no information is presently available to elucidate the ecology and possible pathology of the wide host range species, and inoculation studies would now be required to fully resolve their status as foliar pathogens of banana. The possibility exists that some of the species described here as new have been described previously on hosts other than banana. However, none of the sequences presently in GenBank, or in the MycoBank database, match any known comparable species.
From the data presented in this study, it is clear that the Sigatoka disease complex is caused by a multitude of Mycosphaerella species. However, the exact contribution of each of these species to the disease complex remains unclear. The multi-locus DNA sequence data set established in this study can be used to develop species-specific molecular detection tools, which is a good alternative for traditional diagnostics. These tools can subsequently be implemented in disease management programmes.
Acknowledgments
The work of Mahdi Arzanlou was funded by the Ministry of Science, Research and Technology of Iran, which we gratefully acknowledge. Several colleagues from different countries provided material without which this work would not have been possible. Errol Reid from the Windward Islands, Yasmina Jaufeerally-Fakim from Mauritius, Gert Kema from the Netherlands and Randy Christopher Ploetz from Florida, USA. We thank Marjan Vermaas for preparing the photographic plates, and Arien van Iperen for taking care of the cultures. Dr Lute-Harm Zwiers and Prof. dr P.J.G.M. de Wit are thanked for their critical review of this article, and Prof. dr Walter Gams for providing the Latin diagnoses.
REFERENCES
- Aptroot A. 2006. Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodiversity Series 5: 1 – 231 . [Google Scholar]
- Arzanlou M, Abeln ECA, Kema GHJ, Waalwijk C, Carlier J, Vries I de, Guzmán M, Crous PW. 2007a. Molecular diagnostics for the Sigatoka disease complex of banana. Phytopathology 97: 1112 – 1118 . [DOI] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin H-D, Crous PW. 2007b. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57 – 93 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess TI, Barber PA, Sufaati S, Xu D, Hardy GEStJ, Dell B. 2007. Mycosphaerella spp. on Eucalyptus in Asia; new species; new hosts and new records. Fungal Diversity 24: 135 – 157 . [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553 – 556 . [Google Scholar]
- Carlier J, Lebrun MH, Zapater MF, Dubois C, Mourichon X. 1996. Genetic structure of the global population of bananas black leaf streak fungus Mycosphaerella fijiensis. Molecular Ecology 5: 499 – 510 . [Google Scholar]
- Carlier J, Zapater MF, Lapeyre F, Jones DR, Mourichon X. 2000. Septoria leaf spot of banana: a newly discovered disease caused by Mycosphaerella eumusae (anamorph Septoria eumusae). Phytopathology 90: 884 – 890 . [DOI] [PubMed] [Google Scholar]
- Cortinas MN, Crous PW, Wingfield BD, Wingfield MJ. 2006. Multi-gene phylogenies and phenotypic characters distinguish two species within the Colletogloeopsis zuluensis complex associated with Eucalyptus stem cankers. Studies in Mycology 55: 133 – 146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW. 1998. Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. Mycologia Memoirs 21: 1 – 170 . [Google Scholar]
- Crous PW, Aptroot A, Kang JC, Braun U, Wingfield MJ. 2000. The genus Mycosphaerella and its anamorphs. Studies in Mycology 45: 107 – 121 . [Google Scholar]
- Crous PW, Braun U. 2003. Mycosphaerella and its anamorphs. 1. Names published in Cercospora and Passalora. CBS Biodiversity Series 1: 1 – 571 . [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007a. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004a. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19 – 22 . [Google Scholar]
