INTRODUCTION: STEM CELLS AND REGENERATIVE MEDICINE
Regenerative Medicine and the Promise of Stem Cells
The ability to control the regeneration of tissues or organs that do not normally regenerate in humans would have an enormous impact on medical practice as well as on the general quality of human life. The goal of regenerative medicine is to repair or replace damaged or diseased adult tissues or organs. Three general strategies are being investigated for regenerative therapies: 1) cell based; 2) engineered bio-scaffolds seeded with selected cells prior to engraftment; and 3) boosting endogenous repair mechanisms. Small molecule agents can potentially play critical roles as part of all three approaches.
The cell-based approach has grown largely from the successful use of hematopoietic stem cell (HSC) transplants (bone marrow transplants) for more than 50 years now, primarily to treat blood disorders (for review1). Recently, the potential to significantly expand the scope of this approach has gained momentum from the ability to isolate adult multipotent stem cells from virtually all organs as well as from newly discovered capabilities to direct the differentiation of embryonic stem cells (ES) to multiple lineages. The recent advent to derive patient specific induced pluripotent cells (iPS) has further enhanced the potential of this approach by removing both ethical barriers and barriers to immuno-histocompatibility. The third approach has to this point relied heavily on the hope of recapitulating in mammals/humans some of the extremely impressive regenerative capacity of lower species (e.g. reptiles, amphibians, fish, and birds). However, the potential success of this approach is evidenced by the use of agents such as erythropoietin (EPO) and filgastrim (G-CSF) to boost hematopoietic recovery. These represent important clinically validated applications of this type of approach. The potential to expand this strategy and develop pharmacologic therapies to boost endogenous repair mechanisms for a wide array of tissues by overcoming the intrinsic barriers to regeneration in mammals remains an enormously exciting possibility.
In this review, we will discuss the significant potential role that small molecules have to play in gaining a better understanding of stem cell biology, enabling regenerative therapies and treating stem cell based diseases.
Stem Cells
What is a stem cell? In the popular press, they are characterized as cells with a tremendous potential to cure a wide range of diseases or produce new organs for replacement after damage via disease or injury. However, the term stem cell is in fact extremely broad and covers a diverse array of cell types. By definition, a stem cell is a cell that has the capacity to both self-renew (make at least one identical copy of itself at each division) and also has the capacity to differentiate into more mature, differentiated (and less potent) specialized cells. Stem cells can be embryonic, if derived from an embryo, or adult/somatic if derived from tissue. The enormous interest elicited by embryonic stem (ES) cells is based on their key property of pluripotency. Indeed, ES cells possess the rare and precious capacity of generating all the cell types found in embryos, as well as adult developed organisms. This unique property has been lost in other somatic stem cells (SSCs). SSCs are therefore described as being multipotent, i.e. capable of generating multiple differentiated cell types but generally restricted to that of a particular tissue, organ or physiological system (e.g. hematopoietic stem cells, neural stem cells, etc…) in which they reside, as opposed to pluripotent.
Today, the basic molecular pharmacology/signaling networks in ES cells are coming into focus. Considerable effort has been devoted to understanding the maintenance of the pluripotent state. It is believed to rely heavily on the expression of a relatively few transcription factors, such as Oct4 and Nanog 2–7. Oct4 appears to be the most specific and critical gene for the maintenance of pluripotency, as its expression is mandatory for this purpose. However, expanding our knowledge of the biology of ES cell pluripotency, SSC multipotency and the differentiation programs implemented during development is critical. The potential applications of this expanded knowledge however are immense. If, as hypothesized, similar signaling pathways could be harnessed to produce specific cell types on demand (either from pluripotent ES cells or multipotent somatic/adult stem cells) the potential benefits to regenerative medicine would be enormous. Furthermore, these same pathways appear to be corrupted in cancer (and in particular cancer stem cells/tumor initiating cells) to drive malignancies. Understanding how to correct these aberrantly utilized pathways in cancer stem cells will be critical to the effective treatment of malignancies.
