Abstract
BACKGROUND
Protein biomarker candidates from discovery proteomics must be quantitatively verified in patient samples before they can progress to clinical validation. Here we demonstrate that peptide immunoaffinity enrichment coupled with stable isotope dilution mass spectrometry (SISCAPA-MRM) can be used to configure assays with performance suitable for candidate biomarker verification. As proof of principle, we configured SISCAPA assays for troponin I (cTnI), an established biomarker of cardiac injury, and interleukin 33 (IL-33), an emerging immunological and cardiovascular marker for which robust immunoassays are currently not available.
METHODS
We configured individual and multiplexed assays in which peptides were enriched from digested human plasma using antipeptide antibodies. Assay performance was established using response curves for peptides and proteins spiked into normal plasma. We quantified proteins using labeled peptides as internal standards, and we measured levels of cTnI in patients who underwent a planned myocardial infarction for hypertrophic obstructive cardiomyopathy.
RESULTS
Measurement of cTnI and IL-33 proteins from trypsin-digested plasma was linear from 1.5 to 5000 μg/L, with imprecision <13% for both proteins, processed individually or multiplexed. Results correlated well (R=0.89) with a commercial immunoassay.
CONCLUSIONS
We used an established biomarker of cardiac injury and an emerging biomarker to demonstrate how SISCAPA can detect and quantify changes in concentration of proteins present at 1–10 μg/L in plasma. Our results demonstrate that these assays can be multiplexed and retain the necessary precision, reproducibility, and sensitivity to be applied to new and uncharacterized candidate biomarkers for verification of low-abundance proteins in blood.
The critical gap in biomarker discovery and development is the ability to evaluate lengthy lists of candidate biomarker proteins in a statistically relevant number of patient samples with sufficient sensitivity and specificity toward the targeted disease. Currently, clinical validation of biomarkers relies primarily on immunoassays because of their specificity for the target analyte, sensitivity, and high throughput. However, antibody reagents for a clinical-grade immunoassay only exist for few, if any, candidates suggested by discovery experiments. Developing reliable immunoassays for each candidate biomarker is not practical owing to the high cost and long development time for useful ELISAs. Alternative methods are clearly needed to verify candidate protein biomarkers and triage the long lists of potential biomarkers (1, 2).
Multiple reaction monitoring (MRM)4 coupled with stable isotope dilution mass spectrometry (SID-MS) has recently been shown to be well suited for direct quantification of proteins in plasma owing to the high specificity, sensitivity, and precision of the quantitative measurements (3–6). In addition, MRM assays can be highly multiplexed such that many candidate proteins can be simultaneously targeted and measured in the large numbers of patient samples required for verification (7). Currently, up to 50 proteins can be routinely monitored, but higher numbers are possible by targeting known peptide retention times using Scheduled MRM™ (AB Sciex). Sensitivity for unambiguous detection and quantification of proteins by MS-based assays, however, is often constrained by sample complexity, especially in plasma (8, 9). Plasma can be made significantly less complex by depleting abundant proteins and then fractionating the peptides produced by enzymatic digestion of the plasma before SID-MRM-MS. Using this approach, we have demonstrated limits of quantification (LOQ) in the range of 1–20μg/L with CVs of 10%–20% for a range of proteins in plasma (4). Upfront sample processing substantially restricts sample analysis throughput compared to immunoassays, however, and accurate quantification of proteins in the medium to low ng/L range has been inaccessible to date by this approach.
Here we couple the advantages of immunoaffinity enrichment of target peptides with the sensitivity and quantitative capabilities of SID-MRM-MS. The method, first described by Anderson et al. (10) and termed SISCAPA (stable isotope standards capture with antipeptide antibodies), has been shown to achieve more than a thousandfold enrichment for target peptides in a plasma digest and to be useful for quantifying proteins in the μg/L range in plasma with CV <20% (10, 11). In the present study, we successfully configured SISCAPA-MRM assays for 2 clinically relevant protein biomarkers of cardiovascular disease, cardiac troponin I (cTnI) (12, 13) and interleukin-33 (IL-33) (14), the latter currently lacking a well-validated immunoassay. This proof-of-principle study demonstrates, for the first time, that SISCAPA assays can be multiplexed to measure clinically relevant proteins. We applied our SISCAPA-MRM assay to quantify cTnI in plasma of patients who underwent planned myocardial infarction (PMI), a therapeutic intervention for patients with hypertrophic obstructive cardiomyopathy (HOCM). This work further demonstrates the enormous potential of SISCAPA-MRM assays in verification studies to quantify proteins for which immunoassays do not exist.
Materials and Methods
PREPARATION OF STANDARDS AND LC-MRM-MS METHOD
We selected peptides from in vitro digestion of protein standards analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) on a LTQ linear ion trap mass spectrometer (Thermo Fisher). Peptide standards were chosen for synthesis and antipeptide antibody development following the guidelines of Keshishian et al. (4) and further selected in silico as described in Supplemental Methods, which accompanies the online version of this article at www.clinchem.org/content/vol55/issue6. The peptides identified by MS/MS that were both high responding in electrospray LC-MS/MS and sequence unique within the human genome are referred to as “signature peptides” (underlined peptides in online Supplemental Fig. 1). We selected a subset of these peptides for antibody development (blue underlines in online Supplemental Fig. 1). The synthesis of stable isotope peptides, optimization of the triple-quadrupole mass spectrometer (TQ-MS), and a description of the LC-MRM-MS methods are in the online Supplemental Methods. Optimized MRMmethod transition ion masses and voltages are listed in online Supplemental Table 1.
POLYCLONAL ANTIBODY PRODUCTION AND MAGNETIC BEAD PREPARATION
Antibodies were produced and linked to magnetic beads as described in online Supplemental Methods.