- Crous PW, Groenewald JZ, Aptroot A, Braun U, Xavier M, Carlier J. 2003. Integrating morphological and molecular data sets in Mycosphaerella, with specific reference to species occurring on Musa. In: Jacome L, Lepoivre P, Marin D, Ortiz R, Romero R, Escalant JV. (eds), Mycosphaerella leaf spot diseases of bananas: present status and outlook. Proceedings of the Second International Workshop on Mycosphaerella leaf spot diseases of bananas, San José, Costa Rica: 43–57. INIBAP, France . [Google Scholar]
- Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ. 2004b. Cryptic speciation and host specificity among Mycosphaerella spp. occurring on Australian Acacia species grown as exotics in the tropics. Studies in Mycology 50: 457 – 469 . [Google Scholar]
- Crous PW, Kang JC, Braun U. 2001. A phylogenetic redefinition of anamorph genera in Mycosphaerella based on ITS rDNA sequence and morphology. Mycologia 93: 1081 – 1101 . [Google Scholar]
- Crous PW, Mourichon X. 2002. Mycosphaerella eumusae and its anamorph Pseudocercospora eumusae spp. nov.: causal agent of eumusae leaf spot disease of banana. Sydowia 54: 35 – 43 . [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Mohammed C, Himaman W, Groenewald JZ. 2007b. Foliicolous Mycosphaerella spp. and their anamorphs on Corymbia and Eucalyptus. Fungal Diversity 26: 143 – 185 . [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ. 2006a. Eucalyptus microfungi known from culture. 1. Cladoriella and Fulvoflamma genera nova, with notes on some other poorly known taxa. Studies in Mycology 55: 53 – 63 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ. 1997. Colletogloeopsis, a new coelomycete genus to accommodate anamorphs of Mycosphaerella occurring on Eucalyptus. Canadian Journal of Botany 75: 667 – 674 . [Google Scholar]
- Crous PW, Wingfield MJ, Mansilla JP, Alfenas AC, Groenewald JZ. 2006b. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. II. Studies in Mycology 55: 99 – 131 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton FC. 1976. Studies on Cercospora and allied genera. VI. Pseudocercospora Speg., Pantospora Cif. and Cercoseptoria Petr. Mycological Papers 140: 1 – 168 . [Google Scholar]
- Deighton FC. 1979. Studies on Cercospora and allied genera. VII. New species and redispositions. Mycological Papers 144: 1 – 56 . [Google Scholar]
- Farr DF, Bills GF, Chamuris GP, Rossman AY. 1995. Fungi on plants and plant products in the United States APS Press, St Paul, MN, USA: . [Google Scholar]
- Gams W, Verkley GJM, Crous PW. 2007. CBS course of mycology, 5th ed Centraalbureau voor Schimmelcultures, Utrecht, Netherlands: . [Google Scholar]
- Goodwin SB, Dunkle DL, Zismann VL. 2001. Phylogenetic analysis of Cercospora and Mycosphaerella based on the internal transcribed spacer region of ribosomal DNA. Phytopathology 91: 648 – 658 . [DOI] [PubMed] [Google Scholar]
- Groenewald M, Groenewald JZ, Braun U, Crous PW. 2006. Host range of Cercospora apii and C. beticola and description of C. apiicola, a novel species from celery. Mycologia 98: 275 – 285 . [DOI] [PubMed] [Google Scholar]
- Hayden HL, Carlier J, Aitken EAB. 2003. Population differentiation in the banana leaf spot pathogen Mycosphaerella musicola, examined at a global scale. Plant Pathology 52: 713 – 719 . [Google Scholar]
- Hoog GS de, Hijwegen T, Batenburg van der Vegte WH. 1991. A new species of Dissoconium. Mycological Research 95: 679 – 682 . [Google Scholar]
- Hunter GC, Wingfield BD, Crous PW, Wingfield MJ. 2006. A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Studies in Mycology 55: 147 – 161 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SL, Maxwell A, Neumeister-Kemp HG, Dell B, Hardy GEStJ. 2004. Infection, hyperparasitism and conidiogenesis of Mycosphaerella lateralis on Eucalyptus globulus in Western Australia. Australasian Plant Pathology 33: 49 – 53 . [Google Scholar]
- Jones DR. 2000. Diseases of banana, abaca and enset CAB International, Wallingford, Oxon, UK: . [Google Scholar]
