Abstract
Huntington's disease (HD) is an autosomal dominant neurodegenerative disease caused by a polyglutamine expansion in huntingtin. There are no treatments that are known to slow the neurodegeneration caused by this mutation. Mutant huntingtin causes disease via a toxic gain-of-function mechanism and has the propensity to aggregate and form intraneuronal inclusions. One therapeutic approach for HD is to enhance the degradation of the mutant protein. We have shown that this can be achieved by upregulating autophagy, using the drug rapamycin. In order to find safer ways of inducing autophagy for clinical purposes, we previously screened United States Food and Drug Administration-approved drugs for their autophagy-stimulating potential. This screen suggested that rilmenidine, a well tolerated, safe, centrally acting anti-hypertensive drug, could induce autophagy in cell culture via a pathway that was independent of the mammalian target of rapamycin. Here we have shown that rilmenidine induces autophagy in mice and in primary neuronal culture. Rilmenidine administration attenuated the signs of disease in a HD mouse model and reduced levels of the mutant huntingtin fragment. As rilmenidine has a long safety record and is designed for chronic use, our data suggests that it should be considered for the treatment of HD and related conditions.
INTRODUCTION
Huntington's disease (HD) is a devastating neurodegenerative disorder characterized by progressive motor dysfunction, dementia and emotional disturbances (reviewed in 1 and 2). It is inherited in an autosomal dominant manner and its prevalence is 5–10 cases per 100 000. The median age of clinical onset is about 37 years of age and the disease progresses over time and is invariably fatal 15–20 years after onset. Currently, there is no therapy that slows degeneration in humans with this disease.
The HD gene codes for a large highly conserved protein of numerous apparent functions, huntingtin (1). In affected individuals, there is an expanded polyglutamine sequence in the protein due to expansion of a polymorphic trinucleotide repeat sequence (CAGn) near the 5′ end of the gene (3). The HD mutation results from more than 35 CAG repeats (4). An inverse relationship exists between the CAG repeat number (i.e. glutamine residues) and the age of onset of the first symptoms, with higher repeat numbers associated with a younger age of onset.
Proteolysis of mutant huntingtin releases a persistent N-terminal fragment comprising the first 100–150 residues with the expanded polyglutamine sequence. This fragment forms aggregates with itself and other proteins and is believed to confer toxicity via a gain-of-function mechanism (5). Thus, HD pathogenesis is frequently modelled with exon 1 fragments containing expanded polyglutamine repeats, which form aggregates and cause toxicity in cell models and in vivo (6).
The pathological hallmark of HD is the gradual atrophy of the striatum (caudate nucleus and putamen), and it is the medium-sized projection spiny neurons which are most affected within the striatum (7). However, neuronal loss has been identified in many other regions of the brain and cortical degeneration also occurs in early phases (8). Striatal atrophy begins more than a decade before motor symptoms develop and therefore by the time of diagnosis, the striatum may be atrophied by as much as 50% (9,10). Therefore, the ultimate goal is to develop therapies that prevent the onset of clinical symptoms in mutation carriers and slow down disease progression in post-symptomatic individuals.
One way to slow or attenuate the effects of the HD mutation may be to enhance the removal of the mutant protein, because it acts as a toxin. We have previously shown that mutant huntingtin is cleared by (macro)autophagy, a finding that has been subsequently replicated by others (11,12). The autophagy-lysosomal pathway is a major route for protein clearance in eukaryotic cells. It involves the formation of double membrane structures (called autophagosomes) around a portion of cytosol, which then fuse with lysosomes where their contents are degraded. Our data suggest that autophagy induction may represent a therapeutic strategy for neurodegenerative diseases like HD that are caused by intracytoplasmic aggregate-prone proteins (13). We found that the rapamycin analogue CCI-779, which induces autophagy by inhibiting the protein kinase mTOR (mammalian target of rapamycin), improved behavioural performance and decreased aggregate formation in a mouse model of HD (14) and in a mouse model of another polyglutamine disease, spinocerebellar ataxia type 3 (15). As rapamycin has non-trivial side effects, our laboratory screened FDA-approved drugs to identify new autophagy-inducing pathways (16). This screen revealed clonidine and the related compound rilmenidine, which are both used for the chronic treatment of hypertension, as mTOR-independent autophagy inducers. These compounds act on α2-adrenoceptors and imidazoline I1 receptors in the brain and in the periphery (17). Clonidine and rilmenidine are centrally acting antihypertensive agents. They act primarily within the rostral part of the ventrolateral medulla to reduce sympathic outflow to peripheral organs (18). We decided to use rilmenidine in our in vivo trial, since, in comparison to the prototypical compound clonidine, this drug is 30 times more selective for imidazoline receptors than for α2-adrenoceptors and thus causes fewer adverse central side effects like sedation or antinociception (19–21). At equihypotensive doses, rilmenidine causes less bradycardia and reduction in cardiac output, less sedation and little or no antinociceptive action compared with clonidine (17). Rilmenidine is known to get into the brain and mediate its antihypertensive effects in this organ (18). The imidazoline I1 receptor is expressed in regions affected by the HD mutation in both the rodent and human, including the striatum, cerebral cortex, hippocampus, ventrolateral medulla and hypothalamus (22–25).
To examine the effect of rilmenidine in vivo, we used the N171-82Q mice, which express the first 171 amino acids of mutant huntingtin under the control of the mouse prion promoter, which restricts expression of the protein mainly to the brain (26). These transgenic mice show a profound phenotype including loss of motor coordination, tremors, weight loss and premature death.