PEPTIDE STANDARD CURVE PREPARATION
Corresponding [12C] forms of the heavy peptide internal standards were added to digested plasma from a healthy female donor and diluted to generate a 7-point response curve with concentrations equivalent to those described in the protein standard curve (see online Supplemental Table 2). Samples were stored at −80 °C until assayed.
PROTEIN STANDARD CURVE PREPARATION
cTnI and IL-33 standard proteins were added to nondigested plasma from a healthy female donor and diluted to generate a 7-point response curve with final concentrations of 0, 0.1, 0.5, 2, 20, 200, and 5000μg/L. Samples were stored at −80 °C until aliquots of 50 μL were reduced, alkylated, and digested with porcine trypsin (Promega) as described in online Supplemental Methods. Digested samples were stored dry at −80 °C until assayed.
PATIENT SAMPLE COLLECTION PROTOCOL
In this study, we included 5 patients undergoing PMI using alcohol septal ablation for the treatment of symptomatic HOCM, inclusion criteria to receive alcohol ablation treatment as described (15, 16). The protocol for drawing blood and preparing plasma is described in online Supplemental Methods. The protocol for obtaining blood from patients was approved by the Massachusetts General Hospital institutional review board, and all subjects gave written informed consent.
PEPTIDE IMMUNOAFFINITY ENRICHMENT AND SID-MRM QUANTIFICATION
We reconstituted digested plasma samples in 50 μL of 1× PBS. Two microliters of magnetic beads bound with 2μg polyclonal antibody and 50 fmol [13C] internal standard peptide were added to each concentration point, diluted 1:5 with PBS, and rotary mixed at 4 °C. After overnight incubation, we isolated the magnetic beads using a magnet and removed the unbound material. The beads were washed twice with 200 μL PBS and once with 200μL 50mMammonium bicarbonate, pH 7.8. Peptides were eluted with 2 volumes of 30 μL 5% acetic acid. The recovered fractions were pooled and desalted using a pipette tip packed with C18 Empore material (3M) (17). The purified mixture of enriched peptides was collected with 40 μL 90% acetonitrile: 0.1% formic acid and vacuum-centrifuged to dryness. Enriched samples were stored dry at −80 °C until analyzed by LC-MRM-MS as described in online Supplemental Methods.
DATA ANALYSIS AND PROTEIN QUANTIFICATION
We integrated extracted ion chromatograms (XICs) of all transition ions and calculated peak area ratios (PARs) using MultiQuant™ (AB Sciex) version 1.1.0.2. Analyte:internal standard PAR for the most intense transition ion was entered into the equation (PAR)(5 nmol/L)(protein molecular weight)(analysis volume)/process volume/1000 to calculate the concentration of protein in the original sample, reported in μg/L. We plotted observed concentration against the theoretical concentration for each curve and average protein concentration against time for the PMI patient time course. We calculated the SD of peptide curves, uniplex protein curves, and PMI patient samples from 3 injections of 1 processed sample and the SD of the multiplex protein curves using 3 injections of 3 processed samples. No outlier values were excluded for the analyses. We calculated imprecision (CV) and percent recovery at a concentration within the linear range of each curve and the limit of detection (LOD) and LOQ according to the method described by Currie (18).
MEASUREMENT OF cTnI BY IMMUNOASSAY
We measured the concentration of cTnI in PMI patients using the antibody plate and protein standards supplied with immunoassay kit (Alpco Diagnostics) according to the manufacturer’s instructions and calibrated using Liquichek™ Cardiac Markers Control (Bio-Rad). This in vitro diagnostic (IVD) EIA kit specifies a functional sensitivity of 0.75 μg/L with interassay CV <10%, a correlation coefficient of 0.86, and a slope of 1.04 compared to the Abbott MEIA for cTnI. The functional sensitivity of the kit in our hands was 1 μg/L when the cTnI protein response curve samples were assayed. The straight line summarizing the relation between immunoassay and SISCAPA concentrations was derived using Deming linear regression (19).
Results
SISCAPA ASSAY DEVELOPMENT AND CHARACTERIZATION
We prepared peptide and protein response curves for cTnI and IL-33 in healthy female plasma to evaluate the performance of the SISCAPA assay (online Supplemental Table 2). Analyte peptide standards spiked directly (Fig. 1A) or peptides derived from the spiked proteins (Fig. 1B) were subsequently captured using bead-bound polyclonal antibodies raised against the signature peptide. After enrichment, we analyzed antibody bead eluates using SID-LC-MRM-MS on the TQ-MS. We calculated protein concentrations from the analyte to internal standard PAR as described in Materials and Methods.
Fig. 1.
Strategy for generation of peptide (A) and protein (B) standard addition curves in plasma for SISCAPA assay development of cTnI and IL-33.
SELECTION OF PEPTIDES FOR SISCAPA ASSAY DEVELOPMENT
We selected high-responding peptides for cTnI and IL-33 based on detectability and response in LC-MS/MS analyses of tryptic digests of isolated and purified or recombinant versions of the target proteins (online Supplemental Fig. 1). The selected signature peptides were synthesized with an N-terminal cysteine, conjugated to keyhole limpet hemocyanin (KLH) for immunization, spotted onto microtiter plates for ELISA, and conjugated to affinity media to purify the reactive antisera. For cTnI, we raised an antibody against the signature peptide NITEIADLTQK, and we purified rabbit antisera against 2 of the IL-33 signature peptides, TDPGVFIGVK (Pep-1) and VLLSYYESQHPSNESGDGVDGK (Pep-2).
