Abstract
Coniothyrium-like fungi are common wood and soil inhabitants and hyperparasites on other fungi. They belong to different fungal genera within the Pleosporales. Several isolates were obtained on wood of different Prunus species (plum, peach and nectarine) from South Africa, on Actinidia species from Italy and on Laurus nobilis from Turkey. Morphological and cultural characteristics as well as DNA sequence data (5.8S nrDNA, ITS1, ITS2, partial SSU nrDNA) were used to characterise them. The isolates belonged to three species of the recently established genus Paraconiothyrium. This is the first report of Paraconiothyrium brasiliense on Prunus spp. from South Africa. Two new species are described, namely Paraconiothyrium variabile sp. nov. on Prunus persica and Prunus salicina from South Africa, on Actinidia spp. from Italy and on Laurus nobilis from Turkey, and Paraconiothyrium africanum sp. nov. on Prunus persica from South Africa. Although other known species of Paraconiothyrium commonly produce aseptate conidia, those of P. africanum and P. hawaiiense comb. nov. are predominantly two-celled.
Keywords: Coniothyrium, Microdiplodia, Paraphaeosphaeria, phylogeny, Pleosporales, systematics
INTRODUCTION
Coniothyrium-like fungi are known as biological control agents (Finch-Savage et al. 2003), potential bioremediators (Sasaki et al. 2006), producers of metabolites inhibiting influenza virus replication (Fukami et al. 2000), and as producers of substances with potential anticancer activity (Turbyville et al. 2006). Tsuda et al. (2003) described metabolites of a Paraconiothyrium isolate from a marine horse mussel with antagonistic and antifungal abilities. On the other hand, Coniothyrium species are involved in human skin infections (Guarro et al. 1999, Miele et al. 2002). Ascomycetous fungi with coniothyrium-like anamorphs are common colonisers of wood and leaves of woody host plants, for example Leptosphaeria coniothyrium (anamorph: Coniothyrium fuckelii) on stems of Rubus spp., Microsphaeropsis olivacea on twigs and branches of Cytisus, Hedera, Laurus, Lycium and Sambucus and Thyridaria rubronotata (anamorph: Cyclothyrium juglandis) as plurivorous species (Ellis & Ellis 1985). Paraconiothyrium sporulosum (≡ C. sporulosum) is common in soil and P. minitans (≡ C. minitans) is almost exclusively known from fungal sclerotia (Whipps & Gerlagh 1992). However, P. minitans can also cause wood rot of birch and pine (Nilsson 1973), while P. sporulosum can cause core rot of apples in California (Michailides et al. 1994). Coniothyrium cerealis and P. sporulosum are able to degrade wood (Haider & Domsch 1969, Nilsson 1973). Readeriella zuluensis (≡ C. zuluense; Crous et al. 2007) causes cankers on Eucalyptus (Cortinas et al. 2006), while Coniothyrium species were also associated with stem necroses of Fraxinus excelsior (Przybyl 2002).
The delimitation of genera and species of these fungi is hampered by the variability in characters such as mode of conidiogenesis and conidial morphology. Fungi with brown, 1(–2)-celled conidia that are formed on simple conidiogenous cells in brown pycnidia are generally referred to as Coniothyrium species. Since the type species of the genus, C. palmarum, forms annellidic conidiogenous cells, Sutton (1980) included in Coniothyrium only species with annellides and brown, thick-walled, 0–1-septate, verruculose conidia, species with thin-walled, 1-celled conidia and phialides were referred to as Microsphaeropsis. Other coniothyrium-like species separated from Coniothyrium s.s. by Sutton (1980) are Cyclothyrium and Cytoplea. Verkley et al. (2004) established a new genus, Paraconiothyrium, to accommodate some species with Coniothyrium anamorphs, including C. minitans and C. sporulosum, and described four new species. Within this genus, they observed both phialidic and percurrent (annellidic) conidiogenesis. Only a few species with coniothyrium-like anamorphs have known teleomorphs, belonging in the ascomycete genera Cucurbidothis, Leptosphaeria, Massarina, Paraphaeosphaeria, Pleospora, Thyridaria (anamorph: Cyclothyrium) (Sivanesan 1984), Neophaeosphaeria, Phaeosphaeriopsis (Camara et al. 2001, 2003), and Readeriella (Teratosphaeria) (Crous et al. 2007). Paraconiothyrium spp., decribed by Verkley et al. (2004), did not produce teleomorph states, but were shown to belong to Paraphaeosphaeria s.s. (Pleosporales) based on their SSU phylogeny. Coniothyrium palmarum, Microsphaeropsis olivacea and Cyclothyrium juglandis, the type species of Coniothyrium, Microsphaeropsis and Cyclothyrium, respectively, as well as Cytoplea spp. also grouped in the Pleosporales, but were distant from each other and from Paraphaeosphaeria/Paraconiothyrium spp.
Microsphaeropsis olivacea (≡ C. olivaceum) has been isolated from many woody hosts including Prunus persica (Sutton 1980) and Laurus spp. (Ellis & Ellis 1985). In a study of Buck et al. (1998), fungi in the form genus Coniothyrium belonged to the most prevalent epiphytic fungi from peach bark and were found more often on young bark than on scaffold bark, and more often on smooth bark than on lenticels. Buck & Traquair (1998) showed some of these isolates (identified as C. olivaceum) to produce siderophores and to have antagonistic abilities against other fungi. Additional coniothyrium-like species described from Prunus spp. include: C. cerasi on branches of Prunus cerasi in Italy, C. insitivum f. syringae on branches of Prunus padi in Italy and France (Saccardo 1884) and C. pruni on leaves of Prunus armeniaca and Prunus domestica and in mature fruits of Prunus armeniaca in Australia (Saccardo 1906). However, these species have not been recollected since their original description and are presently not known from culture. Therefore, it remains uncertain which coniothyrium-like genus they belong to. Paraconiothyrium sporulosum has been reported from Actinidia sp. in New Zealand, identified by CBS in 1999 (PDD 70683; http://nzfungi.landcareresearch.co.nz/html/search_collections.asp). In South Africa, several coniothyrium-like species are known as causal organisms of leaf spot diseases of Eucalyptus spp. and Proteaceae (Crous et al. 2000), though several have been shown to belong to Readeriella (Crous et al. 2007). Although C. fuckelii is associated with stem cankers of apple trees (Doidge et al. 1953) and roses (Pole-Evans 1928) in South Africa, no coniothyrium-like fungi have been reported from Prunus spp.