RESULTS
Rilmenidine increases autophagy in tissue from wild-type mice
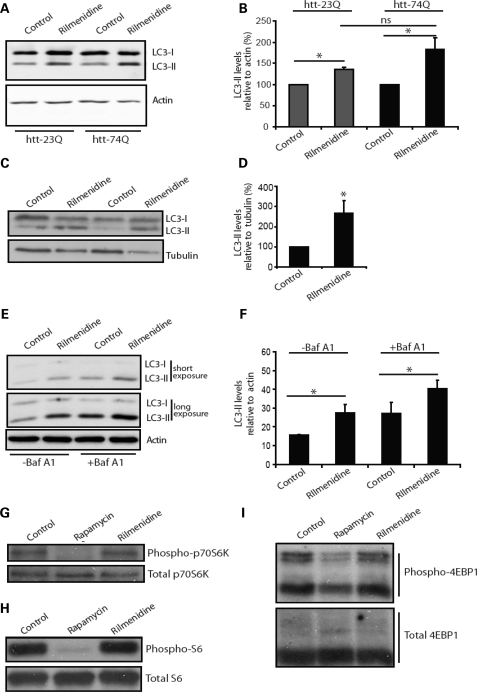
Our previous studies have demonstrated the ability of rilmenidine to induce autophagy and clear model aggregate-prone proteins such as mutant huntingtin exon 1 (16). In order to further confirm the suitability of rilmenidine as a potential therapeutic for use in HD, we showed that its ability to induce autophagy was not compromised by the presence of mutant huntingtin. We assessed autophagosome numbers using the microtubule-associated protein 1 light chain (LC3) (27). LC3 is processed post-translationally into LC3-I, then converted to LC3-II, the only known protein that specifically associates with autophagosome membranes (28). LC3-positive vesicle numbers or LC3-II levels (versus loading control) correlate with autophagosome numbers (27). Stable inducible PC12 cells expressing either wild-type (23Q) or expanded polyglutamine (74Q) huntingtin both showed a significant increase in LC3-II when treated with rilmenidine (Fig. 1A and B).
Figure 1.
Rilmenidine enhances autophagy in wild-type mice. (A) Endogenous LC3-II levels were measured in stable inducible PC12 cells 48 h after switching on expression of either huntingtin exon 1 with 23 polyglutamine repeats (htt-23Q) or 74 polyglutamine repeats (htt-74Q) in the presence or absence of rilmenidine (for the final 24 h). Actin was used as a loading control. (B) LC3-II levels were measured using fluorescent intensity of the bands by Li-Cor Odyssey. Results are shown as a percentage of control in each individual cell line (n = 5, *P < 0.05 by t-test). The increase in LC3-II levels in rilmenidine treated, htt-74Q is not significantly greater than in htt-23Q treated cells, and possible differences may be due to variations between the clonal cell lines used. (C) LC3-II levels in muscle lysates from rilmenidine-treated and placebo-treated wild-type mice after 24 weeks of treatment. Western blots were also probed for tubulin as a loading control (D) Densitometric analysis of LC3-II-levels relative to tubulin. Control condition is set to 100%. Error bars show SEM (*P = 0.036, t-test, n = 4 for rilmenidine, n = 5 for control). (E) In cultured primary cortical neurons, LC3-II levels were assessed by western blot. Two exposures are shown to allow comparison of weaker bands in non-bafilomycin A1-treated lanes (−Baf A1) and stronger bands in bafilomycin A1-treated lanes without saturation. (F) Densitometric quantification of LC3-II levels relative to actin in triplicate experiments. (*P < 0.05 by t-test). Effect of rilmenidine treatment on phosphorylation of downstream mTOR targets was investigated by western blotting, (G) phosphorylated p70 S6 kinase levels, (H) phosphorylated S6 ribosomal protein and (I) phosphorylated 4EBP1. In these experiments, rapamycin treatment was used as a control for the inactivation of mTOR where the effects of treatment can be clearly seen.
In order to investigate the in vivo effects of rilmenidine on autophagy, we measured LC3-II levels in muscle samples from rilmenidine-treated wild-type mice. As shown in Figure 1C and D rilmenidine significantly increased LC3-II levels. Not unexpectedly, we saw no obvious change in LC3-II levels in the brains of treated mice. Previous studies have demonstrated that the detection of increased LC3-II levels in the brain is very difficult, as neurons appear to clear autophagosomes very efficiently (29). We therefore used primary neuronal culture to show that treatment with rilmenidine increases LC3-II levels (Fig. 1E and F). This also allowed us to demonstrate that this increase in LC3-II levels results from an increase in synthesis rather than a decrease in degradation of autophagosomes, as increased levels were also seen in the presence of bafilomycin A1, an autophagy inhibitor that blocks autophagosome-lysosome fusion and therefore LC3-II degradation (27) (Fig. 1E and F).
Further analysis of the effects of rilmenidine on primary neurons confirmed that rilmenidine does not act to upregulate autophagy via inactivation of the mTOR pathway. Unlike rapamycin, which has previously been identified as a possible therapeutic due to its upregulation of autophagy, rilmenidine had no effect on phosphorylation of p70 S6 kinase (Fig. 1G), S6 protein (Fig. 1H) or 4E-BP1 (Fig. 1I), all downstream substrates of mTOR kinase activity.
Rilmenidine improves motor phenotype in HD mice
We have studied the effects of rilmenidine in the N171-82Q mice, which we have previously used to study rapamycin, thus allowing a rough comparison of the two drugs. We chose this mouse model as it does not have the very early onset of disease seen in the R6/2 mice. Also the N171-82Q transgene is driven by the mouse prion protein promoter which results in expression predominantly in the brain, compared with the R6/2 mice where the transgene is expressed widely in the peripheral tissues, such as muscle which may effect behaviour on various motor tasks (30). The N171-82Q mice show obvious motor signs on the rotarod and grip-strength tests, wire-manoeuvre task and increased tremors from about 12 weeks of age. We pre-tested the mice at 4 weeks of age to estimate the baseline motor performances, in order to ensure that the randomly assigned treatment and placebo groups (controlled by litter) were not significantly different in sex ratio or in their abilities to perform any of the tasks.
Rilmenidine treatment then started at 5 weeks of age. Behavioural testing began at the age of 12 weeks, when N171-82Q mice show a clear difference in motor performances compared with non-transgenic littermates, and mice were then tested every 2 weeks until death or euthanasia due to reaching the humane endpoint.
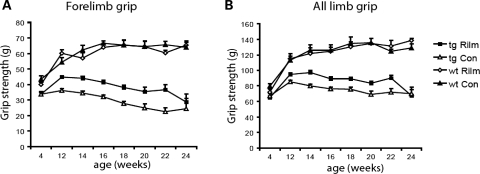
Grip strength
Grip strength was quantified using a grip strength meter from 12 to 24 weeks. Rilmenidine-treated 171-82Q mice displayed significant improved forelimb grip strength (Fig. 2A) and all limbs grip strength (Fig. 2B) from 12 to 22 weeks of age. There was no influence of rilmenidine treatment on the performance of wild-type mice. Also there was no difference between the two groups in forelimb or all limb grip strength before treatment commenced at 4 weeks of age.