EVALUATION OF SISCAPA ASSAY PERFORMANCE USING SYNTHETIC PEPTIDES
Although quantification of proteins in plasma is the primary objective, peptide response curves are useful to define the maximum assay performance possible. Many potential sources of sample loss and interference resulting from reduction, alkylation, and tryptic digestion of the protein in plasma are not factors when response curves are generated using peptides. Therefore the assay accuracy, CV, LOD, LOQ and overall peptide recoveries measured under these conditions provide a useful estimate of the maximum that can be achieved. We generated response curves for peptides derived from cTnI and IL-33 by adding 0.16–210 pmol [12C] analyte peptides together (online Supplemental Table 2) with a fixed amount of internal standard peptide into 50 μL reduced, alkylated, and trypsin-digested plasma. As summarized in Table 1 under the Peptide column, the response for cTnI protein (as defined by the peptide response) was linear from 0.6 to 5000 μg/L, with a recovery of 94.4% and intraassay imprecision of 3.2% at 200 μg/L. The response for IL-33 protein (based on Pep-1) was linear from 2.5 to 5000 μg/L, with a recovery of 129% and CV of 0.8% at 200 μg/L. IL-33 Pep-2 had a smaller linear response range (1–200 μg/L), a recovery of 122%, and CV 1.4% at 200 μg/L.
Table 1.
Summary of SISCAPA assay in plasma using peptide and protein standards.
| cTnI | IL-33 Pep-1 | IL-33 Pep-2 | ||||
|---|---|---|---|---|---|---|
| Parameter | Peptide | Protein | Peptide | Protein | Peptide | Protein |
| LOD (0 + 3SD), μg/L | 0.6 | 1.2 | 2.5 | 3.4 | 1.0 | 2.6 |
| LOQ (0 + 10SD), μg/L | 0.9 | 2.8 | 2.8 | 5.2 | 1.5 | 4.7 |
| Linearity, R2 | 1.00 | 1.00 | 1.00 | 1.00 | 0.91 | 0.99 |
| Range of linearity, μg/L | 0.6–5000 | 1.2–5000 | 2.5–5000 | 3.4–16 000 | 1–200 | 2.6–600 |
| Recovery at 200 μg/L, % | 94.4 | – | 129 | – | 122 | – |
| Intraassay imprecision at 200 μg/L, CV | 3.2 | – | 0.8 | – | 1.4 | – |
| Recovery at 20 μg/L (cTnI) or 65 μg/L (IL-33), % | 46–53 | 80–99 | a | |||
| Interassay imprecision at 20 (2) μg/L (cTnI) or 65 (6.5) μg/L (IL-33), % | 12.9 (20.5) | 9.4 (15.8) | a | |||
Results for analysis of IL-33 using Pep-2 antibody not included because of poorer performance in the peptide response curves.
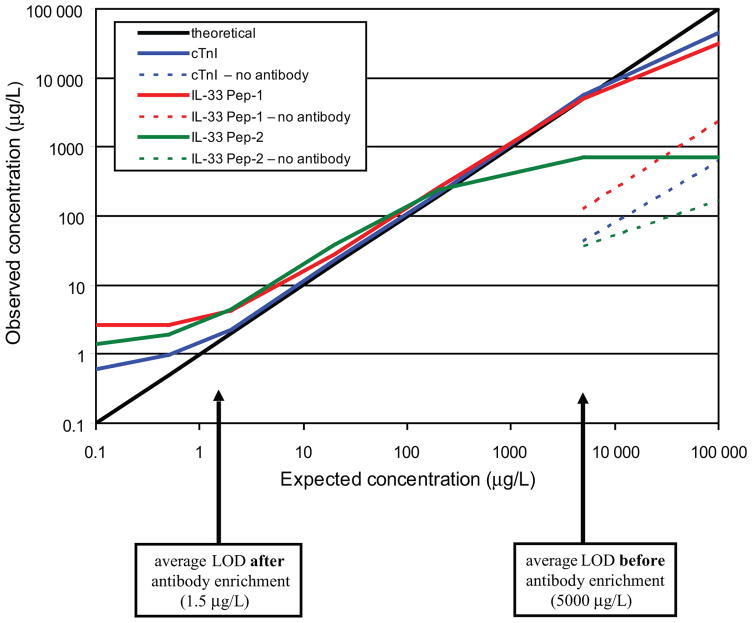
At concentrations <1 μg/L, the response for all 3 peptides became nonlinear owing to background ion current in the product-ion channels monitored (Fig. 2). The origin of this background signal has not been identified, but presumably arises from peptide or other small molecules associated with the antibody and/or tosyl activated magnetic beads. Investigations are underway to see how the antibody preparation, magnetic beads, peptide standards, or input plasma contribute a background signal at lower concentrations. These background signals account for the higher-than-theoretical concentration values observed in the cTnI and IL-33 Pep-1 curves. The higher-than-theoretical values obtained for IL-33 Pep-2 persisted even after accounting for this background signal, suggesting overaddition of this peptide based on original volumes or concentration of the peptide standards. Because it is unlikely that these background signals reflect the presence of true endogenous levels of analyte, it is necessary to include them into the calculation of LOD and LOQ. The LOD and LOQ for cTnI, when measured in undiluted plasma, were 0.6 μg/L (25 pmol/L) and 0.9 μg/L (38 pmol/L), respectively. The LOD was 2.5 μg/L (83 pmol/L) for IL-33 Pep-1 and 1.0 μg/L (33 pmol/L) for IL-33 Pep-2. The LOQ was 2.8 μg/L (93 pmol/L) for IL-33 Pep-1 and 1.5 μg/L (50 pmol/L) for IL-33 Pep-2 (Table 1, Peptide column). At concentrations of IL-33 Pep-2 >1000 μg/L, the response curve flattens, indicating that the binding capacity for this antibody has been exceeded. The apparent lower binding capacity of antibody to IL-33 Pep-2 compared with the other 2 antipeptide antibodies in this assay format may be due to steric hindrance of the larger IL-33 Pep-2 peptide or a lower population of high-affinity antibody in this purified polyclonal mixture. Based on these performance data, IL-33 Pep-1 is the preferred antibody for measuring this target protein in plasma by SISCAPA.