STEM CELLS COME IN VARIOUS FLAVORS
Human Embryonic Stem Cells
In the past decade, since the first human embryonic stem (hES) cells were described8, a number of additional lines have been derived. These lines, which are derived from the inner cell mass (ICM) of blastocyst-stage embryos, can be cultured on feeder layers of mouse embryonic fibroblasts (MEFs) in the presence of serum and/or basic fibroblast growth factors (bFGF), where they maintain self-renewal and pluripotent capacity9, (for a recent review please see10). However, protocols to cleanly and completely differentiate hES cells into specific cell types have not been successfully developed, thereby hindering the use of these derivatives in transplantation therapies. A better understanding of the molecular programs governing lineage commitment is desperately needed to achieve homogeneous enrichment for cell types of interest. Furthermore, in addition to important ethical issues, many anticipated pitfalls in the use of hES cells remain to be addressed; including concerns regarding rejection of the transplanted cells, as well as the possibility of ES cell-derived tumor formation. Although hES cells are relatively immunologically inert compared to normal somatic cells, blocking natural rejection is still required. Currently available immunosuppressive drugs are far from ideal for long-term use and have been associated with numerous complications, making the development of alternative solutions mandatory. One potential solution to the rejection problem would be the creation of individualized patient on demand specific “self” ES cells. Nuclear transfer technology, involving the fusion of the nucleus from a patient’s somatic cell with an enucleated oocyte11 is one method to achieve this strategy. Although such patient-derived hES cells could provide a valuable resource to better understand diseases and for drug testing, this expensive and time-consuming technique is undermined by its requirement for the destruction of an oocyte and additionally brings us one step closer to human cloning.
Induced Pluripotent Stem (iPS) Cells
One way to circumvent some of the ethical and technical issues discussed above is to generate ES-like cells directly from somatic cells. As demonstrated by nuclear reprogramming technology, the somatic cell nucleus, when exposed to the correct cellular environment, can be reprogrammed to provide pluripotent cells. Several years ago, Yamanaka and colleagues screened for combinations of factors that could induce the reprogramming of somatic cells (for recent reviews please see12,13). This groundbreaking study led to a potential major breakthrough for cell therapy. They demonstrated that the forced expression of only four transcription factors (i.e. Oct4, Sox2, Kfl4 and c-Myc) is sufficient to de-differentiate mouse embryonic and adult tail-tip fibroblasts into pluripotent stem-like cells14 (referred to as iPS, for a recent review please see13). Oct415 and Sox216 are known to be key representatives of the core transcriptional apparatus, which synergistically upregulate “stemness genes” while repressing differentiation pathways. However, Oct4 and Sox2 appear to operate only in a permissive environment, i.e. an environment that is not provided in differentiated cells such as fibroblasts, but is created by epigenetic modifications by the transcription factors Klf4 and c-Myc14, or Lin28 and Nanog17. In conclusion, iPS derivation may provide an appropriate method to generate both patient and disease specific pluripotent cell lines, without raising ethical barriers.
Despite these advances, major limitations remained, as initially iPS derivation was based on retroviral infections to maintain the expression of the 4 key factors. This protocol raised concerns for increased risk of tumorigenicity due to multiple viral integration events and the overexpression of c-Myc, a well known proto-oncogene (for review13). To address this first point, alternative methods have already been developed, using non-integrating adenovirus18, repeated transfection of plasmids19, non-integrating episomal vectors20 or protein expression21, providing integration-free and/or xeno-free conditions. However, these methods remain relatively inefficient. Most recently, the use of the piggyBac transposon/transposase22,23 or Cre/LoxP24 systems have been reported to efficiently induce pluripotency in both mouse and human embryonic fibroblasts, while being virtually traceless. With regard to the second point, omission of c-Myc in reprogramming protocols has been reported, albeit with decreased efficiency, thereby considerably reducing the risk of tumorigenicity25. For example, the absence of c-Myc can be compensated for by Wnt3a treatment26.
In conclusion, although tremendous progress has already been made, the direct reprogramming process is still relatively inefficient and slow, as well as based on a genetic selection, thereby limiting its use for patient specific purposes at present. However, this is an extremely active area of research. Ultimately, the goal is to identify combinations of transient cues to replace retroviral infection and the genetic selection strategies used to establish iPS cell lines, as well as to develop serum-free chemically defined media to culture pluripotent cells and specifically trigger a given lineage commitment pathway.