Figure 2.
Rilmenidine improved grip strength in a transgenic mouse model of HD. N171-82Q mice were given rilmenidine ip injections four times a week (tg Rilm: 4, 12 and 14 weeks, n = 20; 16 weeks, n = 19; 18 weeks, n = 16; 20 weeks, n = 12; 22 weeks, n = 6 and 24 weeks, n = 2) or ip injections with the carrier substance (tg Con: 4, 12, 14 and 16 weeks, n = 20; 18 weeks, n = 17; 20 weeks, n = 12; 22 weeks, n = 6 and 24 weeks, n = 2) from 5 weeks of age. Wild-type mice were given rilmenidine or placebo injections with the same frequency (wt Rilm and wt Con, n = 5) (A) Forelimb grip strength in N171-82Q mice at 4 weeks (P = n.s.), 12–18 weeks (P < 0.0001), 20 weeks (P = 0.01), 22 weeks (P = 0.0057) and 24 weeks (P = n.s.) (by t-test). Rilmenidine treatment had no effect on grip strength in wild-type mice. (B) All limb grip strength in N171-82Q mice at 4 weeks (P = n.s.), 12 weeks (P = 0.002), 14 weeks (P < 0.0001), 16 weeks (P = 0.0015), 18 weeks (P = 0.0001), 20 weeks (P = 0.01), 22 weeks (P = 0.0129) and 24 weeks (P = n.s.) (by t-test). Error bars represent SEM.
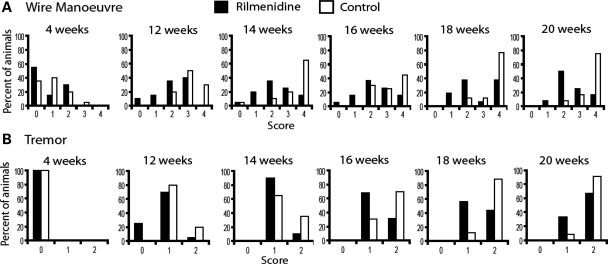
Wire manoeuvre
The wire manoeuvre tests the capacity of the mice to climb back on a horizontal wire when hung on the wire by their forelimbs. Rilmenidine-treated 171-82Q mice performed significantly better on the wire manoeuvre task from 12 until 20 weeks of age (Fig. 3A), and again there was no difference between the two groups before the start of treatment at 4 weeks. Also there was no influence of rilmenidine treatment on the performance of wild-type mice (data not shown).
Figure 3.
Rilmenidine treatment improved overall performance at the wire manoeuvre task as well as the severity of tremors at certain time points in N171-82Q mice. (A) Mice were scored on their ability to perform the wire manoeuvre, score: 0, active grip with hind legs; 1, difficulty grasping with hind legs; 2, unable to lift hind legs; 3, falls within 30 s; 4, falls immediately. The percentage of animals obtaining each score is shown in the graphs, black bars represent rilmenidine-treated animals and white bars represent control, placebo-treated animals. Significant differences between treatment groups were seen at 12 weeks (P = 0.002), 14 weeks (P = 0.017), 16 weeks (P = 0.0246), 18 weeks (P = 0.0211) and 20 weeks (P=0.0056) of age (Mann–Whitney U test). No significant differences were seen pre-treatment at 4 weeks as well as at 22 weeks (P = 0.1495) of age. (B) The severity of tremors in N171-82Q mice was significantly improved by rilmenidine treatment at 16 weeks (P = 0.0403) and 18 weeks (P = 0.0293) of age, and a trend towards an improvement was also evident at the age of 12 weeks (P = 0.0583) in comparison to the control group. No significant differences were seen at 14 weeks (P = 0.1762), 20 weeks (P = 0.2987) and 22 weeks (P = 0.3367) of age. Score: 0, no tremor; 1, mild tremor; 2, severe tremor.
Tremors
The severity of tremors was significantly improved in rilmenidine-treated 171-82Q mice at 16 and 18 weeks of age, and a trend towards an improvement was also seen at the age of 12 weeks (Fig. 3B), in comparison to the control group. Wild-type mice do not demonstrate tremors at these ages, and rilmenidine treatment did not induce tremors in wild-type mice (data not shown).
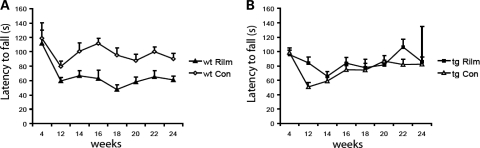
Accelerating rotarod apparatus
The accelerated rotarod apparatus is a measure of motor coordination and measures the ability of a mouse to maintain balance on a rotating cylinder (rotating towards the mouse). As shown in Fig. 4A, we found a significant influence of rilmenidine treatment on the behaviour of wild-type mice in this test. Rilmenidine-treated wild-type mice spent significantly less time on the rotarod at 14–24 weeks of age, and even at 12 weeks a trend towards a diminished performance was visible. This may be due to the reported sedative effects of the compound (31). However, despite the decline in rotarod performance seen in wild-type mice, rilmenidine-treated 171-82Q mice stayed significantly longer on the rotarod than control mice at 12 weeks of age (Fig. 4B). At later time points (14–24 weeks), we did not observe a decline in the performance of rilmenidine-treated 171-82Q mice as observed in the wild-type mice. Rather, the performance of rilmenidine-treated transgenic mice was similar to these mice treated with the carrier substance. At 22 weeks of age, there was again a trend towards an improved performance of the rilmenidine-treated animals. Thus, despite the negative effects of this drug on wild-type performance (probably due to its known sedative effects) (31), a ‘net improvement’ by rilmenidine in our 171-82Q mice was observed.
Figure 4.
