Abstract
The dynamin-related GTPase OPA1 is mutated in autosomal dominant optic atrophy (DOA) (Kjer type), an inherited neuropathy of the retinal ganglion cells. OPA1 is essential for the fusion of the inner mitochondrial membranes, but its mechanism of action remains poorly understood. Here we show that OPA1 has a low basal rate of GTP hydrolysis that is dramatically enhanced by association with liposomes containing negative phospholipids such as cardiolipin. Lipid association triggers assembly of OPA1 into higher order oligomers. In addition, we find that OPA1 can promote the protrusion of lipid tubules from the surface of cardiolipin-containing liposomes. In such lipid protrusions, OPA1 assemblies are observed on the outside of the lipid tubule surface, a protein-membrane topology similar to that of classical dynamins. The membrane tubulation activity of OPA1 is suppressed by GTPγS. OPA1 disease alleles associated with DOA display selective defects in several activities, including cardiolipin association, GTP hydrolysis and membrane tubulation. These findings indicate that interaction of OPA1 with membranes can stimulate higher order assembly, enhance GTP hydrolysis and lead to membrane deformation into tubules.
INTRODUCTION
Three large GTPases are required for mitochondrial fusion, which involves the coordinated fusion of both outer and inner membranes (1). Outer membrane fusion requires the mitofusins Mfn1 and Mfn2, integral membrane proteins of the mitochondrial outer membrane (2). Inner membrane fusion requires the inner membrane protein OPA1 (2), which shows strong sequence similarity to dynamins. OPA1 is proteolytically processed into inner membrane-bound long isoforms and soluble short isoforms. A combination of both long and short isoforms is necessary for mitochondrial fusion (3,4). In the absence of OPA1, outer membrane fusion occurs without inner membrane fusion (2). Similar results are observed with the yeast OPA1 ortholog Mgm1p (5). In addition to mitochondrial fusion, OPA1/Mgm1p is also required for the maintenance of cristae (5–9), extensive infoldings of the mitochondrial inner membrane. However, Δmgm1 yeast cells have normal cristae structure if mitochondrial fission is also disrupted (9).
Mutations in OPA1 cause dominant optic atrophy (DOA, Kjer type), a degeneration of the optic nerve caused by loss of retinal ganglion cells (10–12). Over 200 mutations in OPA1 have been associated with DOA, but the effects of these mutations on OPA1 remain poorly understood, in part because little is known about the biochemistry of OPA1. OPA1 belongs to the dynamin family, which consists of large GTPases that are centrally involved in several important membrane remodeling processes (13–15). Classical dynamins, exemplified by dynamin-1, act as mechanochemical enzymes that mediate the severing of endocytic vesicles from the cell membrane. In the late stages of clathrin-mediated endocytosis, dynamin-1 assembles on the outer surface of the collar of an invaginating vesicle and facilitates its fission from the cell membrane. In contrast, OPA1 mediates the topologically opposite process of membrane fusion.
To better understand OPA1 function, we purified recombinant OPA1 and developed assays to study its GTP hydrolysis activity and association with lipids. In addition, we studied whether these activities were perturbed by mutations in OPA1 that cause DOA. Our results show that several OPA1 activities—including lipid association, GTP hydrolysis and membrane tubulation—are disrupted in DOA disease alleles.
RESULTS
Cardiolipin enhances GTPase hydrolysis by OPA1
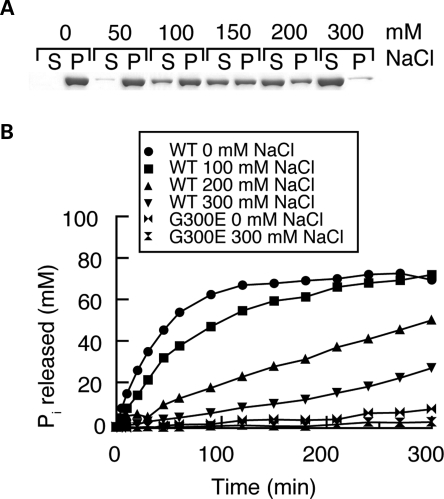
To study the biophysical properties of OPA1, we expressed the short isoform of human OPA1 (splice form 1; termed hereafter as OPA1-S1) with an N-terminal hexahistidine tag in bacteria. Recombinant OPA1-S1 is highly soluble in 300 mm NaCl (Fig. 1A) but shows low GTP hydrolysis activity, with a turnover number of 0.06 GTP/min (Fig. 1B). As the concentration of NaCl is reduced, OPA1-S1 shows a modest increase in GTP hydrolysis activity, reaching a maximum turnover number of 0.6 GTP/min at 0 mm salt (Fig. 1B). No GTP hydrolysis activity was detected with OPA1-S1 containing the mutation G300E in the G1 motif of the GTPase domain, indicating that the low levels of GTP hydrolysis detected are authentic. Concomitant with the 10-fold increase in GTPase activity with decreasing salt, we found OPA1-S1 could be progressively sedimented by a high-speed spin (100 000g). Whereas little OPA1-S1 is sedimented at 300 mm NaCl, OPA1-S1 was found almost entirely in the pellet fraction at 50 and 0 mm salt (Fig. 1A). We examined OPA1 under various salt conditions by negative staining electron microscopy and found that the protein aggregates induced under low salt had no specific or ordered morphology (Supplementary Material, Fig. S1A). However, as shown later, these aggregates are reversible and can functionally assemble onto lipid membranes.
