Abstract
Autosomal recessive limb-girdle muscular dystrophy type 2G (LGMD2G) is an adult-onset myopathy characterized by distal lower limb weakness, calf hypertrophy and progressive decline in ambulation. The disease is caused by mutations in Tcap, a z-disc protein of skeletal muscle, although the precise mechanisms resulting in clinical symptoms are unknown. To provide a model for preclinical trials and for mechanistic studies, we generated knockout (KO) mice carrying a null mutation in the Tcap gene. Here we present the first report of a Tcap KO mouse model for LGMD2G and the results of an investigation into the effects of Tcap deficiency on skeletal muscle function in 4- and 12-month-old mice. Muscle histology of Tcap-null mice revealed abnormal myofiber size variation with central nucleation, similar to findings in the muscles of LGMD2G patients. An analysis of a Tcap binding protein, myostatin, showed that deletion of Tcap was accompanied by increased protein levels of myostatin. Our Tcap-null mice exhibited a decline in the ability to maintain balance on a rotating rod, relative to wild-type controls. No differences were detected in force or fatigue assays of isolated extensor digitorum longus (EDL) and soleus (SOL) muscles. Finally, a mechanical investigation of EDL and SOL indicated an increase in muscle stiffness in KO animals. We are the first to establish a viable KO mouse model of Tcap deficiency and our model mice demonstrate a dystrophic phenotype comparable to humans with LGMD2G.
INTRODUCTION
Tcap [previously termed telethonin (1)] is a muscle-specific titin-capping protein at the z-disc thought to exert both regulatory and structural roles in skeletal muscle (2–6). In cultured skeletal muscle cells, Tcap knockdown resulted in a marked decrease in the expression of myogenic regulatory factors, MyoD and myogenin, suggesting a regulatory role of Tcap during muscle growth (4). Tcap provides structural support to the sarcomere by linking the N-terminus of titin to other z-disc proteins. In addition to structural support, Tcap is thought to function in the z-disc protein complex as a stretch sensor by closely interacting with titin (7) and responding to titin-generated tension. Recent experiments in Tcap-deficient zebrafish suggest that mechanical forces can regulate Tcap transcription during development (8). How mechanical forces might change within the sarcomere in response to Tcap deficiency have not previously been studied in mammals.
In humans, autosomal recessive mutations disrupting the carboxy terminus of Tcap result in clinical pathology termed limb-girdle muscular dystrophy type 2G (LGMD2G) (9). Although the disease is rare, LGMD2G exists worldwide (10) and may escape specific diagnosis (11). Affected patients develop weakness in the distal lower limb muscles, calf hypertrophy and progressive decline in ambulation; histopathology of affected muscles shows fiber size variation with central nucleation (10). The mechanisms responsible for clinical decline and muscle pathology in LGMD2G patients are generally unknown. Lack of translational research progress for this condition is due, in part, to the lack of available animal models of the disease. Although a Tcap-deficient zebrafish model has been reported (8), no mammalian Tcap knockout (KO) models have been described. To provide a new model for preclinical trials and mechanistic studies, we generated mice homozygous for a null mutation in the Tcap gene (Tcap).
In this article, we present the first results of a molecular and phenotypic characterization of skeletal muscle in a novel Tcap KO mouse. Our objective was to generate Tcap-null mice and to characterize their skeletal muscle phenotype. Targeted deletion of the mouse Tcap gene resulted in a mild dystrophic phenotype with histopathological features similar to those described from muscle biopsies of LGMD2G patients. Analysis of a Tcap binding partner, myostatin, revealed increased protein levels of this pro-hypotrophic factor in skeletal muscle. Functional analysis revealed that Tcap deficiency leads to an early decline in motor ability. Surprisingly, we found that skeletal muscles of Tcap-null mice maintain their force-generating capacity but display an increase in passive muscle stiffness. Together, our results provide a framework for future translational and mechanistic studies in a novel mammalian model of Tcap deficiency.
RESULTS
Generation of Tcap KO mice
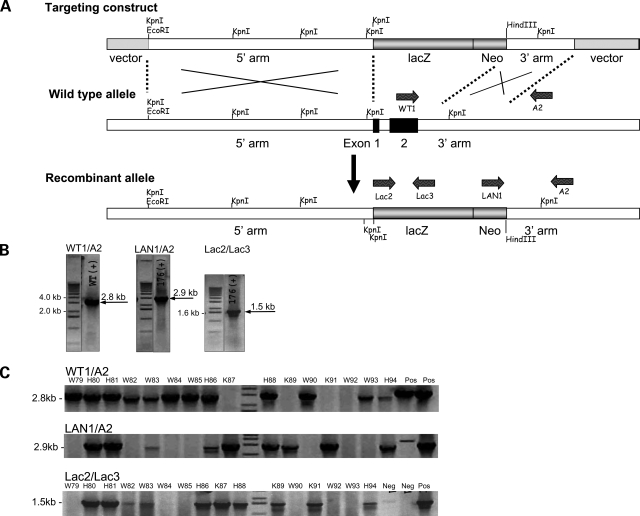
The Tcap gene was disrupted in embryonic stem (ES) cells by replacing the putative promoter region and exons 1 and 2 with a LacZ/Neo cassette (Fig. 1A). ES cells were selected by antibiotic resistance and surviving clones were expanded for PCR analysis to identify recombinant ES clones (Fig. 1B). Clones were further confirmed by sequencing (not shown) prior to microinjection into blastocyst-stage embryos (3.5 dpc). Blastocyst injection yielded viable F1 KO mice. Genomic DNA of F1 mice was probed with high-fidelity PCR primers (Fig. 1C) for the recombinant (KO) allele and to confirm the presence of the LacZ cassette. Deletion of the Tcap gene product was further confirmed in the skeletal and cardiac muscles of mice by immunoblot using anti-Tcap polyclonal and monoclonal antibodies (Fig. 2). Tcap was not detected in immunoblots of liver, kidney or brain lysates (not shown).
Figure 1.
Targeted disruption of the Tcap gene locus and establishment of mouse lines expressing normal and mutant Tcap. (A) Schematic diagram illustrating the targeting construct, the WT Tcap allele, and the predicted recombinant allele with exons 1 and 2 replaced by a lacZ/Neo cassette. The 15.4-kb targeting construct included the plasmid vector, the long 5′ and short 3′ homology arms, a lacZ reporter gene and neomycin-resistance (Neo) cassette. Homologous recombination resulted in replacement of exons 1 and 2 of the Tcap gene locus with a DNA fragment containing a lacZ/Neo gene. (B) PCR primers (LAN1/A2) external and internal (Lac2/Lac3) to the targeting construct (as shown in A) were used to characterize the targeted locus in ES cells. The 3′ homology arm of the targeting construct was amplified by high-fidelity PCR from genomic DNA of ES cells using the primers LAN1 and A2. LAN1 anneals to the recombinant allele inside the Neo cassette and A2 anneals 3′ to the SA, outside the region used to create the targeting construct. LAN1/A2 and Lac2/3 primer sets amplify fragments within the KO allele measuring 2.9 and 1.5 kb, respectively; WT1/A2 amplifies a 2.8 kb fragment within the WT allele. (C) Detection of the recombinant gene using PCR primers (shown in A) of mouse tail DNA from each of the founder lines. Neg, negative control; Pos, positive control, W, wild-type; H, heterozygote; K, knockout.
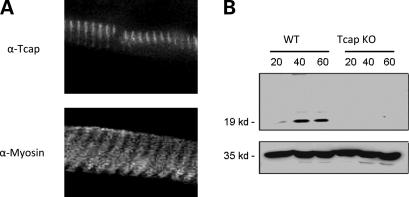
Figure 2.
Immunohistochemistry and western blot of WT and Tcap KO mouse hindlimb muscles. (A) Staining pattern in longitudinal thin muscle sections reveals thin bands (top panel), consistent with Tcap's z-disc localization, whereas thick bands are observed with myosin staining (bottom panel). Fluorescence imaging was accomplished using primary monoclonal antibodies against myosin heavy chain (MF-20) and full-length Tcap. Bar = 50 µm. (B) Representative western immunoblot of WT and Tcap KO muscle lysates. Gastrocnemius lysate from 2-month-old mice was loaded at 20, 40 and 60 µg per lane (top panel). Anti-GAPDH antibody was used as a loading control (bottom panel).
Immunohistochemical staining of thin longitudinal wild-type (WT) muscle sections probed with anti-Tcap antibodies revealed a regular repeating pattern of thin bands whereas a repeating pattern of thick bands was observed when probed with anti-myosin antibodies (Fig. 2A). Sections from brain, liver and kidney did not display Tcap staining (not shown). Together these findings demonstrate the specificity of the Tcap antibody for the z-disc of WT skeletal muscle and demonstrate the absence of Tcap staining in KO muscles.
Skeletal muscles of Tcap KO mice develop normally
Size and mass of mice were regularly assessed during the first 6 months, and relative to their WT littermates, KO mouse muscles developed normally. Necropsy of several hindlimb muscles [quadriceps, gastrocnemius, anterior tibialis, extensor digitorum longus (EDL)] did not reveal gross muscle atrophy during the first 12 months (not shown). These findings indicate that Tcap protein is not critical for normal skeletal muscle growth or development in mice.
