Abstract
Mutations in CHMP2B cause frontotemporal dementia (FTD) in a large Danish pedigree, which is termed FTD linked to chromosome 3 (FTD-3), and also in an unrelated familial FTD patient. CHMP2B is a component of the ESCRT-III complex, which is required for function of the multivesicular body (MVB), an endosomal structure that fuses with the lysosome to degrade endocytosed proteins. We report a novel endosomal pathology in CHMP2B mutation-positive patient brains and also identify and characterize abnormal endosomes in patient fibroblasts. Functional studies demonstrate a specific disruption of endosome–lysosome fusion but not protein sorting by the MVB. We provide evidence for a mechanism for impaired endosome–lysosome fusion whereby mutant CHMP2B constitutively binds to MVBs and prevents recruitment of proteins necessary for fusion to occur, such as Rab7. The fusion of endosomes with lysosomes is required for neuronal function and the data presented therefore suggest a pathogenic mechanism for FTD caused by CHMP2B mutations.
INTRODUCTION
Frontotemporal dementia (FTD) is the third most common form of primary degenerative dementia, accounting for up to 20% of young onset dementia (1). FTD linked to chromosome 3 (FTD-3) is an autosomal dominant presenile dementia, associated with a large pedigree originating from the Jutland region of Denmark (2,3).
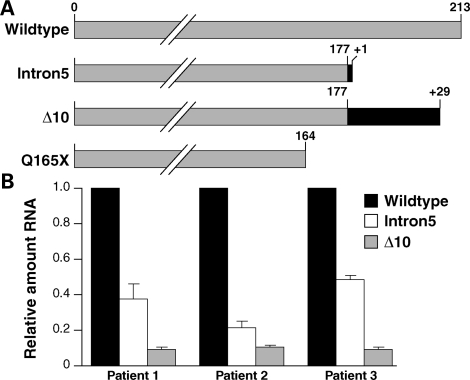
Analysis of the Jutland pedigree identified a disease segregating mutation in CHMP2B (4,5). This G>C transition in the 5′ acceptor splice site of exon 6 produces two aberrant transcripts: CHMP2BIntron5 and CHMP2BΔ10 (Fig. 1A). The proteins predicted from these splice variants are both C-terminal truncations of 36 amino acids (4). A CHMP2B nonsense mutation has been found in an unrelated familial FTD patient (6). This mutation (C493T) predicts replacement of a glutamine residue with a stop codon (CHMP2BQ165X) causing a C-terminal truncation of 49 amino acids (Fig. 1A).
Figure 1.
Mutant CHMP2B transcripts are expressed in patient brain. (A) Schematic diagram showing CHMP2BWildtype and the C-terminal truncations predicted by expression of the mutant transcripts. Grey indicates conservation of the wild-type protein, black indicates novel amino acids. Notably CHMP2BΔ10 includes a 29 amino acid C-terminal nonsense sequence. (B) Quantitative real-time PCR analysis of CHMP2B transcripts in three FTD-3 patient brains. CHMP2BIntron5 is expressed at ∼36% and CHMP2BΔ10 at ∼10% of CHMP2BWildtype levels.
CHMP2B is a component of the endosomal sorting complex required for transport-III (ESCRT-III), which is involved in endocytic trafficking of proteins (7). Membrane proteins that are targeted for degradation are internalized, or endocytosed, into the cell in vesicles which converge on endosomes. A subset of endosomes mature to form multivesicular bodies (MVBs), which are characterized by the presence of intraluminal vesicles (ILVs) formed by inward budding of the endosomal membrane. MVBs undergo fusion with lysosomes, where their contents are degraded. ESCRT proteins, including CHMP2B, are recruited to the endosomal outer membrane, where they provide a cellular machinery for the sorting of proteins destined for degradation into ILVs (8).
Here we report a novel endosomal pathology in FTD-3 patient brains and fibroblast cell lines. Using cellular overexpression models and patient fibroblast cell lines, we show that CHMP2B mutants disrupt a late stage of endosomal trafficking: the fusion of endosomes with lysosomes, while protein sorting into ILVs is intact. We further demonstrate that deficient fusion may be due to impaired ability of CHMP2BIntron5-positive endosomes to recruit Rab7, a GTPase required for endosome–lysosome fusion.
RESULTS
Mutant CHMP2B expression in patient brains
We used a quantitative real-time PCR assay to determine the relative levels of CHMP2BIntron5 and CHMP2BΔ10 compared with CHMP2BWildtype transcripts in the frontal cortex of three FTD-3 patient brains (Fig. 1B). This analysis showed that CHMP2BIntron5 was present at ∼36% and CHMP2BΔ10 at ∼10% of CHMP2BWildtype levels. This suggests that CHMP2BIntron5 is present at sufficient levels to play a role in pathogenesis. As CHMP2BΔ10 is expressed at lower levels than CHMP2BIntron5 but does not have a greater effect in the functional assays described below, CHMP2BIntron5 may be more relevant to the disease process.