Effect of rilmenidine treatment on rotarod performance of wild-type and transgenic N171-82Q mice. (A) Wild-type mice were given rilmenidine ip injections (wt Rilm; n = 5) or ip injections with the carrier substance (wt Con; n = 5) four times a week from 5 weeks of age. Performance on the accelerated rotarod was significantly impaired in rilmenidine-treated wild-type mice at 14 weeks (P = 0.0455), 16 weeks (P = 0.0068), 18 weeks (P = 0.0042), 20 weeks (0.0265), 22 weeks (P = 0.0117) and 24 weeks (P = 0.0141) of age (by t-test). Also a trend towards a diminished rotarod performance was seen at an age of 12 weeks (P = 0.067). Overall effect from all treatment points, P = 0.0007 (repeated measures ANOVA). (B) N171-82Q mice were given rilmenidine ip injections four times a week (tg Rilm; 4, 12 and 14 weeks, n = 20; 16 weeks, n = 19; 18 weeks, n = 16; 20 weeks, n = 12; 22 weeks, n = 6 and 24 weeks, n = 2) or ip injections with the carrier substance (tg Con; 4, 12, 14 and 16 weeks, n = 20; 18 weeks, n = 17; 20 weeks, n = 12; 22 weeks, n = 6 and 24 weeks, n = 2) from 5 weeks of age. Rilmenidine-treated N171-82Q mice stayed significantly longer on the rotarod than carrier-treated mice at 12 weeks of age (P = 0.0027). However, there were no significant differences at 14,16,18,20 and 24 weeks of age, at 22 weeks there was a trend towards an improvement with rilmenidine treatment (P = 0.092) (by t-test).
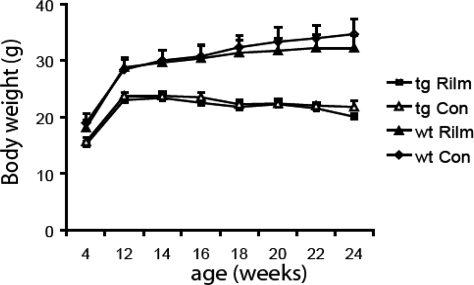
Rilmenidine does not prevent weight loss or premature death in HD mice
Weight loss is a feature in the N171-82Q mice, possibly due to a hypermetabolic state (32). Transgenic N171-82Q mice weighed significantly less than wild-type mice from 12 weeks of age and rilmenidine treatment did not have an effect on body weight in transgenic or wild-type mice (Fig. 5). The 171-82Q mouse model of HD displays premature death between 16 and 24 weeks of age, the reason for which is unknown (26). Rilmenidine treatment had no significant effect on the lifespan of our transgenic 171-82Q mice. Rilmenidine-treated transgenic mice showed an average lifespan of 148.27 ± 4.62 days (mean ± SD), whereas mice treated only with the carrier substance survived 145.74 ± 4.24 days. Our survival analysis, however, may be confounded by the fact that the mice had to be euthanized when their disease severities exceeded defined humane endpoints to prevent the disease severity exceeding the UK home office moderate category (no interaction with peers, no righting reflexes, major loss of weight). Thus, we do not know how long the mice would have survived beyond this point. Furthermore, the cause of death in these transgenic mice is unknown and is probably more dependent on metabolic disturbances than on neuronal pathology (33).
Figure 5.
Rilmenidine treatment did not prevent the disease-associated weight loss in 171-82Q transgenic mice (tg Rilm versus tg Con). The drug also did not have any influence on the body weight of wild-type or transgenic mice (wt Rilm versus wt Con).
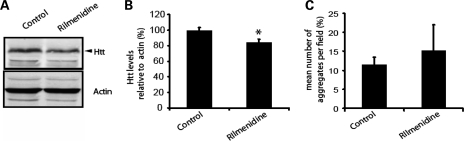
Rilmenidine decreases levels of mutant huntingtin
In order to investigate the effect of rilmenidine treatment on levels of mutant huntingtin in the brains of transgenic, mice we measured the levels of the soluble htt-exon1 fragment using quantitative western blotting methods. A small change in soluble huntingtin levels was observed after a 6-week period of drug treatment (Fig. 6A and B). This is compatible with a biologically significant enhancement of transgene clearance, since this transgene has a long half-life (11).
Figure 6.
Rilmenidine treatment decreases soluble huntingtin levels, but not aggregates. (A and B) Quantification of soluble huntingtin levels was carried out by western blotting with IRDye conjugated secondary antibodies and measurement of fluorescence intensity of the bands corresponding to mutant huntingtin and actin, as a loading control. Representative lanes from the western blot are shown in (A) and quantification is shown in (B) (*P < 0.05 by t-test, n = 7 for control, placebo-treated mice and n = 6 for rilmenidine-treated mice, error bars represent SEM). (C) Aggregate number was counted in the motor cortex of control, placebo-treated and rilmenidine-treated 171-82Q transgenic mice. Aggregates were counted on three sections from three mice in each group (P = n.s. by t-test).
One of the neuropathological hallmarks of HD is the presence of aggregates, which are predominantly intranuclear, in the striatum and deeper layers of the cortex. Immunohistochemistry was performed on coronal sections from rilmenidine-treated and control HD mice. Aggregates were counted in three different regions of the brain, the piriform cortex, motor cortex and hippocampus. The number of aggregates seen was very variable, particularly in the rilmenidine treated brains, and no alterations in aggregate number were observed between treatment groups (Fig. 6C and data not shown). Aggregate size in these sections was also assessed and the diameter of aggregates showed no change, 0.63 ± 0.04 µm in placebo versus 0.67 ± 0.04 µm in rilmenidine-treated mice. The lack of a decrease in aggregate number or size in brains may be due to the relative availability of the soluble protein compared with the aggregated protein. As the aggregates seen in these mice were almost exclusively nuclear they will be less accessible for degradation by autophagy, which is a cytoplasmic process, unlike the soluble, cytoplasmic protein (34). Of relevance, it appears that the large inclusions visible by light microscopy may be comparatively protective sequestrations of the more toxic and more soluble oligomeric species of mutant huntingtin (35). The marked improvement in behavioural outcome associated with the decrease in soluble mutant huntingtin seen in these mice is consistent with this view.