Fig. 2. Peptide response curves before and after antibody enrichment. Peak area ratios of analyte (12C-peptide) and internal standard (13C-peptide) were determined from the XIC of the most abundant transition ions (cTnI [12C]623.3/1018.5, [13C]626.3/1024.5; IL-33 Pep-1 [12C]516.8/816.5, [13C]519.3/821.5; IL-33 Pep-2 [12C]794.4/1084.9, [13C]796.0/1087.5).
The observed concentration value was plotted against the expected concentration value for each individual response curve before (dashed line) and after (solid line) antibody enrichment. The average LOD is labeled at the bottom of the linear range of each antibody–peptide curve. Enrichment factors between 2000 (IL-33 Pep-2) and 8000 (cTnI) were calculated as the ratio of LOD observed before and after antibody enrichment.
These results can be used to calculate an enrichment factor for SISCAPA of these target peptides in plasma. We determined the enrichment factor by comparing the LOD to a separate aliquot analyzed by SID-MRM-MS before SISCAPA (Fig. 2). The ratio of the LOD before and after antibody enrichment was 8333:1 (cTnI), 2000:1 (IL-33 Pep-1), and 5000:1 (IL-33 Pep-2).
EVALUATION OF SISCAPA ASSAY PERFORMANCE USING PROTEINS
To establish the ability of these antibodies to measure protein directly in plasma, we prepared protein response curves by adding protein into plasma before reduction, alkylation, and trypsin digestion to final concentrations of 0, 0.1, 0.5, 2, 20, 200, and 5000 μg/L. These amounts were equivalent to the molar amounts of peptide added at each level (Fig. 1 and online Supplemental Table 2). As shown in Table 1 in the Protein column, the linear range between the peptide assay and the protein assay was similar, differing only in the lower range owing to a slight increase in the LOD of the protein curve (cTnI 1.2–5000 μg/L, IL-33 Pep-1 3.4–16 000 μg/L, and IL-33 Pep-2 2.6–600 μg/L). The LOQs observed for these 3 peptides were 2.8 μg/L (cTnI), 5.2 μg/L (IL-33 Pep-1), and 4.7 μg/L (IL-33 Pep-2).
Peptide recovery from the protein digests was near theoretical for IL-33 but lower than theoretical for cTnI (Table 1). The lower than 100% recovery for the cTnI peptide is most likely caused by incomplete trypsin digestion and/or less-than-quantitative recovery of this peptide from the C18 cartridge used to desalt the digested spiked plasma samples before antibody capture of the signature peptide. Glycosylation of this peptide, which contains an N-linked glycosylation consensus site, could also result in lower recovery before antibody enrichment. However, we have not found any reference to glycosylation of this tissue-derived protein in the literature.
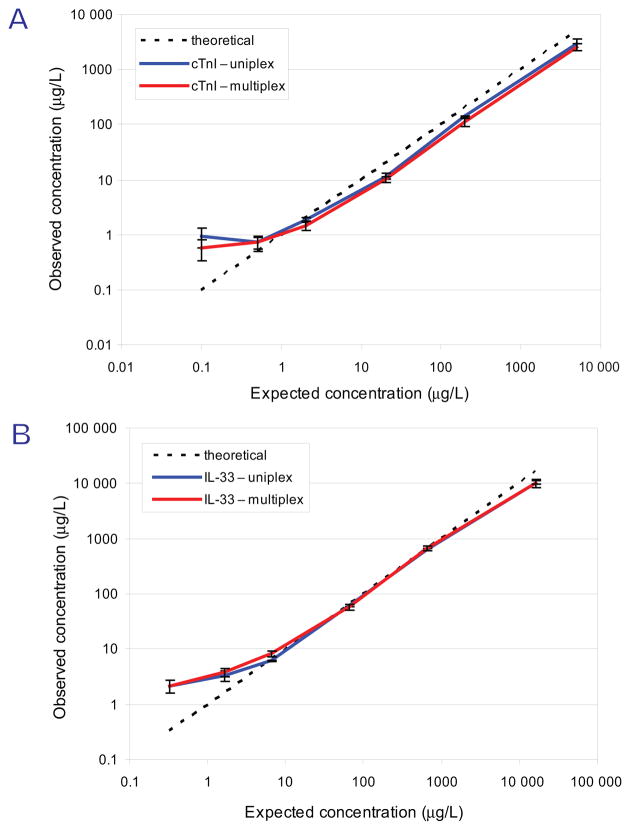
We performed 4 process replicates of the protein curve to determine the interassay reproducibility (Fig. 3). One process replicate was performed using a single antibody bead type per sample. Three additional process replicates were performed using both bead types simultaneously as a multiplexed assay. Only minor differences between the plotted curves were observed (Fig. 3A, cTnI; B, IL-33), indicating minimal deterioration in assay performance when >1 antibody bead is added to 1 aliquot of plasma. As shown in Table 1 in the Protein column, CVs were conserved between each analysis, with a demonstrated overall interassay imprecision of 12.9% at 20 μg/L and 20.5% at 2 μg/L for cTnI and 9.4% at 65μg/L and 15.8% at 6.5μg/L for IL-33 Pep-1. As observed in the peptide curves, the interference of background signal produced a nonlinear trend in concentrations <1 μg/L (for cTnI) and <10 μg/L (for IL-33).
Fig. 3. Reproducibility of multiplexed SISCAPA assay for cTnI and IL-33.
Four independent protein standard addition curves for the proteins are shown. Target peptides were enriched once in separate tubes (uniplex) or 3 times in 1 tube (multiplex). The average protein concentration of 3 injections (uniplex) or 12 injections (multiplex) for cTnI (A) and IL-33 Pep-1 (B) was plotted against the theoretical protein concentration.