Figure 1.
Stimulation of OPA1-S1 GTPase activity by low-salt and liposome binding. (A) Sedimentation of OPA1-S1 under varying salt concentrations. OPA1-S1 was dialyzed against the indicated concentration of NaCl and centrifuged at 100 000g. The supernatant and pellet were analyzed by SDS–PAGE. S, supernatant; P, pellet. (B) GTP hydrolysis activity of wild-type and G300E OPA1 under varying salt concentrations. GTP hydrolysis was measured at 100 μm GTP with the indicated NaCl concentration.
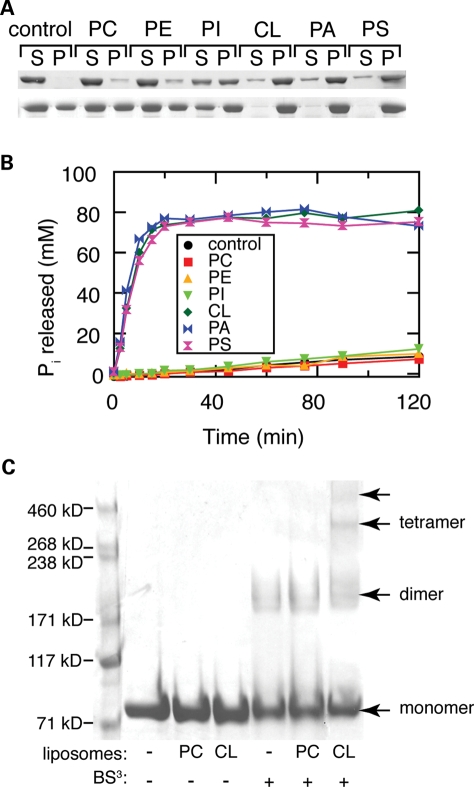
Because OPA1 is associated with the mitochondrial inner membrane, we tested the effect of lipids on OPA1-S1 behavior. Using a liposome co-sedimentation assay, we found no interaction of OPA1-S1 with liposomes composed of phosphatidylcholine (POPC), or a mixture of POPC and phosphatidylethanolamine (POPE) (Fig. 2A). A unique feature of mitochondrial inner membranes is the high concentration of cardiolipin, a negatively charged lipid that constitutes almost 20% by weight of the total lipid (16). We found that recombinant OPA1-S1 co-sediments robustly with liposomes containing 25% cardiolipin (Fig. 2A). This association had a profound effect on the GTP hydrolysis rate of OPA1, resulting in a 100-fold increase in the turnover number to 8 GTP/min (Fig. 2B). The association of OPA1 with cardiolipin-containing liposomes also correlated with its assembly into higher order oligomers that can be detected with the homobifunctional crosslinker bis(sulfosuccinimidyl)suberate (Fig. 2C).
Figure 2.
Association of OPA1-S1 with artificial lipsomes. (A) Selective binding of OPA1-S1 to liposomes. OPA1-S1 was incubated with the indicated liposomes, and the mixture was centrifuged at 100 000g to sediment the liposomes. The sedimentation of OPA1-S1 was analyzed in 300 (top panel) and 150 mm NaCl (lower panel) by SDS–PAGE. S, supernatant; P, pellet; PC, 100% POPC; PE, 78% POPC, 22% POPE; PI, 70% POPC, 22% POPE, 8% PI; CL, 45% POPC, 22% POPE, 8% PI, 25% cardiolipin; PA, 45% POPC, 22% POPE, 8% PI, 25% POPA; PS, 45% POPC, 22% POPE, 8% PI, 25% POPS. Liposome binding was analyzed by centrifugation and SDS–PAGE. (B) Enhancement of GTP hydrolysis activity of wild-type OPA1 by liposomes. GTP hydrolysis was assayed at 100 μm GTP and 0.2 mg/ml liposome. All experiments were performed with 0.2 mg/ml OPA1-S1 at pH 7.0. (C) Higher order oligomers of OPA1 form upon interaction with cardiolipin-containing liposomes. The composition of the liposomes and the presence of the crosslinker (BS3) are indicated at the bottom. Arrows indicate distinct species of OPA1 resolved by SDS–PAGE.