Central nucleation, abnormal nicotinamide adenine dinucleotide, reduced staining and hypotrophic myofibers in Tcap-deficient muscle
Histology
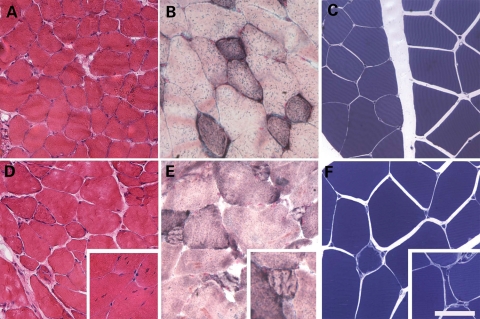
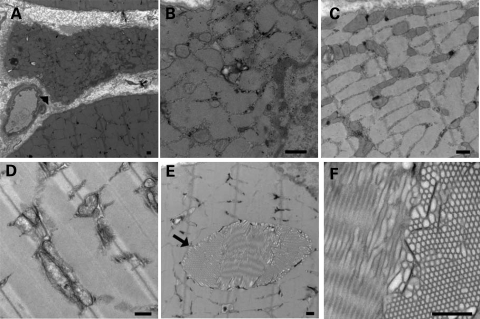
Hematoxylin and eosin-stained cross sections of gastrocnemius muscle were analyzed according to the guidelines suggested by the TREAT-NMD consortium (12). Scattered small atrophic fibers in the KO mice were observed with coarse basophilic inclusions in some small fibers (Fig. 3A and D), resulting in a 33% decrease in minimum fiber diameter when comparing myofibers from WT (19.6 µm) and KO (13.0 µm) mice. No significant differences between WT and TCAP KO animals were detected with respect to mean or maximum fiber diameter. KO mice also demonstrated a significantly increased number of centrally nucleated fibers (8.5 ± 2.0%, P < 0.05) in comparison to WT animals (1.1 ± 0.7%). There was no observed increase in endomysial fibrosis, and necrotic fibers were not present. Nicotinamide adenine dinucleotide, reduced (NADH) staining (Fig. 3B and E) shows that some small fibers in KO muscles have areas of increased NADH staining surrounded by areas devoid of staining. Fibers with this pattern were not seen in WT animals. Toluidine blue staining of Epon-embedded quadriceps muscle further illustrated the presence of scattered atrophic fibers in comparison to WT animals (Fig. 3C and F). Additionally, focal areas of disrupted cytoplasmic architecture were observed in myofibers in areas void of contractile filaments or mitochondria (not shown). Electron microscopy of KO gastrocnemius muscles revealed slightly enlarged and prominent mitochondria in atrophic fibers (Fig. 4A–C) whereas contractile filaments were appropriately organized (Fig. 4D). Tubular aggregates in KO gastrocnemius muscles (Fig. 4E and F), were less numerous in the quadriceps muscles (not shown). Aggregates were also seen in the gastrocnemius muscles of some age-matched WT animals but were not seen in small, hypotrophic fibers of KO mice. These findings demonstrate that KO mouse muscles develop a skeletal myopathy comparable to findings reported in patients with LGMD2G (9–11).
Figure 3.
Tcap KO mouse muscles reveal atrophic scattered fibers with central nucleation. Isopentane-frozen gastrocnemius muscles from WT (A–C) and Tcap KO (D–F) mice were stained with hematoxylin and eosin (A, D) or NADH (B, E), showing scattered atrophic fibers in the Tcap KO mice, with coarse basophilic inclusions in some small fibers and an increase in the number of centrally nucleated fibers (D, inset). Toluidine blue staining of Epon-embedded quadriceps muscle also reveals atrophic fibers in Tcap KO mice (C, F). Scale Bar = 100 µm in A and D, and 200 µm in B, C, E and F.
Figure 4.
Tcap KO mouse muscles display ultrastructural abnormalities. Electron microscopy of Tcap KO muscles revealed prominent mitochondria in atrophic fibers (arrowhead) (A–C) whereas contractile filaments are appropriately organized (D). Tubular aggregates (arrow) were also present in numerous muscle fibers in the Tcap KO muscles (E, F). Scale bar = 500 nm.
Microarray analyses in Tcap-deficient muscle
To test the hypothesis that genes differentially expressed in cultured C2C12 muscle cells after siRNA-mediated Tcap knockdown (4) are similarly expressed in Tcap KO muscles, we performed microarray analysis (Table 1) of both WT and KO gastrocnemius muscle RNA. RNA derived from biological replicates in each group [WT (n = 3) and KO (n = 3)] was used to query mouse whole-genome microarrays containing probes representing 41 000 genes and transcripts. Statistical analysis with the student's t-test, using a significance cut-off of P ≤ 0.05, led to the identification of 136 significantly changed transcripts between the two groups (Table 1). Of these, none corresponded to the genes we previously identified in an earlier transcript analysis Tcap knockdown in cultured C2C12 cells. However, in the present study, several genes associated with pathogenic mechanisms in muscular dystrophies (cardiac ryanodine receptor 2 (14), nitric oxide synthase 2 (13,26) ankyrin repeat (22) and nuclear factor-kappa B (NFkB) repressing factor genes) were significantly (P < 0.05) upregulated in Tcap KO muscles (Table 1).
Table 1.
mRNAs differentially expresseda in age-matched Tcap KO versus WT gastrocnemius muscle
| Ratio | Direction | P-value | Gene identifier | Gene name |
|---|---|---|---|---|
| 3.55 | Up | 0.032 | AK043675 | Ryanodine receptor 2, cardiac |
| 1.7 | Up | 0.049 | NM_010927 | Nitric oxide synthase 2, inducible, macrophage |
| 1.6 | Up | 0.047 | AK171682 | Ankyrin repeat, family A (RFXANK-like), 2 |
| 1.55 | Up | 0.039 | NM_029891 | NF-kappaB repressing factor |
| 1.25 | Down | NS | NM_010834 | Myostatin |
| 1.05 | Up | NS | NM_008046 | Follistatin |
| 1.02 | Down | NS | NM_008047 | Follistatin-like 1 |
| 1.08 | Up | NS | NM_031380 | Follistatin-like 3 |
| 1.26 | Down | NS | NM_177059 | Follistatin-like 4 |
| 1.07 | Up | NS | AK047518 | Follistatin-like 5 |
| 1.04 | Up | NS | NM_178673 | Follistatin-like 5 |
| 1.06 | Down | NS | NM_010514 | IGF-2 |
| 1.27 | Up | NS | NM_010866 | MyoD1 Myogenic Differentiation 1 |
| 1.01 | Up | NS | NM_031189 | Myogenin |
| 1.04 | Up | NS | NM_134092 | Mdm2, transformed 3T3 cell double minute p53 binding protein |
| 1.01 | Up | NS | NM_178939 | P53 and DNA damage regulated 1 |
| 1.21 | Up | NS | NM_011035 | P21 (CDKN1A)-activated kinase 1 |
| 1.09 | Up | NS | NM_177326 | P21 (CDKN1A)-activated kinase 2 |
| 1.29 | Down | NS | NM_008778 | P21 (CDKN1A)-activated kinase 3 |
| 1.15 | Down | NS | NM_027470 | P21 (CDKN1A)-activated kinase 4 |
| 1.22 | Up | NS | AK028788 | P21 (CDKN1A)-activated kinase 6 |
| 1.57 | Up | NS | NM_172858 | P21 (CDKN1A)-activated kinase 7 |
| 1.06 | Up | NS | NM_175246 | Smad nuclear interacting protein 1 |
| 1 | Up | NS | NM_001038627 | SMAD specific E3 ubiquitin protein ligase 1 |
| 1.21 | Up | NS | NM_025481 | SMAD specific E3 ubiquitin protein ligase 2 |
| 3.03 | Up | 0.029 | AK016833 | Transcribed locus, strongly similar to XP_204338.2 PREDICTED: hypothetical protein LOC71159 isoform 1 [Mus musculus] |
| 3.01 | Up | 0.035 | AK014613 | Amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| 2.95 | Up | 0.023 | AK077587 | CDNA clone IMAGE:3491339 |
| 2.87 | Up | 0.046 | NM_177742 | Tripartite motif family-like 1 |
| 2.78 | Up | 0.030 | AK053064 | gb|Mus musculus 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930038O08 product:unclassifiable, full insert sequence [AK053064] |
| 2.72 | Up | 0.016 | NM_177093 | Leucine rich repeat containing 58 |
| 2.65 | Up | 0.048 | AK054366 | Zinc finger, MIZ-type containing 1 |
| 2.56 | Up | 0.042 | NM_177568 | Phospholipase C, beta 2 |
| 2.48 | Up | 0.027 | NM_175696 | RIKEN cDNA C530028O21 gene |
| 2.43 | Up | 0.012 | NM_181858 | CD59b antigen |
| 2.42 | Up | 0.040 | NM_146780 | Olfactory receptor 715 |
| 2.42 | Up | 0.016 | BB161836 | Transcribed locus |
| 2.42 | Down | 0.024 | NM_001025607 | Predicted gene, EG382275 |
| 2.41 | Up | 0.027 | AK044518 | RIKEN cDNA A930016P21 gene |
| 2.37 | Up | 0.029 | NM_021885 | Tubby candidate gene |
| 2.22 | Up | 0.017 | NM_011164 | Prolactin |