During a survey of Prunus wood from South Africa, several fungal strains forming coniothyrium-like anamorphs were isolated. These isolates included three different Paraconiothyrium species. Two of these species could not be assigned to described species. One of them proved to be conspecific with isolates obtained from Actinidia spp. in Italy and from Laurus nobilis in Turkey, which had earlier been identified as undescribed species of Paraconiothyrium. The aim of the present study is to describe these taxa morphologically and to elucidate their phylogenetic relationships.
MATERIAL AND METHODS
Isolates
Branches of trees with dieback or necrotic symptoms, as well as pruning debris, were sampled from stone fruit (Prunus spp.) orchards in the Western Cape and the Limpopo Provinces of South Africa. Strains from necrotic Prunus tissue were isolated according to the protocols of Damm et al. (2007). Single-conidial isolates were obtained from sporulating pycnidia in these cultures and on the bark of pruning debris by transferring germinating conidia from 2 % tap water agar onto potato-dextrose agar (2 % PDA, Biolab, Midrand, South Africa). The isolate from Laurus nobilis was obtained as described by Göre & Bucak (2007). Isolates from Actinidia chinensis in Italy were obtained from necrotic wood tissue of plants with leader die-back disease (Riccioni et al. 2007). Reference strains are maintained in the culture collections of the Department of Plant Pathology, University of Stellenbosch (STE-U), Stellenbosch, South Africa, the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands, and the CRA - Centro di Ricerca per la Patologia Vegetale (CRA-PAV), Rome, Italy. Isolates used for morphological and sequence analyses are presented in Table 1.
Table 1.
Names, accession numbers and collection details of isolates studied.
| Species | Accession no.1 | Host | Location | Collector | Isolated from | GenBank accessions | |
|---|---|---|---|---|---|---|---|
| ITS | SSU | ||||||
| Paraconiothyrium africanum | STE-U 6316/CBS 121166* | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295650 | EU295654 |
| Paraconiothyrium brasiliense | STE-U 6300, CBS 121426 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295632 | EU295651 |
| STE-U 6301 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295638 | ||
| STE-U 6303, CBS 121165 | Prunus persica var. nucipersica | Mookgopong, South Africa | U. Damm | Necrotic wood | EU295635 | ||
| STE-U 6302, CBS 121427 | Prunus salicina | Paarl, South Africa | U. Damm | Necrotic wood | EU295633 | ||
| STE-U 6304 | Prunus salicina | Mookgopong, South Africa | U. Damm | Necrotic wood | EU295637 | ||
| STE-U 6305 | Prunus salicina | Mookgopong, South Africa | U. Damm | Necrotic wood | EU295636 | ||
| STE-U 6306 | Prunus salicina | Mookgopong, South Africa | U. Damm | Necrotic wood | EU295634 | ||
| Paraconiothyrium hawaiiense | CPC 12265/CBS 120025* | Sophora chrysophylla | Hawaii, USA | W. Gams & Y. Degawa | Stems | DQ885897 | EU295655 |
| CPC 12268 | Sophora chrysophylla | Hawaii, USA | W. Gams & Y. Degawa | Stems | DQ885896 | EU295656 | |
| Paraconiothyrium variabile | ER 1380, CBS 119486 | Actinidia chinensis | Latina, Italy | L. Riccioni | Necrotic wood | EF055359 | |
| ER 1386, CBS 120014 | Actinidia chinensis | Latina, Italy | L. Riccioni | Necrotic wood | EF055360 | ||
| CBS 119633, Det 303-2005 nr 41 | Laurus nobilis | Günlüce/Marmaris, Turkey | M.E. Göre & C. Bucak | Leaves | EU295649 | ||
| STE-U 5848 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295645 | ||
| STE-U 6308 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295646 | ||
| STE-U 6309, CBS 121164 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295640 | ||
| STE-U 6310 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295647 | ||
| STE-U 6311, CBS 121163* | Prunus persica | Paarl, South Africa | U. Damm | Necrotic wood | EU295639 | EU295653 | |
| STE-U 6312 | Prunus persica | Paarl, South Africa | U. Damm | Pycnidia on bark | EU295642 | ||
| STE-U 6315 | Prunus persica | Paarl, South Africa | U. Damm | Necrotic wood | EU295644 | ||
| STE-U 6307 | Prunus salicina | Stellenbosch, South Africa | U. Damm | Necrotic wood | EU295641 | ||
| STE-U 6313, CBS121754 | Prunus salicina | Paarl, South Africa | U. Damm | Necrotic wood | EU295648 | EU295652 | |
| STE-U 6314 | Prunus salicina | Paarl, South Africa | U. Damm | Necrotic wood | EU295643 |
1 STE-U: Culture collection of the Department of Plant Pathology, University of Stellenbosch, South Africa; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; ER: Culture collection of CRA – Centro di Ricerca per la Patologia Vegetale (CRA-PAV), Rome, Italy, * ex-type cultures.