Because cardiolipin is an acidic phospholipid, we tested other acidic phospholipids (Fig. 2A). We found that OPA1-S1 associates strongly with liposomes containing 25% phosphatidic acid (POPA) or 25% phosphoserine (POPS), and these interactions also enhance GTP hydrolysis (Fig. 2B). These results suggest that OPA1-S1 associates with negatively charged phospholipids. However, given that cardiolipin is by far the major negatively charged lipid in the mitochondrial inner membrane (PI constitutes 5% and POPS <0.1%) (16), cardiolipin may be the physiological lipid that regulates OPA1 activity.
Membrane tubulation by OPA1
Given the association of OPA1 with artificial liposomes and the ability of dynamin family members to deform membranes, we tested whether this association resulted in changes in membrane structure. We prepared large multilamellar liposomes that contain a lipid composition (45% POPC, 22% POPE, 8% PI, 25% cardiolipin) approximating that of the mitochondrial inner membrane. To visualize the liposomes, 3,3′-dioctadecyloxacarbocyanine (DiO) and biotin were incorporated, and the fluorescent liposomes were immobilized via biotin–streptavidin interactions within a flow cell. Liposome morphology was monitored with and without the addition of OPA1-S1 into the flow cell.
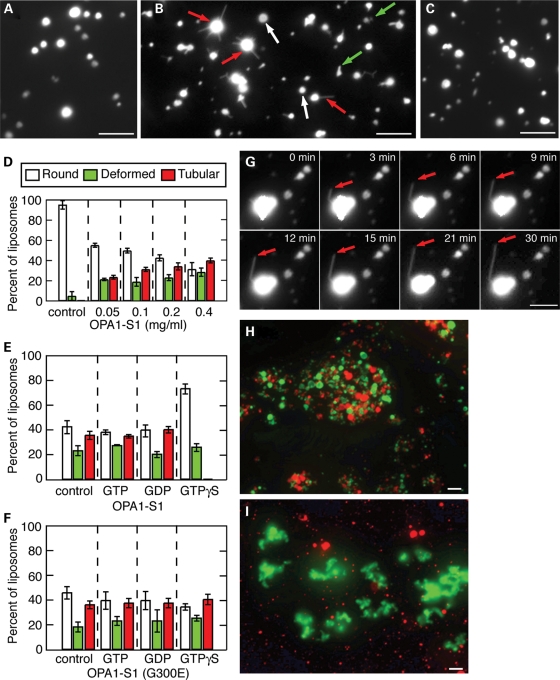
Remarkably, addition of OPA1-S1 in the absence of guanine nucleotides resulted in the protrusion of multiple, thin lipid tubules from the liposome surface (Fig. 3B and D). Such protrusions were often >5 μm in length. In addition to liposome tubulation, we also observed an increase in the number of liposomes with deformed surfaces with the addition of OPA1-S1 (Fig. 3B and D and Supplementary Material, Fig. S3). Because liposome tubulation is a more clearly defined morphological category, we consider it to be a more definitive measure of OPA1 activity. By morphologically scoring liposome reactions, we could quantify the ability of OPA1 to tubulate membranes (Fig. 3D). Using time-lapse microscopy, we observed real-time extension of long tubules from liposome surfaces for up to 30 min (Fig. 3G and Supplementary Material, Video S1). Liposome deformation and tubulation events were not observed in the absence of OPA1-S1 (Fig. 3A) or with neutrally charged liposomes and OPA1-S1 (Fig. 3C).
Figure 3.