Enlarged endosomes in patient brains
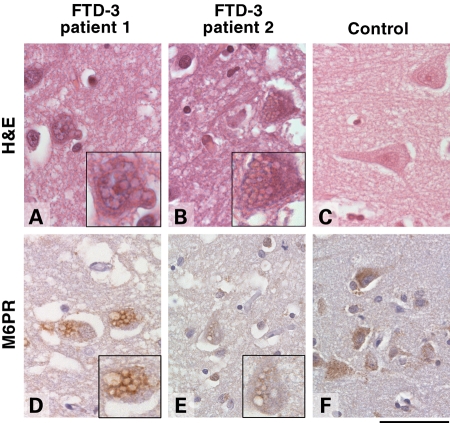
We have previously shown that overexpression of CHMP2BIntron5 and CHMP2BQ165X produce enlarged late endosomes in human neuroblastoma cells (6). If this endosomal phenotype is relevant to neurodegeneration, we would predict a similar finding in brain tissue. Frontal cortex sections from four FTD-3 patients were immunostained for mannose-6 phosphate receptor (M6PR), a marker of late endosomes. Anti-M6PR labelled large vacuoles in the cytoplasm of cortical neurons which were also visible by haematoxylin and eosin staining (Fig. 2). The vacuolar pathology was most abundant in the deep layers of the cortex. To assess their frequency, we counted the number of neurons that contained enlarged M6PR-positive vacuoles per high power field of cortical layers III to V in three FTD-3 brains. There was a range of 0–15 neurons with vacuolar pathology per high power field (an area of 0.249 mm2) with an average of 2.95 per field (average range 2–4.85). We then determined the distribution of vacuolar pathology in one FTD-3 brain by examining 14 brain regions (Table 1). Vacuoles were found in all cortical areas examined, with the exception of the cerebellar cortex. This is consistent with the generalized cortical atrophy observed on post-mortem (9) and with imaging data showing that atrophy is widespread in the FTD-3 cortex (10,11). The vacuoles were not observed in non-cortical grey matter regions such as the thalamus, substantia nigra, locus coeruleus and medulla. To determine whether these vacuoles were specific to FTD-3, we analysed brains from three Alzheimer's disease patients, six familial FTD patients (four with MAPT mutations, two with GRN mutations), 12 prion disease patients and nine age-matched controls (Supplementary Material, Fig. S1 and Table S1). None of these cases showed enlarged M6PR-positive vacuoles suggesting that this pathology is a result of the CHMP2B mutation.
Figure 2.
Enlarged endosomal phenotype in patient brain. Frontal cortex sections from FTD-3 (A, B, D, E) and age-matched control brains (C, F). Representative images of haematoxylin and eosin staining (H&E, A–C) and immunohistochemical staining against M6PR (D–F) are shown. Cytoplasmic vacuoles are visible in frontal cortical neurons of FTD-3 brains only, and these are positive for M6PR, a marker of late endosomes. Insets show enlargements of affected neurons. Scale bar = 60 µm.
Table 1.
Distribution of enlarged vacuoles in FTD-3 patient brain
| Brain area | Presence of vacuoles |
|---|---|
| Frontal cortex | + |
| Insula | + |
| Parietal cortex | + |
| Occipital cortex | + |
| Temporal cortex | + |
| Hippocampus | + |
| Basal ganglia | − |
| Thalamus | − |
| Substantia nigra | − |
| Locus coeruleus | − |
| Raphe nuclei | − |
| Pons | − |
| Olive | − |
| Cerebellar cortex | − |
Enlarged endosomes in patient fibroblasts
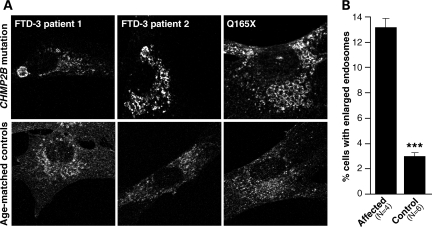
To investigate whether a similar morphological phenotype was present in peripheral tissue, we derived fibroblast cell lines from patient skin biopsies and immunostained them for CD63 to visualize late endosomes. Enlarged CD63-positive vesicles were observed; sometimes one very enlarged structure was present, or more typically, clusters of enlarged endosomes were seen when compared with age-matched controls (Fig. 3A).
Figure 3.
Enlarged endosomal phenotype in patient fibroblasts. Enlarged endosomal structures are observed at a higher frequency in fibroblast cell lines from mutation positive patients than in age-matched controls. (A) Representative images of CD63 staining showing enlarged structures and normal endosomal staining. (B) Blind counts revealed a highly significant difference between FTD-3 and control cell lines in the percentage of cells showing the enlarged endosomal phenotype (P < 0.001). Error bars show the SEM of three independent experiments.
Four patient (three FTD-3 patients and the CHMP2BQ165X patient) and six age-matched control cell lines were examined by an investigator blind to conditions. Six hundred cells per line were counted across three independent experiments, with cells matched for passage number (Fig. 3B). A significantly greater proportion of patient fibroblast cell lines had enlarged endosomes when compared with controls (t-test, P < 0.0001).
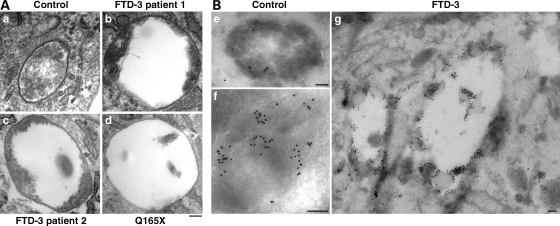
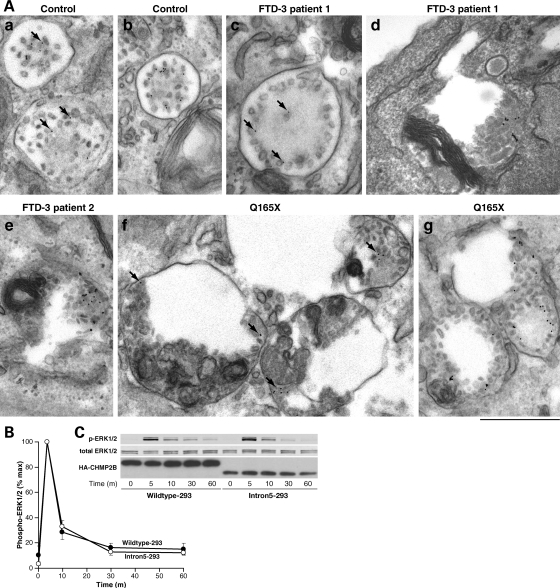
Examination of the ultrastructure of these enlarged endosomes by electron microscopy suggested that they were dysmorphic MVBs. Unusual vesicular structures present in three patient cell lines, including the CHMP2BQ165X patient, were not seen in age-matched controls (Fig. 4A). These structures were typically enlarged vacuoles with aberrant membrane deformation at the periphery, or ILVs which did not fill the lumen. In contrast, normal MVBs, which were seen in both patient and control cells, were smaller and packed with ILVs. Immunolabelling showed that these characteristic structures were likely to be late endosomes as they were positive for CD63, which also labelled normal degradative compartments and MVBs in patient and control cells (Fig. 4B).