CORRELATION OF cTnI QUANTIFICATION BY SISCAPA WITH IMMUNOASSAY USING THE PROTEIN CURVE AND CLINICAL SAMPLES
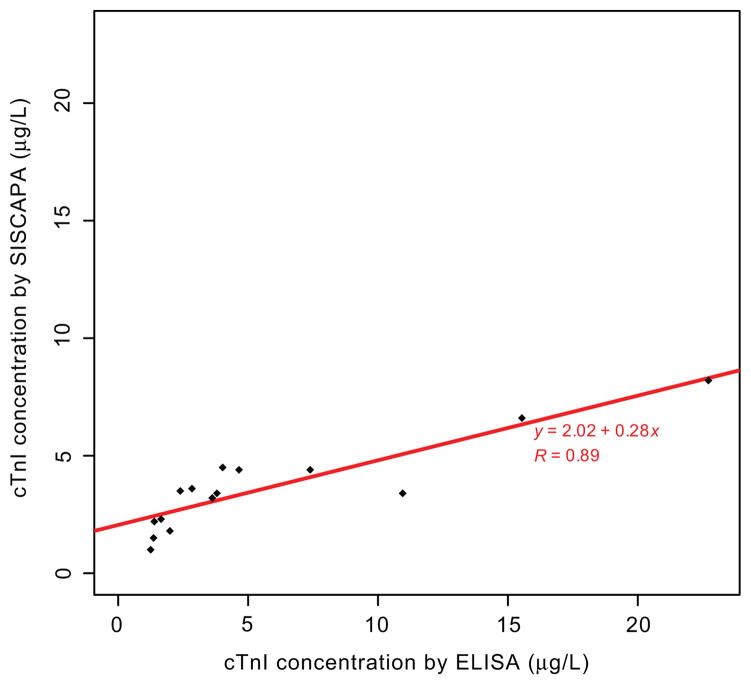
We next compared the results for PMI patient samples determined by SISCAPA with an established immunoassay (Fig. 4). Using the data points (n = 15) with a reported cTnI concentration >1 μg/L, the working LOD for both the cTnI-ELISA and cTnI-SISCAPA, the Pearson correlation coefficient (R) was 0.89 over the concentration range of 1 to 24 μg/L in 5 PMI patients. The Deming regression equation had a slope of 0.28 and an intercept of 2.02.
Fig. 4. Correlation of cTnI concentrations determined by SISCAPA and immunoassay in PMI patient plasma.
The linear fit obtained using Deming regression of the correlation between commercially available immunoassay kit and SISCAPA of cTnI concentration on PMI patient time course samples measured >1 μg/L by both assays. The equation of the robust linear regression line is displayed with the Pearson coefficient of correlation (R) calculated for the comparison of these 2 methods. At concentrations <1 μg/L for cTnI, the interference of background signal produced a nonlinear trend; these data points were therefore excluded from the comparison.
TEMPORAL MEASUREMENTS OF cTnI IN CARDIOVASCULAR PATIENT PLASMA
We measured the plasma concentration of cTnI in patients undergoing therapeutic planned heart attack for the disease hypertrophic cardiomyopathy (20). In this procedure, approximately 1mL alcohol is injected into the septal branch of the left anterior descending artery. This causes endothelial denudation and thrombosis of the vessel and infarction of the heart muscle downstream of the occlusion. The standard biochemical metrics of myocardial infarction, creatine kinase and troponin, rise and fall as with spontaneous myocardial infarction. Because blood can be sampled before and after injury, the clinical scenario provides a unique opportunity to test the robustness of the analytical method. We measured cTnI by SISCAPA at baseline (preinjury) and 10min, 1 h, 2 h, 4 h, and 24 h after onset of injury (Fig. 5). In 4 of 5 patients, the cTnI concentration was 0.6 (0.1) μg/L 1 h after the treatment. Over the next 2–4 h, the plasma concentration of cTnI increased and ranged between 2 and 5 μg/L. Three PMI patients monitored had slightly higher cTnI after 24 h (3–8 μg/L), whereas the concentration of cTnI decreased slightly or leveled off in 2 patients after 4 h. Although all patients experienced large infarctions overall, the heterogeneous response is entirely consistent with clinical observations, since the amount of heartmuscle subserved by the vessels being occluded varies from patient to patient. We also measured IL-33 concentrations in these patient samples using our SISCAPA assay. In all cases, the levels measured were below the LOD for this assay.
Fig. 5. cTnI concentrations determined by SISCAPA in PMI patient plasma.
Plasma concentration of cTnI was measured in 5 patients undergoing PMI. Six time points were sampled over the course of the procedure (0, 0.2, 1, 2, 4, and 24 h). Each measurement is an average of 3 injections; error bars represent SD. A nonlinear scale was used for the time axis.
Discussion
Until recently, only ELISA analyses have been able to detect and quantify cardiac troponins in the low μg/L range in plasma. Other approaches, such as immunoaffinity depletion/limited fractionation coupled to MRM, have been successful in quantifying low-abundance proteins in plasma with comparable precision, reproducibility, and multiplexing capability (4). However, this methodology involves multistep plasma processing, which limits overall sample throughput. Here we demonstrated that SISCAPA can be multiplexed to quantify low-abundance cTnI and IL-33 in plasma with CV <15% in a linear range of 1.2 μg/L to 5 mg/L, a range where many protein biomarkers are expected (8). This is also the first demonstration that SISCAPA MRM–based assays can quantify relative changes in concentration of cTnI pre- and postinjury in clinical samples. Similar trends in cTnI concentration were observed using SISCAPA and a commercial immunoassay in PMI patients in the 1–10 μg/L range, with an excellent correlation (R = 0.89). Our results are consistent with the assay performance and LOQ recently described by Hoofnagle et al. for thyroglobulin (21).