| 2.19 | Up | 0.042 | NM_015728 | Solute carrier family 33 (acetyl-CoA transporter), member 1 |
| 2.16 | Up | 0.041 | AF175408 | MAD homolog 9 (Drosophila) |
| 2.08 | Up | 0.026 | NM_019508 | Interleukin 17B |
| 2.07 | Up | 0.042 | NM_147061 | Olfactory receptor 691 |
| 2.04 | Up | 0.006 | AK054195 | gb|Mus musculus 2 days pregnant adult female oviduct cDNA, RIKEN full-length enriched library, clone:E230027A21 product:purinergic receptor P2X-like 1, orphan receptor, full insert sequence [AK054195] |
| 2.02 | Up | 0.023 | AK029221 | Transcribed locus |
| 2.01 | Down | 0.026 | NM_019743 | RING1 and YY1 binding protein |
| 2 | Up | 0.030 | NM_013912 | Apelin |
| 1.98 | Down | 0.049 | NM_001001884 | Riken cDNA C230021P08 gene |
| 1.98 | Up | 0.010 | NM_009520 | Wingless related MMTV integration site 2b |
| 1.98 | Up | 0.042 | NM_053271 | Regulating synaptic membrane exocytosis 2 |
| 1.96 | Down | 0.050 | NM_031388 | Ubiquitin specific peptidase 26 |
| 1.95 | Up | 0.043 | AK166269 | Hedgehog interacting protein-like 1 |
| 1.94 | Down | 0.034 | NM_030218 | RIKEN cDNA 9130017N09 gene |
| 1.94 | Down | 0.016 | AK049328 | gb|Mus musculus ES cells cDNA, RIKEN full-length enriched library, clone:C330024F03 product:unclassifiable, full insert sequence [AK049328] |
| 1.94 | Up | 0.009 | AK084222 | gb|Mus musculus 12 days embryo eyeball cDNA, RIKEN full-length enriched library, clone:D230010D07 product:unclassifiable, full insert sequence [AK084222] |
| 1.93 | Up | 0.025 | NM_008260 | Forkhead box A3 |
| 1.93 | Up | 0.033 | BC045114 | CDNA sequence BC016201 |
| 1.92 | Up | 0.036 | AK048563 | Transcribed locus, strongly similar to NP_891550.1 ADAM metallopeptidase with thrombospondin type 1 motif, 9 preproprotein [H] |
| 1.92 | Up | 0.036 | TC1744477 | tc|Q9Y649_HUMAN (Q9Y649) GW128 (YWHAB protein), partial (16%) [TC1744477] |
| 1.91 | Up | 0.016 | AK033743 | Lipocalin 6 |
| 1.88 | Up | 0.031 | AK034064 | Dedicator of cytokinesis 10 |
| 1.87 | Down | 0.043 | NM_029881 | RIKEN cDNA C030003D03 gene |
| 1.87 | Up | 0.018 | NM_019694 | Leucine zipper-EF-hand containing transmembrane protein 1 |
| 1.87 | Up | 0.022 | NM_001099631 | SH2 domain-containing 5 |
| 1.8 | Up | 0.019 | AK041796 | Transcribed locus, strongly similar to NP_006339.2 component of oligomeric golgi complex 5 isoform 1 [Homo sapiens] |
| 1.77 | Up | 0.019 | AK016421 | RIKEN cDNA 4931400O07 gene |
| 1.76 | Down | 0.005 | NM_001112729 | CDNA sequence BC019943 |
| 1.75 | Down | 0.022 | NM_027893 | Poliovirus receptor-related 4 |
| 1.75 | Up | 0.036 | TC1650944 | tc|Q3TLP8_MOUSE (Q3TLP8) Mammary gland RCB-0527 Jyg-MC(B) cDNA, RIKEN full-length enriched library, clone:G930005L08 product:RAS-related C3 botulinum substrate 1, full insert sequence, partial (9%) [TC1650944] |
| 1.75 | Up | 0.042 | NM_145959 | DNA segment, Chr 15, ERATO Doi 621, expressed |
| 1.74 | Up | 0.008 | AK142481 | Transcribed locus, weakly similar to XP_001479644.1 PREDICTED: hypothetical protein [Mus musculus] |
| 1.74 | Up | 0.015 | ENSMUST00000092674 | ens|HBS1-like protein [Source:Uniprot/SWISSPROT;Acc:Q69ZS7] [ENSMUST00000092674] |
| 1.74 | Up | 0.004 | NM_001039655 | RAS protein-specific guanine nucleotide-releasing factor 1 |
| 1.73 | Up | 0.045 | NM_029663 | Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| 1.73 | Up | 0.042 | AK050374 | Tocopherol (alpha) transfer protein |
| 1.72 | Down | 0.022 | NM_026390 | UBX domain-containing 2 |
| 1.72 | Up | 0.038 | NM_001081012 | RIKEN cDNA 4930473A06 gene |
| 1.71 | Up | 0.039 | AK028249 | Ubiquitin specific peptidase 27, X chromosome |
| 1.68 | Down | 0.012 | AK031662 | gb|Mus musculus 13 days embryo male testis cDNA, RIKEN full-length enriched library, clone:6030474N09 product:unclassifiable, full insert sequence [AK031662] |
| 1.65 | Up | 0.035 | NM_147032 | Olfactory receptor 705 |
| 1.64 | Up | 0.036 | NM_001008231 | Dishevelled associated activator of morphogenesis 2 |
| 1.64 | Up | 0.013 | AK081940 | Transcribed locus |
| 1.64 | Up | 0.038 | NM_011218 | Protein tyrosine phosphatase, receptor type, S |
| 1.63 | Up | 0.046 | BC037468 | gb|Mus musculus cDNA clone IMAGE:4223409 [BC037468] |
| 1.63 | Up | 0.043 | AK046200 | gb|Mus musculus adult male corpora quadrigemina cDNA, RIKEN full-length enriched library, clone:B230350K09 product:unclassifiable, full insert sequence [AK046200] |
| 1.62 | Down | 0.037 | NM_022007 | FXYD domain-containing ion transport regulator 7 |
| 1.62 | Up | 0.010 | NM_173427 | Kelch domain-containing 7A |
| 1.6 | Up | 0.042 | AK053298 | gb|Mus musculus 0 day neonate eyeball cDNA, RIKEN full-length enriched library, clone:E130008E24 product:surfactant associated protein B, full insert sequence [AK053298] |
| 1.6 | Up | 0.022 | AK005365 | G protein-coupled receptor kinase-interactor 2 |
| 1.6 | Up | 0.044 | NM_181400 | WD repeat domain 47 |
| 1.6 | Up | 0.034 | AK080744 | PIH1 domain-containing 2 |
| 1.6 | Up | 0.030 | NM_026563 | Serologically defined colon cancer antigen 3 |
| 1.59 | Down | 0.049 | NM_176835 | RIKEN cDNA 2810451A06 gene |
| 1.59 | Down | 0.048 | AK046930 | Transcribed locus |
| 1.57 | Up | 0.010 | NM_010154 | V-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) |
| 1.56 | Up | 0.039 | NM_031199 | Transforming growth factor alpha |
| 1.55 | Up | 0.041 | AK079838 | Glutaminase |
| 1.55 | Up | 0.047 | AK133637 | Protocadherin 17 |
| 1.54 | Up | 0.031 | NM_009215 | Somatostatin |
| 1.54 | Up | 0.049 | NM_009288 | Serine/threonine kinase 10 |
| 1.54 | Up | 0.014 | AK139092 | CDNA clone IMAGE:574497 |
| 1.53 | Up | 0.047 | NM_001081182 | Atpase, class I, type 8B, member 2 |
| 1.51 | Down | 0.041 | AK054138 | SUMO/sentrin specific peptidase 6 |
| 1.5 | Up | 0.048 | NM_207683 | Phosphatidylinositol 3-kinase, C2 domain-containing, gamma polypeptide |
Changes (ratio) are relative to control where control = 1.00; Up, Up-regulation; Down, Down-regulation. Student's t-test values are shown, n = 6 microarrays (three per group).
aMuscle regulatory factors and cell cycle regulators of special interest that were assayed but not significantly different (NS) between KO and WT tissue are included.
Elevated myostatin protein expression in Tcap-deficient muscle
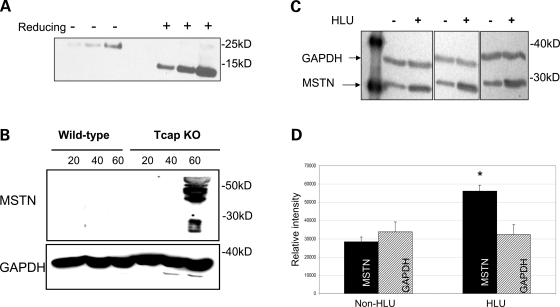
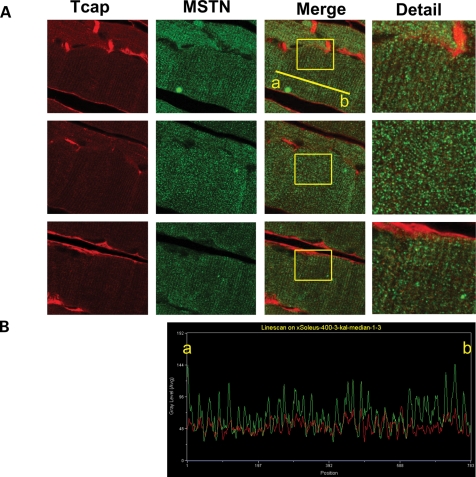
Differential expression of myostatin and follistatin from microarray results (Table 1) led us to address the question of whether Tcap deficiency might result in altered myostatin protein levels. Myostatin immunoblots performed in KO gastrocnemius muscle lysates revealed marked 45- and 26-kDa bands that were not detected in WT lysates (Fig. 5B). These findings indicate that 45-kDa precursor (17) and 26-kDa processed myostatin (17) levels are increased in KO mouse muscle relative to WT controls. To further investigate a potential association between Tcap and myostatin in normal skeletal mouse muscle, thin longitudinal sections of WT muscle were double-stained with anti-Tcap and anti-myostatin antibodies (Fig. 6A). Line-scan analysis (Fig. 6B) indicated that the Tcap and myostatin stains overlapped 20.5 ± 0.9%. Together, our immunoblot and immunohistochemical findings suggest an association between Tcap and myostatin, and indicate that Tcap deficiency influences myostatin protein expression in skeletal muscle.
Figure 5.