Morphology
To enhance sporulation, autoclaved filter paper and double-autoclaved pine needles were placed onto the surface of synthetic nutrient agar medium (SNA, Nirenberg 1976), and incubated for 2–4 wk at 25 °C under near-ultraviolet (nuv) light. Measurements, photographs of characteristic structures and vertical sections through conidiomata were made according to Damm et al. (2007). Radial growth rates and cultural characteristics were determined on oatmeal (OA, Gams et al. 2007), cornmeal (CMA, Gams et al. 2007) and 3 % malt extract (MEA, Oxoid) agars. Plates were incubated in the laboratory under diffuse daylight at 20 °C, or in an incubator under nuv light (12 h light, 12 h dark) at 15 °C. Colony colours were rated according to Rayner (1970). Growth characteristics were studied on MEA plates incubated in the dark at temperatures ranging from 5–35 °C, in 5 °C intervals.
DNA isolation, amplification and analyses
Genomic DNA of all isolates was extracted from fungal mycelium grown on PDA plates following the protocol of Damm et al. (in press). The 5.8S ribosomal gene with the two flanking internal transcribed spacers (ITS1 and ITS2) and the 18S rDNA gene (SSU) were amplified and sequenced using the primer pairs ITS1F (Gardes & Bruns 1993) and ITS4 (White et al. 1990) or ITS5 (White et al. 1990) and ITS4, NS1 and NS8 (White et al. 1990), as well as the primers NS2, NS3, NS4, NS5 (White et al. 1990) and NS24 (Gargas & Taylor 1992). The sequences were added to the outgroup (ITS: Helminthosporium velutinum AF145704 and Helminthosporium solani AF163089, SSU: Peziza echinospora AF006309) and sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov). GenBank accession numbers and corresponding taxon names are given in Fig. 1 & 2. The alignments were assembled and manually adjusted using Sequence Alignment Editor v. 2.0a11 (Rambaut 2002). Phylogenetic analyses were performed using PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2003). All characters were unordered and of equal weight. Characters with insertions/deletions, ambiguous position homology as well as constant characters were excluded from the ITS analyses. Gaps, uninformative and constant characters were excluded from the SSU analyses. Maximum parsimony analyses were performed using the heuristic search option with 100 random sequence additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm. The robustness of the trees obtained was evaluated by 1 000 bootstrap replications (Hillis & Bull 1993) with 100 random sequence additions. Tree length, consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for the resulting trees. Sequences derived in this study were lodged at GenBank (Table 1), and the alignments in TreeBASE (TreeBASE: 12345).
Fig. 1.
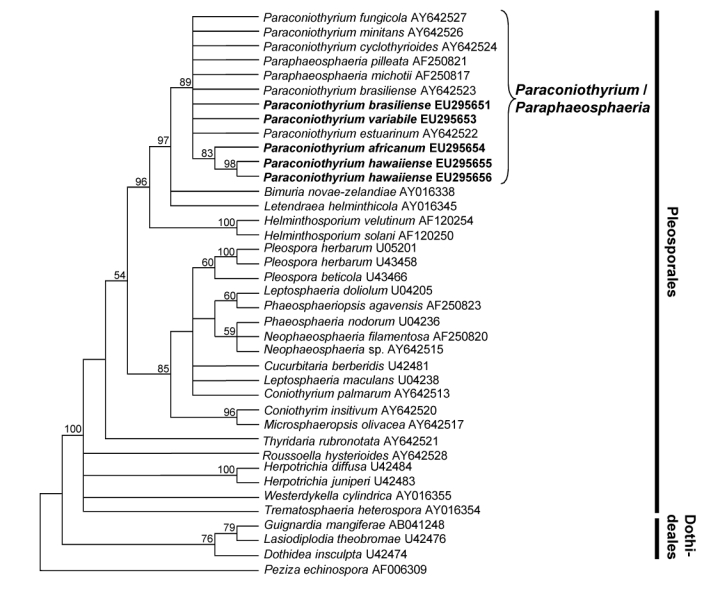
Strict consensus tree obtained from SSU sequence data of Pleosporales and Dothideales (Length: 353 steps, CI: 0.552, RI: 0.819, RC: 0.452, HI: 0.448). Bootstrap support values (1 000 replicates) above 50 % are shown at the nodes. Peziza echinospora AF006309 was used as outgroup. Isolates analysed in this study are emphasized in bold.
Fig. 2.
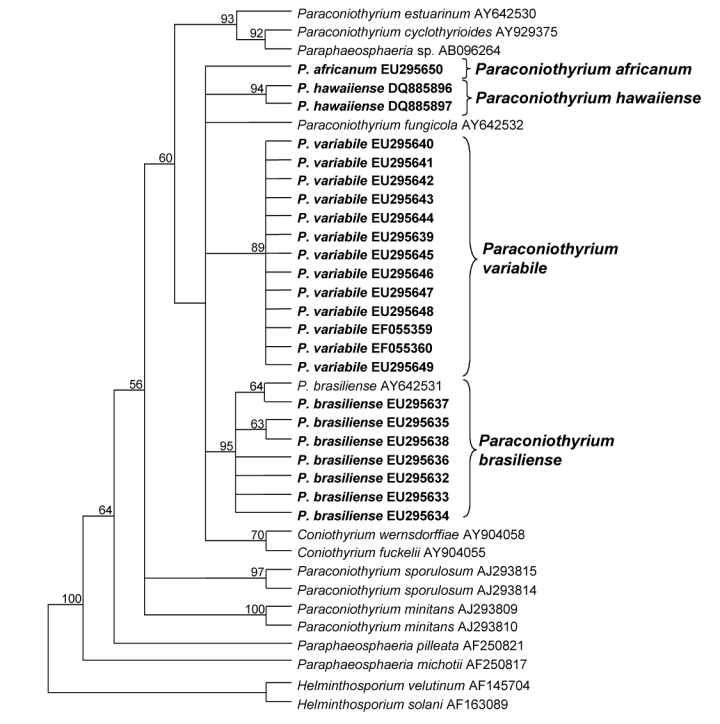
Strict consensus tree obtained from ITS sequence data of Parasphaeosphaeria/Paraconiothyrium species (Length: 164 steps, CI: 0.768, RI: 0.877, RC: 0.674, HI: 0.232). Bootstrap support values (1 000 replicates) above 50 % are shown at the nodes. Helminthosporium velutinum AF145704 and Helminthosporium solani AF163089 were used as outgroup. Isolates analysed in this study are emphasized in bold.