Somatic stem cells (SSCs)
Another option worthy of significant consideration is the use of endogenous SSCs either via isolation/amplification or alternatively via endogenous stimulation. The first type of SSC to be isolated and utilized therapeutically was the HSC in the form of bone marrow for transplantation therapy. More than a half century later, their self-renewal and multilineage differentiation capacity has been fully demonstrated. More precisely, an entire hierarchy of progenitors has been identified and isolated (for review1). These studies have been the template for the isolation of a number of other SSC populations (e.g. Neural Crest SC,27). Adult SSCs, although frequently present in only limited numbers, are believed to be the source for naturally occurring tissue regeneration and repair in adult tissues, as already demonstrated in the lung (for review28), in the heart after myocardial infarction (for review29) or the adult CNS for therapeutic approaches to stroke and neurodegenerative disorders (for review30). Nevertheless, a great deal of work remains to isolate factors that either can mobilize and/or modulate endogenous stem cell populations to efficiently regenerate damaged tissue or that can allow for the routine expansion in vitro, without differentiation, of the SSC populations for cell therapy. One caveat to this approach is that although many tissues appear to have SSCs, to date, not all tissues have been proven to contain adult SSC populations31. Fortunately, in the past decade, reports have indicated that some populations of adult SSCs may possess greater plasticity than originally believed. This has led to the concept that these adult SSC populations could be utilized as multipotent progenitors, which could be directed to a diverse array of lineages. Subsequently, significant effort has been devoted to deciphering the molecular mechanisms that regulates such plasticity and developing ways to exploit it for therapeutic purposes. Mesenchymal Stem Cells/Multipotent Stromal Cells (MSCs) are one of the most widely studied adult stem cell. They can be readily derived from many adult tissues and have been shown to give rise to many cell types (for review32). However, such trans-differentiation has been controversial and has not been consistently reproduced in vitro or in vivo, pointing out that significantly more work will be required to understand and control this process. One point of consternation with the introduction of ex vivo cultured stem cells is their ability to home to and engraft in the correct site and subsequently incorporate properly into their new environment, as this has been proven to be generally not a very efficient process. For example, MSCs have been shown to have the capacity of promoting endogenous myelin repair and modulating the immune response (for short review33), despite a low level of engraftment and/or trans-differentiation. Since MSCs are known to produce a variety of cytokines and adhesion molecules that regulate various aspects of hematopoiesis and the immune response34 it has been suggested that alterations in the tissue microenvironment (more than trans-differentiation) may be responsible for the positive outcome35. This modulation could enhance the proliferation and the differentiation of endogenous stem-like progenitors found in many tissues and decrease the inflammatory, immune and fibrotic response, which often limits endogenous regeneration. Furthermore, the complexity and heterogeneity of MSC populations leaves room for the possibility that only a minority subpopulation is actually responsible for the therapeutic effect of MSCs. Identification, isolation and optimization of factors capable of positively altering the tissue microenvironment is of primary importance to understand the underlying mechanism and increase its efficiency. Further knowledge how to enhance engraftment and subsequent differentiation of injected stem/progenitor cells is also essential.
Cancer stem cells (CSCs)
The term cancer stem cell (CSC) or tumor initiating cell (TIC) refers to cells that propagate the tumor phenotype, hence their name. Like their normal counterparts (SSCs), CSCs exhibit self-renewal capacity and differentiation potential, albeit an aberrant and incomplete differentiation potential. Since the initial isolation of CSCs in leukemia, their existence in a wide variety of other cancers has been successfully demonstrated (for review36). It is worth noting that the term CSCs does not necessarily imply that these cells were directly derived from mutations to normal tissue SSCs. Alternatively, CSC/TIC may be derived from progenitor populations that have acquired mutations that have essentially reprogrammed the cell to a “stem-like” status37. Therefore, the term TIC may be generically more appropriate. Traditional therapies, targeting rapidly proliferating cell populations, often with cytotoxic agents, may kill the bulk of the tumor, yet are inefficient at targeting the CSC/TICs. For this reason, alternative therapies, which directly target CSCs are needed to complement classic approaches. A significant caveat to this approach is that given the similarities between endogenous SSC and CSC/TIC, the potential to damage the endogenous normal population is ever present. To successfully accomplish this approach, we will need to improve our understanding of stem cell biology: understanding the molecular pathways that regulate normal SSC versus CSC activation versus quiescence in tissues. This may be the key to successful cancer chemotherapy, as most cancers probably originate from an inappropriate perturbation of the finely tuned balance of stem cell quiescence versus activation and subsequent terminal differentiation. Normal and cancer somatic stem cell biology and the mechanisms that govern their maintenance and differentiation are undoubtedly closely entwined.
