Abstract
In biological systems, membrane fusion is mediated by specialized proteins. Although soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptors (SNAREs) provide the minimal molecular machinery required to drive membrane fusion, the precise mechanism for SNARE-mediated fusion remains to be established. Here, we used atomic force microscope (AFM) spectroscopy to determine whether the pulling force generated by interacting SNAREs is directly coupled to membrane fusion. The mechanical strength of the SNARE binding interaction was determined by single molecule force measurements. It was revealed that the forced unbinding of the SNARE complex formed between opposing (trans) bilayers involves two activation barriers; where the steep inner barrier governs the transition from the bound to an intermediate state and the outer barrier governs the transition between the intermediate and the unbound state. Moreover, truncation of either SNAP-25 or VAMP 2 reduced the slope of the inner barrier significantly and, consequently, reduced the pulling strength of the SNARE complex; thus, suggesting that the inner barrier determines the binding strength of the SNARE complex. In parallel, AFM compression force measurements revealed that truncated SNAREs were less efficient than native SNAREs in facilitating hemifusion of the apposed bilayers. Together, these findings reveal a mechanism by which a pulling force generated by interacting trans-SNAREs reduces the slope of the hemifusion barrier and, subsequently, facilitates hemifusion and makes the membranes more prone to fusion.
Introduction
Membrane fusion is mediated by a specialized family of proteins, soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP) receptors (SNAREs). Since their introduction as the minimal machinery for membrane fusion,1 SNARE proteins have been extensively used in model membrane systems to investigate membrane fusion.2-5 The SNARE hypothesis postulates that the interaction of cognate SNAREs between opposing membranes drives triggered, as well as, constitutive membrane fusion.6-8 The neuronal v-SNARE, vesicle-associated membrane protein (VAMP 2, also referred to as synaptobrevin 2) and the plasma membrane t-SNAREs, syntaxin 1A and SNAP-25 (synaptosomal-associated protein of 25 kD) are expressed in the presynaptic terminal and mediate vesicle fusion with the plasma membrane during neurotransmitter release. Interaction between cognate v- and t-SNAREs in the opposite (trans) membranes forms a ternary core complex that contributes significantly to the energy required for membrane fusion.9,10 Although numerous studies investigating the interactions amongst SNAREs or between SNAREs and other proteins have been carried out,11-13 the precise mechanism of SNARE-mediated membrane fusion remains to be established.
It has been suggested that the oblique angle of insertion of the transmembrane domain (TMD) of VAMP 2 promotes lipid mixing without formation of the SNARE complex.14 This is consistent with the role of tilted peptides,15 where the 36° angle of the TMD of VAMP, relative to the normal of the lipid bilayer, destabilizes the membrane and promotes fusion. Moreover, most SNAREs are anchored in the membrane at their C-terminal end and the assembly of the SNARE complex has been suggested to occur in the N- to the C-terminal direction.16-18 Consequently, the membranes in which the SNAREs are anchored are pulled together into a close proximity that is favorable for membrane fusion. However, rather than merely providing apposition between the membranes to promote fusion, emerging evidence suggests that the SNARE interaction plays an additional role in destabilizing the bilayers through movement of the SNARE transmembrane segments.19-22
In order to address the question of how interacting trans-SNAREs act upon opposing membranes to mediate their fusion, we have established an experimental system that permitted the detection of induced hemifusion and fusion events between apposed floating lipid bilayers using atomic force microscope (AFM) force spectroscopy.23 This experimental approach was used to measure the compression force required to induce the hemifusion of the phospholipid bilayers (Fig. 1). We have shown previously that membrane fusion is facilitated when v- and t-SNAREs are present in the opposing bilayers, and that the inhibition of the trans-SNARE interaction by the soluble cytoplasmic domain of VAMP eliminated the observed facilitation of membrane fusion.21 Since apposition of the bilayers in our experimental system is accomplished by the AFM-applied compression, the above findings suggested that the mere presence of SNARE proteins in the bilayers is insufficient to promote their fusion, and that the interactions of the cytoplasmic domains of trans-SNAREs are required to facilitate hemifusion of the bilayers and subsequently membrane fusion. Here, we demonstrate that the trans-interaction of the SNARE cytoplasmic domains provides the necessary pulling force to destabilize the lipid bilayers and facilitate hemifusion of the membranes.
Fig. 1.
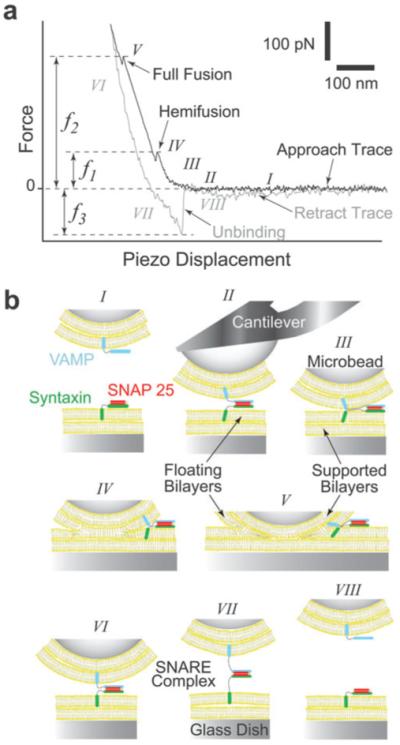
AFM force vs. piezo displacement measurement of interactions between opposing lipid bilayers and SNAREs. (a) A typical AFM force scan measurement showing hemifusion and full fusion of the compressed lipid bilayers and the unbinding of the SNARE complex during approach and retraction of the cantilever, respectively.21 The compression force required to induce bilayer hemifusion (f1) and fusion (f2) is measured at the onset of each event. Here, we generically refer to f1 as the fusion force. Alternatively, the unbinding force (f3) is measured during the sharp transition in the retraction trace as the SNARE complex dissociates under pulling. (b) Cartoon of our experimental system (not to scale) depicting the different steps (roman numerals) during the force scan measurement shown in (a). Lipid bilayers were formed on the glass dish and glass microbead attached to the cantilever tip. Upon approach of the cantilever toward the substrate, SNAREs embedded in the opposing bilayers form a complex (II) and the bilayers hemifuse (IV) and fully fuse (V) under compression. During the retraction phase, the SNARE complex is extended (VII) before it dissociates (VIII) under pulling.