Figure 4.
Ultrastructural analysis of patient fibroblasts. (A) Aberrantly formed multivesicular bodies (MVBs) were found in fibroblast cell lines from two FTD-3 patients (b and c) and the Q165X patient (d). In contrast to normal MVBs (a), these were typically enlarged and showed unusual peripheral membrane deformations or sparsity of ILVs. Scale bar = 100 nm. (B) These aberrant structures were positive for CD63, which also decorated normal MVBs and heavily decorated lysosomes in patient and control cell lines. (e) Normal MVB from a control cell line; (f) normal lysosome from a control cell line; (g) abnormal compartments from an FTD-3 cell line. Anti-CD63, 10 nm gold; scale bars = 100 nm.
CHMP2B mutations impair fusion of endosomes with lysosomes
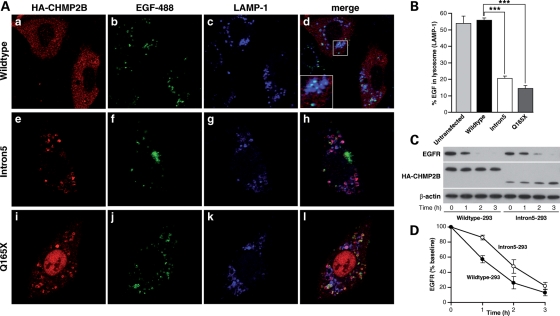
We next investigated the function of these abnormal endosomes. Transfected cells were treated with EGF-488 in the presence of leupeptin, an inhibitor of lysosomal proteases. A 2 h chase period was sufficient to deliver most EGF-488 to the lysosome, as visualized with anti-LAMP1, where it remained undegraded. Therefore, colocalization of EGF-488 with LAMP-1 was taken as a measure of successful endosome–lysosome fusion (Fig. 5A). Fifty cells from three independent experiments were counted for each condition, and the percentage of EGF-488 which colocalized with LAMP-1 was measured. The degree of endosome–lysosome fusion was significantly higher in cells expressing CHMP2BWildtype compared with CHMP2BIntron5 (t-test, P < 0.0001) or CHMP2BQ165X (t-test, P < 0.0001). CHMP2BWildtype expressing cells were comparable to untransfected cells (Fig. 5B).
Figure 5.
Delayed fusion of endosomes with lysosomes in CHMP2B mutant cells. (A) SK-N-SH cells were treated with leupeptin to inhibit lysosomal proteases, followed by feeding of AlexaFluor-488 labelled EGF. Trafficking to the lysosome can be confirmed by co-localization of EGF (green) with the lysosomal marker LAMP-1 (blue). Co-localization (cyan) of EGF with lysosomes occurs when CHMP2BWildtype is expressed (a–d), however when CHMP2BIntron5 (e–h) or CHMP2BQ165X (i–l) is expressed, there is less co-localization, indicating that EGF containing endosomes have not fused with lysosomes. (B) ∼55% of EGF is colocalized with LAMP-1 in CHMP2BWildtype expressing cells, which is comparable to untransfected cells. In contrast, only 15–20% colocalization is seen in cells expressing CHMP2BIntron5 (P < 0.0001) or CHMP2BQ165X (P < 0.0001). (C) Wildtype-293 and Intron5-293 cells were stimulated with EGF for 0, 1, 2 or 3 h before lysis and western blotting with antibodies against EGFR and HA-tag. (D) Detected protein was quantitated using ImageJ. EGFR was corrected against β-actin and the level of EGFR plotted as a percentage of the baseline level (i.e. that at time 0). Wildtype-293 cells, filled symbols; Intron5-293 cells unfilled symbols; error bars represent the SEM of three experiments.
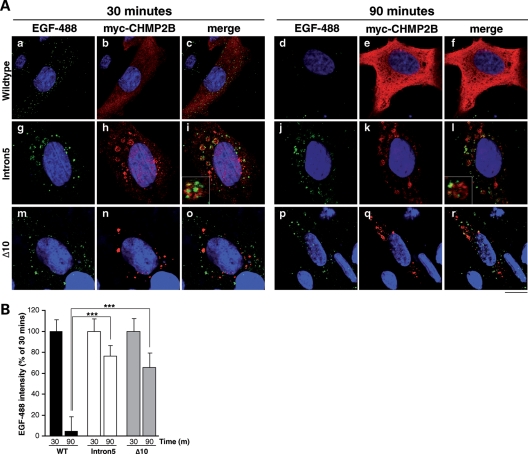
Impaired fusion was confirmed by delayed degradation of EGF and its receptor epidermal growth factor receptor (EGFR). Tetracycline-inducible HEK-293 cell lines expressing HA-tagged CHMP2BWildtype (Wildtype-293) or CHMP2BIntron5 (Intron5-293) showed downregulation of EGFR in response to stimulation with EGF, a process requiring delivery of EGFR from the MVB to the lysosome. However, this process was delayed in Intron5-293 cells when compared with Wildtype-293 cells (Fig. 5C and D), also suggesting that fusion is impaired. Impairment of EGF-488 degradation was also shown in transfected SK-N-SH cells. Levels of cellular EGF, as measured by fluorescence intensity, were recorded after 30 and 90 min chase (Fig. 6B). In CHMP2BWildtype expressing cells, ∼90% of the EGF signal present at 30 min was degraded at 90 min. In contrast, in cells expressing CHMP2BIntron5 or CHMP2BΔ10, much less (25–30%) degradation of EGF was observed. Interestingly, there was partial colocalization of EGF with CHMP2BIntron5-positive compartments at 90 min (Fig. 6A, inset), suggesting that the failure of degradation may be due to EGF being trapped in these aberrant compartments.
Figure 6.