Membrane tubulation and liposome aggregation by OPA1-S1. (A–C) Images of fluorescently labeled liposomes. Immobilized cardiolipin-containing liposomes were incubated with buffer (A) or OPA1-S1 (B) and imaged by fluorescence microscopy after 30 min. In (C), PC liposomes were incubated with OPA1-S1. In (B), the arrows indicate examples of the three categories of liposome morphologies. White arrows indicate normal round liposomes; green arrows indicate liposomes with deformed borders but not clear tubulation; red arrows indicate liposomes with clear membrane tubulation. The concentration of OPA1-S1 and liposomes were both 0.2 mg/ml. (D) Quantification of OPA1-S1-mediated membrane tubulation at the indicated concentrations of OPA1-S1 and in the absence of guanine nucleotides. In the control, no protein was added. The ‘tubular’ category was defined as liposomes that contained one or more thin tubular extensions of at least 1 μm in length. This category therefore consisted of liposomes that showed unambiguous tubulation. The ‘deformed’ category was defined as liposomes that contained surface protrusions, but not meeting the tubular criteria. The ‘round’ category consisted of spherical liposomes with no surface protrusions. An electron micrograph showing tubular and deformed liposomes is shown in Supplementary Material, Figure S3. In each of three independent experiments, 200 liposomes were scored; error bars indicate standard deviations. (E) Effect of guanine nucleotides on liposome tubulation. OPA1 was incubated with Mg2+ (control) or Mg2+ and the indicated guanine nucleotide prior to the addition to cardiolipin-containing liposomes. In each of three independent experiments, more than 100 liposomes were scored; error bars indicate standard deviations. (F) Effect of guanine nucleotides on the GTPase mutant G300E was analyzed at in (E). (G) Time-lapse observation of OPA1-S1-mediated membrane tubulation from a cardiolipin-containing liposome. Arrows indicate extension of a >5 μm lipid tubule over the course of 30 min. The corresponding movie is shown in Supplementary Material, Video S1. (H and I) Liposome aggregation by OPA1-S1. OPA1-S1 was added to a mixture of DiO-labeled cardiolipin-containing liposomes (green) and DiI-labeled cardiolipin-containing liposomes (red) (H), or a mixture of DiO-labeled cardiolipin-containing liposomes and DiI=1,1'-dioctadecyl-3,3,3'3'-tetramethylindocarbocyanine perchlorate-labeled PC liposomes (I). In (I), note that only the cardiolipin-containing liposomes aggregate. The concentration of OPA1-S1 and liposomes were both 0.2 mg/ml. All scale bars are 5 μm.
We found that guanine nucleotides had a profound effect on the ability of OPA1-S1 to tubulate cardiolipin-containing liposomes. Pre-incubation of OPA1-S1 with GTPγS strongly inhibited liposome tubulation (Fig. 3E), reducing the percent of tubulated liposomes from 36 to <1% (P-value < 0.0001, unpaired t-test). As expected, this effect was dependent on the presence of Mg2+ (data not shown). No effect of OPA1-mediated membrane tubulation was found with GDP or GTP. We also found that the GTPase mutant G300E showed liposome tubulation activity (Fig. 3F). In contrast to wild-type OPA1, however, this mutant, which is not expected to bind guanine nucleotides, was unaffected by incubation with GTPγS. Taken together, these results indicate that GTP hydrolysis is not necessary for membrane tubulation by OPA1 and that the GTPγS-trapped form of OPA1 is incompetent to tubulate membranes. We found that guanine nucleotides modestly increased the solubility of OPA1 under intermediate salt conditions (150 mm NaCl) and had no effect on the ability of OPA1 to bind to liposomes (Supplementary Material, Fig. S2).
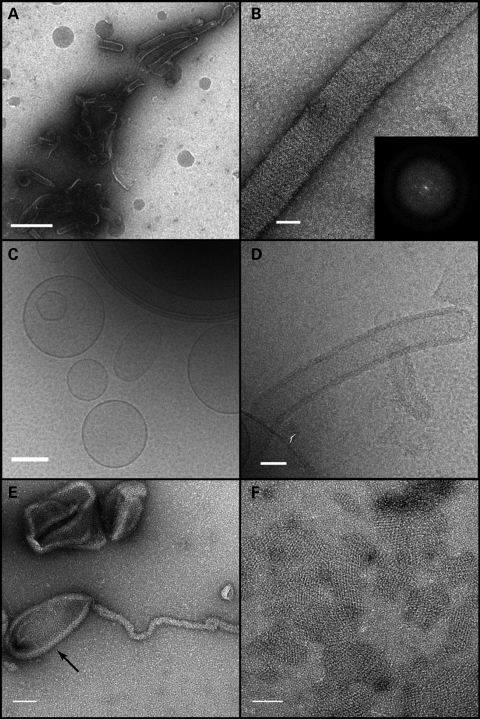
We further examined the properties of these lipid tubules by negative staining electron microscopy and cryo-electron microscopy (Fig. 4). The lipid tubules were of variable diameter, ranging from 50 to >100 nm. OPA1 was present on the outer surface of the lipid tubules and assembled there into ordered structures as judged by Fourier analysis (Fig. 4B). OPA1 assembled not only in the tubulated portions of the liposomes, but also on the non-tubulated portion (Fig. 4E). In these reactions, patches of ordered protein, apparently free of membranes, were also readily observed (Fig. 4F).
Figure 4.