Materials and methods
Plasmids and protein purification
Plasmids to generate recombinant full-length VAMP 2 (pTW2),1 and the full-length t-SNARE heterodimer (syntaxin 1A and SNAP-25; pTW34),1 were provided by J. E. Rothman (Columbia University, New York, NY). cDNA encoding full-length syntaxin 1A24 (provided by R. H. Scheller, Genentech; San Francisco, CA) was sub-cloned into a pTrc-His vector (Invitrogen Life Technologies; Carlsbad, CA), resulting in a N-terminal His6-tag. We generated the SNAP-25 truncations that mimic the BoNT/A or BoNT/E (mut2-SNAP-25 or mut1-SNAP-25, respectively) cleavage products by placing a stop codon after amino acid position 197 or 180, respectively, using standard PCR methods and pTW34 as the parent construct.25 Proteins were expressed and purified as described.3,25 Briefly, bacterial pellets were resuspended in 25 mM HEPES-KOH, 400 mM KCl, 20 mM imidazole and 5mM β-mercaptoethanol. Bacterial extracts were mixed with Ni-NTA agarose (Qiagen; Valencia, CA) for 2 hours at 4 °C. Beads were washed twice in wash buffer (25 mM HEPES-KOH, 400 mM KCl, 20 mM imidazole and 5 mM β-mercaptoethanol, 1 mM MgCl2) plus 10 μg ml−1 DNase and RNase (Roche Applied Science; Indianapolis, IN) to remove residual RNA/DNA. Two more washes were carried out in the resuspension buffer. Proteins were eluted from the beads in resuspension buffer with 500 mM imidazole, 10% glycerol (w/v) and 1% n-octylglucoside (Research Products International Corp.; Philadelphia, PA). SNAP-25 was purified as above except that n-octylglucoside was omitted from the elution buffer, and soluble protein was dialyzed using dialysis tubing from Spectrum Labs (Rancho Dominguez, CA) against 25 mM HEPES-KOH, 100 mM KCl, 10% glycerol (w/v) and 1 mM DTT.
Reconstitution of SNAREs into vesicles and bilayer formation
Chicken egg l-α-phosphatidylcholine (egg PC) was purchased from Avanti Polar Lipids (Alabaster, AL). Reconstitution of v-SNARE or t-SNARE vesicles was carried out as previously described3,25 with modifications. In brief, v-SNARE or binary t-SNAREs were reconstituted in separate populations of 100% egg PC vesicles by the detergent depletion method followed by a floatation step on an Accudenz density gradient, as previously described.3,25 Protein concentrations were adjusted to yield an average of 25 copies of protein molecules per vesicle (1 : 1000 protein to lipid ratio). SNARE-free vesicles were prepared as described previously3,25 but using 100% egg PC. For each experiment, floating bilayers were formed by vesicle adsorption and fusion to hydrophilic surfaces as previously described21 with modification. In brief, SNARE-free and v- or t-SNARE vesicles were adsorbed separately to the cleaned glass dish and glass microbead attached to the cantilever tip for 1 h at 4 °C. After adsorption, the bilayers were gently washed, while always in solution, three times with Tris-buffered saline (TBS; 10 mM Tris–100 mM NaCl, PH 7.2) to remove excess vesicles. The AFM was assembled next, and necessary temperature adjustments were performed before initiation of the experiments.
Atomic force microscope
We used a custom built AFM, in which the lateral and vertical scans are decoupled.26 In brief, the sample sits on an X–Y stage which can be adjusted relative to the cantilever mounted on a stacked piezo-electric transducer (Physik Instrumente L.P.; Auburn, MA). The piezo-electric transducer provides the necessary vertical movement (0 to ~10 μm range) to approach and retract the cantilever from the stationary substrate (dish).
AFM cantilevers were purchased from Veeco (model MLCT-AUHW, part 00-103-0925; Woodbury, NY), and the largest V-shaped cantilever with a nominal spring constant of 10 mN m−1 was used in all experiments after attachment of the glass microbead (~50 μm diameter; Polysciences, Inc., Warrington, WA) using micromanipulation. The microbead and custom glass dishes provided hydrophilic substrates for bilayer formation after extensive cleaning. Custom software was used to calibrate the cantilever tip based on thermal noise analysis,27 and to control the position of the piezo-electric transducer and timing during force scan measurements. A dry Peltier element was positioned underneath the glass dish to maintain desired temperatures ±0.3 °C.
Substrate preparation for bilayer formation
Glass dishes and stainless steel utensils were boiled in distilled water containing ~10% RBS 35 detergent (Pierce; Rockford, IL) and ethanol, and rinsed extensively with distilled water. Cantilevers with attached glass microbeads were soaked in 1% n-octyl β-d-glucopyranoside, then in 100% ethanol followed by ultraviolet (UV) irradiation with extensive rinsing in nanopure water (18 MΩ cm; Barnstead; Dubuque, IA) in between. Finally and immediately before vesicle adsorption and bilayer formation, cleaned cantilevers and glass dishes were further treated for 5 min in a nitrogen-plasma cleaner (Harrick Plasma; Ithaca, NY).
AFM force measurement of SNARE-mediated bilayer hemifusion and the unbinding of the SNARE complex
During an AFM force scan measurement, the approach and retract traces correspond to the movement of the cantilever tip toward and away from the substrate, respectively. As the cantilever is lowered and pressed against the substrate during approach, the cantilever is subjected to forces that result in its upward bending. Deflection of the cantilever is monitored by the position of a laser beam reflected off the coated back-side of the cantilever tip onto a two-segment photodiode. Down or upward deflection in the cantilever signifies an attractive or repulsive interaction, respectively. The change of the laser position on the photodiode is calibrated based on the force causing the cantilever deflection and the resulting force scan represents the interaction force vs. the displacement of the piezo-electric transducer (Fig. 1(a)). It should be noted that the piezo displacement largely results in the deflection (bending) of the AFM cantilever and a negligible compression of the apposed bilayers. All force measurements were performed at ~25 °C in TBS and forces were corrected for hydrodynamic drag on the cantilever and the attached bead (drag coefficient 20 pN s μm−1).
During approach in the presence of opposing (trans) v- and t-SNARE bilayers, interaction between VAMP 2 and binary syntaxin 1A/SNAP-25 leads to the formation of a trans-SNARE complex (illustration II of Fig. 1(b)). Continued approach leads to further compression (illustration III) of the bilayers, which results in an increase in the force subjected to the cantilever. With further approach, the contacting bilayers hemifuse (illustration IV) and fully fuse (illustration V) under compression.21 Typically, we increased the applied compression force from ~300 to ~1000 pN and the contact time from 0.5 to 3 s with increasing scan velocity. Hemifusion and fusion are detected during the instability (jump) in the force scan. The jump is due to the sudden displacement of the cantilever tip toward the substrate.