Delayed degradation of EGF in CHMP2B mutant cells. (A) SK-N-SH cells were transiently transfected with CHMP2BWildtype (a–f), CHMP2BIntron5 (g–l) or CHMP2BΔ10 (m–r), and used 24 h later for an EGF-488 trafficking assay. Cells were fed 100 ng/ml EGF-488 (green) and fixed at either 30 (left panel) or 90 min (right panel). Fixed cells were immunostained with an anti-myc antibody to visualize myc-CHMP2B transfected cells (red) and the nuclei counterstained with Hoecsht (blue). At 30 min, all cells show punctate localization of EGF-488. In CHMP2BWildtype-expressing cells at 90 mins (d–f), the EGF-488 signal is degraded. However, in cells overexpressing CHMP2BIntron5 (j–l) or CHMP2BΔ10 (p–r), the majority of EGF-488 remains undegraded. Insets show magnification of EGF-488 colocalizing with CHMP2BIntron5-positive compartments. Scale bar = 20 µm. (B) LSM 510 Meta software was used to quantify this effect. ∼95% EGF-488 is degraded in CHMP2BWildtype expressing cells, compared with only 25–35% in cells expressing CHMP2BIntron5 or CHMP2BΔ10 (t-tests, both P < 0.0001).
Sorting of EGFR onto ILVs of MVBs is not affected by CHMP2B mutations
Given the evidence for disruption of endosome–lysosome fusion and resultant impaired degradation of EGF and EGFR, we also tested whether CHMP2B mutants impair the sorting of EGFR onto the intralumenal vesicles of the MVB, since the ESCRT-III complex has been implicated in this process. Undegraded EGFR associated with MVBs may be internalized normally into these structures or remain unsorted on the outer endosomal membrane. If retained on the outer membrane, the signalling domain of EGFR will still be active in the cytoplasm, allowing the activation of downstream signalling molecules, such as phosphorylation of ERK1/2. Sorting onto ILVs will sequester EGFR in the MVB, effectively silencing its downstream signalling (8,12).
EGF-induced phosphorylation of ERK1/2 was not affected by the expression of CHMP2BIntron5. Both Wildtype-293 and Intron5-293 cells showed a rapid peak in phosphorylation after 5 min, which rapidly returned to baseline levels (Fig. 7B and C). This suggests that EGF-induced signalling was silenced in both lines, due to the sequestration of EGFR in the ILVs. To determine whether EGFR was sequestered on ILVs in patient tissue, FTD-3 patient fibroblasts and age-matched control fibroblasts were incubated with 100 ng/ml EGF and a gold-conjugated anti-EGFR antibody. The EGFR–gold complexes are endocytosed in response to the EGF stimulation and can be used to monitor endosomal trafficking by electron microscopy (13). Sorting of EGFR onto internal vesicles of both normal and abnormal MVBs was observed in three patient fibroblasts lines: two FTD-3 patients and one CHMP2BQ165X patient (Fig. 7). This suggests that impaired degradation of EGFR is due to decreased fusion with lysosomes rather than impaired sorting onto ILVs.
Figure 7.
Intralumenal vesicle formation and sequestration of the EGF receptor is intact in CHMP2B mutant cells. (A) FTD-3 and control fibroblast cell lines were stimulated with 100 ng/ml EGF with anti-EGFR-gold for 2–4 h. EM analysis of control cells showed that EGFR moved onto the internal vesicles of MVBs (a and b) within this time frame. EGFR was also observed on ILVs within normal MVBs and abnormal structures in two different FTD-3 patients (c–e), and the Q165X patient (f, g). (c) A relatively normal MVB with peripheral ILVs associated with gold particles; (d, e) Abnormal structures containing unusual membranes, but also ILVs associated with gold particles. (f) Enlarged structures containing membranes and ILVs associated with gold particles. (g) EGFR–gold is clearly identifiable on ILVs in MVBs. Anti-EGFR, 10 nm gold; examples of gold particles indicated by arrows; scale bar = 500 nm. (B) Wildtype-293 and Intron5-293 cells were not stimulated (0) or stimulated with EGF for 5 min and then chased for 5, 10, 30 or 60 min. Western blotting with antibodies against phosphorylated ERK1/2 and total ERK1/2 (C) was quantitated using LI-COR Odyssey software. Phosphorylated ERK1/2 was corrected against total ERK1/2 and a time course of ERK1/2 activation was plotted as a percentage of the respective phosphorylation intensities after 5 min of EGF stimulation and 5 min of chase. Wildtype-293 cells, filled symbols; Intron5-293 cells unfilled symbols; error bars represent the SEM of three experiments.
Recruitment of Rab7 onto endosomes is impaired by expression of mutant CHMP2B
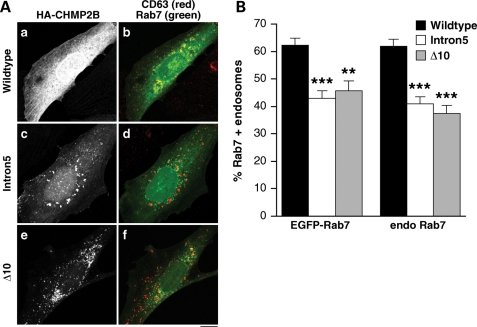
We hypothesised that the impaired fusion of endosomes with lysosomes may be due to a failure of CHMP2BIntron5-positive endosomes to recruit proteins necessary for heterotypic fusion. Endosome–lysosome fusion requires recruitment of Rab7, a recognized marker of late, fusion-competent endosomes (14). SK-N-SH cells were co-transfected with EGFP-Rab7 and CHMP2BWildtype, CHMP2BIntron5 or CHMP2BΔ10 and then immunostained for CD63 to visualize late endosomes (Fig. 8A). There was an approximate one-third reduction in the recruitment of EGFP-Rab7 onto endosomes in CHMP2BIntron5 cells (t-test, P < 0.0001) and an approximate one-quarter reduction in CHMP2BΔ10 cells (test, P < 0.001) when compared with cells expressing CHMP2BWildtype. This assay was repeated using an anti-Rab7 antibody to show endogenous Rab7 rather than using overexpression (Supplementary Material, Fig. S2). Both CHMP2BIntron5 and CHMP2BΔ10 cells showed at least a one-third reduction in recruitment of endogenous Rab7 onto endosomes compared with CHMP2BWildtype cells (t-tests, both P < 0.0001) (Fig. 8B). There were no significant differences when comparing between experiments, i.e. CHMP2BIntron5 or CHMP2BΔ10 cells showed a similar reduction in both EGFP-Rab7 and endogenous Rab7 recruitment (Fig. 8B).