SMALL MOLECULE PHENOTYPIC SCREENING, CHEMICAL GENOMICS AND REGENERATIVE MEDICINE
The promise of small molecules for use in regenerative medicine depends on identifying small molecules, which activate, synergize, inhibit or modulate complex natural pathways required for self-renewal and/or differentiation, while having minimal side effects. This perspective will briefly review recent developments in the use of small molecules to: (a) control self-renewal of embryonic and adult stem cells; (b) control the balance between pluripotency/multipotency and lineage commitment; (c) enhance the derivation of iPS cells and (d) pharmacologically enhance endogenous repair mechanisms, to enable the goals of regenerative medicine.
As described below, the majority of studies published to date have focused the selection process for candidate small molecules on phenotypic screening. However these studies often omit the identification and validation of the molecular target(s) or pathway(s) responsible for the activity of the identified hits. From our perspective, this is how we definitionally distinguish “Small Molecule Phenotypic Screening” from “Chemical Genomics”. While inherently not problematic from a process perspective, small molecule phenotypic screening alone, limits the ability to enhance our knowledge concerning the fundamental networks that control “stemness” and lineage commitment.
Maintenance of embryonic stem cell pluripotency
Although embryonic stem cells can be grown in culture essentially indefinitely, this has traditionally required the use of fibroblast feeders and serum. Current conditions result not only in sub-optimal maintenance of pluripotency and proliferation, thereby increasing the complexity of biological studies, but also prohibiting those cells from being utilized in patient cell replacement therapy for safety reasons. However, to be readily translated clinically for therapy/transplantation, there exists a need to develop feeder-free, chemically defined media to culture embryonic stem cells.
Mouse ES cells
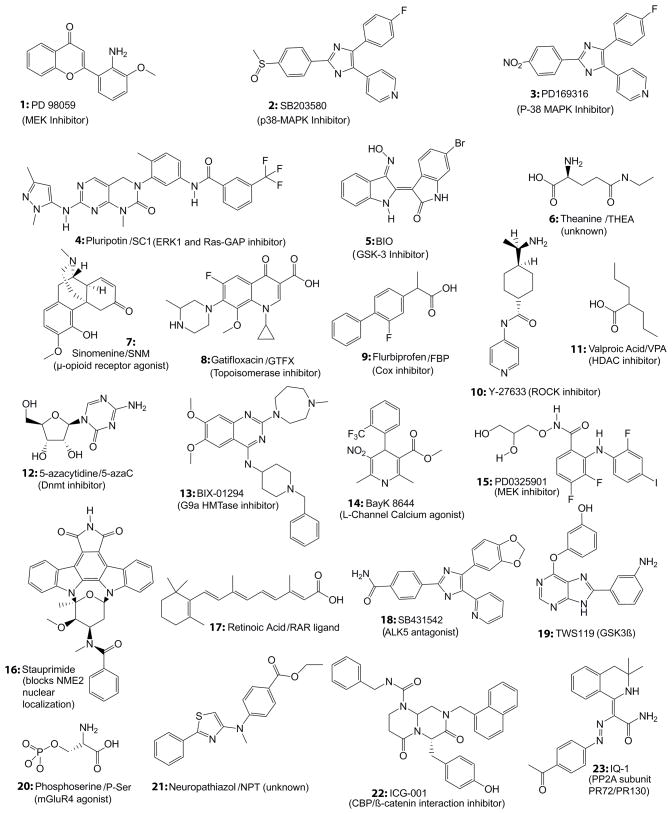
Mouse ES (mES) cells have been derived and maintained for decades often by co-culture on a feeder layer of mitotically inactivated MEFs, in the presence of Leukemia inhibitory factor (LIF) and serum. Such a complex and only partially defined environment provides a “cocktail” of signaling molecules synergistically sustaining pluripotency and proliferation of mES cells. Smith and collaborators demonstrated that mES cell self-renewal capacity, which is generally enabled by activation of the LIF/STAT3 pathway, can also be maintained solely by stimulation of BMP signaling5. The presence of BMPs in the media triggers the synthesis of Inhibitors of Differentiation (Id) transcription factors thereby blocking differentiation programs, acting in synergy with the stimulation of pluripotency and self-renewal activity sustained by LIF signaling. The maintenance of pluripotency of mES cells by LIF has been further distilled down to (i) activation of the STAT3 pathway to maintain self-renewal and (ii) inhibition of ERK signaling to suppress differentiation6,38. Taken together, this simple distillation of the biology of mouse ES cells allowed for a direct search for specific regulators of ‘stemness’ and laid the groundwork for development of chemically defined media sustaining mES cell pluripotency and proliferation. Inhibitors of MEK, such as PD98059 (Figure 1, structure 1), and of p38 MAPK, such as SB203580 (Figure 1, structure 2) or PD169316 (Figure 1, structure 3), have also been shown to enhance self-renewal and decrease LIF withdrawal triggered apoptosis, thereby potentially improving ES cell maintenance39,40. Recently, combinations of LIF with MEK inhibitor, LIF with GSK3 inhibitor, or inhibitors of MEK/GSK3/FGFR, have been used to sustain self-renewal and the derivation of various mES cell lines under chemically defined conditions41.