Myostatin protein expression is increased in Tcap KO mouse muscles. Immunoblots were performed on muscle lysates from WT and Tcap KO gastrocnemius muscles probed with an anti-myostatin antibody. (A) To evaluate the myostatin antibody, recombinant purified human myostatin protein was subjected to both non-reducing (−) and reducing (+) conditions as indicated. One hundred and twenty-five, 250 and 500 ng purified myostatin protein were loaded in each condition. Bands at ∼26 and ∼13 kDa were clearly seen, indicating the sensitivity of the antibody to detect intact and cleaved C-terminal dimer myostatin proteins, respectively. (B) Both the 45-kDa precursor and 26-kDa processed myostatin bands were observed, but only in Tcap KO muscle lysates and not in WT muscles. (C) To assess the sensitivity of myostatin antibody in skeletal muscle tissue, female rats (n = 3) were suspended by their tails (HLU) for four weeks, SOL muscles were subsequently removed and muscle lysates were probed with anti-myostatin antibody. Intensity of myostatin bands (∼26 kDa) increased relative to GAPDH bands (∼36 kDa) during HLU compared with non-HLU controls. Densitometry analysis (D) of MSTN bands demonstrated significantly greater intensity (P < 0.05) relative to GAPDH bands in the HLU group compared with non-HLU controls. Equal protein loading of lanes was confirmed by intensity of GAPDH bands.
Figure 6.
Tcap and myostatin associate in mouse skeletal muscle tissue. HLU was used to increase myostatin expression in skeletal muscle. Adult female mice (n = 2) were suspended by their tails (HLU) for 2 weeks. (A) Three independent thin sections of SOL muscles were stained with primary antibodies directed against either Tcap or myostatin and conjugated to red or green fluorescent secondary antibodies, respectively, as indicated. In merged images (second column from the right) the relative amount of color overlap was determined with computer software. The mean (±SD) percent overlap between Tcap and myostatin was 20.5 ± 0.9%, suggesting a biological association between the two proteins. The far-right column shows detail of the area indicated by the box. (B) Representative line-scan analysis of fluorescent intensity. Points along the line drawn on the image connecting two points, labeled ‘a’ and ‘b’, were sampled to generate an intensity profile of two fluorescent colors, red (Tcap) and green (myostatin). Peak intensity for both Tcap and myostatin line scans correlate with the regular repeating z-disc striations observed in longitudinal thin sections of muscle.
Impaired rotor rod performance in Tcap KO mice
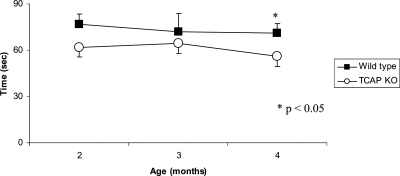
To measure the impact of Tcap deficiency on in vivo muscle performance in mice, we performed a rotor rod test at various times in young mice. The ability to maintain balance on a rotating rod was compared between WT and KO mice by measuring the length of time mice stayed on the rod before falling. Performance was assessed over 90 days beginning at 2 months of age (Fig. 7). KO mice fell from the rod earlier than WT mice at two (62 ± 6 versus 77 ± 7 s; P = 0.08), three (65 ± 6 versus 72 ± 12 s; P = 0.30) and 4 months of age (56 ± 6 versus 71 ± 7 s; P = .04). These findings indicate that Tcap deficiency impairs motor function in young mice.
Figure 7.
Tcap KO mice exhibit a decline in motor performance by 4 months of age. The ability to maintain balance on a rotating rod was compared between WT and Tcap KO mice by measuring the time (in seconds) to fall from a Rotor Rod. Performance was assessed longitudinally, and compared with WT controls, Tcap KO mice (open circles) exhibited impaired performance relative to controls at 2 months (n = 32, 62±6 versus 77 ± 7 s), 3 months (n = 32, 65 ± 6 versus 72 ± 12 s) and 4 months (n = 30, 56 ± 6 versus 71 ± 7), respectively. Data indicate mean ± SE.
Normal force and fatigue profiles in Tcap-deficient EDL and soleus muscles
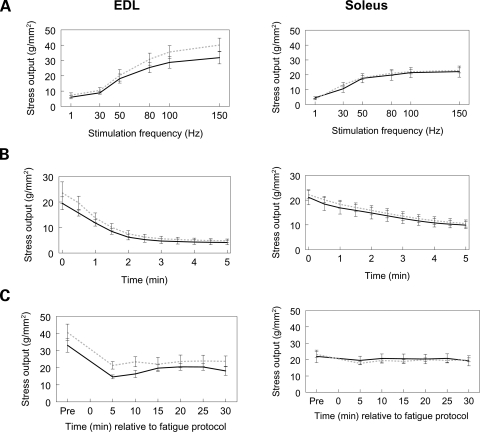
We next set out to determine if Tcap deficiency would alter the force-generating capabilities of either a fast-twitch (e.g. EDL) or a slow-twitch [e.g. soleus (SOL)] mouse muscle. To address this question we conducted ex vivo muscle assays at 1 year of age. Morphological results demonstrated that there were no differences detected in overall body mass nor were there differences detected in mass, length or cross-sectional area between either the two muscles or between the two genotypes (Table 2). However, compared with the SOL muscle, the EDL was longer and contracted with greater force independent of genotype (Table 2, P < 0.05). In addition, between genotypes both EDL and SOL muscles exhibited similar stress-frequency profiles (i.e. the force produced at a given stimulation frequency; Table 2; Fig. 8A). Furthermore, in response to a fatigue challenge and subsequent recovery, no significant differences were detected in stress profiles between genotypes for either EDL or SOL muscles (Fig. 8B and C). These findings suggest that Tcap deficiency does not impair the ability of mouse muscles to generate maximum force, nor does Tcap deficiency predispose the muscles to fatigue.
Table 2.
Morphological and stress output data
| Tcap KO | WT | |
|---|---|---|
| Body mass was not affected by genotype (n = 9 mice/genotype) | ||
| Body mass (g) | 35.3 ± 2.3 | 34.6 ± 2.4 |
| Neither muscle mass, length, nor cross-sectional area were affected by genotype (n = 17–18 muscles/genotype) | ||
| EDL mass (mg) | 12.1 ± 0.5 | 10.8 ± 0.4 |
| EDL length (mm) | 13.3 ± 0.3* | 12.8 ± 0.3* |
| EDL XSA (g/mm2) | 0.86 ± 0.03 | 0.80 ± 0.04 |
| SOL mass (mg) | 11.0 ± 0.6 | 10.0 ± 0.4 |
| SOL length (mm) | 11.2 ± 0.2 | 10.7 ± 0.2 |
| SOL XSA (g/mm2) | 0.95 ± 0.07 | 0.88 ± 0.05 |
| Maximal twitch and tetanic (at 150 Hz) stress outputs were not affected by genotype (n = 16–18 muscles/group) | ||
| EDL twitch stress (g/mm2) | 7.19 ± 0.66* | 7.12 ± 0.82* |
| EDL tetanic stress (g/mm2) | 38.97 ± 3.24* | 40.97 ± 3.58* |
| SOL twitch stress (g/mm2) | 4.09 ± 0.53 | 4.05 ± 0.39 |
| SOL tetanic stress (g/mm2) | 24.62 ± 2.62 | 25.14 ± 1.61 |
*P < 0.05.
Independent of genotype (i.e. values pooled from both genotypes), EDL muscles had a greater length (13.1 ± 0.2 versus 11.0 ± 0.2 mm), greater twitch (7.2 ± 0.5 versus 4.1 ± 0.3 g/mm2) and greater tetanic (40.0 ± 2.4 versus 24.9 ± 1.5 g/mm2) stress values compared with SOL muscles.
Figure 8.
Stress profiles of EDL and SOL muscles. Absence of the Tcap protein was not associated with a change in the stress frequency (A), fatigue (B) or recovery (C) profiles of EDL or SOL muscles. Solid black: WT, dashed gray: Tcap KO. n = 7–9 muscles per group.
Tcap-deficient muscles are stiffer than normal
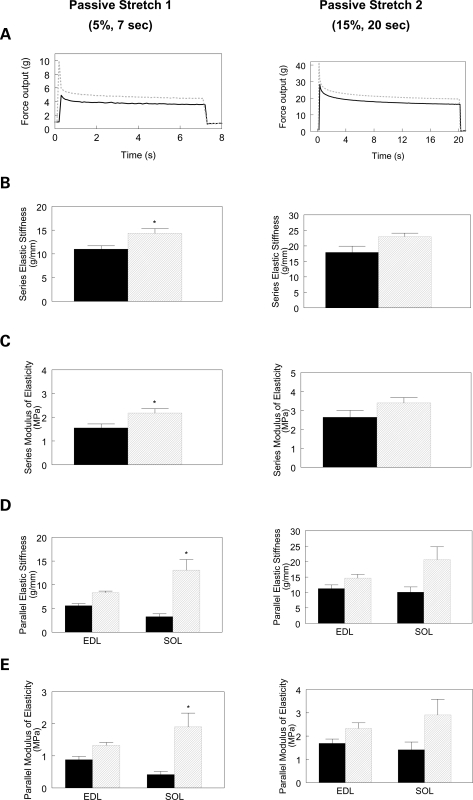
Finally, we performed mechanical studies to carefully assess stiffness profiles in isolated EDL and SOL muscles in mice 1 year of age. Muscles were stretched using passive stretch protocols to generate stress-relaxation profiles. From the stress-relaxation profile induced by the short (5%) length change, independent of muscle type (i.e. main effect of genotype), passive series elastic stiffness (Fig. 9B) and modulus of elasticity (Fig. 9C) were greater in KO muscles compared with age-matched WT controls (14.4 ± 1.0 versus 10.9 ± 0.7 g/mm; P = 0.0039; and 2.2 ± 0.2 versus 1.6 ± 0.2 MPa; P = 0.0046). Passive parallel elastic stiffness (Fig. 9D) and modulus of elasticity (Fig. 9E) were greater in KO SOL (13.1 ± 2.2 versus 3.3 ± 0.6 g/mm; P = 0.0037 and 1.9 ± 0.4 versus 0.4 ± 0.1 MPa; P = 0.0246), but not EDL muscles (Fig. 9D, P > 0.05). When the muscles were stretched at long (15%) lengths and held for 20 s, similar patterns were evident for each stiffness parameter, but differences were not significant. These findings indicate that Tcap deficiency increases passive stiffness in slow-twitch skeletal muscles of the mouse.
Figure 9.