Figure 8.
Impaired recruitment of Rab7 onto endosomes in mutant CHMP2B expressing cells. (A) SK-N-SH cells were co-transfected with EGFP-Rab7 (green), HA-CHMP2BWildtype (a, b), HA-CHMP2BIntron5 (c, d) or CHMP2BΔ10 (e, f) and then immunostained for CD63 to visualise endosomes (red) and HA. All CD63-positive endosomes within each of 40–50 cells per condition were scored for colocalization of EGFP-Rab7 by an experimenter blind to conditions. (B) There was a significant reduction in the percentage of endosomes that recruited EGFP-Rab7 in cells expressing CHMP2BIntron5 (P < 0.0001) or CHMP2BΔ10 (P < 0.001). Recruitment of endogenous Rab7 was similarly impaired (P < 0.0001 for both CHMP2BIntron5 and CHMP2BΔ10). Scale bar = 10 µm.
DISCUSSION
We have shown that FTD-specific CHMP2B mutant proteins cause enlarged late endosomes and disrupt endosome–lysosome fusion in cell culture models. Aberrant endosomes were also observed in patient brains and fibroblasts supporting the relevance of our in vitro overexpression model. We further dissect the function of the endocytic pathway to show that protein sorting is intact, indicating a potential pathogenic event downstream, at the level of fusion with lysosomes.
Cellular degradation of EGF, which is dependent on endosomal trafficking to the lysosome, was significantly decreased within the normal time course of this process. Furthermore, the amount of EGF reaching the lysosome was significantly reduced even after a 2 h chase. Western blotting analysis showed delayed degradation of the EGF receptor in response to EGF stimulation, this effect was marked over 2 h but less discernible at 3 h, suggesting that this process is impaired but not completely abrogated.
Sorting of EGFR onto the ILVs of MVBs is not functionally impaired, as shown by the normal time course of EGF-induced ERK1/2 activation and the presence of immunogold-labelled EGFR on the ILVs of abnormal endosomes in patient fibroblast cells. Knockdown of the ESCRT-III protein CHMP3 similarly prevents fusion of endosomes and lysosomes but not the deactivation of EGFR (15). CHMP3 knockdown cells could sequester EGFR into endosomal structures, even though they formed fewer ILVs than control cells. Similarly, abnormal structures present in FTD-3 patient fibroblasts internalized EGFR despite some abnormal membrane deformation and sparsity of ILVs within the enlarged lumen. In CHMP5 knockout mice, ILV formation is also intact despite impairment of lysosomal degradation (16). Taken together, this evidence suggests uncoupling of these two processes, and that some CHMP proteins may act downstream of ILV formation at the level of endosome–lysosome fusion. ILV formation, which includes deformation of the outer endosomal membrane and scission, is driven by the ESCRT-III complex (17–20). In vitro assays using the yeast homologues of the CHMP proteins, where one ESCRT-III subunit was omitted at a time, showed VPS2 (the homologue of CHMP2) to be unnecessary for both membrane deformation and scission (20). Instead, Vps2 was shown to be necessary for recruiting the AAA+ ATPase VPS4, required for disassembly and recycling of ESCRT-III subunits.
The negatively charged C terminus of CHMP proteins functions as an auto-inhibitory domain, interacting with the positively charged N terminus to keep the protein in a ‘closed’ conformation which masks the membrane binding properties of the N terminus (21–24). Deletion of the C terminus would therefore be predicted to produce a constitutively active protein which is likely to accumulate on endosomal membranes. Furthermore, the C terminus of CHMP2B is necessary for binding VPS4 (25,26); and CHMP2B truncation mutants lose this VPS4 binding domain so would be predicted to fail to dissociate from the endosomal membrane. Dissociation-incompetent CHMP2B may allow ILVs to form normally but also sequester other ESCRT-III subunits at the endosomal membrane so that fewer ILVs can be formed over time.
We propose a model where the accumulation of constitutively bound CHMP2B on endosomes may render them incapable of fusion with lysosomes. This latter process is not well understood, but it may be necessary for ESCRT-III to dissociate before fusion occurs, and/or the constitutive presence of ESCRT-III may mean that the endosomal membrane is unable to recruit factors required for fusion. We demonstrate impaired recruitment of Rab7 onto endosomes in CHMP2B mutant cells. This may be relevant to the neurodegenerative process as mutations in RAB7 cause the motor and sensory neuropathy Charcot–Marie–Tooth type 2B (27,28). Rab7 is a GTPase marking fusion-competent MVBs, which is recruited by the HOPS (homotypic vacuole fusion and vacuole protein sorting) and CORVET (class C core vacuole/endosome tethering) complexes (14,29). The proportion of endosomes that recruit Rab7 is reduced by approximately one-third in CHMP2B mutant cells but not completely diminished. This may explain why fusion is decreased and degradation delayed instead of completely abrogated. Interestingly, overexpression of EGFP-Rab7 did not rescue this deficit; the reduction in recruitment was the same for EGFP-Rab7 and endogenous Rab7. This is consistent with a model where mutant CHMP2B prevents a proportion of late endosomes from recruiting Rab7 and a greater concentration of Rab7 is therefore redundant to these recruitment-incompetent endosomes.
That cortical neurons are selectively vulnerable in the time course of disease remains a conundrum; although it may suggest that these cells are particularly dependent on some aspect of endosomal function for their survival. Normal trafficking of growth factors and their receptors is one possible candidate. Here we showed that epidermal growth factor remained undegraded over the normal time course within CHMP2B-positive enlarged compartments which failed to fuse with lysosomes. However, owing to its rapid sorting into the ILVs of MVBs, there was no prolongation of EGF signalling. Whereas EGF is rapidly internalized and activates its downstream signalling pathways over a short time course (30), other neuronal growth factor-receptor complexes, such as BDNF-TrkB, are known to signal from the endosomal membrane over a longer period (31–33). Therefore, further studies examining how these signalling endosomes are affected in neuronal models of CHMP2B mutations will be of great interest. The ESCRT complex is also involved in the endosomal–lysosomal degradation of neurotransmitter receptors (34,35), so impairment of receptor homeostasis and excitotoxic insult is another potential neurotoxic mechanism that could be mediated by mutant CHMP2B proteins. Finally, studies in Drosophila have recently shown that mutant CHMP2B can activate the Toll-like receptor pathway (36): a process which has been implicated in neurodegeneration (37).