RESULTS
Phylogenetic analysis
The SSU alignment of 39 taxa contained 2088 characters, including the alignment gaps and the 350 bp intron of isolate STE-U 6316, of which 160 parsimony-informative characters were included in the maximum parsimony analysis. After a heuristic search 112 most parsimonious trees were retained (Length: 353 steps, CI: 0.552, RI: 0.819, RC: 0.452, HI: 0.448), of which the strict consensus tree is shown in Fig. 1. There were two main clades in the SSU phylogeny, one formed by members of the Pleosporales (100 % bootstrap) and another by members of the Dothideales (76 %). All isolates analysed in this study are situated within the Pleosporales clade. They formed a cluster with Paraphaeosphaeria/Paraconiothyrium isolates (89 %). Within this clade, only P. africanum and P. hawaiiense formed a well-supported group (83 %).
The ITS dataset contained 38 taxa and 639 characters, including the gaps, of which 105 characters were parsimony-informative and included in the maximum parsimony analysis. After a heuristic search, three most parsimonious trees were retained (Length: 164 steps, CI: 0.768, RI: 0.877, RC: 0.674, HI: 0.232), of which the strict consensus tree is shown in Fig. 2. All isolates analysed in this study clustered with the majority of the taxa within the phylogeny (60 % bootstrap), representing different species of the genus Paraphaeosphaeria/Paraconiothyrium. Seven isolates grouped with the type strain of P. brasiliense AY642531 (95 %). The 13 isolates of P. variabile from Prunus, Actinidia and Laurus formed a well-supported sister group (89 %) to P. brasiliense. Their ITS sequences showed variations in only a single nucleotide. Both P. africanum EU295650 and P. hawaiiense DQ885896 and DQ885897 (94 %) did not group with any other Paraconiothyrium species.
Taxonomy
The 21 strains isolated from wood of Prunus, Actinidia and Laurus, could be assigned to three Paraconiothyrium spp. based on the DNA sequence data and their morphology. Two species proved distinct from known species and are newly described below, and one species, formerly belonging to Microdiplodia, is newly assigned to the genus Paraconiothyrium.
Paraconiothyrium variabile Riccioni, Damm, Verkley & Crous, sp. nov. — MycoBank MB511290; Fig. 3
Fig. 3.
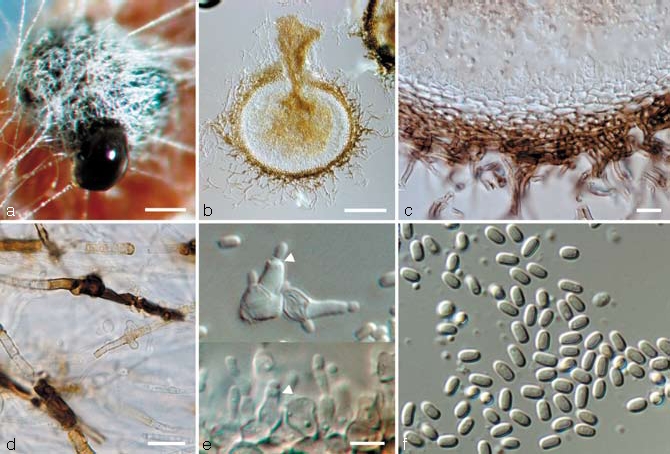
Paraconiothyrium variabile. a. Conidia oozing from pycnidia; b. longitudinal section through a pycnidium; c. pycnidial wall with conidiophores; d. dark- walled hyphae on OA; e. conidiophores (arrow heads indicate percurrent proliferations and periclinical wall thickening); f. conidia; all from CBS H-20004 (holotype); a: DM, b–f: DIC. — Scale bars: a, b = 100 μm; c, d = 10 μm; e = 5 μm; e applies to e & f.
Conidiomata pycnidialia 300–600 μm diam, conidiophora uni-ad tricellularia, 3–15 × 2.5–6 μm. Cellulae conidiogenae conicae ad subulatae vel subcylindricae, doliiformes, late vel elongate ampulliformes variantes, phialidicae tunica sursum periclinaliter incrassata, vel semel ad repetite percurrentes, 2.5–5 × 3–7 μm. Conidia primum hyalina demum pallide brunnea, subcylindrica usque ellipsoidea, utrinque obtusa, continua, pariete tenui glabro vel minute verruculoso, (2.5–)3–4(–5) × 1–2(–2.5) μm, 2.2 × longiora quam lata.
Etymology. Named after the variable shape of the conidiogenous cells (variabilis Lat. = variable).
Conidiomata pycnidial, produced on pine needles on SNA in 2–4 wk, solitary, subglobose, 1–3 ostioles, dark brown, superficial to semi-immersed, 300–600 μm diam, wall consisting of 5–8 cell-layers (25–50 μm) of thick-walled dark brown textura angularis, becoming hyaline and thin-walled towards the inside of the pycnidium, that is surrounded by brown hyphal appendages. Conidiophores lining the inner conidiomatal cavity, hyaline, 1–3-celled, 3–15 × 2.5–6 μm. Conidiogenous cells variable in shape, conical to subulate or subcylindrical, doliiform, broadly or elongated ampulliform, sometimes with a long neck, phialidic with periclinical wall thickening or with one or more percurrent proliferations near the apex, 2.5–5 × 3–7 μm. Conidia initially hyaline, mature conidia pale brown, subcylindrical to ellipsoidal, both ends obtuse, 1-celled, smooth-walled to fine verruculose and thin-walled, sometimes with two small polar droplets, (2.5–)3–4(–5) × 1–2(–2.5) μm, mean ± SD = 3.3±0.6 × 1.5±0.3 μm, average L/W ratio = 2.2. Vegetative hyphae 1.5–5 μm wide, hyaline to pale brown, septate, smooth, chlamydospores absent. On OA parts of the hyaline vegetative hyphae are transformed to very dark-walled hyphal pieces (delimited by septa), which can become locally swollen or accumulate amorphous brown material on the outer wall surface (Fig. 3d).