On the other hand, during retraction the force decreases progressively toward zero as the cantilever returns to its relaxed position. However, in the presence of adhesion which was observed only in the presence of SNARE in the bilayers, downward deflection of the cantilever leads to an increase in the force in the opposite direction as the SNARE complex is extended (illustration VI) until de-adhesion (i.e., unbinding of the SNARE complex) takes place (illustration VII). The force required to induce hemifusion (f1) or fusion (f2) and the unbinding (f3) of the SNARE complex are derived from the product of the spring constant (κ) of the cantilever and the extent of its deflection.
Data analysis and interpretation
Custom data analysis programs were written in IGOR software to detect the jump events during the approach phase of the AFM force scans and to measure the associated compression forces. We typically carried out 300–400 force scans for each compression/loading rate. Different compression and loading rates were accomplished by varying the AFM scan velocity during approach and retraction of the cantilever, respectively. A minimum of triplicate experiments for each condition were performed on different days. The compression rate was calculated by multiplying the cantilever spring constant by the rate of approach of the cantilever. Alternatively, the loading rate was obtained from the product of the rate of retraction of the cantilever by the system spring constant, which is derived from the slope of the force–displacement curve during step VII of the force scan in Fig. 1(a). 15–20% of the force scans produced jump events, from which fusion force measurements were obtained. More than 60% of the selected force scans with jump events displayed simultaneous unbinding events. Therefore, the simultaneous presence of jump and unbinding events during force scan measurements was considered as a signature for SNARE-mediated membrane fusion. Moreover, given the SNARE surface density in our lipid bilayers, a frequency of ~20% of unbinding events was detected which insured ≥ 85% probability that the unbinding events were mediated by single SNARE complexes.28,29 Both measured fusion and unbinding forces were grouped in histograms (10 pN bin size) and the most probable force was obtained from the Gaussian fit to histograms and plotted against the logarithm of the corresponding compression (loading) rate to generate the dynamic force spectra (DFS). The DFS were analyzed in the context of the Bell–Evans model,30,31 which postulates that an applied force f along the reaction coordinate x contributes a mechanical potential fx toward the suppression of the potential of membrane fusion or the SNARE complex dissociation. Each linear regime in the force vs. log(compression/loading rate) of the DFS is interpreted as evidence for a prominent activation barrier in the fusion and molecular unbinding processes. In the Bell–Evans model, each of the activation barriers is characterized by two parameters xβ(ϕ) and , where the indices β and ϕ refer to molecular unbinding and membrane fusion, respectively. Experimentally, xβ(ϕ) and were derived from fitting the following equation:
| (1) |
to the acquired DFS, where is the most probable unbinding (fusion) force of the SNARE complex (membrane fusion) at the loading (compression) rate rf. Using the derived energy barrier parameters, we can estimate the relative difference (δΔG) in the height of the energy barrier between two similar systems or between the inner and outer barriers of the same system by:
| (2) |
where the indices β1 and β2 refer, respectively, to the inner and outer activation barriers during forced unbinding of the SNARE complex, and ϕ and ϕ’ refer to the rate of membrane fusion in the compared systems.
For a two energy barrier process such as during the unbinding of the SNARE complex, the dissociation rate under an applied pulling force (f) can be derived using the following equation:
| (3) |
where is the overall dissociation rate of the SNARE complex as a function of a pulling force f, and the indices β1 and β2 refer to the inner and outer energy barriers, respectively.
Results
SNAREs facilitate membrane hemifusion
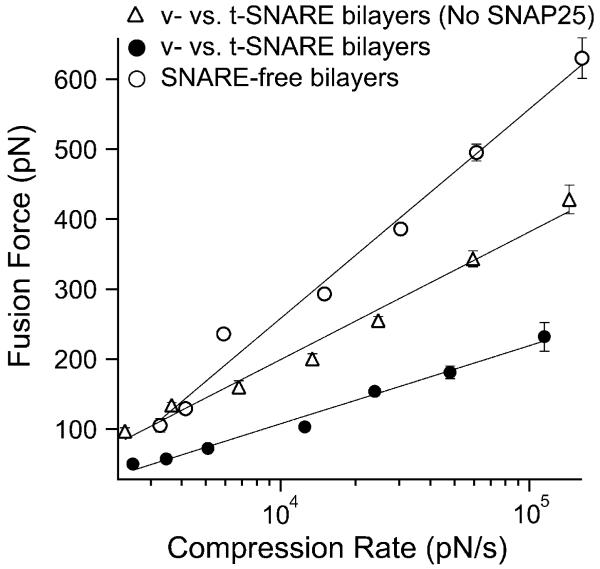
AFM compression experiments of opposing v- and t-SNARE bilayers reconstituted with VAMP 2 and binary syntaxin 1A/SNAP-25, respectively, were carried out as illustrated in Fig. 1. The compression force required to induce hemifusion of the bilayers (i.e., fusion force) was measured at different compression rates (see Methods). The plot of the most probable fusion force vs. the logarithm of the compression rate, also referred to as the dynamic force spectrum (DFS), was derived from these measurements (Fig. 2). The fusion DFS of the v- and t-SNARE bilayers revealed a significant reduction in the fusion force as compared to the fusion of SNARE-free bilayers. These measurements confirmed our earlier finding21 that the presence of SNARE proteins in the lipid bilayers facilitates their hemifusion by lowering the energy requirements.
Fig. 2.
Dynamic force spectrum (DFS) of bilayer hemifusion. With SNAREs in the opposing bilayers, the compression (fusion) force was significantly reduced as compared to SNARE-free bilayers. This revealed facilitation of membrane fusion due to the interaction of VAMP (v-SNARE) with syntaxin/SNAP-25 (v-SNAREs) in the opposite bilayers. Omission of SNAP-25 from the t-SNARE bilayers resulted in partial fusion facilitation due to the direct weaker binary interaction of VAMP with syntaxin. Lines are fits of eqn (1) to the data points. Force values at the different compression rates were derived from distribution histograms of force measurements that were acquired in triplicate experiments on different days (see Materials and methods section). Error bars represent the standard error of the mean (s.e.m.) of all the force values in the distribution histograms of the same compression rates (n; 50 < n < 312).