Transient moderate side effects of rilmenidine
Approximately 10 min after ip injection of rilmenidine, we observed moderate side effects probably related to the known hypotensive effect of this drug. The mice showed decreased activity, unsteady gait and some also presented with hunched posture and piloerection. These side effects lasted for about 2 h and disappeared completely after 3 h. We routinely checked the mice 1.5 h post-injection to ensure that the side effects remained in the moderate severity category (as defined by the UK Home Office) and then again 3 h post-injection to ensure that all mice had fully recovered. Also we gave transgel as an additional fluid source to ensure that the mice were always properly hydrated. Nevertheless, one rilmenidine-treated transgenic mouse did not completely recover from its side effects and had to undergo euthanasia after 10 weeks of treatment, additionally, two mice treated with rilmenidine for measurement of soluble protein levels had to be euthanized for the same reason. Another two mice (one transgenic and one wild-type) had to undergo euthanasia due to problems related to the repeated intraperitoneal injections (boils, abdominal signs of infection).
DISCUSSION
The autophagy–lysosomal and ubiquitin–proteasome pathways are the major routes for protein degradation in eukaryotic cells. Whereas the narrow pore of the proteasome barrel precludes clearance of large protein complexes and intracytoplasmic aggregate-prone proteins like mutant huntingtin, mammalian lysosomes can degrade these by autophagy. Autophagy is regulated by a number of protein kinases, the best characterized is mTOR. However, this kinase controls several cellular processes besides autophagy, which probably contributes to the complications seen with long-term use of its inhibitor rapamycin (36). A screen of FDA-approved drugs revealed the Gi signalling activator clonidine as an autophagy inducer acting via an mTOR-independent pathway (16). We also found that the related compound rilmenidine increased autophagy in a dose-dependent manner in cell culture.
We report here that rilmenidine, a drug known to act in the brain, which is currently used for the chronic treatment of hypertension, improves behavioural performances in a variety of domains in a transgenic animal model of HD. The transgene expression is driven by the mouse prion protein promoter and results in predominantly central nervous system expression with phenotypes that are widely believed to be the consequence of neuronal dysfunction. We have assessed the phenotypes that we have validated as showing clear differences between the transgenic mice and wild-type littermates and which all improved with rapamycin treatment in a previous study (11). We did not assess neuronal cell death as this does not occur to any overt extent in this mouse model (26). Also, measurements of brain volume will be difficult to interpret with autophagy inducers, which might affect this parameter, since the induction of autophagy can reduce cell size in various cellular and in vivo systems (37,38). The improvement of multiple behavioural parameters throughout the rilmenidine treatment period was also associated with a decrease in the levels of soluble mutant huntingtin. The reduction in the load of the mutant protein is consistent with at least some of its effects being due to autophagy. However, the mechanism of action of rilmenidine could also include other effects apart from inducing autophagy. Imidazoline drugs, independent of their affinity for imidazoline receptors, also interact (in the micromolar range) with several cation channels, including N-methyl-d-aspartate (NMDA) receptors. Some researchers have attributed the neuroprotective effects of imidazolines against NMDA-induced neuronal death and hypoxic insults in vitro to a voltage dependent, fast and fully reversible blockade of NMDA receptors at a phencyclidine-like site (36,39).
Unfortunately, there are no ways we can directly show increased autophagosome synthesis in the brains of mice treated with rilmenidine in vivo. This is because steady state levels of autophagosomes in the brain could either increase, decrease or stay the same, if we induced autophagy (40). The only way one could show increased autophagosome synthesis would be to clamp autophagosome degradation, something that is impossible in vivo. However, in order to do this robustly we have studied the effects of rilmenidine on primary neuronal culture. The proposed effect on autophagy is also consistent with the reductions in soluble levels of the mutant protein we observed in the brains of the treated mice.
The treated mice did show moderate side effects like reduced activity, unsteady gait and sometimes piloerection and hunched posture, which lasted for roughly 2 h. This is probably due to the relatively high doses used in this study (10 mg/kg intraperitoneally four times a week). Rilmenidine does bind, albeit with a lower affinity than clonidine, to alpha2-adrenoceptors and therefore side effects via this receptor cannot be excluded at higher concentrations (41). It is important to point out that our previous data suggest that agonists acting on either imidazoline or α2-adrenoceptors will have the same effects on autophagy. Both receptors are Gi coupled, and agonists of either receptor will reduce cAMP levels, which we have found to induce autophagy (16). The deleterious effect of rilmenidine on the rotarod performance of wild-type mice was not unexpected, as this is seen with the prototypical compound clonidine in mice (42). Despite this likely sedative effect observed in rilmenidine-treated wild-type mice, the rilmenidine-treated 171-82Q mice did not perform worse than the placebo group on the rotarod, indicating a net improvement in this test (improvement by the drug was counteracted by the drug's sedative effect).
The side effects related to alpha-2 adrenergic agonism may be tractable in the context of treating HD. First, it is possible that current doses of rilmenidine that are well tolerated for treating hypertension, may be sufficient to have benefit over the many decades it takes for HD to evolve in humans. Indeed, the excellent tolerability of this drug (reviewed in 43) makes it a suitable candidate drug for delaying the onset of HD. Since HD is a highly penetrant, autosomal dominant disease, most cases will have a family history and the mutation can be easily detected at the DNA level. Thus, it would be possible to identify HD mutation carriers who are still asymptomatic in order to delay disease onset. Second, compounds acting exclusively at the imidazoline I1 receptor have been synthesized (44), and since these are devoid of any α2-adrenoceptor binding, they may perhaps be more suited for the long-term treatment of neurodegenerative diseases with fewer side effects.
In conclusion, our data provide proof-of-principle that rilmenidine, a centrally acting, well-tolerated drug, can alleviate the severity of signs in an HD mouse model. The extent of improvement on grip strength, tremors and wire manoeuvre seen in HD mice treated with rilmenidine is comparable with that seen in our previous studies with the autophagy enhancing drug rapamycin (14). Although there was no major improvement in rotarod performance in the HD mice treated with rilmenidine, this can be explained by the side effect of rilmenidine actually impairing rotarod performance in wild-type mice. A comparison between the wild-type and HD mice treated with rilmenidine is compatible with an overall improvement in rotarod performance mediated by this drug in HD mice. Thus, we believe that these data and its very favourable benefit-to-risk profile, suggest that rilmenidine or drugs acting on the same targets as rilmenidine, may be strong candidates to consider for human trials in HD patients.
MATERIALS AND METHODS
Mouse model
We used HD-N171-82Q mice (B6C3F1/J-Tg(HD82Gln)81Dbo/J, Jackson Laboratory, Bar Harbour, ME, USA) backcrossed on a C57BL/6J background for more than 10 generations. These mice carry an N-terminal fragment expressing the first 171 amino acids of human huntingtin with 82 glutamine repeats under the mouse prion promotor (26).