Cultural characteristics — Colonies on OA reaching 32 mm after 7 d, 46–53 mm after 14 d (15 °C, nuv, 12 h light : 12 h dark; 43 mm in 7 d at 20 °C, in diffuse daylight), flat, with an even to slightly ruffled colourless and glabrous margin, immersed mycelium slowly developing, at first honey, then isabelline to somewhat greenish olivaceous pigmentation, sometimes also with conspicuous brick to cinnamon sectors, aerial mycelium absent or consisting of sparse, scattered white to greyish tufts; reverse concolourous, entire edge. Colonies on CMA reaching 45–50 mm diam in 14 d (15 °C, nuv; 44 mm in 7 d at 20 °C in diffuse daylight), as on OA, but without or very sparse greenish olivaceous aerial mycelium. Colonies on MEA reaching 25–35 mm diam in 7 d, 43–45 mm in 14 d (15 °C, nuv; 42 mm in 7 d at 20 °C in diffuse daylight), low convex, with an even to slightly ruffled, glabrous and colourless margin, most of the colony surface covered by dense woolly-floccose, first whitish or pale primrose, then smoke-grey to grey-olivaceous to olivaceous aerial mycelium; reverse ochreous to fulvous, with some darker cinnamon to isabelline patches or entire centre cinnamon to isabelline or olivaceous, entire edge. Conditions for growth: max 35 °C, opt 20–25 °C.
Hosts — Actinidia chinensis, Actinidia deliciosa, Laurus nobilis, Prunus persica, Prunus salicina.
Distribution — South Africa, Italy, Turkey.
Specimens examined. South Africa, Western Cape Province, Paarl, from discoloured wood of Prunus persica close to a pruning wound, 10 June 2004, U. Damm, CBS H-20004 holotype, culture ex-type CBS 121163 = STE-U 6311; from small reddish brown V-shaped necrosis under cracked bark of Prunus salicina, 10 June 2004, U. Damm, CBS 121754 = STE-U 6313. – Italy, Latina, from discoloured wood of decaying vine of Actinidia chinensis, 16 June 2005, L. Riccioni, CBS 119486 = ER1380; from discoloured wood in trunk, of Actinidia chinensis, 5 May 2006, L. Riccioni, CBS120014 = ER1386. – Turkey, Western Anatolia, Günlüce/Marmaris, from leaves of Laurus nobilis, M.E. Göre & C. Bucak, CBS 119633 = Det 303-2005 nr 41.
Notes — Conidia of P. variabile are similar to those of P. estuarinum, but smaller than those of most other Paraconiothyrium species, and longer and narrower than conidia of P. cyclothyrioides (Camara et al. 2001, Verkley et al. 2004, Domsch et al. 2007). However, P. variabile grows more slowly than P. estuarinum and the shape of the conidiogenous cells is more variable. On OA, P. variabile has paler colours and forms no or only restricted aerial mycelium as well as dark-walled hyphal pieces (Verkley et al. 2004).
Paraconiothyrium africanum Damm, Verkley & Crous, sp. nov. — MycoBank MB511291; Fig. 4
Fig. 4.
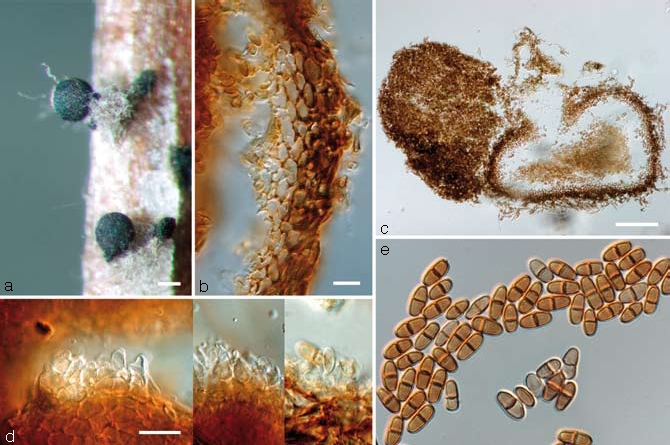
Paraconiothyrium africanum. a. Conidia oozing from pycnidia; b. pycnidial wall; c. longitudinal section through a pycnidium; d. conidiophores; e. conidia; all from CBS H-19847 (holotype); a: DM, b–e: DIC. — Scale bars: a = 100 μm; b = 10 μm; c = 100 μm; d = 10 μm; d applies to d & e.
Conidiomata pycnidialia 100–600 μm diam, conidiophora cellulis conidiogenis reducta. Cellulae conidiogenae ampulliformes ad doliiformes, phialidicae vel semel ad repetite percurrentes, 3–8 × 2–6 μm. Conidia primum hyalina demum brunnea, cylindrica, ellipsoidea vel ovoidea, utrinque obtusa vel basi truncata, plerumque continua vel 1-septata, raro 2–3-septata, septum atrobrunneum crassum, pariete verruculoso, (4–)6.5–9.5(–12) × (2.5–)3–4(–5) μm, 2.3 × longiora quam lata.
Etymology. Named after the continent of origin, Africa.