FIGURE 1.
structures and described modes of action of small molecules that have been shown to regulate stem cell biology and are potentially useful for regenerative medicine.
The use of small molecules with known molecular targets highlights the significant role that small molecules can play in controlling the complex regulatory circuits involved in stem cell biology. However, a limitation of this target-based method is the need a priori for reasonable knowledge of the molecular pathways involved in stem cell biology and ready access to specific inhibitors of the molecular pathways that one is attempting to manipulate. This is particularly problematic in stem cell biology where many of the biological mechanisms are still not completely understood. Given our current partial understanding about the control of stem cell fate, unbiased phenotypic screens of chemical libraries holds great promise for the generation of desired cell types in a controlled manner as well as for providing useful tools to study the underlying mechanisms involved in stem cell regulation. A number of high throughput, cell based screens of chemical libraries have been performed on mES cells (for review42). Assessment of “stemness” is usually based on GFP expression as a readout based upon Oct4-promoter activity (lost after only 4–6 days in vitro without feeders and/or LIF), or simply on alkaline phosphatase (AP) activity, a marker of ES cell pluripotency. Using this type of approach, Ding and collaborators demonstrated that the small molecule SC1 (pluripotin; Figure 1, structure 4) is sufficient to replace both feeder cells and LIF, by apparently inhibiting both ERK1 and Ras-GAP-dependent signaling, thereby promoting self-renewal43.
Human ES cells
hES cells appear, at least at first blush, to share very few characteristics with their mouse counterparts44, besides the key “pluripotency transcription factors” i.e. Oct4, Nanog and Sox2, which they have in common45–47. The extrinsic factors regulating hES cell maintenance and early differentiation events seem to differ from mES cells and to date are poorly understood. It is clear that LIF signaling does not play a significant role in maintaining hES cell self-renewal capacity, despite the ability of LIF to similarly activate the STAT pathway in hES cells as it does in mES48. Furthermore, BMP signaling seems to increase the differentiation of hES cells49 as opposed to maintaining pluripotency as previously described in mES cells. Wnt signaling, which in some reports has been demonstrated to have positive effects on the maintenance of mES cells, appears to also facilitate the maintenance of the undifferentiated phenotype and maintain expression of critical “pluripotency” transcription factors in hES cells48. More precisely, Brivanolou and colleagues described the positive, although limited-time frame effects of BIO (Figure 1, structure 5), a known small molecule inhibitor of the Ser/Thr kinase GSK-3, on both mouse and human ES cell maintenance, while preserving a normal differentiation program capacity after withdrawal. However, there is some controversy regarding this report50. In an unbiased high throughput screen, Desbordes and colleagues identified 4 small molecules capable of replacing bFGF signaling in maintaining hES cells, based on Oct4 expression. A more detailed analysis showed that theanine (THEA, Figure 1, structure 6), sinomenine (SNM, Figure 1, structure 7), gatifloxacin (GTFX, Figure 1, structure 8), and flurbiprofen (FBP, Figure 1, structure 9) sustain both Oct4 and Nanog expression in at least two different hES cell lines (H1 and H9)51. Although these compounds all appear to aid in the maintenance of pluripotency, they are used at extremely high concentrations (approximately 100 μM), and their lack of structural similarities leads one to conclude that they may act through various targets/pathways.