Electron microscopy of liposome tubulation. (A and B) Negative stain of OPA1-S1 reconstituted with liposomes mimicking the mitochondrial inner membrane lipid. (A) A low magnification view showing OPA1-S1 tubular structures with diameters in the range of 50 to 150 nm. In (B), a higher magnification view shows an individual membrane tubule with a periodic arrangement of OPA1-S1 on the lipid substrate. The inset represents the Fourier transform of a segment of the displayed tube indicating molecular order mainly along the long axis of the tube. (C) Cryo-TEM image of vesicle preparations prior to the addition of OPA1-S1. Frozen–hydrated vesicles appear as a mixture of unilamellar and multilamellar vesicles of varying diameters. (D) Cryo-TEM of the vesicle preparation after the addition of OPA1-S1. OPA1-S1 reconstitutes into tubular structures with OPA1-S1 molecules decorating the surface of the lipid tubules. (E) Negative stain of OPA1 mixed with liposomes, showing that OPA1 assembles on the tubular as well as non-tubular (arrow) portions of the liposome. (F) Negative-stained preparation of an OPA1/liposome reaction, showing patches of OPA1 arrays. Scale bars: (A) 500 nm; (B and D) 50 nm; (C) 200 nm; (E and F) 100 nm.
We also observed that OPA1 can dynamically change conformation upon interaction with membranes. As noted earlier, under low-salt conditions, OPA1 forms protein aggregates that have no specific morphology (Supplementary Material, Fig. S1A). However, addition of cardiolipin-containing liposomes to such aggregates leads to the assembly of OPA1 onto the liposome surface and membrane tubulation (Supplementary Material, Fig. S1B). After the addition of liposomes, few of the amorphous protein aggregates can be found, indicating that such aggregates are reversible and competent for lipid binding, assembly into arrays and membrane tubulation.
We also tested whether OPA1-S1 could mediate the fusion of liposomes. When OPA1-S1 was added to a mixture of two distinctly labeled cardiolipin-containing liposome preparations, the liposomes were found to undergo extensive aggregation, but no fusion was found either in the presence and or in the absence of GTP (Fig. 3H). This aggregation was dependent on lipid composition. No aggregation was found when one of the liposome populations contained only POPC, which does not associate with OPA1-S1 (Fig. 3I). The ability of OPA1 to aggregate liposomes may reflect its ability to form intermolecular complexes in trans, a property that has been proposed to be important for its biological activity (5,6).
Distinct functions of OPA1 are disrupted in DOA disease alleles
Our biochemical assays indicate that OPA1 has a low basal rate of GTP hydrolysis activity that is substantially enhanced by association with cardiolipin. In addition, OPA1-S1 is sufficient to mediate membrane tubulation. To uncover whether these properties are physiologically important, we studied a panel of eight mutant alleles that cause DOA. Four of these alleles are associated with classical DOA (G300E, N728K, Q785R, L939), whereas the other four (G439V, S545R, R445H, V910D) are associated with the more severe form termed ‘OPA1 plus’ DOA, which includes multiple mtDNA deletions in addition to the classical optic atrophy (17,18).
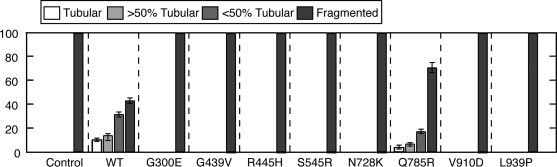
All eight alleles have reduced function in vivo when expressed in OPA1-null mouse embryonic fibroblasts (MEFs). OPA1-null MEFs have completely fragmented mitochondria due to loss of mitochondrial fusion in the face of ongoing mitochondrial fission (2,4). Re-introduction of the wild-type OPA1 gene (splice form 1), which produces both a long and a short isoform (4), results in tubular mitochondria in >50% of the cells (Fig. 5). In contrast, seven of the eight disease alleles are completely defective in inducing any tubular mitochondria, indicating a severe loss of mitochondrial fusion activity. The mutant Q785R shows a partial loss-of-function phenotype, resulting in some mitochondrial tubulation in <30% of expressing cells.
Figure 5.
Mitochondrial fusion activity of OPA1 disease alleles. The indicated alleles of OPA1 were expressed in OPA1-null MEFs by retroviral transduction, and mitochondrial morphology was scored using mitochondrially targeted DsRed. For each mutant, 100 cells were scored in each of three independent experiments; error bars indicate standard deviations. Control cells were not transduced with retrovirus.
To understand the molecular basis of these severe in vivo defects, we studied these alleles in vitro using the biochemical assays detailed earlier (Figs 6 and 7 and Supplementary Material, Fig. S4). Two alleles (V910D and L939P) were found to be almost completely insoluble when expressed in bacteria (data not shown), in contrast to wild-type OPA1-S1. These two alleles likely have a folding defect and were not studied further.
Figure 6.