Conidiomata pycnidial, produced on pine needles on SNA after 2–4 wk, solitary, subglobose, ampulliform or flattened, brown, 100–600 μm diam, semi-immersed, immersed or superficial, 1–2-locular, central ostiole, wall consisting of 4–10 layers (15–30 μm) of dark brown textura angularis, becoming pale brown to hyaline towards the inside of the pycnidium, that is surrounded by brown, verruculose hyphal appendages. Conidiophores reduced to conidiogenous cells. Conidiogenous cells lining the inner conidiomatal cavity, inconspicuous, hyaline to pale brown, ampulliform to doliiform, phialidic, but also proliferating percurrently, 3–8 × 2–6 μm. Conidia initially hyaline, mature conidia medium brown, cylindrical, ellipsoidal or ovoid, generally both ends obtuse, sometimes one end truncate, mainly (1–)2-celled, rarely 3- or 4-celled, septum dark and thick-walled, verruculose, (4–)6.5–9.5(–12) × (2.5–)3–4(–5), mean ± SD = 8.1±1.5 × 3.5±0.6 μm, average L/W ratio = 2.3. Vegetative hyphae: 2–6 μm, hyaline to brown, verrucose.
Cultural characteristics — Colonies on OA reaching 24 mm after 7 d, 60–64 mm after 14 d (15 °C, nuv, 12 h light : 12 h dark; 44 mm in 7 d at 20 °C, in diffuse daylight), flat, with an even, colourless and glabrous margin, immersed mycelium honey to olivaceous buff, aerial mycelium absent or consisting of sparse, white to greyish tufts in the centre; reverse concolourous, entire edge. Colonies on CMA reaching 23 mm after 7 d, 56–62 mm diam after 14 d (15 °C, nuv; 38 mm after 7 d at 20 °C in diffuse daylight), as on OA, flat, with an even, colourless and glabrous margin, immersed mycelium honey, olivaceous buff to greenish olivaceous, aerial mycelium absent or consisting of sparse, white to greyish tufts in the centre; reverse concolourous, entire edge. Colonies on MEA reaching 22 mm diam after 7 d, 52 mm after 14 d (15 °C, nuv; 40 mm after 7 d, 70 mm after 14 d at 20 °C in diffuse daylight), flat, with an even, glabrous and colourless margin, most of the colony surface covered by felty floccose, white to pale smoke-grey aerial mycelium, immersed mycelium buff or honey to greenish olivaceous, reverse buff or honey to greenish olivaceous or olivaceous, entire edge. Conditions for growth: min < 5 °C, max 30 °C, opt 20 °C.
Host — Prunus persica.
Distribution — Paarl (South Africa, Western Cape Province).
Specimen examined. South Africa, Western Cape Province, Paarl, from pycnidia on the bark of Prunus persica, 10 June 2004, U. Damm, CBS H-19847 holotype, culture ex-type CBS 121166 = STE-U 6316.
Notes — The conidia of P. africanum are (1- or) 2-celled, resembling those of Paraconiothyrium hawaiiense (Microdiplodia hawaiiensis) (Crous & Groenewald 2006). However, conidia of P. hawaiiensis are generally much larger, (10–)12–13 × (4–)5(–5.5) μm. Paraconiothyrium fungicola produces only occasionally 2-celled conidia with similar size (6–8 × 4.5–5.2 μm) (Verkley et al. 2004). Other known Paraconiothyrium species only produce aseptate, smaller conidia (Verkley et al. 2004). Conidia of Coniothyrium palmarum are the same size, but are 0- or 1-euseptate, and the conidiogenous cells are exlusively annellidic (Sutton 1980). According to SSU phylogeny, C. palmarum is not closely related to P. africanum (Fig. 1).
Paraconiothyrium hawaiiense (Crous) Damm, Crous & Verkley, comb. nov. — MycoBank MB511292
Basionym. Microdiplodia hawaiiensis Crous, Fungal Planet 7 (2006).
Holotypus. CBS H-19778, ex-type culture: CBS 120025 = CPC 12265.
DISCUSSION
After a first attempt to reveal the phylogenetic relationship of coniothyrium-like fungi and establishing the genus Paraconiothyrium to accommodate new as well as well-known species by Verkley et al. (2004), we found this to be a commonly occurring fungal genus. Species were frequently isolated from wood and leaves of Prunus, Actinidia and Laurus, and two additional species, P. africanum and P. variabile could be distinguished based on their DNA sequence data and unique morphological characteristics.
Paraconiothyrium brasiliense was recently described from a fruit of Coffea arabica in Brazil (Verkley et al. 2004). According to DNA sequence data deposited in GenBank, P. brasiliense also occurs endophytically in Ginkgo biloba (DQ094168, unidentified fungus) and Pinus tabulaeformis (AY546076, unidentified fungus), in leaves of Picea glauca in Canada (AY561200, AY566890, unidentified fungi), Alliaria petiolata in the USA (EF432267), in a marine fish (Pennahia argentata) in China (AJ619957, identified as Myrothecium) and in surface water in wetland in Japan (AB303550). The fungus has also been isolated from discoloured wood of a living tree of Platanus × acerifolia in Rome, Italy (M. Pilotti, CRA-PAV, Rome, Italy, pers. comm.). In our study, however, P. brasiliense was isolated from peach, nectarine and plum trees in two different areas in South Africa. This is the first report of this fungus on Prunus, as well as from South Africa. This fungus was isolated both from wood with necrotic symptoms found on living trees of Prunus salicina and Prunus persica var. nucipersica, as well as from pycnidia on the bark of pruning debris of Prunus persica collected on the orchard floor. The species seems to be widespread (different countries and continents) and common on a wide range of host plants and other habitats.
The novel species, P. variabile, could be distinguished based on DNA sequence data and morphology, and fits well in the concept of the genus Paraconiothyrium by producing smooth-walled to verruculose, pale brown, 1-celled conidia from inconspicuous phialides with periclinal thickening or percurrent proliferations (Camara et al. 2003, Verkley et al. 2004). The fungus was frequently isolated from wood necroses and pycnidia on the bark of Prunus persica in South Africa, and from wood of Prunus salicina, in association with wood necrosis symptoms. Additionally, P. variabile was frequently isolated from wood of Actinidia chinensis and A. deliciosa in Italy in association with trunk or vine disorders and from necrotic wood under pruning cut surfaces (Riccioni et al. 2007), and from leaves of Laurus nobilis in Turkey (Göre & Bucak 2007). This indicates that it may also have a broad host range, including several distantly related host plants and a wide geographical distribution. However, this species has so far only been found in and on wood and leaves of woody hosts.