Passive stiffness of EDL and SOL muscles. (A) Representative EDL passive stretch profiles for the two stress-relaxation protocols. (B, C) Independent of muscle type, absence of Tcap was associated with increased series elastic stiffness and series modulus of elasticity for the 5%, 7 s passive stretch protocol but not the 15%, 20 s passive stretch condition. Independent of genotype, series elastic stiffness and modulus of elasticity were greater in EDL (13.8 ± 0.6 g/mm and 2.2 ± 0.1 MPa, respectively) than SOL (11.4 ± 1.2 g/mm and 1.5 ± 0.2 MPa, respectively; P < 0.05) muscles in the 5%, 7 s passive stretch condition, but not the 15%, 20 s condition (data not shown). (D, E) SOL parallel elastic stiffness and modulus of elasticity were greater in the KO muscles compared with WT muscles stretched 5% for 7 s, but not different when stretched 15% for 20 s. Solid black: WT, dashed gray: Tcap KO; * different from WT; n = 7–9 muscles per group (P < 0.05).
DISCUSSION
We present the first description of a Tcap KO mouse model with histological features comparable to findings reported in the muscles of LGMD2G patients. The human disorder has been described as a muscular pathology caused by the absence of Tcap (9), a 19 kDa protein expressed in adult skeletal and cardiac muscle, and a recent report suggests expression in gastrointestinal smooth muscle (14). Until now, research in LGMD2G has been limited by the lack of an available animal model (4). Results in our KO mouse model indicate that Tcap deficiency leads to motor impairment and stiff skeletal muscles that can generate near-normal levels of maximum force. How Tcap deficiency leads to muscle weakness in LGMD2G patients remains unclear.
Functions of Tcap in muscle development and force-generating capacity
Our earlier work (4) hypothesized that Tcap is crucial for normal skeletal muscle development. In the present work, we generated KO mice to provide an in vivo model for further study. We previously demonstrated that siRNA-mediated knockdown of Tcap impaired normal myogenic differentiation in cultured muscle precursor (C2C12) cells, accompanied by decreased expression of myogenic regulatory factors and insulin-like growth factor 2 (IGF-2). In contrast to our previous in vitro findings, our present study indicates that KO mice develop normally and maintain near-normal muscle strength, and this was reflected by our microarray data which revealed transcriptional changes in a distinctly different subset of genes. This indicates that Tcap is not crucial for normal skeletal muscle development in mice, suggesting that the previously reported transcriptional changes may represent secondary effects in vitro under conditions where Tcap deficiency prevents full differentiation in culture. An a priori prediction was that, similar to our previous work, we would observe differential expression of MyoD, myogenin and IGF-2 in Tcap KO muscle transcripts. However, none of these myogenic regulatory factors, nor many of the genes encoding for cell cycle progression proteins, were significantly differentially expressed in KO muscle. This discordance between in vitro and in vivo findings is not novel or unexpected (15,16,18). The in vivo environment offers rescue pathways not always possible to reproduce in vitro (19). For example, in the mdx mouse model of Duchenne muscular dystrophy, compensatory pathways, such as utrophin expression, rescue some of the pathology caused by absence of dystrophin (20). Furthermore, the lack of significant change in expression for many genes in a dystrophic animal model is not without precedent. In a screen of over 12 000 mRNAs in mdx mice, only 137 were differentially regulated (21). In the present study, we noted significant up-regulation of genes such as ryanodine receptor 1 (RYR1), inducible nitric oxide synthase, ankyrin repeat and NFkB repressing factor. Altered expression of these genes was postulated to have clinical consequences in centronucleation myopathies and muscular dystrophies (22–26): for example, mutations in the RYR1 gene are accompanied by abundant central nuclei, and increased activation of the NFkB pathway is noted in mdx mice. Further study will be required to understand the clinical implications of altered gene expression in Tcap-deficient muscles.
Tcap influences myostatin expression
Previous in vitro studies indicate that Tcap interacts with myostatin, a key regulator of myoblast proliferation (27–31) and differentiation (27,32,33). The finding that myostatin negatively regulates myoblast proliferation and muscle growth (34), together with a potential binding interaction between Tcap and myostatin (28), prompted us to address the question of whether Tcap deficiency might result in altered myostatin protein levels in KO muscles. In preliminary experiments, we validated the myostatin antibody used in the present study by assessing its ability to recognize purified recombinant human myostatin protein (encoding 111 amino acids thought to be the mature protein; Fig. 5A). In immunoblots performed under non-reducing and reducing conditions, with increasing amounts of this recombinant protein, we observed a proportionate increase in the intensity of bands at ∼26 and ∼13 kDa. In other experiments, this same antibody was used to probe for myostatin in hindlimb muscles subjected to hindlimb unweighting (HLU), a ground-based method used to mimic weightlessness and to increase the level of intramuscular myostatin (35,36). Consistent with previous reports (17,32), increases in a ∼26 kDa band were observed on immunoblots (Fig. 5C and D) of HLU tissue relative to non-HLU controls. Results indicated that myostatin protein levels increased following four weeks of HLU, and that the myostatin antibody used in the present study faithfully detects the biologically active carboxy-terminal dimer at ∼26 kDa. Fluorescent images of HLU SOL muscle sections were used for line-scan analysis (Fig. 6B), which further suggested that Tcap and myostatin associate at the z-disc in limb skeletal muscle, consistent with the localization of intracellular myostatin (37).
Our observations of elevated myostatin expression in Tcap KO muscle are noteworthy in light of an earlier report that Tcap and myostatin interact (28). Our data suggest that alterations in Tcap expression might directly influence intracellular myostatin content, and offers a plausible explanation for the muscle atrophy observed in LGMD2G patients. However, we did not observe muscle atrophy or marked weakness in Tcap KO mice. These observations suggest that the mouse may compensate by modulating some other aspect of the myostatin pathway, such as follistatin expression, or perhaps that myostatin may act not to promote atrophy per se, but to control muscle hypertrophy (38). Whether or not myostatin levels are altered in LGMD2G are not presently known. Because myostatin is subject to complex transcriptional, translational and post-translational regulation (39,40) further study will be required to elucidate the clinical significance of myostatin expression in Tcap-deficient muscle.
Dystrophic phenotype in muscles from Tcap KO mice
In KO muscles, we observed atrophy and increased central nucleation in scattered fibers, with an abnormal distribution of mitochondria observed on NADH staining. Our histopathological findings resemble the abnormalities reported in a patient with LGMD2G carrying a Tcap mutation (10). The presence of scattered atrophic fibers in the absence of fiber type grouping is a non-specific finding that can be seen in myopathic or early neuropathic disease. Mild increases in the number of centrally nucleated fibers are most commonly seen in the context of myopathic changes or locally within areas of recent myofiber regeneration. The presence of abnormal mitochondrial staining in Tcap-deficient mice may be due to focal increases in mitochondrial number within fibers, or due to a cytoplasmic inclusion that disrupts the normal localization of mitochondria. Ultrastructural evaluation of muscles in our Tcap-deficient mice revealed large, prominent aggregates of mitochondria within some small fibers, which may account in part for the abnormal NADH staining seen in some small fibers. Additionally, myofibers from Tcap-deficient mice contained large tubular aggregates that distorted the myofiber architecture and might have resulted in large areas of decreased mitochondrial staining. We also observed these aggregates in occasional gastrocnemius muscle fibers in one of the WT animals. Tubular aggregates are generally considered a non-specific finding in human myopathic disease and can be seen in mouse models of myopathic disease and in aging muscle (41). Our ultrastructural examination revealed no organizational abnormalities of the contractile apparatus, a finding similar to a report by Vainzof et al. (42), who found no ultrastructural abnormalities of sarcomeric organization in LGMD2G patients.
Functions of Tcap in muscle passive parallel and series stiffness
Stiffness is defined as the change in muscle stress for a given change in muscle length. The series stiffness represents the stiffness associated with all elements in series from tendon to tendon of the muscle (i.e. the sarcomere, including titin). The parallel stiffness represents the stiffness of elements in parallel with the sarcomeres, such as the sarcolemmal membrane and connective tissues. Stiffness observed when the muscle is not activated (i.e. when the muscle is not receiving neural signals) is referred to as passive stiffness.
Passive parallel stiffness
Recently, Tcap mutations were reported to result in hypertrophic cardiomyopathy (HCM) and abnormal cardiac wall stiffness (43), a finding attributed to stronger Tcap binding interactions between titin, muscle LIM protein and calsarcin-1. This finding led to the hypothesis that stronger binding could increase passive tension and also augment calcium sensitivity of muscle contraction (43), a phenomenon observed in HCM-dependent sarcomeric protein mutations (43). Of note, the study above is difficult to compare with ours, first because cardiac rather than skeletal muscle was studied, and second because mutated Tcap proteins were studied, rather than complete absence of Tcap. Here we report that Tcap deletion results in a differential effect of stiffness (specifically, parallel stiffness) on slow-twitch (SOL) but not fast-twitch (EDL) muscles.
Passive series stiffness
Tissue specificity of passive series stiffness has been previously observed (44), and is thought to be related to differential splicing of titin's PEVK sequences. This region is generally considered to the principal component responsible for myofibrillar passive tension and elasticity. Because Tcap directly associates with titin (45), a protein contributing to passive stiffness properties of skeletal muscle fibers (46), we examined both passive parallel and series elastic stiffness. We determined parallel and series passive stiffness from a stress-relaxation profile (47) at two different amplitudes and durations (1: 5%, 7 s; 2: 15%, 20 s). We found increased series elastic stiffness in KO muscles for the short-amplitude, short-duration stretch, but failed to detect increased passive series or parallel elastic stiffness for the higher-amplitude, longer-duration step. This finding suggests that absence of Tcap yields a modest increase in passive series stiffness, potentially through changes in the PEVK sequences of titin; however, the increased stiffness appears dependent on either the amplitude and/or the velocity of stretch.