Lastly, hES cells are known to be relatively difficult to culture, due to slow growth and hypersensitivity to stresses such as detachment and complete dissociation. Screening for caspase inhibitors, growth factors or kinase inhibitors, which could potentially improve the yield of survival upon hES cell manipulation, Watanabe and colleagues isolated the Rho-Kinase/ROCK inhibitor (Y-27633, Figure 1, structure 10), which could significantly decrease dissociation and detachment-triggered apoptosis52. This was an important discovery to aid in single cell passaging (hence clonal isolation) as well as suspension culture (hence EB differentiation). This result has been recently confirmed and extended in a study that also suggested a positive role of PKC inhibitors on hES cell survival53.
In vivo modulation/activation of stem cells
The ability to modulate/activate endogenous somatic stem cells to repair damaged tissues or organs would overcome many of the limitations previously outlined for the use of ex vivo cultured stem cells. However, with the exception of the remarkable regenerative capacity of the adult liver54, adult stem cells have an apparently limited replacement capacity. Unquestionably, there is an overall decline in tissue regenerative capacity with age. However, these changes apparently are not due to gradual depletion of the stem cells (for example with HSC see55) as in fact there is an apparent increase in SSC number with age. Therefore, finding molecules capable of increasing the regenerative capacity of SSCs would be of significant value for therapeutic protocols. As an example, promising studies have already reported molecules capable of endogenously stimulating self-renewal of HSC. Bug and collaborators56 have shown that valproic acid (VPA, Figure 1, structure 11), a non-selective HDAC inhibitor, previously known to contribute to inducing differentiation or apoptosis of leukemic blasts, also stimulates proliferation and self-renewal of normal HSC. More recently, North and colleagues57 performed a small molecule screen for molecules modulating SC content in the Zebrafish aorta-gonad-mesonephros region. Many chemicals, that affected HSC numbers in this screen54, were modulators of prostaglandin (PG) E2 synthesis, the main effector prostanoid produced in Zebrafish. The authors demonstrated a conserved role of PGE2 in the regulation of vertebrate HSC homeostasis, concluding that modulation of the PG pathway may facilitate the expansion of HSC number for therapeutic purpose. It is interesting to note that PGE2 through the EP2 and EP4 receptors can also activate the Wnt signaling cascade58. In general, better characterization of the stem cell niche and mechanisms by which SSC can be activated would improve our understanding of stem cell homeostasis and our capacity to appropriately stimulate it when needed.
Reprogramming: generation of iPS cells
As described earlier, direct reprogramming of somatic cells is a slow and inefficient process, the molecular mechanism of which remains poorly understood. Recently, Mikkelsen and colleagues59 systematically compared the genetic and epigenetic profile of ES cells, iPS and partially reprogrammed cell lines, in order to better understand the key elements underlying the establishment of pluripotency. This study offered important insights for the development of safer and more efficient reprogramming strategies. They concluded that establishment of a pluripotent state requires (i) repression of lineage-specific genes, (ii) establishment of an “open” chromatin stage and (iii) reactivation of endogenous pluripotency-related genes. These 3 elements are limiting factors for direct reprogramming, hence a direct approach to each of these elements is required for improving the efficiency of iPS derivation.