In the current work, we investigated the contribution of the different components of the SNARE complex in facilitating membrane fusion; in particular, the roles of SNAP-25 and VAMP 2. Although SNAP-25 was originally thought of as an essential component of the fusion machinery,32,33 recent studies revealed that it might not be required for membrane fusion under certain conditions.2,34 To further examine this issue, we carried out compression force measurements between v- and t-SNARE bilayers that were devoid of SNAP-25. In the absence of SNAP-25, the fusion force was partially reduced to an intermediate level between that of SNARE-free and v- vs. t-SNARE bilayers (Fig. 2). We next substituted the full-length SNAP-25 in the binary syntaxin/SNAP-25 complex in the t-SNARE bilayers with truncated mutant forms (mut1-SNAP-25 or mut2-SNAP-25), which have been truncated to mimic the cleavage of SNAP-25 by botulinum neurotoxin (BoNT) E or A, respectively.25
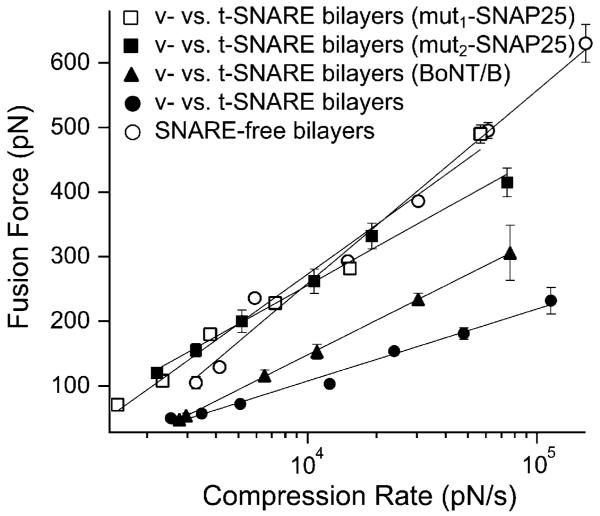
In the presence of either mut1-SNAP-25 or mut2-SNAP-25, an upward shift in the fusion force spectrum toward that of SNARE-free bilayers was observed, which is indicative of suppression in the fusion facilitation that was observed in presence of full-length SNAP-25 (Fig. 3).
Fig. 3.
SNARE perturbations suppress the observed facilitation of membrane fusion. An upward shift in the fusion force spectrum (DFS) was observed upon cleavage of VAMP 2 in the v-SNARE bilayers or substitution of SNAP-25 with mutant forms in the t-SNARE bilayers. Such shift in the DFS is due to the increased compression force that is required to induce bilayer hemifusion and is indicative of the increased overall energy requirements for membrane fusion pursuant to the SNARE perturbations in our experimental system. Lines are fits of eqn (1) to the data points. Error bars are the s.e.m. (33 < n < 289).
To investigate the role of VAMP 2 in the facilitation of membrane fusion, we conducted compression experiments after neurotoxin treatment of the v-SNARE bilayers. Chlostridial neurotoxins are Zn2+-dependent endopeptidases and are known to completely inhibit exocytosis.35,36 In our experimental system, BoNT/B treatment of v-SNARE bilayers (250 nM + 250 μM Zn2+ for 1 h at 37 °C) resulted in an upward shift in the fusion force spectrum especially at high compression rates (Fig. 3). The observed partial reduction in the compression force is due to the remaining portion of the cytoplasmic domain of VAMP 2 after BoNT/B treatment (see Discussion). Repeating the same experiments without the addition of Zn2+ resulted in force values similar to those measured before BoNT/B treatment, thus, confirming the specific effect of BoNT/B on VAMP.
Dynamic strength of the SNARE complex
It has been proposed that formation of the SNARE complex occurs via an N- to C-terminal zippering of the helices of the SNARE cytoplasmic domains.18,37,38 This process may involve overcoming multiple activation barriers corresponding to different conformational states. Recent studies have suggested that pulling force generated during the association of the SNARE molecules is directly coupled to the destabilization of the lipid bilayers and subsequent membrane fusion.19,22 We have carried out single-molecule AFM force measurements in order to characterize the energy landscape of the SNARE complex. In performing these experiments, our assumption is that the trajectory of the forced unbinding of the SNARE complex in the AFM experiment is equal and opposite to that of the SNARE complex formation during membrane fusion. With this assumption, the slope of the inner most barrier of the energy landscape provides an estimate of the maximal pulling force produced during complex formation.
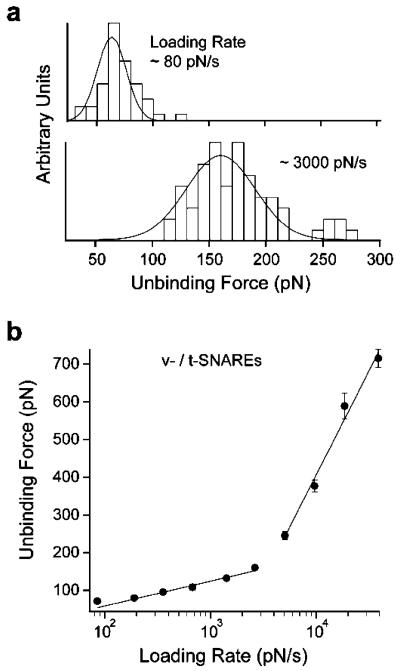
Unbinding force measurements of the unitary SNARE interaction were acquired by AFM force spectroscopy using purified v- and t-SNAREs reconstituted in lipid bilayers (see Methods section). Fig. 4(a) shows representative histograms of unbinding forces acquired at different loading rates. The plot of the most probable unbinding force vs. the logarithm of the loading rate (DFS) for the interaction between VAMP 2 and binary syntaxin 1A/SNAP-25 is presented in Fig. 4(b). The DFS has two force loading regimes, a slow and a fast one. The presence of two loading regimes reveals at least two energy barriers in the dissociation pathway of the v-/t-SNARE complex; an inner and an outer barrier at fast and slow loading rates, respectively. During the slow loading regime (~60 to ~3000 pN s−1), the unbinding force ranged from ~50 to ~180 pN. A sharp transition in the force occurred at loading rate ~5000 pN s−1, where the slope of the force spectrum increased significantly during the fast loading regime; the unbinding force ranged from ~250 to ~750 pN between loading rates of ~5000 and ~30 000 pN s−1.
Fig. 4.