Conclusions
The pervasiveness of Tcap transcripts in skeletal muscle, its highly conserved nature among mice and humans, its association in vivo with z-disc proteins of the sarcomere, and the resulting physical disability due to its deficiency in patients with LGMD2G suggest that Tcap is an important therapeutic target in the treatment of patients with limb-girdle muscular dystrophy. Until now, no mammalian model of Tcap deficiency was available for translational research. In this report, we present a novel mouse model carrying a constitutive deletion in the Tcap gene, and our model mice display dystrophic features characteristic of patients with LGMD2G.
MATERIALS AND METHODS
Gene targeting
The targeting construct is shown in Figure 1. The total size of the targeting construct (including vector backbone and LacZ/Neo cassette) was 15.4 kb. The 11.4 kb region used to construct the targeting vector was first sub-cloned from a B6 clone using a homologous recombination-based technique. The region was designed such that the short homology arm (SA) extended 2.4 kb to 3′ of exon 2. The long homology arm (LA) consisted of a 7.5 kb fragment upstream of Tcap exon 1. The LacZ/Neo cassette was inserted upstream of the start codon for exon 1 and replaced 1.5 kb of the Tcap gene sequence including exons 1 and 2. The targeting vector was confirmed by restriction analysis after each modification step and by sequencing with primers designed to read from the LacZ/Neo cassette into the 5′ end of the SA and the 3′ end of the LA, or from primers that annealed to the vector sequence (Fig. 1). Ten micrograms of the targeting vector was linearized and transfected by electroporation of hybrid (C57Bl/6×129) ES cells. After selection in G418, surviving clones were expanded for PCR analysis to identify recombinant ES clones. Primers were designed downstream (3′) to the SA outside the region used to generate the targeting construct. PCR reactions using primers at the 5′ end of the Neo cassette amplified 2.7, 2.8 and 2.9 kb fragments, respectively. The control PCR reaction amplified a region 5′ of the SA inside the region used to create the targeting construct resulting in a band of 2.4 kb.
Generation and genotyping of mice
All animal procedures were approved by the institutional Animal Care and Use Committee (ACUC) and adhered to principles outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Mice were generated by standard techniques of homologous recombination in ES cells. Four individual ES clones identified above were microinjected into blastocysts from C57Bl/6 mice. Primer sets A2 and LAN1 (Fig. 1) were used to screen F1 mice. LAN1 anneals inside the Neo cassette and A2 anneals 3′ to the SA, outside the region used to create the targeting construct. A2/LAN1 amplifies a fragment that is 2.9 kb in length. The expanded positive ES cell clone was used as a positive control. The F1 mice above were set up for mating to generate F2 mice; following this, strategies were designed to screen F2 mice for the KO allele (A2/LAN1) and the WT allele (A2/WT1) and to confirm the presence of the LacZ cassette (Lac 2/3). Offspring from were genotyped by PCR analysis of tail snip DNA using the following primers: KO allele 2.9 kb (A2) forward (A2Fw), 5′-AGCAAGGTACCAGGACTCCTC-3′; KO (LAN1) reverse (koRv), 5′-CCAGAGGCCACTTGTGTAGC-3′; WT allele (2.75 kb): A2Fw: 5′-AGCAAGGTACCAGGACTCCTC-3′; (WT1rev): 5′-CTAGCAGGCCAGCTGGCTTCAAG-3′; LacZ cassette (1.5 kb): (LAC2fw): 5′-AGCTGGCGTAATAGCGAAGAG-3′ (LAC3rv): 5′-TTAGCGAAACCGCCAAGACTG-3′. PCR thermocycling parameters were 94°C 20 s, 62°C 60 s, 72°C 180 s, for 35 cycles. A schematic diagram showing the locations of the screening primers, representative gel and an overview of PCR genotyping methods is shown in Figure 1. Heterozygous (Tcap+/−) mice were intercrossed to generate homozygous Tcap-null (Tcap−/−) mice. During establishment and maintenance of the colony, PCR genotyping was performed on tail snips collected when pups were weaned.
Microarray/gene expression profiling
RNA was isolated from diced skeletal muscle (gastrocnemius) stored at −20°C following preservation in ≥5 volumes RNAlater (Applied Biosystems/Ambion). Tissue was sonicated in the presence of TriReagent (Molecular Research Center, Inc., Cincinnati, OH, USA) according to the method of Chomcynski and Sacchi (48). Total RNA was purified using RNeasy Minelute columns and reagents (Qiagen, Valencia, CA, USA) and eluted in nuclease-free water. The quantity and purity of the extracted RNA was evaluated using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and its integrity measured using an Agilent Bioanalyzer. For microarray hybridizations, 500 ng of total RNA from each sample was labeled with a fluorescent dye (Cy3) using the Low RNA Input Linear Amplification Labeling kit (Agilent Technologies, Palo Alto, CA, USA) following the manufacturer's protocol. The amount and quality of the fluorescently labeled cRNA was assessed using a NanoDrop ND-1000 spectrophotometer and an Agilent Bioanalyzer. According to manufacturer specifications, 1.6 µg of Cy3-labeled cRNA was hybridized to the Agilent Whole Mouse Genome Oligo Microarray (Agilent Technologies, Inc., Palo Alto, CA, USA) for 17 h, prior to washing and scanning. Data were extracted from scanned images using Feature Extraction Software (Agilent Technologies, Inc., Palo Alto, CA, USA).
Microarray data analysis
Six mouse whole-genome microarrays were queried with RNA from six biological samples derived from muscle tissue from WT (n = 3) and KO (n = 3) animals. For these single color experiments, gProcessedSignal values from Agilent's Feature.
Extraction software were generated following background subtraction and include correction for multiplicative surface trends. Normalized microarray data representing gProcessedSignal values for every feature were analyzed with GeneSifter™ data analysis suite. For pairwise comparisons, Student's t-tests were performed on median-normalized, log-transformed data, using a cut-off of P ≤ 0.05.
Motor performance on a rotor rod (49)
We followed the method of Gifondorwa for functional muscle testing in mice, with minor modifications. Briefly, following 2 weeks' habituation (six familiarization sessions) to the apparatus (San Diego Instruments), mice completed two trials per session at 2, 3 and 4 months of age, and latency to fall was recorded. Each trial lasted a maximum of 180 s. The mean of the two trials was calculated and used in statistical analysis. The terminal speed of the rod was 10 rpm.
Isolated EDL and SOL whole muscle assay
Muscle preparation
EDL and SOL muscles were surgically excised from euthanized mice and secured via 4–0 suture to a dual-mode servomotor (Aurora Scientific: Aurora, ON, Canada) as previously described (47,50).
Experimental protocol
Muscles were equilibrated in an oxygenated (95% O2–5% CO2) physiological salt solution [previously described (37)] bath for 10 min prior to data collection. Following the equilibration period, muscles were subjected to three twitches and two tetani, each separated by 1 min. After a 5 min rest, one EDL and one SOL from each mouse was subjected to a stretch protocol whereas the remaining muscles were subjected to force frequency and fatigue and recovery protocols.
Stretch protocol
Muscles were subjected to two passive stretch protocols. During the first protocol, muscles were stretched to 105% of their optimal length (L0) for 7 s, rested for 2 min and then for the second protocol, were stretched to 115% L0 for 20 s. After the passive stretch protocols, muscles rested for 5 min, were subjected to one twitch and one tetanus (separated by 1 min) and again rested for 5 min.
Force frequency protocol
Muscles were stimulated for 800 ms at 1, 30, 50, 80, 100 and 150 Hz, each separated by 1 min. After a 5 min rest, muscles were subjected to one twitch and one tetanus (separated by 1 min) and rested for 6 min.
Fatigue and recovery protocol
Muscles were stimulated at 60 Hz (EDL) or 100 Hz (SOL) for 1 s every 5 s for 5 min (total of 60 stimulations) then allowed to rest for 5 min. Force recovery was determined every 5 min for 30 min by stimulating the muscle at 60 Hz for 800 ms, resting for 1 min and stimulating the muscle at 150 Hz for 800 ms. For all protocols, stress output (g/mm2) was calculated as the force output (g) for a given stimulation normalized to the estimated cross-sectional area (mm2) of the muscle. Cross-sectional area of each muscle was determined by the following equation: muscle cross-sectional area = muscle mass in mg/(1.056 g/mm3 × muscle length in mm).
Histology and electron microscopy
Cross sections (8 µm) of isopentane-frozen gastrocnemius muscle were stained with hematoxylin and eosin and evaluated using a Nikon Eclipse 50i (Melville, NY, USA) microscope. For NADH staining, frozen sections were incubated with nitro-blue tetrazolium (1 mg/ml, Sigma) and beta-nicotinamide adenine dinucleotide (0.4 mg/ml, Sigma) in 50 mm Tris–HCl, pH 7.3, at 25°C for 30 min. Light microscopic images were captured using a SPOT Insight 4 Meg FW Color Mosaic camera and SPOT 4.5.9.1 software (Diagnostic Instruments, Sterling Heights, MI, USA). Fiber diameter measurements were performed through point-to-point measurements using NIS Elements AR software (Nikon instruments, Melville, NY, USA). Central nuclei were quantified by a direct count of centrally nucleated fibers on intermediate power (200× magnification) micrographs of areas with acceptable histological quality. The number of fibers counted per specimen ranged from 95 to 267. For electron microscopy studies, tissue was fixed in 5% glutaraldehyde, 2.5% paraformaldehyde, 0.06% picric acid in 0.2 m cacodylate buffer (pH 7.4) followed by osmication and uranyl acetate staining, dehydration in alcohols, and was then embedded in Taab epon (Marivac Ltd, Nova Scotia, Canada). Subsequently, 1 µm scout sections were stained with toluidine blue and were evaluated and photographed as described above. Areas of interest were cut at 95 nm thickness with a Leica ultracut microtome, picked up on 100 m formvar coated Cu grids, stained with 0.2% lead citrate, and viewed and imaged under the Philips Technai BioTwin Spirit Electron Microscope. To address biological association between Tcap and myostatin, muscle tissue was obtained from control and hindlimb-unloaded (HLU) adult female CD-1 mice. Following 2 weeks HLU, SOL and gastrocnemius muscles were harvested with mice under inhaled anesthetic (isoflurane) and stored in 10% formalin for immunofluorescence (IF) microscopy. For IF experiments, muscles were embedded in paraffin wax, sectioned longitudinally at 4 µm, and transferred to slides. Slides were dewaxed, and prepared as described previously (4). The percent color overlap of three independent thin sections of HLU SOL was determined using MetaMorph software (Downingtown, PA, USA). In this experiment, Tcap was labeled red and myostatin was labeled green.