In their study, Mikkelsen and colleagues59 first demonstrated that chromatin demethylation can lower the kinetic barrier to direct reprogramming. When they inhibited endogenous DNA methyltransferase activity either by treatment with a specific inhibitor, 5-aza-cytidine (AZA, Figure 1, structure 12), or by RNA interference of Dnmt1, both methods led to a significant increase in reprogramming efficiency. This improvement can be further enhanced by repressing the expression of endogenous lineage specific transcription factors. The importance of epigenetic modulation for the control of cellular potential was further demonstrated using VPA, which improves the efficiency of mouse fibroblast reprogramming with the four required transcription factors by more than 100 fold. It also allowed for the abandonment of c-Myc in the reprogramming cocktail60. Melton and collaborators60 demonstrated that VPA also improves reprogramming of human fibroblasts and enables reprogramming to take place with only exogenous Sox2 and Oct4 expression. Similarly, a 2000 compound screen identified BIX (Figure 1, structure 13), an inhibitor of the G9a HMTase, for its positive impact on MEF reprogramming efficiency. 13 allows reprogramming under Oct4 and Klf4 forced expression only, although efficiency is greatly affected61. Taken together, these studies shed light on the importance of epigenetic modifications for the control of cellular potential and its key role in cellular reprogramming.
However, these treatments only render cells more permissive to reprogramming and cannot in any way be considered as having a specific impact on the regulation of gene expression. As described by Mikkelsen and colleagues 59, lineage-specific transcription factors need to be down-regulated, whereas pluripotency genes need to be upregulated to achieve complete reprogramming. Obviously, such genetic reorganization can be triggered by the forced expression of the four reprogramming factors: Oct4, Sox2, Klf4 and c-Myc. It is of primary importance to find ways to recapitulate this effect, while avoiding the use of oncogenes and viral vectors. One promising approach is to reprogram cells that already express high endogenous levels of some of those genes. Neural Stem Cells (NSCs) for example, which are potentially available from human biopsies, are known to express high levels of Sox2. Recently, Schoeler and collaborators have shown that NSCs also express c-Myc, Klf-4, as well as AP and SSEA-1, two markers of ES cell pluripotency. Starting from this population, pluripotency has been achieved merely by overexpressing Oct4 and Klf-462,63 or even more recently by overexpressing Oct-4 alone64. Nevertheless, despite the fact that such alternative reprogramming protocols rely on less genetic manipulations, they are also less efficient and NSCs are not nearly as readily obtainable as fibroblasts. Ideally, one would like to identify small molecules capable of triggering or replacing Oct4 and Sox2 upregulation in fibroblasts. BayK8644 (Bayk, Figure 1, structure 14) appears to possess some of these characteristics 61.14, an L-channel calcium agonist, was isolated in a small molecule screen of 2000 compounds to improve reprogramming efficiency. 14 treated MEFs also treated with 13 after Oct4 and Klf4 overexpression could be reprogrammed. Since 14 does not show any effects on its own, it is believed that 14 could actually impact more specifically at the cell signaling/ transduction level, in the permissive cellular environment created by 13 treatment and Oct4 and Klf4 overexpression. Despite the importance of this finding, Oct4 appears strictly necessary for reprogramming. Isolating a small molecule sufficient to replace Oct4 overexpression remains an important challenge in this field.
iPS selection
Finally, another major limitation to reprogramming protocols developed so far is the fact that they rely on genetic selection to unveil the iPS colonies. This cannot lead in any case to applicable cell therapeutic strategies for obvious reasons. In that regard, Ding and collaborators have described a chemical selection technique for iPS colonies62. The MEK inhibitor PD0325901 (Figure 1,structure 15), when applied at the later stages of reprogramming, inhibits the growth of non-iPS colonies and promotes the growth of reprogrammed iPS colonies. MEK is believed to be necessary for cell cycle progression in somatic cells, whereas mouse ES cells are insensitive. MEK inhibition also prevents mouse ES cell differentiation. As a consequence, the cultures contain larger and more homogeneous Oct4-GFP+ colonies, expressing higher levels of Oct4. Use of a small molecule, such as 15, to select and also maintain iPS colonies in culture may be important in achieving a safer and more efficient reprogramming strategy.