Dynamic force spectrum (DFS) of the forced unbinding of the SNARE complex. (a) Distribution histograms of unbinding forces measured at the specified loading rates. The most probable unbinding force is derived from the Gaussian fit to the histogram. It is evident that the unbinding force increases with increasing loading rate. (b) Two force loading regimes were revealed in the DFS, which indicated the presence of two energy barriers in the dissociation pathway of the SNARE complex. Lines are fits of eqn (1) to the data points. Similar to the compression force experiments, the error bars are the s.e.m. of all the unbinding force values measured at the corresponding loading rates (40 < n < 283). The DFS did not fit well to a single barrier model with either a harmonic or cubic potential.39,40
To investigate the effect of the mechanical strength of the SNARE complex on the hemifusion of the lipid bilayers, we performed unbinding force measurements pursuant to perturbations to either VAMP 2 or SNAP-25 that were present in the opposing v- and t-SNARE bilayers, respectively. The measured unbinding force was significantly reduced after treatment of VAMP 2 with BoNT/B, especially, during the fast loading regime (Fig. 5); at loading rate ~30 000 pN s−1, we measured an unbinding force of ~320 pN whereas a force of ~710 pN was measured for intact SNAREs. Similarly, substitution of full-length SNAP-25 in the binary syntaxin/SNAP-25 complex with either mut1-SNAP-25 or mut2-SNAP-25 resulted in a significant reduction of the unbinding force of the SNARE complex (Fig. 5). Moreover, t-SNARE bilayers containing syntaxin 1A, but not SNAP-25, interacted specifically with opposing v-SNARE bilayers containing VAMP 2, though the unbinding forces were lower than those in the presence of full-length SNAP-25 (Fig. 5).
Fig. 5.
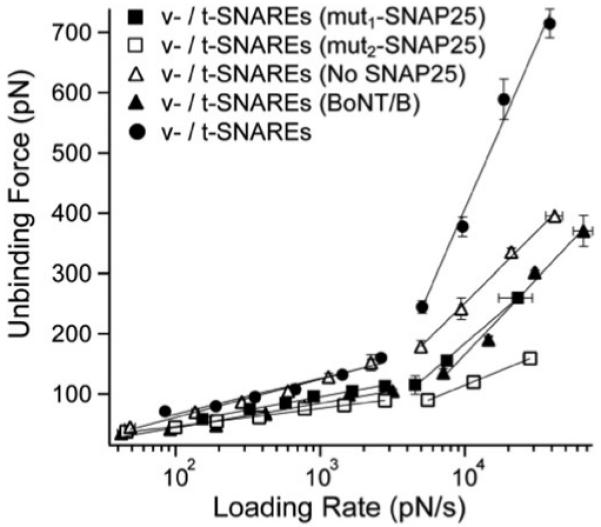
SNARE perturbations interfere with the mechanical strength of the SNARE complex. The unbinding force of the SNARE complex is significantly reduced upon cleavage of VAMP 2 in the v-SNARE bilayers with BoNT/B or omission of SNAP-25 from the t-SNARE bilayers. A further reduction in the SNARE binding strength was observed in presence of mut1-SNAP-25 or mut2-SNAP-25 in the t-SNARE bilayers as compared to the absence of SNAP-25. This was interpreted as the result of the interference of truncated SNAP-25 mutants with the direct binary interaction of VAMP with syntaxin; hence, a weaker SNARE interaction takes place in presence of mut1-SNAP-25 or mut2-SNAP-25 as compared to the complete absence of SNAP-25. However, the strongest v-/t-SNARE interaction takes place in presence of the full-length SNAP-25 in the a binary syntaxin/SNAP-25 complex in the t-SNARE bilayers. Error bars are the s.e.m. (16 < n < 139).
Conclusions
It has been suggested that the formation of the SNARE complex exerts a pulling force on the transmembrane domains and subsequently promotes membrane fusion.19,20,22 Here, we examined how the strength of the SNARE interaction affects hemifusion of model membranes. Using AFM force spectroscopy, we characterized the strength of the SNARE interaction by measuring the pulling force required to unbind the SNARE complex. The experimental conditions were optimized to yield single interactions by using low surface density of SNAREs21 and controlling the AFM scanning parameters.41 The measured unbinding forces were consistent with reported values for single SNARE interactions42,43 and the persistent detection of unbinding events throughout the experiments (6–8 h) ruled out the possibility that SNAREs were extracted from the lipid bilayer during the course of the experiments. It should be noted that such unbinding events were not detected with SNARE-free bilayers.21 Therefore, the exclusive presence of the unbinding events only when SNAREs are present in the bilayers and the change in the unbinding force pursuant to SNARE perturbations confirmed the specificity of the SNARE interaction.
Using a dynamic force approach, we acquired unbinding force spectra for the SNARE complex. As shown in Fig. 4(b), the force spectrum reveals two loading regimes, where the unbinding force increased linearly with the logarithm of the loading rate. This is in contrast to previous studies which have shown a single loading regime during forced unbinding of the SNARE complex.42,43 We have attempted to fit our measurements to a single barrier model with either a harmonic or cubic potential in the context of the Dudko–Hummer model.39,40 Neither the harmonic or cubic potential model was able to adequately describe our force measurements. In earlier studies, purified SNAREs were immobilized directly to solid supports, whereas, they are embedded in lipid bilayers in our experimental system. Perhaps, immobilization of SNAREs to the surfaces interfered with the full assembly of the SNARE complex. The observation of two loading regimes in our results is consistent with an unbinding process whereby the fully assembled SNARE complex must overcome two prominent activation barriers; first, an inner barrier and subsequently an outer barrier before the v- and t-SNAREs are dissociated. In the context of the Bell–Evans model,30,31 a pulling force across the SNARE complex tilts the energy landscape, suppressing the outer barrier to a greater extent than the inner barrier. At large pulling forces, the outer barrier is completely suppressed, allowing the inner barrier to govern the dissociation of the complex. Parameters characterizing the dissociation rate () and barrier width (xβ) of both the inner and outer barriers (Table 1) were obtained by fitting the Bell–Evans model (see Materials and methods section) to the AFM force measurements from the fast and slow loading regimes of the force spectrum, respectively.44
Table 1.
Energy barrier parameters for the fusion of the bilayers and the dissociation of the SNARE complex
| Bilayer | xβ1/Å | /s−1 | xβ2/Å | /s−1 | xϕ/Å | /s−1 |
|---|---|---|---|---|---|---|
| SNARE-free | N/A | N/A | N/A | N/A | 0.31 | 10.57 |
| v-/t-SNAREs | 0.17 | 7.65 | 1.59 | 0.29 | 0.85 | 22.5 |
| v-/t-SNAREs (No SNAP25) | 0.40 | 8.47 | 1.70 | 0.39 | 0.52 | 10.22 |
| v-/t-SNAREs (mut1-SNAP25) | 0.97 | 15.6 | 3.18 | 0.21 | 0.37 | 7.74 |
| v-/t-SNAREs (mut2-SNAP25) | 0.46 | 14.3 | 2.17 | 0.34 | 0.47 | 5.97 |
| v-/t-SNAREs (BoNT/B) | 0.37 | 20.1 | 2.32 | 0.46 | 0.53 | 19.14 |
k° is the dissociation rate of the activation barrier for fusion (ϕ) and unbinding (β), and the indices, 1 and 2, refer to the inner and outer barrier, respectively. x describes the width of the energy barrier during fusion (ϕ) and unbinding (β).