In the present study, we have focused on endosomal dysfunction as the human pathology data so far support a major role for this system in the pathogenesis of FTD-3. It is of great interest that the endosome–lysosome pathway described here also converges with the autophagosomal degradation pathway (38–40) and that other cellular studies of CHMP2B overexpression have shown a deficit in the fusion of autophagosomes with lysosomes (41–43). Basal autophagy is required in neuronal cells to degrade organelles and long lived proteins which might otherwise accumulate with deleterious effects for the cell (44,45). It is likely that both of these highly related processes contribute to the pathogenesis of FTD-3. It is thought that similar mechanisms mediate autophagosome–lysosome and endosome–lysosome fusion (reviewed in 29), so data presented here for a putative role for impaired Rab7 recruitment may also be of relevance to the reported deficits in autophagy.
The findings presented here strongly support a model where C-terminal truncations of CHMP2B are pathogenic by a toxic gain-of-function which disrupts endocytic trafficking, producing deleterious effects for neuronal cells. Defects in endosomal trafficking have been implicated in a number of neurodegenerative diseases and the novel phenotypic and functional assays described here may aid diagnosis and the eventual screening and development of therapeutics.
MATERIALS AND METHODS
Materials
Antibodies used were: rat anti-HA tag (Roche, 1:500), mouse anti-CD63 (University of Iowa DSHB, 1:400), mouse anti-LAMP-1 (University of Iowa DSHB, 1:100), anti-rat-Alexa Fluor 546 (1:500, Invitrogen), anti-mouse-Alexa Fluor 546 (1:1000, Invitrogen), anti-mouse Alexa Fluor 633 (1:500, Invitrogen), anti-M6PR (1:100, Affinity Bioreagents), 1B5 anti-CD63 (a gift from Prof. Mark Marsh, UCL, 1:1000). EGF-488 was from Invitrogen. All cell culture reagents were from Invitrogen and all other reagents from Sigma-Aldrich unless otherwise stated. Human tissues were obtained at autopsy with consent for use in research. This study was approved by the UCL Institute of Neurology and National Hospital for Neurology and Neurosurgery Local Research Ethics Committee.
Plasmids
CHMP2B DNA constructs were used as previously described (6) or were N-terminally tagged with the HA epitope by PCR and TOPO® subcloned into the pcDNA3.1+ or the pTREx-DEST30 vectors (Invitrogen), and sequence verified. EGFP-Rab7 plasmid was a kind gift from Dr G. Schiavo, Cancer Research UK.
Cell lines
The SK-N-SH cell line (ECACC) and fibroblast cell lines were cultured in DMEM-4500 mg/ml glucose, 10% foetal bovine serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 100 U/ml penicillin and 100 µg/ml streptomycin. The TREx-293™cell line (Invitrogen) was additionally cultured with 5 µg/ml blasticidin. Mixed inducible-stable cell lines were generated by transfection with HA-tagged CHMP2BWildtype or CHMP2BIntron5 constructs in the pTREx-DEST30 vector using Lipofectamine 2000. Cells were then selected in 1 mg/ml geneticin and 5 µg/ml blasticidin. To induce CHMP2B expression, cells were incubated with 0.1 µg/ml tetracycline for 48 h prior to assay. Fibroblast cells lines were derived from forearm skin biopsies, tissue was dissected into small pieces and cultured in DMEM-4500 mg/ml glucose, 15% foetal calf serum, 2 mm l-glutamine, 50 U/ml penicillin and 50 µg/ml streptomycin.
Quantification of RNA transcripts
RNA was extracted from frozen FTD-3 frontal cortex using TRIzol (Invitrogen), treated with TURBO DNase (Ambion) and purified using an RNAeasy Mini Column (Qiagen). cDNA was generated using the Omniscript Reverse Transcription Kit (Qiagen). Relative levels of each CHMP2B transcript (Wildtype, Intron5 and Δ10) were determined using a CHMP2B Taqman probe for quantitative real-time PCR and allele-specific primers (Supplementary Material, Table S2). Within each sample, the amount of each transcript was determined by comparing to standard curves and then normalizing to the level of wild-type CHMP2B.
Western blotting
For the EGFR degradation assay, cells were starved for 1 h in serum-free medium with 1 µg/ml cyclohexamide, then 100 ng/ml EGF were added for the timepoints indicated. For the phospho-ERK1/2, cells were starved for 4 h, pulsed with 50 ng/ml EGF for 5 min, washed three times with pre-warmed PBS and pulsed with serum-free medium for the timepoints indicated. Cell lysates were western blotted as described previously (6). Quantitation was with ImageJ (NIH) or LeiCor Odyssey software.
Immunohistochemistry
Immunohistochemistry on formalin-fixed brain tissue was performed as described previously (46). Haematoxylin and eosin staining was done with conventional methods using Harris' haematoxylin.