- Jones DR. 2003. The distribution and importance of the Mycosphaerella leaf spot diseases of banana. In: Jacome L, Lepoivre P, Marin D, Ortiz R, Romero R, Escalant JV. (eds), Mycosphaerella leaf spot diseases of bananas: present status and outlook. Proceedings of the Second International Workshop on Mycosphaerella leaf spot diseases of bananas, San José, Costa Rica: 25–42 INIBAP, France: . [Google Scholar]
- Leach R. 1941. Banana leaf spot Mycosphaerella musicola, the perfect stage of Cercospora musae Zimm. Tropical Agriculture (Trinidad) 18: 91 – 95 . [Google Scholar]
- Li KN, Rouse DI, German TL. 1994. PCR primers that allow intergeneric differentiation of ascomycetes and their application to Verticillium spp. Applied and Environmental Microbiology 60: 4324 – 4331 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A, Jackson SL, Dell B, Hardy GEStJ. 2005. PCR-identification of Mycosphaerella species associated with leaf diseases of Eucalyptus. Mycological Research 109: 992 – 1004 . [DOI] [PubMed] [Google Scholar]
- Morelet M. 1969. Micromycetes du var et d’ailleurs (2ème Note). Annales de la Societe des Sciences Naturelles et d’Archèologie de Toulon et du Var 21: 104 – 106 . [Google Scholar]
- Mourichon X, Fullerton RA. 1990. Geographical distribution of the two species Mycosphaerella musicola Leach (Cercospora musae) and M. fijiensis Morelet (C. fijiensis), respectively agents of Sigatoka disease and black leaf streak disease in bananas and plantains. Fruits 45: 213 – 218 . [Google Scholar]
- Mulder JL, Stover RH. 1976. Mycosphaerella species causing banana leaf spot. Transactions of the British Mycological Society 67: 77 – 82 . [Google Scholar]
- Park RF, Keane PJ, Wingfield MJ, Crous PW. 2000. Fungal diseases of eucalypt foliage. In: Keane PJ, Kile GA, Podger FD, Brown BN. (eds), Diseases and pathogens of eucalypts: 153–259 CSIRO Publishing, Collingwood, Australia: . [Google Scholar]
- Pons N. 1987. Notes on Mycosphaerella fijiensis var. difformis. Transactions of the British Mycological Society 89: 120 – 124 . [Google Scholar]
- Pont W. 1960. Three leaf speckle diseases of the banana in Queensland. Queensland Journal of Agricultural Science 17: 271 – 309 . [Google Scholar]
- Pretorius MC, Crous PW, Groenewald JZ, Braun U. 2003. Phylogeny of some cercosporoid fungi from Citrus. Sydowia 55: 286 – 305 . [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society; Kew, UK: . [Google Scholar]
- Rhodes PL. 1964. A new banana disease in Fiji. Commonwealth Phytopathological News 10: 38 – 41 . [Google Scholar]
- Rivas GG, Zapater MF, Abadie C, Carlier J. 2004. Founder effects and stochastic dispersal at the continental scale of the fungal pathogen of bananas Mycosphaerella fijiensis. Molecular Ecology 13: 471 – 482 . [DOI] [PubMed] [Google Scholar]
- Stover RH. 1963. Leaf spot of bananas caused by Mycosphaerella musicola: associated ascomycetous fungi. Canadian Journal of Botany 41: 1481 – 1485 . [Google Scholar]
- Stover RH. 1969. The Mycosphaerella species associated with banana leaf spots. Tropical Agriculture (Trinidad) 46: 325 – 331 . [Google Scholar]
- Stover RH. 1977. A non-virulent benomyl tolerant Cercospora from leaf spots caused by Mycosphaerella fijiensis var. difformis and M. musicola. Transactions of the British Mycological Society 69: 500 – 502 . [Google Scholar]
- Stover RH. 1980. Sigatoka leaf spots of banana and plantain. Plant Disease 64: 750 – 755 . [Google Scholar]
- Stover RH. 1994. Mycosphaerella musae and Cercospora non-virulentum from Sigatoka leaf spots are identical. Fruits 49: 187 – 190 . [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other Methods). Version 4 Sinauer Associates, Sunderland, Massachusetts: . [Google Scholar]
- Verkley GJM, Starink-Willemse M. 2004. A phylogenetic study of some Septoria species pathogenic to Asteraceae based on ITS ribosomal DNA sequences. Mycological Progress 3: 315 – 322 . [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR Protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, California, USA: . [Google Scholar]