Paraconiothyrium africanum was isolated from the bark of Prunus persica in South Africa. Conidia of P. africanum resemble those of Microdiplodia hawaiiensis, a species recently described on stems of Sophora chrysophylla in Hawaii (Crous & Groenewald 2006), and shown to be phylogenetically distinct from similar anamorphs in the Botryosphaeriaceae (Crous et al. 2006). In both species, the conidia, formed on phialidic as well as percurrently proliferating conidiogenous cells, are mainly 2-celled, but conidia of P. africanum are much smaller than those of M. hawaiiensis. Microdiplodia hawaiiensis was shown to belong to the genus Paraconiothyrium by means of ITS and SSU sequence data and is therefore renamed as Paraconiothyrium hawaiiense. This shows, that the genus Paraconiothyrium comprises not only species with pigmented 1-celled conidia, but also species with pigmented 2-celled conidia that would be considered as Microdiplodia species. There are 349 records listed in Index Fungorum (http://www.speciesfungorum.org) under Microdiplodia, most of them are not connected to any teleomorph genus and of uncertain position within the Ascomycetes. However, there is no type species of Microdiplodia designated (Sutton 1977) nor any authentic cultures, and thus its taxonomic position remains uncertain.
Within the genus Paraconiothyrium, there are closely related species with mainly 1-celled conidia, that formerly belonged to or would have been addressed as Coniothyrium or Microsphaeropsis, as well as 2-celled conidia, that formerly would have been regarded as Microdiplodia. One species, P. fungicola, mainly forms 1-celled, but occasionally also 2-celled, conidia. According to this study, the concept of the genus Paraconiothyrium should be amended to also accommodate species with predominantly 2-celled conidia. While comparing Coniothyrium and Microsphaeropsis, Morgan-Jones (1974) emphasised that conidia of C. palmarum are septate at maturity, while those of M. olivacea are always unicellular. However, since conidia of C. eucalypticola are unicellular also, Morgan-Jones (1974) considered the presence of a septum as a less significant feature within the genus Coniothyrium, as can now also be concluded with regards to Paraconiothyrium.
Confirming observations of Buck et al. (1998), we also found coniothyrium-like fungi to be common on peach bark. Furthermore, we found Paraconiothyrium species in necrotic wood of peach, plum and nectarine wood with necrotic symptoms. It is, however, not known whether the Paraconiothyrium species found here have antagonistic activities against other fungi as shown for P. minitans (Whipps & Gerlagh 1992), and for Coniothyrium olivaceum isolated from peach bark (Buck & Traquair 1998). It has also not yet been determined whether the Paraconiothyrium species isolated from Prunus wood could cause disease on these hosts.
Acknowledgments
The authors acknowledge the University of Stellenbosch, National Research Foundation, THRIP, Winetech and the Deciduous Fruit Producer’s Trust for financial support. Dr E. Göre (Plant Protection Institute, Bornova-Ýzmir, Turkey) is kindly thanked for providing isolates, and Dr J.Z. Groenewald (CBS, the Netherlands) for providing the SSU sequences of Paraconiothyrium hawaiiense. Dr M. Valvassori and Mr. G. Di Giambattista (CRA-PAV, Rome, Italy) are thanked for their technical assistance.
REFERENCES
- Buck JW, Lachance MA, Traquair JA. 1998. Mycoflora of peach bark: population dynamics and composition. Canadian Journal of Botany 76: 345 – 354 . [Google Scholar]
- Buck JW, Traquair JA. 1998. Influence of iron depletion on interactions between peach bark fungi in vitro. Mycologia 90: 947 – 953 . [Google Scholar]
- Camara MPS, Palm ME, Van Berkum P, Stewart EL. 2001. Systematics of Paraphaeosphaeria: a molecular and morphological approach. Mycological Research 105: 41 – 56 . [Google Scholar]
- Camara MPS, Ramaley AW, Castlebury LA, Palm ME. 2003. Neophaeosphaeria and Phaeosphaeriopsis, segregates of Paraphaeosphaeria. Mycological Research 107: 516 – 522 . [DOI] [PubMed] [Google Scholar]
- Cortinas MN, Burgess T, Dell B, Xu DP, Crous PW, Wingfield BD, Wingfield MJ. 2006. First record of Colletogloeopsis zuluense comb. nov., causing a stem canker of Eucalyptus in China. Mycological Research 110: 229 – 236 . [DOI] [PubMed] [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ. 2006. Microdiplodia hawaiiensis. Fungal Planet No. 7 . [Google Scholar]
- Crous PW, Phillips AJL, Baxter AP. 2000. Phytopathogenic fungi from South Africa Department of Plant Pathology Press, University of Stellenbosch Printers; Stellenbosch, South Africa: . [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Phillips AJL, Alves A, Burgess T, Barber P, Groenewald JZ. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235 – 253 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2007. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora spp. nov. Mycologia: 99: 664 – 680 . [DOI] [PubMed] [Google Scholar]
- Damm U, Crous PW, Fourie PH. In press Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Mycological Research: in press . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidge EM, Bottomley AM, Van der Plank JE, Pauer GD. 1953. A revisited list of plant diseases in South Africa. Union of South Africa, Department of Agriculture, Science Bulletin 346: 1 – 122 . [Google Scholar]
- Domsch KH, Gams W, Anderson T-H. 2007. Compendium of soil fungi. 2nd ed IHW-Verlag, Eching, Germany: . [Google Scholar]