Our primary goal in these studies is not to replace existing immunoassays for proteins like cTnI that are already being measured in clinical practice (22, 23), but rather to leverage the unique advantages of targeted MS-based assay methods (10, 11) to configure assays for target proteins where high-quality antibodies suitable for an immunoassay are not available. We developed and characterized a SISCAPA assay for IL-33, a novel candidate marker of cardiovascular disease (14) for which an immunoassay does not yet exist. Of note, IL-33 is the ligand for ST-2, a protein that is highly upregulated by myocardial cells subjected to mechanical stress (24) and an emerging biomarker of acute and chronic myocardial injury (25). The present study therefore paves the way for future investigation testing the additional clinical utility of simultaneous measurements of this ligand–receptor pair.
Sandwich immunoassays suffer from a variety of nonspecific interference that can be difficult to recognize and correct (26–28). In SISCAPA, plasma proteins are denatured and enzymatically digested in the first processing step. These steps destroy specific and nonspecific protein–protein interactions that may produce false positives and interfere with the integrity of the measurements (21). Because proteins are quantified by measuring peptides produced by the digestion of intact protein, MRM and SISCAPA-MRM assays can be used to measure truncated or degraded forms of the full-length protein, provided that the epitope recognized by the SISCAPA antipeptide antibody has not been compromised. Proteolytic degradation of cTnI in serum can cause measurement inaccuracies for ELISA assays that SISCAPA could, in principle, sidestep (29). SISCAPA assays are also easier to multiplex than ELISA or sandwich immunoassays because off-target interactions are less likely to occur with peptides than proteins, and if they do occur the mass spectrometer can distinguish their presence and identity. In addition, MS-based SISCAPA assays are less expensive to develop than immunoassays, as only 1 antibody rather than a carefully matched pair is required because the mass spectrometer functions as the secondary antibody.
MS-based quantification is based on measuring the ion-counts for the fragment ions from the peptide analyte. Even slight differences in the composition of the matrix (digested plasma) at the time of elution and detection of the analyte can significantly alter the absolute signal of the analyte observed. This is why internal calibrants are routinely employed in LC-MS/MS–based assays (30). In our assays, we employ the ideal internal standard, a 13C and/or 15N-labeled version of the same peptide that has identical physiochemical properties, including retention time, and differs from the analyte only in its parent and fragment masses. Any matrix constituent that causes suppression of the signal from the analyte also affects the internal standard equivalently, and therefore these effects are compensated for in the measurement. Whereas external calibration is typically used in immunoassay, internal calibration methods are beginning to be used, with clear benefits to assay precision being realized (31).
Antiprotein antibody enrichment before SID-MRM-MS has also been used to establish high-quality assays (32–34). These assays require the existence of affinity purification-grade antibodies that are not always available for the majority of proteins. Although antiprotein antibodies are widely used for rapid enrichment of proteins from complex mixtures for characterization by MS (35), quantitative analyses require that the immunoprecipitation (IP) efficiency does not vary from sample to sample. Because the exogenous labeled peptide standard is added after protein IP, interassay reproducibility cannot be internally controlled. Addition of a suitably labeled protein standard (36) could be used to correct for this variability in a manner similar to the use of stable isotopically labeled peptides described here. This approach could also be used to normalize for incomplete protein digestion. However, this method would be practically applicable only to cases in which a very small number of high-quality targets are to be measured.
There are limitations of SISCAPA-MRM assays worth noting. First, as demonstrated here, peptides derived from the same protein can have different assay performance. This may be caused by differences in the antibody affinity, the MS response of the respective peptides, or the recovery of the peptides owing to differences in the efficiency of release by digestion or differential loss upon desalting of the digest. Adding the internal standard peptide earlier in the process would account for losses that occur during desalting before antibody enrichment. Second, the results may not correlate well with immunoassay. The proportional bias observed here (slope = 0.28) may become the rule rather than the exception, since the immunoassay is measuring protein in its native state whereas SISCAPA is measuring a peptide recovered from that protein. More importantly, each assay was developed with separately qualified standards and calibrants. The LOQ of SISCAPA assays, presently in the low μg/L range for proteins in plasma, is limited by interferences from nonspecific binding of peptides to the antibody and resin, as well as traces of target peptide that remain bound to the antibody postpurification (21). Investigations are underway using other types of magnetic beads and elution conditions to reduce nonspecific binding.
The LOD for our MS-based cTnI assay is well above the LOD of current commercial immunoassays (37). Consistent with the expected literature levels of IL-33 in the blood (38), IL-33 was not detected by this assay in patient samples. However, we expect that, as was the case for immunoassays, the sensitivity of the SISCAPA assays will improve over time. Provided that sample is not limiting, increasing the volume of plasma processed from 50μL used here to 500μL should provide a 10-fold increase in sensitivity without altering the assay format. Coupling immunoaffinity depletion to SISCAPA could also potentially improve the overall LOD (39, 40). Specific or nonspecific loss of target proteins can occur on depletion columns, however, and should be defined as part of assay configuration. For example, we have evaluated the recovery of cTnI following depletion of abundant proteins with both the MARS-7 and Genway-12 immunodepletion columns and found that about 90% of this protein was lost to the depletion columns (data not shown). Use of monoclonal antibodies in place of polyclonal antibodies could also increase capture efficiency and translate into an increase in sensitivity. It would also provide a continually renewable source of identical antibody. With higher affinities, less antibody and fewer magnetic beads would be required for the assay. As a result, nonspecific binding would be reduced and more target peptide could be loaded per analysis, ultimately increasing the signal and sensitivity of the SISCAPA assay. Regardless of the current sensitivity limitations, we have achieved the primary goal of clearly establishing that SISCAPA assays can multiplexed and used to measure proteins of clinical interest in the absence of commercial antibodies suitable for immunoaffinity/ELISA assays.