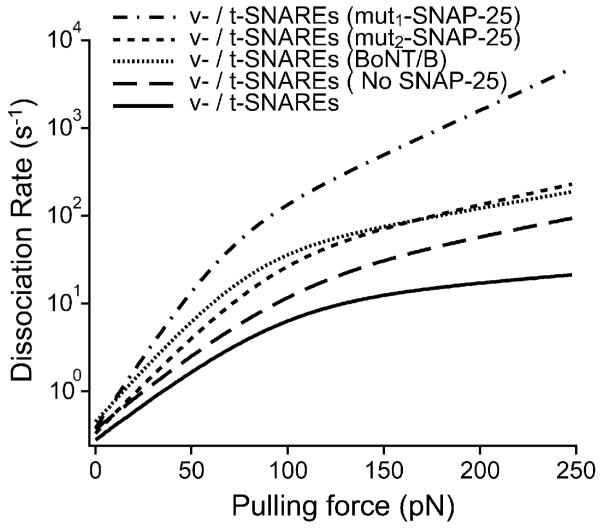
To modify the strength of the SNARE interaction, we substituted full-length SNAP-25 with truncated mutant forms. Fig. 5 shows that these perturbations resulted in a significant decrease in the pulling force required to dissociate the v-/t-SNARE complex. Analysis of the force spectra revealed that the reduction in unbinding force for both SNAP-25 mutants can be attributed to widening of both the inner and outer barriers of the energy landscape rather than to changes in the height of the activation barriers (see Materials and methods section and Table 1). Widening of the activation barriers results in reduction of their slope and consequently leads to acceleration of the dissociation of the SNARE complex in response to a given pulling force. This is evident in the kinetic profiles (Fig. 6) for the unbinding of the SNARE complex under the different conditions. In parallel AFM compression measurements, we noted that the SNAP-25 mutants completely eliminated the facilitation of membrane fusion (Fig. 3), confirming earlier results using bulk fusion assays.25 Moreover, omitting SNAP-25 from the t-SNARE bilayers resulted in a greater interaction strength of the binary VAMP/syntaxin complex as compared to the v-/t-SNARE complex with SNAP-25 mutants in the t-SNARE bilayers (Fig. 5) and introduced some facilitation of membrane fusion (Fig. 3). This observation is consistent with recent reports which revealed that SNAP-25 is not required for membrane fusion under certain experimental conditions.2,34
Fig. 6.
The kinetic profile for the dissociation of the SNARE complex reveals increased dissociation kinetics pursuant to SNARE perturbations. Based on the energy barrier parameters under the different experimental conditions (Table 1), the overall dissociation rate () of the SNARE complex was derived using eqn (3). The slope of the kinetic profile reflects the dissociation kinetics of the complex; a steeper profile indicates fast dissociation kinetics and a weak interaction. Thus, the native v-/t-SNARE complex has the slowest dissociation kinetics which corresponds to a strong binding interaction. The minimal change in the dissociation rate of the SNARE complex above ~100 pN, where the inner energy barrier dominates the SNARE dissociation process indicates that the inner barrier chiefly determines the mechanical strength of the SNARE binding interaction.
In the ternary SNARE complex, SNAP 25 binds to both syntaxin and VAMP via the conserved SNARE domains sn1 and sn2, respectively, and acts as a “bridge” between the two.8,45 The sn1 domain is located in the N-terminal region of SNAP-25, while the sn2 domain is located in the C-terminal region where binding with VAMP is mediated. The SNAP-25 mutants (mut1-SNAP-25 and mut2-SNAP-25) are truncated from the C-terminal end to mimic cleavage by BoNT/A or E, respectively.10,46 Hence, mut1-SNAP-25 or mut2-SNAP-25 binding with VAMP is compromised, which yielded weak binding interactions of the SNARE complex under these conditions (Fig. 5). However, since mut1-SNAP-25 and mut2-SNAP-25 retain their ability to bind with syntaxin, they interfered with the direct binding of VAMP with syntaxin in the opposite bilayers and, hence, they eliminated the modest facilitation in membrane fusion observed in the absence of SNAP-25 (Fig. 2).
Furthermore, treatment of VAMP 2 in the v-SNARE bilayers with BoNT/B reduced the binding strength of the SNARE interaction (Fig. 5) and increased the compression force required to induce hemifusion (Fig. 3). BoNT/B cleaves VAMP 2 in its cytoplasmic domain between residues Gln76 and Phe7736 and approximately 30% of the SNARE binding domain remains after cleavage. With BoNT/B treatment, the reduced unbinding frequency and lower unbinding and fusion forces confirm that the partial fusion facilitation is due to interaction of the remaining portion of the cytoplasmic domain of VAMP with the t-SNAREs in the opposing bilayer rather than inadequate VAMP 2 cleavage. Thus, only the interaction between the native (intact) v- and t-SNAREs in the opposite bilayers will yield the strongest binding interaction and mediate the most facilitation of membrane fusion in our experimental system.
Taken together, the above results reveal an inverse relationship between the strength of the SNARE interaction and the compression force required to induce fusion of the membranes (Fig. 7(a)). To gain a general understanding of this relationship, it is important to first establish parameters that properly describe the pulling force generated during SNARE complex formation and the extent of facilitation of membrane fusion. Our interpretation of the energy landscape of the unbinding process is that the two activation barriers correspond to two conformational states of the bound SNARE complex, where the outer barrier governs the SNARE binding affinity toward each other while the steep inner barrier determines the mechanical strength of their interaction and provides the pulling force necessary to pin the membranes together during SNARE-mediated fusion.
Fig. 7.