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis and immunoblot analyses
Surgically excised gastrocnemius muscles from WT and Tcap KO mice were snap frozen. Tissue was subjected to liquid nitrogen grinding and homogenization, followed by lysis in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1:10 protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Protein concentration of the muscle homogenate was determined using the Bio-Rad Dc protein assay. Varying amounts of total protein for all samples (20, 40 and 60 µg) were separated on 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis, transferred to a PVDF membrane (Millipore Corporation, Bedford, MA, USA), and probed with appropriate dilutions of respective antibodies against Tcap and myostatin proteins overnight, followed by corresponding secondary IgG, labeled with horseradish peroxidase. Chemiluminescent detection was performed using Supersignal West Dura Extended Duration Substrate (Pierce Biotechnology, Inc., Rockford, IL, USA) or Western blotting Luminol Reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Images were obtained using LAS3000 Imager. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to ensure equal loading of total protein. GST-tagged Tcap recombinant protein was used as a positive control. Antibody against Tcap was obtained commercially from BD Biosciences (Cat# 612328) or from the laboratory of G. Faulkner. Custom-made GST-tagged Tcap was obtained from ProteinTech Group, Inc (Chicago, IL, USA). Anti-myostatin antibody and purified myostatin protein were obtained from Millipore Corporation (Bedford, MA, USA; Cat# ab3239) and ProSpec (Rehovot, Israel; Cat# CYT-418), respectively. All antibodies were diluted in appropriate ranges in 5% w/v skim milk/1× PBST (0.5% v/v Tween-20). GAPDH and HRP-conjugated secondary antibodies were obtained from Abcam Inc. (Cambridge, MA, USA; Cat# ab9484) and Pierce Biotechnology, Inc., respectively. Prestained broad range molecular marker (Bio-Rad Laboratories, Hercules, CA, USA) and prestained biotinylated protein ladder (Cell Signaling Technology, Danvers, MA, USA) were used to determine molecular masses of the bands. The specificity and sensitivity of all antibodies used were optimized in pilot studies. The band density was quantitated using the Multi Gauge program and was normalized against GAPDH bands. The densitometry bar graphs were created using GraphPad Prism and data are reported as mean normalized intensity ± SE. One-way ANOVA analysis was performed, followed with a Newman–Keuls post hoc test. A P-value of <0.05 was considered significant.
Muscular performance statistical analysis
For EDL and SOL whole muscle assays, and rotor rod performance, Student's t-tests were used for comparison between groups (P ≤ 0.05). For muscle length, mass and cross-sectional area, twitch and tetanic stress, and passive and active stiffness measures a two-way ANOVA (factors: genotype and muscle type) were used for comparison. Mixed-model ANOVAs with repeated measures were used for comparison of stress frequency (factors: genotype and stimulation frequency), and fatigue and recovery (factors: genotype and time) profiles. Tukey HSD was used for post hoc analyses when ANOVAs indicated significance. For all measures, significance was set at P ≤ 0.05. Data are presented as mean ± SE.
FUNDING
This work was supported by the Departments of Physical Medicine & Rehabilitation, School of Medicine, University of Missouri and the Department of Neurology, School of Medicine, Wake Forest University Health Sciences as well as by NIH R01 AR044345, P50 NS040828, and the Lee and Penny Anderson Family Foundation.
ACKNOWLEDGEMENTS
We thank Masood Machingal, Charles Childers and Devin Odom for technical assistance, G. Faulkner for sharing of antibody, and S.-J. Lee for supplying myostatin-knockout mice. MF-20 (myosin) antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Dept. of Biological Sciences, Iowa City, IA 52242.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Valle G., Faulkner G., De Antoni A., Pacchioni B., Pallavicini A., Pandolfo D., Tiso N., Toppo S., Trevisan S., Lanfranchi G. Telethonin, a novel sarcomeric protein of heart and skeletal muscle. FEBS Lett. 1997;415:163–168. doi: 10.1016/s0014-5793(97)01108-3. doi:10.1016/S0014-5793(97)01108-3. [DOI] [PubMed] [Google Scholar]
- 2.Faulkner G., Pallavicini A., Comelli A., Salamon M., Bortoletto G., Ievolella C., Trevisan S., Kojic S., Dalla Vecchia F., Laveder P., et al. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 2000;275:41234–41242. doi: 10.1074/jbc.M007493200. doi:10.1074/jbc.M007493200. [DOI] [PubMed] [Google Scholar]
- 3.Furukawa T., Ono Y., Tsuchiya H., Katayama Y., Bang M.L., Labeit D., Labeit S., Inagaki N., Gregorio C.C. Specific interaction of the potassium channel beta-subunit minK with the sarcomeric protein T-cap suggests a T-tubule-myofibril linking system. J. Mol. Biol. 2001;313:775–784. doi: 10.1006/jmbi.2001.5053. doi:10.1006/jmbi.2001.5053. [DOI] [PubMed] [Google Scholar]
- 4.Markert C.D., Ning J., Staley J.T., Heinzke L., Childers C.K., Ferreira J.A., Brown M., Stoker A., Okamura C., Childers M.K. TCAP knockdown by RNA interference inhibits myoblast differentiation in cultured skeletal muscle cells. Neuromuscul. Disord. 2008;18:413–422. doi: 10.1016/j.nmd.2008.03.010. doi:10.1016/j.nmd.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Mayans O., van der Ven P.F., Wilm M., Mues A., Young P., Furst D.O., Wilmanns M., Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. doi:10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 6.Mues A., van der Ven P.F., Young P., Furst D.O., Gautel M. Two immunoglobulin-like domains of the Z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett. 1998;428:111–114. doi: 10.1016/s0014-5793(98)00501-8. doi:10.1016/S0014-5793(98)00501-8. [DOI] [PubMed] [Google Scholar]
- 7.Epstein N.D., Davis J.S. Sensing stretch is fundamental. Cell. 2003;112:147–150. doi: 10.1016/s0092-8674(03)00037-0. doi:10.1016/S0092-8674(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang R., Yang J., Zhu J., Xu X. Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere-membrane interaction, not sarcomere assembly. Hum. Mol. Genet. 2009;18:4130–4140. doi: 10.1093/hmg/ddp362. doi:10.1093/hmg/ddp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreira E.S., Wiltshire T.J., Faulkner G., Nilforoushan A., Vainzof M., Suzuki O.T., Valle G., Reeves R., Zatz M., Passos-Bueno M.R., et al. Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene encoding the sarcomeric protein telethonin. Nat. Genet. 2000;24:163–166. doi: 10.1038/72822. doi:10.1038/72822. [DOI] [PubMed] [Google Scholar]
- 10.Olive M., Shatunov A., Gonzalez L., Carmona O., Moreno D., Quereda L.G., Martinez-Matos J.A., Goldfarb L.G., Ferrer I. Transcription-terminating mutation in telethonin causing autosomal recessive muscular dystrophy type 2G in a European patient. Neuromuscul. Disord. 2008;18:929–933. doi: 10.1016/j.nmd.2008.07.009. doi:10.1016/j.nmd.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanin M., Nascimbeni A.C., Aurino S., Tasca E., Pegoraro E., Nigro V., Angelini C. Frequency of LGMD gene mutations in Italian patients with distinct clinical phenotypes. Neurology. 2009;72:1432–1435. doi: 10.1212/WNL.0b013e3181a1885e. doi:10.1212/WNL.0b013e3181a1885e. [DOI] [PubMed] [Google Scholar]
- 12.Willmann R., Possekel S., Dubach-Powell J., Meier T., Ruegg M.A. Mammalian animal models for Duchenne muscular dystrophy. Neuromuscul. Disord. 2009;19:241–249. doi: 10.1016/j.nmd.2008.11.015. doi:10.1016/j.nmd.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Froehner S.C. Just say NO to muscle degeneration? Trends Mol. Med. 2002;8:51–53. doi: 10.1016/s1471-4914(02)02275-x. doi:10.1016/S1471-4914(02)02275-X. [DOI] [PubMed] [Google Scholar]
- 14.Morel J.L., Dabertrand F., Fritz N., Henaff M., Mironneau J., Macrez N. The decrease of expression of ryanodine receptor subtype 2 is reversed by gentamycin sulfate in vascular myocytes from mdx mouse. J. Cell. Mol. Med. 2009;13:3122–3130. doi: 10.1111/j.1582-4934.2009.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson S., Luffer-Atlas D., Knadler M.P. Predicting circulating human metabolites: how good are we? Chem. Res. Toxicol. 2009;22:243–256. doi: 10.1021/tx8004086. doi:10.1021/tx8004086. [DOI] [PubMed] [Google Scholar]
- 16.Fujii K., Nagai J., Sawada T., Yumoto R., Takano M. Effect of PEGylation of N-WASP181-200 on the inhibitory potency for renal aminoglycoside accumulation. Bioconjug. Chem. 2009;20:1553–1558. doi: 10.1021/bc900094g. [DOI] [PubMed] [Google Scholar]