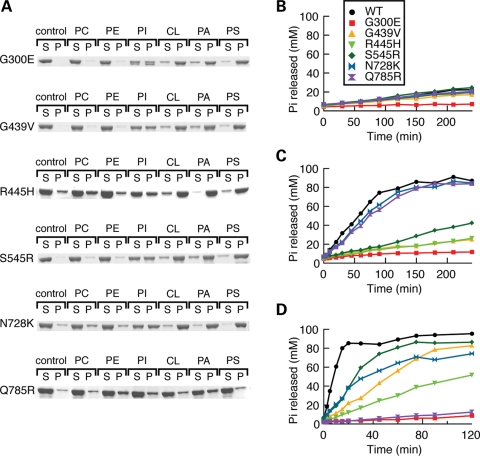
Effects of DOA disease mutations in liposome-binding and GTP hydrolysis activity. (A) Liposome binding was measured by a liposome co-sedimentation assay, as in Fig. 2A. (B–D) GTP hydrolysis activity was measured at 300 mm NaCl (B), 0 mm NaCl (C) and 300 mm NaCl with cardiolipin-containing liposomes (D).
Figure 7.
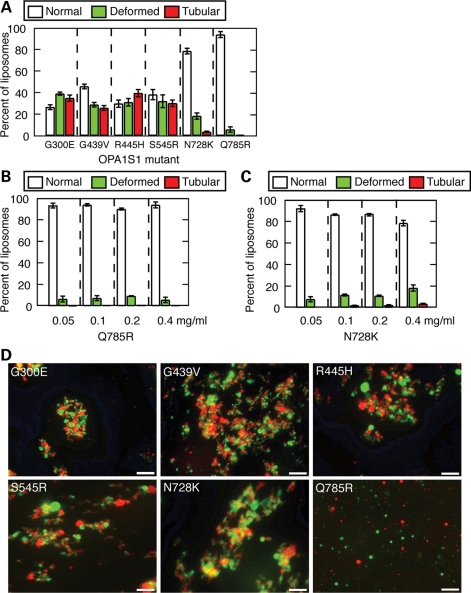
Effects of DOA disease mutations on membrane tubulation and liposome aggregation. (A) Quantification of membrane tubulation by mutant alleles at 0.4 mg/ml, scored as in Fig. 3D. (B) Quantification of membrane tubulation by Q785R at varying concentrations. (C) Quantification of membrane tubulation by N728K. In (A–C), 200 liposomes were scored in each of three independent experiments, and error bars indicate standard deviations. (D) Images of DiO-labeled cardiolipin-containing liposomes (green) and DiI-labeled cardiolipin-containing liposomes (red) in the presence of OPA1-S1 mutant alleles, assayed as in Fig. 3H. Scale bars are 10 μm.
In the remaining six disease alleles, we uncovered three overlapping classes. The first class consists of the single mutant Q785R, which has a severe defect in the binding of lipids, including cardiolipin. This defect is reflected in its inability to sediment with liposomes containing cardiolipin, POPA, POPS or PI (Fig. 6A). Associated with this binding defect, Q785R shows a complete loss of GTPase enhancement by cardiolipin-containing liposomes (Fig. 6D). Importantly, however, this mutant showed normal levels of basal GTPase activity and normal levels of stimulation by assembly under low-salt conditions, indicating that its GTPase domain is intact (Fig. 6B and C). Consistent with its defect in lipid binding, Q785R was unable to either tubulate or aggregate liposomes (Fig. 7A, B and D).
The second and largest class of OPA1 mutants shows defects in GTP hydrolysis. One mutant, G300E, has a complete loss of GTP hydrolysis activity. It shows no basal GTPase activity and no stimulation by low salt or lipids (Fig. 6B–D). The other alleles with mutations in the GTPase domain (G439V, S545R, R445H) show reduced levels of GTP hydrolysis under low-salt and lipid-containing conditions (Fig. 6C and D). All the GTPase mutants show no detectable mitochondrial fusion activity when expressed in OPA1-null MEFs (Fig. 5).
The third class of mutants shows a defect in membrane tubulation activity. As mentioned earlier, Q785R has a complete defect in membrane tubulation secondary to loss of lipid binding. In addition, N728K shows reduced membrane tubulation activity (Fig. 7A and C), even though this mutant retains association with liposomes (Fig. 6A) and could aggregate liposomes (Fig. 7D). Taken together, these studies suggest that lipid binding, regulated GTP hydrolysis and membrane tubulation are all likely to be important aspects of OPA1 function.