Immunofluoresence
SK-N-SH cells were transfected with Lipofectamine 2000 (Invitrogen) according to standard protocols. For the EGF-488 degradation assay, 24 h post-transfection cells were starved in serum-free DMEM for 30 min, before the addition of 100 ng/ml EGF-488 (Molecular Probes) for the timepoints indicated. For the endosome–lysosome fusion assay, 24 h post-transfection cells were starved in serum-free DMEM with 100 µM leupeptin (Sigma) for 30 min, before the addition of 100 ng/ml EGF-488 for 30 min. Cells were then chased with leupeptin-DMEM containing 100 ng/ml unlabelled EGF for 2 h. Following any cellular assays, coverslips were processed for confocal analysis as previously described (6). Standard settings were used to allow comparisons between cells and between independent experiments. Fluorescence intensity of 12-bit images was assessed using the histogram function of LSM Meta, where all pixel values above background fluorescence were counted. Colocalization was also assessed using the histogram function; threshold fluorescence for each channel was set automatically for the region of interest. CD63-positive endosomes were scored for the presence or absence of endogenous Rab7 or EGFP-Rab7 by eye after thresholding all images to the same levels using Adobe Photoshop. All endosomes were counted within 40–50 cells per condition by an experimenter blind to conditions. For quantification of enlarged endosomes in patient fibroblasts blind counts were performed on 600 cells per line across three independent experiments using cells matched for passage number. A fibroblast was scored as having enlarged endosomes if one or more enlarged endosomes were present, as determined by eye after examination of several hundred normal cells to familiarize the counter with the normal range of endosome sizes. The same researcher performed all the cell counts to ensure consistency.
Electron microscopy
Cells were serum starved for 1 h and then stimulated with human EGF in the presence of 108 (anti-EGFR) gold for the specified length of time. Electron microscopy analysis was performed as described previously (13).
SUPPLEMENTARY MATERIAL
FUNDING
This work was funded by the Medical Research Council, UK, the Novo Nordisk Foundation and the Fund for Scientific Research—Flanders (FWO-V). Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council, UK.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ray Young for help preparing the figures, Professor Mark Marsh for the kind gift of anti-CD63 antibody and Dr Giampietro Schiavo for the kind gift of EGFP-Rab7 plasmid.
Conflict of Interest statement. None declared.
APPENDIX
The FReJA (Frontotemporal Dementia Research in Jutland Association) Consortium: Anders Gade, Tove Thusgaard, Memory Disorders Research Group, Copenhagen University Hospital Rigshospitalet, Copenhagen, Denmark. Susanne Gydesen, Psychiatric Centre Ballerup, Copenhagen University Hospital, Ballerup, Denmark. Elisabet Englund, Department of Pathology, University Hospital, Lund, Sweden. Martin Rossor, Department of Neurodegenerative Disease, UCL Institute of Neurology, London, UK.
REFERENCES
- 1.Snowden J.S., Neary D., Mann D.M. Frontotemporal dementia. Br. J. Psychiatry. 2002;180:140–143. doi: 10.1192/bjp.180.2.140. doi:10.1192/bjp.180.2.140. [DOI] [PubMed] [Google Scholar]
- 2.Lindquist S.G., Braedgaard H., Svenstrup K., Isaacs A.M., Nielsen J.E. Frontotemporal dementia linked to chromosome 3 (FTD-3)—current concepts and the detection of a previously unknown branch of the Danish FTD-3 family. Eur. J. Neurol. 2008;15:667–670. doi: 10.1111/j.1468-1331.2008.02144.x. doi:10.1111/j.1468-1331.2008.02144.x. [DOI] [PubMed] [Google Scholar]
- 3.Gydesen S., Brown J.M., Brun A., Chakrabarti L., Gade A., Johannsen P., Rossor M., Thusgaard T., Grove A., Yancopoulou D., et al. Chromosome 3 linked frontotemporal dementia (FTD-3) Neurology. 2002;59:1585–1594. doi: 10.1212/01.wnl.0000034763.54161.1f. [DOI] [PubMed] [Google Scholar]
- 4.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. doi:10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 5.Momeni P., Rogaeva E., Van D.V., Yuan W., Grafman J., Tierney M., Huey E., Bell J., Morris C.M., Kalaria R.N., et al. Genetic variability in CHMP2B and frontotemporal dementia. Neurodegener. Dis. 2006;3:129–133. doi: 10.1159/000094771. doi:10.1159/000094771. [DOI] [PubMed] [Google Scholar]
- 6.van der Zee J., Urwin H., Engelborghs S., Bruyland M., Vandenberghe R., Dermaut B., De P.T., Peeters K., Santens P., De Deyn P.P., et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum. Mol. Genet. 2008;17:313–322. doi: 10.1093/hmg/ddm309. doi:10.1093/hmg/ddm309. [DOI] [PubMed] [Google Scholar]
- 7.Urwin H., Ghazi-Noori S., Collinge J., Isaacs A. The role of CHMP2B in frontotemporal dementia. Biochem. Soc. Trans. 2009;37:208–212. doi: 10.1042/BST0370208. doi:10.1042/BST0370208. [DOI] [PubMed] [Google Scholar]
- 8.Katzmann D.J., Odorizzi G., Emr S.D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 2002;3:893–905. doi: 10.1038/nrm973. doi:10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 9.Holm I.E., Englund E., Mackenzie I.R., Johannsen P., Isaacs A.M. A reassessment of the neuropathology of frontotemporal dementia linked to chromosome 3. J. Neuropathol. Exp. Neurol. 2007;66:884–891. doi: 10.1097/nen.0b013e3181567f02. doi:10.1097/nen.0b013e3181567f02. [DOI] [PubMed] [Google Scholar]
- 10.Rohrer J.D., Ahsan R.L., Isaacs A.M., Nielsen J.E., Ostergaard L., Scahill R., Warren J.D., Rossor M.N., Fox N.C., Johannsen P. Presymptomatic generalized brain atrophy in frontotemporal dementia caused by CHMP2B mutation. Dement. Geriatr. Cogn. Disord. 2009;27:182–186. doi: 10.1159/000200466. doi:10.1159/000200466. [DOI] [PubMed] [Google Scholar]
- 11.Eskildsen S.F., Ostergaard L.R., Rodell A.B., Ostergaard L., Nielsen J.E., Isaacs A.M., Johannsen P. Cortical volumes and atrophy rates in FTD-3 CHMP2B mutation carriers and related non-carriers. Neuroimage. 2009;45:713–721. doi: 10.1016/j.neuroimage.2008.12.024. doi:10.1016/j.neuroimage.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 12.Eden E.R., White I.J., Futter C.E. Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem. Soc. Trans. 2009;37:173–177. doi: 10.1042/BST0370173. doi:10.1042/BST0370173. [DOI] [PubMed] [Google Scholar]