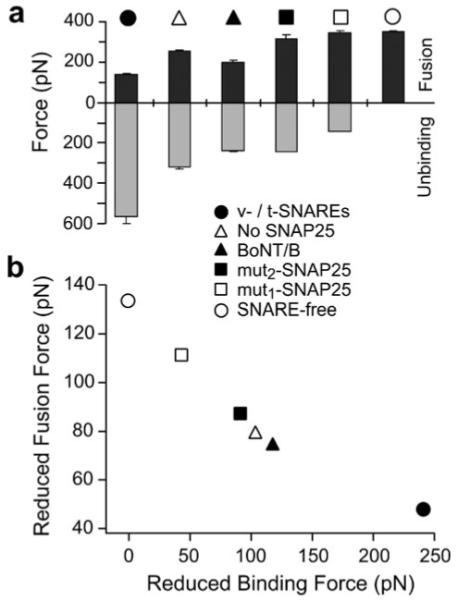
Facilitation of membrane fusion is coupled to the pulling strength of interacting SNAREs in the opposite bilayers. (a) Fusion forces (black) were derived from the DFS (see Fig. 2 and 3) at a compression rate of 20 000 pN s−1 in the compression experiments under the specified experimental conditions. The unbinding force measurements (gray) were derived from the DFS (see Fig. 5) at loading rate of 20 000 pN s−1 in the unbinding experiments under the same conditions. It is evident that an inverse relationship between the fusion force and the SNARE unbinding force exists under these conditions. Error bars are the s.e.m. of either the fusion or the unbinding forces measured at the compression/loading rate of 20 000 pN s−1, respectively. (b) Membrane fusion facilitation quantified by the reduced fusion force (fϕ) correlates with the pulling force of interacting SNAREs as characterized by the reduced unbinding force (fβ). This reveals that the pulling force generated by interacting SNAREs facilitates membrane fusion in a force dependent manner.
To quantify the strength of the interacting SNAREs, we use the reduced unbinding force (fβ) for the inner barrier of the SNARE complex. This parameter corresponds to the force required to suppress the inner activation barrier by 1 kBT during forced unbinding. It is also the pulling force generated as the interacting SNAREs transition from the peak of the inner activation barrier to the fully bound state by an amount equivalent to 1 kBT in the energy landscape. In the context of the Bell–Evans model, fβ equals kBT divided by xβ1, where xβ1 is the molecular distance between the bound and transition state of the inner barrier (i.e., inner barrier width). Experimentally, xβ1 and hence fβ are determined by fitting the Bell–Evans model to the acquired force spectra of the SNARE complexes as described in the Materials and methods section.
The reduced unbinding force of the inner barrier can be used to characterize the pulling force generated by interacting SNAREs, provided that its height does not change significantly among the conditions being compared (Table 1). Analysis of the dissociation rates () for the inner barrier of the different SNARE complexes revealed a 2.6 fold difference in extreme cases; this corresponds to a difference δΔG of <1 kBT (see eqn (2) in Methods section). An estimate of the absolute height of the inner barrier of the SNARE complex can be obtained from the difference between the binding energy of the complex and the energy separation between the inner and outer barriers.23 The binding energy of a partly unstructured SNARE complex was estimated to be ~35 kBT.47 Using the derived values, we obtained a difference of ~3.3 kBT between the outer and inner barriers and, hence, a conservative estimate of ~32 kBT for the height of the inner barrier was obtained. Thus, the variation in the relative height of the inner barrier is small compared to the variation in fβ (~40 to ~250 pN; see Fig. 7(b)) amongst the different SNARE complexes.
Similarly, we use the reduced fusion force (fϕ) to quantify the facilitation of bilayer fusion. This parameter corresponds to the compression force required to overcome 1 kBT of the fusion energy barrier and is obtained from fitting a similar model23 to the force spectra (Fig. 2 and 3) of membrane fusion (also see Methods). As shown in Fig. 6(b), fϕ ranges from ~50 pN for the fusion of native t-/v-SNARE bilayers to ~130 pN for SNARE-free bilayers. A lower value of fϕ indicates that less force is required to induce fusion. Since the relative difference in height (δΔG) of the fusion energy barrier of the different bilayers (Table 1; ) does not change by >1 kBT from an estimated value of ~45 kBT,48 fϕ is a good indicator of how a compression force suppresses the fusion barrier.
Fig. 6(b) reveals a strong correlation between the reduced unbinding force fβ and the reduced fusion force fϕ. We interpret this result as evidence that, in addition to providing critical proximity between the membranes, the pulling force generated by interacting trans-SNAREs facilitates membrane fusion possibly through destabilization of the lipid bilayers. Our results are consistent with a model where the initial interaction of cognate SNAREs brings the opposing bilayers to close proximity. This step corresponds to the transition of the dissociated SNAREs over the outer activation barrier to a transient intermediate bound state between the outer and inner barriers of the complex. In neurons for example, this intermediate state corresponds to the primed state of docked vesicles and might be stabilized by complexin49-51 or synaptotagmin as recently suggested.52 Nonetheless upon stimulation and intracellular Ca2+ increase, release of the constraint on the molecular system occurs53-55 allowing it to cross over the inner activation barrier toward a tightly bound complex. The strong pulling force generated as the system moves down the steep inner barrier induces conformational changes in the juxtamembrane and transmembrane domains of the SNAREs.56 Consistent with the role of tilted peptides15,57 and viral fusion proteins in destabilizing the lipid bilayers and facilitating viral fusion with the host cell membrane,58,59 such pulling force can lead to local destabilization of the lipid bilayers at SNARE anchorage sites21 and, subsequently, to facilitation of membrane fusion at fusion sites.
In summary, using AFM force spectroscopy we established a direct correlation between the strength of the SNARE interaction and SNARE-mediated membrane fusion. We also revealed that the SNARE interaction is governed by two energy barriers, and that the mechanical strength of the SNARE complex stems primarily from the steep inner barrier. Moreover, our findings showed a critical role for trans-SNARE interactions in mediating membrane fusion by pinning opposing membranes at close proximity and, possibly, locally destabilizing the lipid bilayers. Finally, we conclude that the combined effects of SNAREs on the membranes contribute to the overall work performed on the bilayers and subsequently lead to SNARE-mediated membrane fusion.
Insight, innovation, integration.
The current work deals the mechanism of SNARE-mediated membrane fusion which is essential for survival. We obtained nano-force measurements using AFM force spectroscopy to examine the biological interactions at the core of the membrane fusion process. This research is crucial to the understanding of a variety of physiological and pathological conditions that involve vesicle fusion with the plasma membrane. Using an AFM-based experimental system for membrane fusion, we demonstrate for the first time a direct correlation between the pulling strength of the SNARE complex with the SNARE-mediated membrane fusion.
Acknowledgements
We thank Drs J. E. Rothman and R. H. Scheller for protein constructs. E. R. C is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from NIH (GM086808)/NSF (MRI 0722372)/AHA (0855395E) to V. T. M., and NIH (GM56827 and MH61876)/AHA (0440168N) to E. R. C.