In conclusion, SISCAPA-MRM assays have the potential for high sensitivity, ease of automation, ability to be highly multiplexed, and high sample analysis throughput (10, 11, 21). The present work further establishes that SISCAPA is a flexible alternative to sandwich and ELISA immunoassays, with the potential to triage large lists of candidate proteins and thereby help to alleviate the significant bottleneck that exists between unbiased discovery and clinical validation (2, 7).
Supplementary Material
Acknowledgments
Research Funding: M.S. Sabatine, Roche, Singulex, Donald W. Reynolds Foundation, and National Heart, Lung, and Blood Institute grant U01-HL081341; S.A. Carr, National Heart, Lung, and Blood Institute grant U01-HL081341, NIH grant 1U24 CA126476-02 from the National Cancer Institute as part of the NCI’s Clinical Proteomic Technologies Initiative; R.E. Gerszten, National Heart, Lung, and Blood Institute grant U01-HL081341 and Donald W. Reynolds Foundation.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
The authors thank Dick Cook, Rick Schiavoni, and Katie Tone from the Biopolymers Laboratory at Massachusetts Institute of Technology for the synthesis and purification of the heavy labeled peptides, Will Beavers and Shannon Kerivan from Dana Farber for amino acid analysis of all protein and peptide stocks, Laurie Farrell from Massachusetts General Hospital as the clinical coordinator for the PMI samples, Ed Greenfield for sharing his antibody experience, Leigh Anderson from Plasma Proteome Institute and Terry Pearson from University of Victoria for helpful discussions on the properties and applications of antipeptide antibodies, and Stanley Prince at Covance Research Products and Youling Zou and Xiuwen Liu at Epitomics for their insight on rabbit polyclonal antibodies.
Footnotes
Nonstandard abbreviations: MRM, multiple reaction monitoring; SID-MS, stable isotope dilution mass spectrometry; LOQ, limit of quantitation; SISCAPA, stable isotope standards capture with antipeptide antibodies; cTnI, cardiac troponin I; IL-33, interleukin 33; PMI, planned myocardial infarction; HOCM, hypertrophic obstructive cardiomyopathy; LC-MS/MS, liquid chromatography–tandem mass spectrometry; TQ-MS, triple-quadrupole mass spectrometer; XIC, extracted ion chromatogram; PAR, peak area ratio; LOD, limit of detection; IVD, in vitro diagnostic; KLH, keyhole limpet hemocyanin; IP, immunoprecipitation.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest: Employment or Leadership: None declared.
Consultant or Advisory Role: M.S. Sabatine, Singulex.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature Biotechnol. 2006;24:971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Carr SA, Anderson L. Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory? Clin Chem. 2008;54:1749–52. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–88. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–29. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics. 2004;4:1175–86. doi: 10.1002/pmic.200300670. [DOI] [PubMed] [Google Scholar]
- 6.Bondar OP, Barnidge DR, Klee EW, Davis BJ, Klee GG. LC-MS/MS quantification of Zn-alpha2 glycoprotein: a potential serum biomarker for prostate cancer. Clin Chem. 2007;53:673–8. doi: 10.1373/clinchem.2006.079681. [DOI] [PubMed] [Google Scholar]
- 7.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteom Clin Appl. 2008;2:1386–402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Gerszten RE, Accurso F, Bernard GR, Caprioli RM, Klee EW, Klee GG, et al. Challenges in translating plasma proteomics from bench to bedside: update from the NHLBI Clinical Proteomics Programs. Am J Physiol. 2008;295:L16–22. doi: 10.1152/ajplung.00044.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable 1116 Clinical Chemistry 55:6 (2009) isotope standards and capture by anti-peptide antibodies (SISCAPA) J Proteome Res. 2004;3:235–44. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 11.Whiteaker JR, Zhao L, Zhang HY, Feng LC, Piening BD, Anderson L, Paulovich AG. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams JE, 3rd, Bodor GS, Davila-Roman VG, Delmez JA, Apple FS, Ladenson JH, Jaffe AS. Cardiac troponin I: a marker with high specificity for cardiac injury. Circulation. 1993;88:101–6. doi: 10.1161/01.cir.88.1.101. [DOI] [PubMed] [Google Scholar]
- 13.Bodor GS, Porter S, Landt Y, Ladenson JH. Development of monoclonal antibodies for an assay of cardiac troponin-I and preliminary results in suspected cases of myocardial infarction. Clin Chem. 1992;38:2203–14. [PubMed] [Google Scholar]
- 14.Sanada S, Hakuno D, Higgins LJ, Schreiter ER, McKenzie AN, Lee RT. IL-33 and ST2 comprise a critical biomechanically induced and cardioprotective signaling system. J Clin Invest. 2007;117:1538–49. doi: 10.1172/JCI30634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight C, Kurbaan AS, Seggewiss H, Henein M, Gunning M, Harrington D, et al. Nonsurgical septal reduction for hypertrophic obstructive cardiomyopathy: outcome in the first series of patients. Circulation. 1997;95:2075–81. doi: 10.1161/01.cir.95.8.2075. [DOI] [PubMed] [Google Scholar]
- 16.Sigwart U. Non-surgical myocardial reduction for hypertrophic obstructive cardiomyopathy. Lancet. 1995;346:211–4. doi: 10.1016/s0140-6736(95)91267-3. [DOI] [PubMed] [Google Scholar]