All studies and procedures were performed under the jurisdiction of appropriate Home Office Project and Personal animal licenses and with local Ethics Committee approval.
HD genotyping
The litters produced by mating of HD transgenic males with C57BL/6J females were genotyped at an age of 3 weeks by PCR, according to the protocol recommended by the Jackson Laboratory. Two sets of primers were used: IL-2wt-fwd (5′-CTAGGCCA CAGAATTGAAAGATCT-3′), IL-2wt-rvs (5′-GTAGGTGGAAATTCTAGCATCATCC-3′), HD82Gln (5′-GTGGATACCCCCTCCCCCAGCCTAGACC-3′) and HD-591-5′ (5′-GA ACTTTCAGCTACCGAAGAAAGACCGTGT-3′).
Rilmenidine treatment and behavioural tests
Transgenic mice and non-transgenic littermates were compared in the trial and both genders were used. Mice were coded with alpha-numeric identities which provided no clues to their genotype and treated and untreated mice were not housed in separate cages. Thus observers were blind to their treatment and genetic status during testing.
Rilmenidine hemifumarate (Tocris Biosciences, Bristol, UK) was prepared as a stock solution of 10 mg/ml in 20% ethanol and was diluted on the day of experiment to 1 mg/ml in 0.15 m NaCl, 5% Tween-20 and 5% PEG 400 immediately before injection.
We used 20 HD-transgenic mice for rilmenidine treatment as well as the same number of transgenic animals for the control group. For studies of the effects of rilmenidine on behavioural parameters, we included five rilmenidine-treated wild-type mice as well as five control wild-type mice. There was no difference in the male:female ratios of the groups treated with placebo or rilmenidine.
At 4 weeks of age, a pre-assessment of motor performances was performed. There were no significant differences in test performances in the mice assigned to the treatment and placebo groups at 4 weeks and no differences in the sex ratios in the groups.
Rilmenidine treatment was started at an age of 5 weeks. The mice were weighed and received intraperitoneal injections (10 mg/kg body weight rilmenidine) four times a week (Monday, Tuesday, Thursday and Friday). The control group received ip injections with the carrier substance (0.15 m NaCl, 5% Tween-20 and 5% PEG 400 and 2% ethanol). Mice were monitored daily. We chose this dose since 10 mg/kg was reported not to cause overt sedation (18).
We assessed motor performance at 12, 14, 16, 18, 20, 22 and 24 weeks of age with a rotarod apparatus (Accelerating Model, Ugo Basile, Biological Research Apparatus, Varese, Italy). The mice were given training sessions for 2 consecutive days to acclimatize them to the apparatus and on the third day, the definitive testing took place. On the first training day, the mice had three trials at a constant speed of 4 r.p.m. In each trial, the animals were put on the Rotarod for a maximum of 300 s. On the second day, the training took place at a constant speed of 10 r.p.m. (three trials). Day 3 was the testing day, where the speed was accelerated from 3 to 30 r.p.m. in 300 s. A minimum of 10 min break was given between each trial. The latency to fall was taken as the maximum value reached over the three trials.
Grip strength, wire manoeuvre and tremor monitoring was also performed at 12, 14, 16, 18, 20, 22 and 24 weeks of age. Grip strength was monitored quantitatively by using a grip strength meter (Biosep, France). The mice were held above the apparatus grid with their front paws (forelimb grip) or all four paws (all limb grip) grasping the grid, then pulled back by the tail following the axle of the sensor, horizontally and steadily, until they released the grid. The apparatus was used in peak mode, the recorded value corresponding to the maximum force developed by the animal. Forelimb grip strength as well as all paw grip strength was measured three times and the average value was taken. Wire manoeuvre and tremor are part of the SHIRPA battery of behavioural tests (45). For tremors, the mice were placed on a grid in a clear perspex cylinder. We recorded tremor for 2 min and scored the mice as follows: 0, none; 1, mild; 2, marked. For the wire manoeuvre, mice were held above a horizontal wire by the tail and lowered to allow the forelimbs to grip the wire. The mice were held in extension, rotated around the horizontal and released. We scored them as follows: 0, active grip with hind legs; 1, difficulty grasping with hind legs; 2, unable to grasp with hind legs; 3, unable to lift hind legs, falls within 10 s; 4, falls immediately.
The following signs were used as humane endpoints for the mice, which resulted in euthanasia: marked loss of appetite and fluid intake, staring coat, hunched posture and subdued behaviour, or 20% weight loss over a period of less than 3 days.
Cell culture and treatment
For assessment of autophagy in primary cells, cortical neurons were isolated from embryonic day 16.5 Sprague–Dawley rat pups (Charles River). Briefly, pup brains were harvested and placed in ice-cold DMEM medium where the meninges were removed; the cerebral cortices were dissected and then incubated in DMEM containing Accutase (Innovative Cell Technologies, San Diego, CA, USA) for 10 min at 37°C. After mechanical dissociation using sterile micropipette tips, dissociated neurons were resuspended in DMEM medium and centrifuged. Cell count and viability assay was performed using Trypan blue. Viable cells were seeded on poly-d-lysine and laminin coated 6-multiwell (7.5 × 105 cells per well). Cells were maintained in DMEM containing 2 mm glutamine, 2% B27 supplement and 1% PSF (Invitrogen). One-half of the plating medium was changed every third day until treatment. After 7 days in vitro, cultured neurons were treated for 8 h with 1 µm rilmenidine or 0.2 µm rapamycin (Sigma-Aldrich). Where used, a saturating concentration (400 nm) of bafilomycin A1 (Calbiochem) was added to the cells in the last 4 h before harvesting. The stable inducible PC12 cells have been previously described (11), cells were treated with doxycycline to induce huntingtin expression for 24 h at 1 µg/ml with the addition of 1 µm rilmenidine for a further 24 h and bafilomycin A1 as for primary neurons.