DISCUSSION
For mammalian OPA1 and the yeast Mgm1p, simultaneous expression of both long and short isoforms is necessary for mitochondrial membrane fusion (3,4). The only exception in mammalian cells is during stress-induced mitochondrial fusion, which, for unclear reasons, requires only a long OPA1 isoform (19). The long isoforms are constitutively embedded in the mitochondrial inner membrane due to an amino-terminal transmembrane anchor. The long isoforms have been shown to physically interact with short isoforms (5,6), and such interactions in Mgm1 appear essential for fusion (20,21). Our results indicate that the OPA1 short isoforms, which lack a transmembrane anchor, also have important interactions with the mitochondrial inner membrane. We find that OPA1-S1 can assemble on lipid surfaces, and this interaction dramatically activates its GTP hydrolysis activity. Because cardiolipin is the most abundant negatively charged lipid of the mitochondrial inner membrane, it may be the physiological lipid that regulates OPA1 activity, although we cannot rule out that other less abundant lipids may regulate OPA1 within a membrane microdomain. In this respect, OPA1 resembles recently described properties of Mgm1p (20,22). This lipid interaction may serve to regulate OPA1 so that it is activated only when positioned properly with regard to the cardiolipin-rich inner membrane.
In addition, we find that OPA1-S1 alone robustly tubulates liposomes. Electron microscopy clearly indicates that OPA1-S1 assembles on the outer surface of the tubular protrusions, resulting in a membrane topology that is similar to that of classical dynamins. The diameter of the lipid tubules ranges from 50 to 100 nm, larger than dynamin-decorated tubules but in the same range as yeast Dnm1p tubules, which have a diameter of ∼100 nm (23). The similarities of OPA1 to classical dynamin is striking, given that these two proteins mediate topologically opposite reactions—membrane fusion for OPA1 versus membrane fission for dynamin. We speculate that OPA1 may protrude the mitochondrial inner membranes towards each other during the fusion process. In contrast to dynamins, however, the membrane-tubulating activity of OPA1-S1 is inactive in the presence of GTPγS. This observation suggests that the GTPase cycle may regulate the fusion activity of OPA1 by controlling its interaction with the lipid membrane.
Importantly, we find that DOA disease alleles disrupt key biochemical features of OPA1. We uncovered alleles that specifically disrupt lipid binding, GTP hydrolysis and membrane tubulation. These observations suggest that these activities are key to OPA1 function. Further analysis of DOA alleles may further clarify physiologically important features of OPA1.
MATERIALS AND METHODS
Recombinant OPA1-S1 expression and purification
cDNA corresponding to the short S1 isoform of OPA1 was PCR-amplified from the template plasmid pMSCV/OPA1-Isfm1 (4) and subcloned into the pET28a(+) expression vector (Novagen, Madison, WI, USA). Disease alleles were constructed in pET28a/OPA1-S1 through PCR with primers encoding point mutations. Details of OPA1-S1 purification are provided in Supplementary Material, Text S1.
GTPase assay
GTPase reactions were performed with 0.2 mg/ml OPA1-S1 in 50 mm HEPES (pH 7.0), 1 mm dithiothreitol (DTT) and varying NaCl concentrations. In experiments with liposomes, the reactions contained 50 mm HEPES (pH 7.0), 300 mm NaCl, 1 mm DTT and 0.2 mg/ml unilamellar liposomes. GTP hydrolysis was quantified by monitoring the free phosphate concentration using a malachite green assay (24). Reactions were initiated by the addition of GTP to 100 μm and incubation at 37°C. Aliquots (10 μl) at regular time points were quenched with 5 μl of 0.5 M EDTA at regular time points. After the addition of malachite green solution to 750 μm, the free phosphate concentration was monitored by absorbance at 650 nm in a 96-well plate reader (Infinite M200 TECAN, Durham, NC, USA). Details of liposome preparation, as well as the liposome tubulation and aggregation assays are provided in Supplementary Material, Text S1.
Sedimentation assay
OPA1-S1 was dialyzed at 4°C overnight against varying concentrations of NaCl (0 to 300 mm) in 50 mm HEPES (pH 7.0) and 1 mm DTT and diluted to a concentration of 0.2 mg/ml. For the assembly of OPA1-S1 on liposomes, all liposomes were extruded 15 times through a 1 μm polycarbonate filter using a LiposoFast extruder (Avestin, Ottawa, Canada). The liposomes were directly added to the protein solution at a final concentration of 0.2 mg/ml and incubated at room temperature for 30 min. Samples (80 μl) were centrifuged at 100 000g (50 000 rpm in a TLA100 rotor, Beckman Coulter, Inc., Fullerton, CA, USA) for 15 min at 4°C. The supernatant and pellet were analyzed by SDS–PAGE.
Crosslinking assay
OPA1-S1 was dialyzed at 4°C overnight against 50 mm HEPES (pH 7.0), 300 mm NaCl and 1 mm DTT. OPA1-S1 was added at a final concentration of 0.3 to 0.3 mg/ml PC- or cardiolipin-containing liposomes and incubated for 30 min at room temperature. The amine-reactive crosslinker bis(sulfosuccinimidyl)suberate (BS3; Thermo Fisher Scientific, Waltham, MA, USA) was added (50 μm final) to the OPA1-S1–liposome mixture. After a 15 min incubation at room temperature, the crosslinking reaction was quenched with 50 mm Tris (pH 8.0). Crosslinked products were analyzed using a 3–8% Tris–acetate gel (NuPage Novel; Invitrogen, Carlsbad, CA, USA).