- 17.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 18.Currie LA. Limits for qualitative detection and quantitative determination. Anal Chem. 1968;40:586–93. [Google Scholar]
- 19.Linnet K. Evaluation of regression procedures for methods comparison studies. Clin Chem. 1993;39:424–32. [PubMed] [Google Scholar]
- 20.Lewis GD, Wei R, Liu E, Yang E, Shi X, Martinovic M, et al. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–12. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a lowabundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54:1796–804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christenson RH, Duh SH, Apple FS, Bodor GS, Bunk DM, Dalluge J, et al. Standardization of cardiac troponin I assays: round robin of ten candidate reference materials. Clin Chem. 2001;47:431–7. [PubMed] [Google Scholar]
- 23.Khan NA, Hemmelgarn BR, Tonelli M, Thompson CR, Levin A. Prognostic value of troponin T and I among asymptomatic patients with end-stage renal disease: a meta-analysis. Circulation. 2005;112:3088–96. doi: 10.1161/CIRCULATIONAHA.105.560128. [DOI] [PubMed] [Google Scholar]
- 24.Kakkar R, Lee RT. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov. 2008;7:827–40. doi: 10.1038/nrd2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabatine MS, Morrow DA, Higgins LJ, Mac-Gillivray C, Guo W, Bode C, et al. Complementary roles for biomarkers of biomechanical strain ST2 and N-terminal prohormone B-type natriuretic peptide in patients with ST-elevation myocardial infarction. Circulation. 2008;117:1936–44. doi: 10.1161/CIRCULATIONAHA.107.728022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marks V. False-positive immunoassay results: a multicenter survey of erroneous immunoassay results from assays of 74 analytes in 10 donors from 66 laboratories in seven countries. Clin Chem. 2002;48:2008–16. [PubMed] [Google Scholar]
- 27.Kaplan IV, Levinson SS. When is a heterophile antibody not a heterophile antibody? When it is an antibody against a specific immunogen. Clin Chem. 1999;45:616–8. [PubMed] [Google Scholar]
- 28.Katrukha AG, Bereznikova AV, Esakova TV, Pettersson K, Lovgren T, Severina ME, et al. Troponin I is released in bloodstream of patients with acute myocardial infarction not in free form but as complex. Clin Chem. 1997;43:1379–85. [PubMed] [Google Scholar]
- 29.Shi Q, Ling M, Zhang X, Zhang M, Kadijevic L, Liu S, Laurino JP. Degradation of cardiac troponin I in serum complicates comparisons of cardiac troponin I assays. Clin Chem. 1999;45:1018–25. [PubMed] [Google Scholar]
- 30.Zimmer D. Introduction to quantitative liquid chromatography-tandem mass spectrometry (LC-MS-MS) Chromatographia. 2003;57:S325–32. [Google Scholar]
- 31.Nichkova M, Dosev D, Gee SJ, Hammock BD, Kennedy IM. Multiplexed immunoassays for proteins using magnetic luminescent nanoparticles for internal calibration. Anal Biochem. 2007;369:34–40. doi: 10.1016/j.ab.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicol GR, Han M, Kim J, Birse CE, Brand E, Nguyen A, et al. Use of an immunoaffinity-mass spectrometry-based approach for the quantification of protein biomarkers from serum samples of lung cancer patients. Mol Cell Proteomics. 2008;7:1974–82. doi: 10.1074/mcp.M700476-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Berna M, Ott L, Engle S, Watson D, Solter P, Ackermann B. Quantification of NTproBNP in rat serum using immunoprecipitation and LC/MS/MS: a biomarker of drug-induced cardiac hypertrophy. Anal Chem. 2008;80:561–6. doi: 10.1021/ac702311m. [DOI] [PubMed] [Google Scholar]
- 34.Berna MJ, Zhen Y, Watson DE, Hale JE, Ackermann BL. Strategic use of immunoprecipitation and LC/MS/MS for trace-level protein quantification: myosin light chain 1, a biomarker of cardiac necrosis. Anal Chem. 2007;79:4199–205. doi: 10.1021/ac070051f. [DOI] [PubMed] [Google Scholar]
- 35.Labugger R, Simpson JA, Quick M, Brown HA, Collier CE, Neverova I, Van Eyk JE. Strategy for analysis of cardiac troponins in biological samples with a combination of affinity chromatography and mass spectrometry. Clin Chem. 2003;49:873–9. doi: 10.1373/49.6.873. [DOI] [PubMed] [Google Scholar]
- 36.Singh R, Crow FW, Babic N, Lutz WH, Lieske JC, Larson TS, Kumar R. A liquid chromatography-mass spectrometry method for the quantification of urinary albumin using a novel 15N-isotopically labeled albumin internal standard. Clin Chem. 2007;53:540–2. doi: 10.1373/clinchem.2006.078832. [DOI] [PubMed] [Google Scholar]
- 37.Venge P, James S, Jansson L, Lindahl B. Clinical performance of two highly sensitive cardiac troponin I assays. Clin Chem. 2009;55:109–16. doi: 10.1373/clinchem.2008.106500. [DOI] [PubMed] [Google Scholar]
- 38.Sakashita M, Yoshimoto T, Hirota T, Harada M, Okubo K, Osawa Y, et al. Association of serum interleukin-33 level and the interleukin-33 genetic variant with Japanese cedar pollinosis. Clin Exp Allergy. 2008;38:1875–81. doi: 10.1111/j.1365-2222.2008.03114.x. [DOI] [PubMed] [Google Scholar]
- 39.Martosella J, Zolotarjova N, Liu H, Nicol G, Boyes BE. Reversed-phase high-performance liquid chromatographic prefractionation of immunodepleted human serum proteins to enhance mass spectrometry identification of lower-abundant proteins. J Proteome Res. 2005;4:1522–37. doi: 10.1021/pr050088l. [DOI] [PubMed] [Google Scholar]
- 40.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, et al. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5:2167–74. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.