Protein extraction and western blot analysis
Mouse muscle samples from rilmenidine- or placebo-treated wild-type mice (four rilmenidine-treated and five placebo-treated mice) at an age of 28 weeks were homogenized in lysis buffer [10 mm Tris–HCl, pH 6.8, 68.5 nm NaCl, 0.5 mm EGTA, 0.5% Triton X-100, 5% glycerol and protease inhibitor cocktail (Roche Diagnostics)] at 4°C. For brains, the lysis buffer was 50 mm Tris, pH 7.4, 5.5% Triton X-100 and protease inhibitor cocktail. The homogenate was centrifuged at 13 400g at 4°C, the supernatant was removed and used for western blot. Cell pellets were lysed on ice in Laemmli buffer [6.5 mm Tris–HCl pH 6.8, 2% sodium dodecyl sulphate (SDS), 5% β-mercaptoethanol, 10% glycerol, 0.01% Bromophenol blue and protease inhibitors) for 30 min. Proteins were separated on 12% SDS–polyacrylamide gels and transferred onto PVDF membranes (Hybond ECL membrane, Amersham Biosciences). Membranes were blocked by incubation in 5% dried milk in PBS and 0.1% Tween-20, pH 7.6. The primary antibodies used were anti-polyglutamine (1C2, Chemicon), anti-LC3 (Novus), anti-phosphorylated and total 4E-BP1, S6 kinase and S6 protein (all Cell Signaling), anti-tubulin and anti-actin (Sigma). Horseradish peroxidase-conjugated antibodies (Amersham Biosciences, 1:2000) were then added and immunoreactive bands were detected with enhanced chemiluminescence reagent (ECL, Amersham Biosciences). Quantification of western blots was carried out using ImageJ software and band intensities were normalized to actin or tubulin levels. P-values were determined using unpaired t-tests. For quantification of soluble huntingtin levels in rilmenidine-treated mouse brains, protein was extracted from seven placebo-treated mice and six rilmenidine-treated mice as above. For analysis of these levels and those of LC3-II levels in PC12 cells treated with rilmenidine, western blots were probed with secondary antibodies conjugated to IRDye® for detection at 780 or 680 nm (Li-Cor Biosciences) and visualized and quantified using an Odyssey imaging system (Li-Cor Biosciences).
Immunohistochemistry
Thirty micrometer sections of rilmenidine-treated and control transgenic HD mice were analysed for neuronal inclusions according to the protocol of Davies et al. (46). Sections were labelled with anti-huntingtin antibody by free-floating immunohistochemistry (EM48, Chemicon). Staining was performed by peroxidase labelling using Vectastain Avidin:Biotinylated enzyme complex (ABC) kit and visualized with DAB reagent (Vector Laboratories).
Inclusions were counted in the piriform cortex, motor cortex and hippocampus in three fields on at least three sections per animal at a magnification of ×100 (Zeiss Axioskop2, field diameter 0.2 mm). Aggregates were photographed and diameter measured using Zeiss axiovision software.
Statistics
Significance levels for comparisons between groups were determined with t-tests, repeated measure or factorial ANOVA, where appropriate, for parametric data and with Mann–Whitney U tests for non-parametric data, using the STATVIEW software, version 4.53 (Abacus Concepts, Berkeley, CA, USA).
FUNDING
Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
ACKNOWLEDGEMENTS
We are grateful to the MRC (Programme grant to D.C.R. and S.B.), and the Wellcome Trust (Senior Fellowship to DCR) for funding. We thank Sarah Carter and Michelle Stewart (MRCHarwell) for excellent technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. doi:10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 2.Gil J.M., Rego A.C. Mechanisms of neurodegeneration in Huntington's disease. Eur. J. Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. doi:10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 3.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. doi:10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein D.C., Leggo J., Coles R., Almqvist E., Biancalana V., Cassiman J.J., Chotai K., Connarty M., Crauford D., Curtis A., et al. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am. J. Hum. Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- 5.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. doi:10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein D.C. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. doi:10.1016/S0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 7.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P., Jr Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. doi:10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Rosas H.D., Salat D.H., Lee S.Y., Zaleta A.K., Pappu V., Fischl B., Greve D., Hevelone N., Hersch S.M. Cerebral cortex and the clinical expression of Huntington's disease: complexity and heterogeneity. Brain. 2008;131:1057–1068. doi: 10.1093/brain/awn025. doi:10.1093/brain/awn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aylward E.H., Brandt J., Codori A.M., Mangus R.S., Barta P.E., Harris G.J. Reduced basal ganglia volume associated with the gene for Huntington's disease in asymptomatic at-risk persons. Neurology. 1994;44:823–828. doi: 10.1212/wnl.44.5.823. [DOI] [PubMed] [Google Scholar]
- 10.Aylward E.H., Sparks B.F., Field K.M., Yallapragada V., Shpritz B.D., Rosenblatt A., Brandt J., Gourley L.M., Liang K., Zhou H., et al. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- 11.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. doi:10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 12.Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. doi:10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 13.Rubinsztein D.C., Gestwicki J.E., Murphy L.O., Klionsky D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug. Discov. 2007;6:304–312. doi: 10.1038/nrd2272. doi:10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 14.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. doi:10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 15.Menzies F.M., Huebener J., Renna M., Bonin M., Riess O., Rubinsztein D.C. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain. 2010;133:93–104. doi: 10.1093/brain/awp292. doi:10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. doi:10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harron D.W. Distinctive features of rilmenidine possibly related to its selectivity for imidazoline receptors. Am. J. Hypertens. 1992;5:91S–98S. doi: 10.1093/ajh/5.4.91s. [DOI] [PubMed] [Google Scholar]
- 18.Montastruc J.L., Macquin-Mavier I., Tran M.A., Damase-Michel C., Koenig-Berard E., Valet P. Recent advances in the pharmacology of rilmenidine. Am. J. Med. 1989;87:14S–17S. doi: 10.1016/0002-9343(89)90499-3. doi:10.1016/0002-9343(89)90499-3. [DOI] [PubMed] [Google Scholar]