References
- 1.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 2.Liu T, Tucker WC, Bhalla A, Chapman ER, Weisshaar JC. Biophys. J. 2005;89:2458–2472. doi: 10.1529/biophysj.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Nat. Struct. Mol. Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 4.Nickel W, Weber T, McNew JA, Parlati F, Sollner TH, Rothman JE. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parlati F, Weber T, McNew JA, Westermann B, Sollner TH, Rothman JE. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothman JE, Warren G. Curr. Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 7.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 8.Brunger AT. Q. Rev. Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 9.Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. Nat. Struct. Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 10.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 11.Sutton RB, Ernst JA, Brunger AT. J. Cell Biol. 1999;147:589–598. doi: 10.1083/jcb.147.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin RC, Scheller RH. Neuron. 1997;19:1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 13.Chapman ER, Hanson PI, An S, Jahn R. J. Biol. Chem. 1995;270:23667–23671. doi: 10.1074/jbc.270.40.23667. [DOI] [PubMed] [Google Scholar]
- 14.Bowen M, Brunger AT. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas A, Brasseur R. Curr. Protein Pept. Sci. 2006;7:523–527. doi: 10.2174/138920306779025594. [DOI] [PubMed] [Google Scholar]
- 16.Pobbati AV, Stein A, Fasshauer D. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 17.Sorensen JB, Wiederhold K, Muller EM, Milosevic I, Nagy G, de Groot BL, Grubmuller H, Fasshauer D. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwilling D, Cypionka A, Pohl WH, Fasshauer D, Walla PJ, Wahl MC, Jahn R. EMBO J. 2007;26:9–18. doi: 10.1038/sj.emboj.7601467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Sollner TH, Rothman JE. J. Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui TJ, Vites O, Stein A, Heintzmann R, Jahn R, Fasshauer D. Mol. Biol. Cell. 2007;18:2037–2046. doi: 10.1091/mbc.E07-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdulreda MH, Bhalla A, Chapman ER, Moy VT. Biophys. J. 2008;94:648–655. doi: 10.1529/biophysj.107.114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kesavan J, Borisovska M, Bruns D. Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Abdulreda MH, Moy VT. Biophys. J. 2007;92:4369–4378. doi: 10.1529/biophysj.106.096495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett MK, Calakos N, Scheller RH. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 25.Tucker WC, Weber T, Chapman ER. Science. 2004;304:435–438. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 26.Chen A, Moy VT. Methods Cell Biol. 2002;68:301–309. doi: 10.1016/s0091-679x(02)68015-x. [DOI] [PubMed] [Google Scholar]
- 27.Hutter JL, Bechhoefer J. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 28.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Wojcikiewicz E, Moy VT. Biophys. J. 2002;83:2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 31.Evans E, Ritchie K. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YA, Scales SJ, Jagath JR, Scheller RH. J. Biol. Chem. 2001;276:28503–28508. doi: 10.1074/jbc.M103009200. [DOI] [PubMed] [Google Scholar]
- 33.Vikman J, Ma X, Hockerman GH, Rorsman P, Eliasson L. J. Mol. Endocrinol. 2006;36:503–515. doi: 10.1677/jme.1.01978. [DOI] [PubMed] [Google Scholar]
- 34.Bowen ME, Weninger K, Brunger AT, Chu S. Biophys. J. 2004;87:3569–3584. doi: 10.1529/biophysj.104.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wictome M, Rossetto O, Montecucco C, Shone CC. FEBS Lett. 1996;386:133–136. doi: 10.1016/0014-5793(96)00431-0. [DOI] [PubMed] [Google Scholar]
- 36.Evans ER, Sutton JM, Gravett A, Shone CC. Toxicon. 2005;46:446–453. doi: 10.1016/j.toxicon.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Hua SY, Charlton MP. Nat. Neurosci. 1999;2:1078–1083. doi: 10.1038/16005. [DOI] [PubMed] [Google Scholar]
- 38.Duman JG, Forte JG. Am. J. Physiol.: Cell Physiol. 2003;285:C237–249. doi: 10.1152/ajpcell.00091.2003. [DOI] [PubMed] [Google Scholar]
- 39.Dudko O, Hummer G, Szabo A. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudko O, Mathé J, Szabo A, Meller A, Hummer G. Biophys. J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wojcikiewicz EP, Abdulreda MH, Zhang X, Moy VT. Biomacromolecules. 2006;7:3188–3195. doi: 10.1021/bm060559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Montana V, Bai J, Chapman ER, Mohideen U, Parpura V. Biophys. J. 2006;91:744–758. doi: 10.1529/biophysj.105.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yersin A, Hirling H, Steiner P, Magnin S, Regazzi R, Huni B, Huguenot P, De los Rios P, Dietler G, Catsicas S, Kasas S. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8736–8741. doi: 10.1073/pnas.1533137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans E, Leung A, Hammer D, Simon S. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3784–3789. doi: 10.1073/pnas.061324998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poirier MA, Hao JC, Malkus PN, Chan C, Moore MF, King DS, Bennett MK. J. Biol. Chem. 1998;273:11370–11377. doi: 10.1074/jbc.273.18.11370. [DOI] [PubMed] [Google Scholar]
- 46.Breidenbach MA, Brunger AT. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 47.Li F, Pincet F, Perez E, Eng W, Melia T, Rothman J, Tareste D. Nat. Struct. Mol. Biol. 2007;14:890–896. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]
- 48.Cohen F, Melikyan G. J. Membr. Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- 49.Carr C, Munson M. EMBO Rep. 2007;8:834–838. doi: 10.1038/sj.embor.7401051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melia TJ. FEBS Lett. 2007;581:2131–2139. doi: 10.1016/j.febslet.2007.02.066. [DOI] [PubMed] [Google Scholar]
- 51.Bowen M, Weninger K, Ernst J, Chu S, Brunger A. Biophys. J. 2005;89:690–702. doi: 10.1529/biophysj.104.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chicka MC, Hui E, Liu H, Chapman ER. Nat. Struct. Mol. Biol. 2008;15:827–835. doi: 10.1038/nsmb.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaub J, Lu X, Doneske B, Shin Y, McNew J. Nat. Struct. Mol. Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 54.Huntwork S, Littleton J. Nat. Neurosci. 2007;10:1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 55.Tang J, Maximov A, Shin O, Dai H, Rizo J, Südhof T. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 56.McNew JA, Weber T, Engelman DM, Sollner TH, Rothman JE. Mol. Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 57.Crowet JM, Lins L, Dupiereux I, Elmoualija B, Lorin A, Charloteaux B, Stroobant V, Heinen E, Brasseur R. Proteins. 2007 doi: 10.1002/prot.21483. [DOI] [PubMed] [Google Scholar]
- 58.Lau WL, Ege DS, Lear JD, Hammer DA, DeGrado WF. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorin A, Charloteaux B, Fridmann-Sirkis Y, Thomas A, Shai Y, Brasseur R. J. Biol. Chem. 2007;282:18388–18396. doi: 10.1074/jbc.M700099200. [DOI] [PubMed] [Google Scholar]