- 13.Futter C.E., Pearse A., Hewlett L.J., Hopkins C.R. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell. Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. doi:10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. doi:10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 15.Bache K.G., Stuffers S., Malerod L., Slagsvold T., Raiborg C., Lechardeur D., Walchli S., Lukacs G.L., Brech A., Stenmark H. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. doi:10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shim J.H., Xiao C., Hayden M.S., Lee K.Y., Trombetta E.S., Pypaert M., Nara A., Yoshimori T., Wilm B., Erdjument-Bromage H., et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell. Biol. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. doi:10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson P.I., Roth R., Lin Y., Heuser J.E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell. Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. doi:10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saksena S., Wahlman J., Teis D., Johnson A.E., Emr S.D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. doi:10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teis D., Saksena S., Emr S.D. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. doi:10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J.H. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. doi:10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim S., Kimpler L.A., Hanson P.I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. doi:10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 22.Zamborlini A., Usami Y., Radoshitzky S.R., Popova E., Palu G., Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl Acad. Sci. USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. doi:10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y., Kimpler L.A., Naismith T.V., Lauer J.M., Hanson P.I. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. doi:10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 24.Whitley P., Reaves B.J., Hashimoto M., Riley A.M., Potter B.V., Holman G.D. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J. Biol. Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. doi:10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 25.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D.J., Perisic O., Emr S.D., Williams R.L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. doi:10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 26.Stuchell-Brereton M.D., Skalicky J.J., Kieffer C., Karren M.A., Ghaffarian S., Sundquist W.I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. doi:10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 27.Spinosa M.R., Progida C., De Luca A., Colucci A.M., Alifano P., Bucci C. Functional characterization of Rab7 mutant proteins associated with Charcot–Marie–Tooth type 2B disease. J. Neurosci. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. doi:10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhoeven K., De Jonghe P., Coen K., Verpoorten N., Auer-Grumbach M., Kwon J.M., FitzPatrick D., Schmedding E., De Vriendt E., Jacobs A., et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot–Marie–Tooth type 2B neuropathy. Am. J. Hum. Genet. 2003;72:722–727. doi: 10.1086/367847. doi:10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luzio J.P., Pryor P.R., Bright N.A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell. Biol. 2007;8:622–632. doi: 10.1038/nrm2217. doi:10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 30.Wu C., Lai C.F., Mobley W.C. Nerve growth factor activates persistent Rap1 signaling in endosomes. J. Neurosci. 2001;21:5406–5416. doi: 10.1523/JNEUROSCI.21-15-05406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginty D.D., Segal R.A. Retrograde neurotrophin signaling: Trk-ing along the axon. Curr. Opin. Neurobiol. 2002;12:268–274. doi: 10.1016/s0959-4388(02)00326-4. doi:10.1016/S0959-4388(02)00326-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhou P., Porcionatto M., Pilapil M., Chen Y., Choi Y., Tolias K.F., Bikoff J.B., Hong E.J., Greenberg M.E., Segal R.A. Polarized signaling endosomes coordinate BDNF-induced chemotaxis of cerebellar precursors. Neuron. 2007;55:53–68. doi: 10.1016/j.neuron.2007.05.030. doi:10.1016/j.neuron.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hibbert A.P., Kramer B.M., Miller F.D., Kaplan D.R. The localization, trafficking and retrograde transport of BDNF bound to p75NTR in sympathetic neurons. Mol. Cell. Neurosci. 2006;32:387–402. doi: 10.1016/j.mcn.2006.06.001. doi:10.1016/j.mcn.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Kantamneni S., Holman D., Wilkinson K.A., Correa S.A., Feligioni M., Ogden S., Fraser W., Nishimune A., Henley J.M. GISP binding to TSG101 increases GABA receptor stability by down-regulating ESCRT-mediated lysosomal degradation. J. Neurochem. 2008;107:86–95. doi: 10.1111/j.1471-4159.2008.05580.x. doi:10.1111/j.1471-4159.2008.05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kantamneni S., Holman D., Wilkinson K.A., Nishimune A., Henley J.M. GISP increases neurotransmitter receptor stability by down-regulating ESCRT-mediated lysosomal degradation. Neurosci. Lett. 2009;452:106–110. doi: 10.1016/j.neulet.2009.01.011. doi:10.1016/j.neulet.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmad S.T., Sweeney S.T., Lee J.A., Sweeney N.T., Gao F.B. Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc. Natl Acad. Sci. USA. 2009;106:12168–12173. doi: 10.1073/pnas.0903134106. doi:10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okun E., Griffioen K.J., Lathia J.D., Tang S.C., Mattson M.P., Arumugam T.V. Toll-like receptors in neurodegeneration. Brain Res. Rev. 2009;59:278–292. doi: 10.1016/j.brainresrev.2008.09.001. doi:10.1016/j.brainresrev.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordon P.B., Seglen P.O. Prelysosomal convergence of autophagic and endocytic pathways. Biochem. Biophys. Res. Commun. 1988;151:40–47. doi: 10.1016/0006-291x(88)90556-6. doi:10.1016/0006-291X(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky D.J., Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell. Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. doi:10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Tooze J., Hollinshead M., Ludwig T., Howell K., Hoflack B., Kern H. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J. Cell. Biol. 1990;111:329–345. doi: 10.1083/jcb.111.2.329. doi:10.1083/jcb.111.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. ESCRT-III Dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. doi:10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 42.Lee J.A., Gao F.B. Roles of ESCRT in autophagy-associated neurodegeneration. Autophagy. 2008;4:230–232. doi: 10.4161/auto.5384. [DOI] [PubMed] [Google Scholar]
- 43.Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerod L., Fisher E.M.C., Isaacs A., Brech A., Stenmark H., Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell. Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. doi:10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. doi:10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 45.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 46.Isaacs A.M., Powell C., Webb T.E., Linehan J.M., Collinge J., Brandner S. Lack of TAR-DNA binding protein-43 (TDP-43) pathology in human prion diseases. Neuropathol. Appl. Neurobiol. 2008;34:446–456. doi: 10.1111/j.1365-2990.2008.00963.x. doi:10.1111/j.1365-2990.2008.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.