- 19.Chan C.K., Head G.A. Relative importance of central imidazoline receptors for the antihypertensive effects of moxonidine and rilmenidine. J. Hypertens. 1996;14:855–864. doi: 10.1097/00004872-199607000-00008. doi:10.1097/00004872-199607000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Safar M.E. Rilmenidine: a novel antihypertensive agent. Am. J. Med. 1989;87:24S–29S. doi: 10.1016/0002-9343(89)90501-9. doi:10.1016/0002-9343(89)90501-9. [DOI] [PubMed] [Google Scholar]
- 21.Yu A., Frishman W.H. Imidazoline receptor agonist drugs: a new approach to the treatment of systemic hypertension. J. Clin. Pharmacol. 1996;36:98–111. doi: 10.1002/j.1552-4604.1996.tb04174.x. [DOI] [PubMed] [Google Scholar]
- 22.Ernsberger P., Meeley M.P., Mann J.J., Reis D.J. Clonidine binds to imidazole binding sites as well as alpha 2-adrenoceptors in the ventrolateral medulla. Eur. J. Pharmacol. 1987;134:1–13. doi: 10.1016/0014-2999(87)90125-7. doi:10.1016/0014-2999(87)90125-7. [DOI] [PubMed] [Google Scholar]
- 23.Kamisaki Y., Ishikawa T., Takao Y., Omodani H., Kuno N., Itoh T. Binding of [3H]p-aminoclonidine to two sites, alpha 2-adrenoceptors and imidazoline binding sites: distribution of imidazoline binding sites in rat brain. Brain Res. 1990;514:15–21. doi: 10.1016/0006-8993(90)90430-j. doi:10.1016/0006-8993(90)90430-J. [DOI] [PubMed] [Google Scholar]
- 24.De Vos H., Bricca G., De Keyser J., De Backer J.P., Bousquet P., Vauquelin G. Imidazoline receptors, non-adrenergic idazoxan binding sites and alpha 2-adrenoceptors in the human central nervous system. Neuroscience. 1994;59:589–598. doi: 10.1016/0306-4522(94)90179-1. doi:10.1016/0306-4522(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 25.King P.R., Gundlach A.L., Louis W.J. Quantitative autoradiographic localization in rat brain of alpha 2-adrenergic and non-adrenergic I-receptor binding sites labelled by [3H]rilmenidine. Brain Res. 1995;675:264–278. doi: 10.1016/0006-8993(95)00083-3. doi:10.1016/0006-8993(95)00083-3. [DOI] [PubMed] [Google Scholar]
- 26.Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A., et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. doi:10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. doi:10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. doi:10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boland B., Kumar A., Lee S., Platt F.M., Wegiel J., Yu W.H., Nixon R.A. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. doi:10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sathasivam K., Hobbs C., Turmaine M., Mangiarini L., Mahal A., Bertaux F., Wanker E.E., Doherty P., Davies S.W., Bates G.P. Formation of polyglutamine inclusions in non-CNS tissue. Hum. Mol. Genet. 1999;8:813–822. doi: 10.1093/hmg/8.5.813. doi:10.1093/hmg/8.5.813. [DOI] [PubMed] [Google Scholar]
- 31.Bousquet P., Feldman J. Drugs acting on imidazoline receptors: a review of their pharmacology, their use in blood pressure control and their potential interest in cardioprotection. Drugs. 1999;58:799–812. doi: 10.2165/00003495-199958050-00003. doi:10.2165/00003495-199958050-00003. [DOI] [PubMed] [Google Scholar]
- 32.Aziz N.A., van der Burg J.M., Landwehrmeyer G.B., Brundin P., Stijnen T., Roos R.A. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. doi:10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 33.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D., et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. doi:10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Iwata A., Christianson J.C., Bucci M., Ellerby L.M., Nukina N., Forno L.S., Kopito R.R. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. doi:10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. doi:10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov D.D., Ali S.M., Sabatini D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. doi:10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Vellai T., Bicsak B., Toth M.L., Takacs-Vellai K., Kovacs A.L. Regulation of cell growth by autophagy. Autophagy. 2008;4:507–509. doi: 10.4161/auto.5670. [DOI] [PubMed] [Google Scholar]
- 38.Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2007;581:2623–2629. doi: 10.1016/j.febslet.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 39.Milhaud D., Fagni L., Bockaert J., Lafon-Cazal M. Imidazoline-induced neuroprotective effects result from blockade of NMDA receptor channels in neuronal cultures. Neuropharmacology. 2000;39:2244–2254. doi: 10.1016/s0028-3908(00)00085-x. doi:10.1016/S0028-3908(00)00085-X. [DOI] [PubMed] [Google Scholar]
- 40.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. In search of an ‘autophagomometer. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. doi:10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 41.Koenig-Berard E., Tierney C., Beau B., Delbarre G., Lhoste F., Labrid C. Cardiovascular and central nervous system effects of rilmenidine (S 3341) in rats. Am. J. Cardiol. 1988;61:22D–31D. doi: 10.1016/0002-9149(88)90460-2. doi:10.1016/0002-9149(88)90460-2. [DOI] [PubMed] [Google Scholar]
- 42.Young R., Batkai S., Dukat M., Glennon R.A. TDIQ (5,6,7,8-tetrahydro-1,3-dioxolo[4,5-g]isoquinoline) exhibits anxiolytic-like activity in a marble-burying assay in mice. Pharmacol. Biochem. Behav. 2006;84:62–73. doi: 10.1016/j.pbb.2006.04.006. doi:10.1016/j.pbb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Reid J.L. Update on rilmenidine: clinical benefits. Am. J. Hypertens. 2001;14:322S–324S. doi: 10.1016/s0895-7061(01)02239-7. doi:10.1016/S0895-7061(01)02239-7. [DOI] [PubMed] [Google Scholar]
- 44.Bousquet P., Greney H., Bruban V., Schann S., Ehrhardt J.D., Monassier L., Feldman J. I(1) imidazoline receptors involved in cardiovascular regulation: where are we and where are we going? Ann. N. Y. Acad. Sci. 2003;1009:228–233. doi: 10.1196/annals.1304.028. doi:10.1196/annals.1304.028. [DOI] [PubMed] [Google Scholar]
- 45.Rogers D.C., Fisher E.M., Brown S.D., Peters J., Hunter A.J., Martin J.E. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome. 1997;8:711–713. doi: 10.1007/s003359900551. doi:10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- 46.Davies S.W., Sathasivam K., Hobbs C., Doherty P., Mangiarini L., Scherzinger E., Wanker E.E., Bates G.P. Detection of polyglutamine aggregation in mouse models. Methods Enzymol. 1999;309:687–701. doi: 10.1016/s0076-6879(99)09045-x. doi:10.1016/S0076-6879(99)09045-X. [DOI] [PubMed] [Google Scholar]