Transmission electron microscopy and cryo-electron microscopy
OPA1-S1 samples were reconstituted with large multilamellar cardiolipin-containing liposomes at room temperature. For negative stain analysis, aliquots of the reaction mixture were applied to carbon-Formvar coated 400 MESH copper grids and stained with a 2% (w/v) uranyl acetate solution. Images were acquired using a transmission electron microscope (Philips/FEI CM120, Eindhoven, the Netherlands) at 100 kV and recorded on a CCD camera (GATAN, Pleasanton, CA, USA). For cryo-electron microscopy, samples were applied to Quantifoil R 3.5/1 grids (SPI, West Chester, PA, USA), blotted and plunged into liquid ethane using a Vitrobot apparatus (FEI, Eindhoven, the Netherlands). Samples were examined at cryogenic conditions using a Gatan 626 cryo-holder in a CM120 microscope, and images were recorded on a Gatan CCD camera.
Analysis of OPA1 activity in cells
The ability of OPA1 mutants to rescue the fragmented mitochondria of OPA1-null MEFs was performed as described previously (4). Briefly, OPA1-null cells expressing mitochondrial DsRed were infected with retrovirus-expressing mutant OPA1 alleles and plated onto glass coverslips. Mitochondrial phenotypes were scored by fluorescence microcopy into four classes as detailed previously (4).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (grant number GM062967 to D.C.C.) and the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (J.E.H.). T.B. was supported by the Japan Society for Promotion of Science (JSPS) with a Post-Doctoral Fellowship for Research Abroad. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kenichi Morigaki (Kobe University, Japan) for advice on liposome preparation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. doi:10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 2.Song Z., Ghochani M., McCaffery J.M., Frey T.G., Chan D.C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. doi:10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A.S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. doi:10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 4.Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. doi:10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. doi:10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. doi:10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 7.Griparic L., van der Wel N.N., Orozco I.J., Peters P.J., van der Bliek A.M. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. doi:10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- 8.Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. doi:10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 9.Sesaki H., Southard S.M., Yaffe M.P., Jensen R.E. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. doi:10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. doi: 10.1038/79944. doi:10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 11.Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. doi.10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 12.Delettre C., Lenaers G., Pelloquin L., Belenguer P., Hamel C.P. OPA1 (Kjer type) dominant optic atrophy: a novel mitochondrial disease. Mol. Genet. Metab. 2002;75:97–107. doi: 10.1006/mgme.2001.3278. doi:10.1006/mgme.2001.3278. [DOI] [PubMed] [Google Scholar]
- 13.Praefcke G.J., McMahon H.T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. doi:10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 14.Heymann J.A., Hinshaw J.E. Dynamins at a glance. J. Cell Sci. 2009;122:3427–3431. doi: 10.1242/jcs.051714. doi:10.1242/jcs.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pucadyil T.J., Schmid S.L. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 2009;325:1217–1220. doi: 10.1126/science.1171004. doi:10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardail D., Privat J.P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 1990;265:18797–18802. [PubMed] [Google Scholar]
- 17.Amati-Bonneau P., Valentino M.L., Reynier P., Gallardo M.E., Bornstein B., Boissiere A., Campos Y., Rivera H., de la Aleja J.G., Carroccia R., et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. doi:10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- 18.Hudson G., Amati-Bonneau P., Blakely E.L., Stewart J.D., He L., Schaefer A.M., Griffiths P.G., Ahlqvist K., Suomalainen A., Reynier P., et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. doi:10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- 19.Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. doi:10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeVay R.M., Dominguez-Ramirez L., Lackner L.L., Hoppins S., Stahlberg H., Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 2009;186:793–803. doi: 10.1083/jcb.200906098. doi:10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zick M., Duvezin-Caubet S., Schafer A., Vogel F., Neupert W., Reichert A.S. Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 2009;583:2237–2243. doi: 10.1016/j.febslet.2009.05.053. doi:10.1016/j.febslet.2009.05.053. [DOI] [PubMed] [Google Scholar]
- 22.Rujiviphat J., Meglei G., Rubinstein J.L., McQuibban G.A. Phospholipid association is essential for the dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J Biol. Chem. 2009;284:28682–28686. doi: 10.1074/jbc.M109.044933. doi:10.1074/jbc.M109.044933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingerman E., Perkins E.M., Marino M., Mears J.A., McCaffery J.M., Hinshaw J.E., Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. doi:10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard M., Song B.D., Ramachandran R., Schmid S.L. Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. doi:10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.