- Ellis MB, Ellis JP. 1985. Microfungi on land plants: An identification handbook London & Sydney: . [Google Scholar]
- Finch-Savage WE, Clay HA, Budge SP, Dent KC, Clarkson JP, Whipps JM. 2003. Biological control of Sclerotinia pseudotuberosa and other fungi during moist storage of Quercus robur seeds. European Journal of Plant Pathology 109: 615 – 624 . [Google Scholar]
- Fukami A, Nakamura T, Kim Y-P, Shiomi K, Hayashi M, Nagai T, Yamada H, Komiyama K, Omura S. 2000. A new anti-influenza virus antibiotic, 10-norparvulenone from Microsphaeropsis sp. FO-5050. The Journal of Antibiotics 53: 1215 – 1218 . [DOI] [PubMed] [Google Scholar]
- Gams W, Verkley GJM, Crous PW. (eds). 2007. CBS course of mycology Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands: . [Google Scholar]
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113 – 118 . [DOI] [PubMed] [Google Scholar]
- Gargas A, Taylor JW. 1992. Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia 84: 589 – 592 . [Google Scholar]
- Göre ME, Bucak C. 2007. Geographical and seasonal influences on the distribution of fungal endophytes in Laurus nobilis. Forest Pathology 37: 281 – 288 . [Google Scholar]
- Guarro J, Mayayo E, Tapiol J, Aguilar C, Cano J. 1999. Microsphaeropsis olivacea as an etiological agent of human skin infection. Medical Mycology 37: 133 – 137 . [PubMed] [Google Scholar]
- Haider K, Domsch KH. 1969. Abbau und Umsetzung von lignifiziertem Pflanzenmaterial durch mikroskopische Bodenpilze. Archives of Microbiology 64: 338 – 348 . [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182 – 192 . [Google Scholar]
- Michailides TJ, Morgan DP, Mitcham E, Crisosto CH. 1994. Occurrence of mouldy core and core rot of Fuji apple in California. KAC Plant Protection Quarterly 3: 4 – 6 . [Google Scholar]
- Miele PS, Levy CS, Smith MA, Dugan EM, Cooke RH, Light JA, Lucey DR. 2002. Primary cutaneous fungal infections in solid organ transplantations: A case series. American Journal of Transplantation 2: 678 – 683 . [DOI] [PubMed] [Google Scholar]
- Morgan-Jones G. 1974. Concerning some species of Microsphaeropsis. Canadian Journal of Botany 52: 2575 – 2579 . [Google Scholar]
- Nilsson T. 1973. Studies on wood degradation and cellulolytic activity of microfungi. Studia Forestalia Suecica 104: 5 – 40 . [Google Scholar]
- Nirenberg HI. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1 – 117 . [Google Scholar]
- Pole-Evans IB. 1928. Botanical work. In: Annexure to annual report of the Secretary for Agriculture for the year ended 30th June, 1928. Farming in South Africa 3: 1085–1087 . [Google Scholar]
- Przybyl K. 2002. Fungi associated with necrotic apical parts of Fraxinus excelsior shoots. Forest Pathology 32: 387 – 394 . [Google Scholar]
- Rambaut A. 2002. Sequence Alignment Editor. Version 2.0 Department of Zoology, University of Oxford, Oxford, UK: . [Google Scholar]
- Rayner RW. 1970. A mycological colour chart Commonwealth Mycological Institute and British Mycological Society; Kew, Surrey, UK: . [Google Scholar]
- Riccioni L, Manning M, Valvassori M, Haegi A, Casanato S, Spinelli R. 2007. A new disease: leader die-back in Actinidia chinensis Hort16A in Italy. Acta Horticulturae 753: 669 – 676 . [Google Scholar]
- Saccardo PA. 1884. Sphaeropsidearum et Melanconiearum omnium hucosque cognitarum Sylloge Fungorum III: 307 . [Google Scholar]
- Saccardo PA. 1906. Sphaeropsidearum et Melanconiearum omnium hucosque cognitarum. Supplementum universale Sylloge Fungorum XVIII: 308 . [Google Scholar]
- Sasaki K, Matsuda M, Hirajima T, Takano K, Konno H. 2006. Immobilization of Mn(II) ions by a Mn-oxidizing fungus – Paraconiothyrium sp.-like strain at neutral pHs. Materials Transactions 47: 2457 – 2461 . [Google Scholar]
- Sivanesan A. 1984. The bitunicate ascomycetes and their anamorphs Cramer, Vaduz, Lichtenstein: . [Google Scholar]
- Sutton BC. 1977. Coelomycetes, VI. Nomenclature of generic names proposed for Coelomycetes. Mycological Papers 141: 1 – 253 . [Google Scholar]
- Sutton BC. 1980. The coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata Commonwealth Mycological Institute, Kew, England: . [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4 Sinauer Associates, Sunderland, MA, USA: . [Google Scholar]
- Tsuda M, Mugishima T, Komatsu K, Sone T, Tanaka M, Mikami Y, Kobayashi J. 2003. Modiolides A and B, two new 10-membered macrolides from a marine-derived fungus. Journal of Natural Products 66: 412 – 415 . [DOI] [PubMed] [Google Scholar]
- Turbyville TJ, Wijeratne EMK, Liu MX, Burns AM, Seliga CJ, Luevano LA, David CL, Faeth SH, Whitesell L, Gunatilaka AAL. 2006. Search for Hsp90 inhibitors with potential anticancer activity: Isolation and SAR studies of radicicol and monocillin I from two plant-associated fungi of the Sonoran Desert. Journal of Natural Products 69: 178 – 184 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkley GJM, da Silva M, Wicklow DT, Crous PW. 2004. Paraconiothyrium, a new genus to accommodate the mycoparasite Coniothyrium minitans, anamorphs of Paraphaeosphaeria, and four new species. Studies in Mycology 50: 323 – 335 . [Google Scholar]
- Whipps JM, Gerlagh M. 1992. Biology of Coniothyrium minitans and its potential for use in disease biocontrol. Mycological Research 96: 897 – 907 . [Google Scholar]
- White TJ, Bruns T, Lee J, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, California, USA: . [Google Scholar]