- 17.Sharma M., Kambadur R., Matthews K.G., Somers W.G., Devlin G.P., Conaglen J.V., Fowke P.J., Bass J.J. Myostatin, a transforming growth factor-beta superfamily member, is expressed in heart muscle and is upregulated in cardiomyocytes after infarct. J. Cell. Physiol. 1999;180:1–9. doi: 10.1002/(SICI)1097-4652(199907)180:1<1::AID-JCP1>3.0.CO;2-V. doi:10.1002/(SICI)1097-4652(199907)180:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 18.Jespersen B., Thiesson H.C., Henriksen C., Therland K., Falk C., Poulsen T., Fogh B., Madsen K., Walther S., Jensen B.L. Differential effects of immunosuppressive drugs on COX-2 activity in vitro and in kidney transplant patients in vivo. Nephrol. Dial. Transplant. 2009;24:1644–1655. doi: 10.1093/ndt/gfp004. doi:10.1093/ndt/gfp004. [DOI] [PubMed] [Google Scholar]
- 19.Infante J.P., Huszagh V.A. Mechanisms of resistance to pathogenesis in muscular dystrophies. Mol. Cell. Biochem. 1999;195:155–167. doi: 10.1023/a:1006972315739. doi:10.1023/A:1006972315739. [DOI] [PubMed] [Google Scholar]
- 20.Weir A.P., Morgan J.E., Davies K.E. A-utrophin up-regulation in mdx skeletal muscle is independent of regeneration. Neuromuscul. Disord. 2004;14:19–23. doi: 10.1016/j.nmd.2003.09.004. doi:10.1016/j.nmd.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Tseng B.S., Zhao P., Pattison J.S., Gordon S.E., Granchelli J.A., Madsen R.W., Folk L.C., Hoffman E.P., Booth F.W. Regenerated mdx mouse skeletal muscle shows differential mRNA expression. J. Appl. Physiol. 2002;93:537–545. doi: 10.1152/japplphysiol.00202.2002. [DOI] [PubMed] [Google Scholar]
- 22.Bean C., Facchinello N., Faulkner G., Lanfranchi G. The effects of Ankrd2 alteration indicate its involvement in cell cycle regulation during muscle differentiation. Biochim. Biophys. Acta. 2008;1783:1023–1035. doi: 10.1016/j.bbamcr.2008.01.027. doi:10.1016/j.bbamcr.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Kojic S., Medeot E., Guccione E., Krmac H., Zara I., Martinelli V., Valle G., Faulkner G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J. Mol. Biol. 2004;339:313–325. doi: 10.1016/j.jmb.2004.03.071. doi:10.1016/j.jmb.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 24.Sewry C.A., Jimenez-Mallebrera C., Muntoni F. Congenital myopathies. Curr. Opin. Neurol. 2008;21:569–575. doi: 10.1097/WCO.0b013e32830f93c7. doi:10.1097/WCO.0b013e32830f93c7. [DOI] [PubMed] [Google Scholar]
- 25.Siegel A.L., Bledsoe C., Lavin J., Gatti F., Berge J., Millman G., Turin E., Winders W.T., Rutter J., Palmeiri B., et al. Treatment with inhibitors of the NF-kappaB pathway improves whole body tension development in the mdx mouse. Neuromuscul. Disord. 2009;19:131–139. doi: 10.1016/j.nmd.2008.10.006. doi:10.1016/j.nmd.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Wehling M., Spencer M.J., Tidball J.G. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J. Cell. Biol. 2001;155:123–131. doi: 10.1083/jcb.200105110. doi:10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joulia D., Bernardi H., Garandel V., Rabenoelina F., Vernus B., Cabello G. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp. Cell. Res. 2003;286:263–275. doi: 10.1016/s0014-4827(03)00074-0. doi:10.1016/S0014-4827(03)00074-0. [DOI] [PubMed] [Google Scholar]
- 28.Nicholas G., Thomas M., Langley B., Somers W., Patel K., Kemp C.F., Sharma M., Kambadur R. Titin-cap associates with, and regulates secretion of, Myostatin. J. Cell. Physiol. 2002;193:120–131. doi: 10.1002/jcp.10158. doi:10.1002/jcp.10158. [DOI] [PubMed] [Google Scholar]
- 29.Rios R., Carneiro I., Arce V.M., Devesa J. Myostatin regulates cell survival during C2C12 myogenesis. Biochem. Biophys. Res. Commun. 2001;280:561–566. doi: 10.1006/bbrc.2000.4159. doi:10.1006/bbrc.2000.4159. [DOI] [PubMed] [Google Scholar]
- 30.Taylor W.E., Bhasin S., Artaza J., Byhower F., Azam M., Willard D.H., Jr, Kull F.C., Jr, Gonzalez-Cadavid N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am. J. Physiol. Endocrinol. Metab. 2001;280:E221–E228. doi: 10.1152/ajpendo.2001.280.2.E221. [DOI] [PubMed] [Google Scholar]
- 31.Thomas M., Langley B., Berry C., Sharma M., Kirk S., Bass J., Kambadur R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000;275:40235–40243. doi: 10.1074/jbc.M004356200. doi:10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- 32.Langley B., Thomas M., Bishop A., Sharma M., Gilmour S., Kambadur R. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J. Biol. Chem. 2002;277:49831–49840. doi: 10.1074/jbc.M204291200. doi:10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]
- 33.Rios R., Carneiro I., Arce V.M., Devesa J. Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiol. Cell. Physiol. 2002;282:C993–C999. doi: 10.1152/ajpcell.00372.2001. [DOI] [PubMed] [Google Scholar]
- 34.McPherron A.C., Lawler A.M., Lee S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. doi:10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 35.Carlson C.J., Booth F.W., Gordon S.E. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am. J. Physiol. 1999;277:R601–R606. doi: 10.1152/ajpregu.1999.277.2.r601. [DOI] [PubMed] [Google Scholar]
- 36.Jones S.W., Hill R.J., Krasney P.A., O'Conner B., Peirce N., Greenhaff P.L. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- 37.Artaza J.N., Bhasin S., Mallidis C., Taylor W., Ma K., Gonzalez-Cadavid N.F. Endogenous expression and localization of myostatin and its relation to myosin heavy chain distribution in C2C12 skeletal muscle cells. J. Cell. Physiol. 2002;190:170–179. doi: 10.1002/jcp.10044. doi:10.1002/jcp.10044. [DOI] [PubMed] [Google Scholar]
- 38.Greenhaff P.L. The molecular physiology of human limb immobilization and rehabilitation. Exerc. Sport. Sci. Rev. 2006;34:159–163. doi: 10.1249/01.jes.0000240017.99877.8a. doi:10.1249/01.jes.0000240017.99877.8a. [DOI] [PubMed] [Google Scholar]
- 39.Lee S.J. Regulation of muscle mass by myostatin. Annu. Rev. Cell. Dev. Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. doi:10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- 40.Markert C., Childers M.K. Myostatin expression in muscular dystrophies and mitochondrial encephalomyopathies. Pediatr Neurol. 2006;35:300. doi: 10.1016/j.pediatrneurol.2006.08.001. author reply 300. doi:10.1016/j.pediatrneurol.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Agbulut O., Destombes J., Thiesson D., Butler-Browne G. Age-related appearance of tubular aggregates in the skeletal muscle of almost all male inbred mice. Histochem. Cell. Biol. 2000;114:477–481. doi: 10.1007/s004180000211. [DOI] [PubMed] [Google Scholar]
- 42.Vainzof M., Moreira E.S., Suzuki O.T., Faulkner G., Valle G., Beggs A.H., Carpen O., Ribeiro A.F., Zanoteli E., Gurgel-Gianneti J., et al. Telethonin protein expression in neuromuscular disorders. Biochim. Biophys. Acta. 2002;1588:33–40. doi: 10.1016/s0925-4439(02)00113-8. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi T., Arimura T., Itoh-Satoh M., Ueda K., Hohda S., Inagaki N., Takahashi M., Hori H., Yasunami M., Nishi H., et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:2192–2201. doi: 10.1016/j.jacc.2004.08.058. doi:10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 44.Wang K., McCarter R., Wright J., Beverly J., Ramirez-Mitchell R. Regulation of skeletal muscle stiffness and elasticity by titin isoforms: a test of the segmental extension model of resting tension. Proc. Natl. Acad. Sci. USA. 1991;88:7101–7105. doi: 10.1073/pnas.88.16.7101. doi:10.1073/pnas.88.16.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregorio C.C., Trombitas K., Centner T., Kolmerer B., Stier G., Kunke K., Suzuki K., Obermayr F., Herrmann B., Granzier H., et al. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell. Biol. 1998;143:1013–1027. doi: 10.1083/jcb.143.4.1013. doi:10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linke W.A., Ivemeyer M., Olivieri N., Kolmerer B., Ruegg J.C., Labeit S. Towards a molecular understanding of the elasticity of titin. J. Mol. Biol. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. doi:10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- 47.Wolff A.V., Niday A.K., Voelker K.A., Call J.A., Evans N.P., Granata K.P., Grange R.W. Passive mechanical properties of maturing extensor digitorum longus are not affected by lack of dystrophin. Muscle Nerve. 2006;34:304–312. doi: 10.1002/mus.20588. doi:10.1002/mus.20588. [DOI] [PubMed] [Google Scholar]
- 48.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. doi:10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 49.Gifondorwa D.J., Robinson M.B., Hayes C.D., Taylor A.R., Prevette D.M., Oppenheim R.W., Caress J., Milligan C.E. Exogenous delivery of heat shock protein 70 increases lifespan in a mouse model of amyotrophic lateral sclerosis. J. Neurosci. 2007;27:13173–13180. doi: 10.1523/JNEUROSCI.4057-07.2007. doi:10.1523/JNEUROSCI.4057-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grange R.W., Gainer T.G., Marschner K.M., Talmadge R.J., Stull J.T. Fast-twitch skeletal muscles of dystrophic mouse pups are resistant to injury from acute mechanical stress. Am. J. Physiol. Cell Physiol. 2002;283:C1090–C1101. doi: 10.1152/ajpcell.00450.2001. [DOI] [PubMed] [Google Scholar]