Control of lineage commitment
As described above, pluripotent and multipotent cells have been isolated and cultured (ES, iPS cells, MSC etc.), raising the hope that they can be specifically differentiated into any cell type required for cell therapy as well as basic biologic and pharmacologic studies. To date, conventional methods for cellular differentiation of ES and iPS cells (EB formation, co-culture with feeders) are neither efficient nor specific, and thus require an appropriate selection of the cell type needed. Efforts have been made to improve these protocols. For example, Smith and collaborators achieved up to 60% neuronal differentiation, exploiting the default choice of mES cells65. Jessell and collaborators obtained motor neurons (MNs) by the addition of multiple sequential or combinatorial signals to progressively drive the cells toward this phenotype66. Nevertheless, even considering one of the most efficient protocols to date65, still “a minority of cells” differentiate into non-neural cell types and 10–15% persist as clusters of undifferentiated ES cells”, this represents a major risk for cellular therapy. The in vitro differentiation process needs to become much more efficient and specific to be viable for clinical utilization. Again small molecules offer great promise in this regard. Recently, a small molecule, Stauprimide (Figure 1, structure 16) has been isolated from a high-content screen for its capacity to direct mouse and human ES cells toward differentiation process67. 16 could “prime” pluripotent cells for differentiation, which in combination with the appropriate extracellular cues could significantly reduce some of the inherent risks associated with currently available protocols.
Over 2 decades ago, 12 was shown to induce muscle cell differentiation from mesenchymal cells by inducing MyoD expression68. This could be considered amongst the earliest evidence that small molecules could have a major impact on cellular differentiation. Recently, a great deal of effort has been focused on the neuronal differentiation process, with the hope to potentially “prime” transplantable cells such as iPS or ES cells to cure degenerative neurological diseases or injuries. Although retinoic acid (RA, Figure 1, structure 17) has proven to induce differentiation from multiple cell lines, its effects are pleiotropic, so the need for more specific and efficient molecules remains. Adopting a candidate-based strategy, Chambers and colleagues have recently developed a very efficient method to derive neurons69. The critical role of SMAD signaling in preventing neural induction has been previously demonstrated. Starting with adherent cultures of human ES or iPS cells, the authors described the synergistic effect of two upstream inhibitors of SMAD dependent transcription, i.e. the protein noggin and the small molecule ALK5 (TGFβ receptor 1) inhibitor SB431542 (Figure 1, structure 18), that enables an almost complete (>80%) neuronal induction.
In parallel, high throughput phenotypic screens have been performed, leading to the isolation of molecules specifically directing precursors toward a defined phenotype. For example, TWS119 (Figure 1, structure 19) has been shown to induce neuronal differentiation of mouse P19 EC and ES cells, by inhibiting GSK-3β activity and thereby stabilizing β-catenin70. More recently, phospho-serine (p-Ser, Figure 1, structure 20) has been found to induce neuronal differentiation of primary NSC and hES cells71. Also, neuropathiazol (Figure 1, structure 21) was identified for its capacity to inhibit the proliferation of adult hippocampal NSC and induce neuronal differentiation and survival, at the expense of astroglial differentiation72. Interestingly, similar results have been previously found for 11, both in vitro and in vivo73. These are just a few examples among many studies aimed at the discovery of small molecules capable of influencing the fate of pluripotent progenitors (for a recent excellent review please see42). Neurons are obviously not the only cell type of interest. Better control over the production of HSCs, cardiomyocytes, myoblasts, pancreatic islets etc. is necessary to enable cell based therapies for numerous diseases and injuries.
CHEMICAL GENOMICS
Chemical genomics is a powerful method to complement more traditional genetic techniques (i.e. knockout mice, siRNA) for the dissection of complex signaling networks. Small organic molecules that selectively modulate cellular networks that are composed of complex protein/protein interactions and control cellular/organismal phenotypes represent powerful tools to study complex cellular processes. Large-scale forward chemical genomic screens do not bias the choice of molecular target, but rather seek to find any relevant target(s) in a pathway and as such, are well adapted to study complex signaling networks, where all the components and interactions might not be known. The ability to selectively block a subset of protein/protein interactions provides the unique ability to modulate signaling networks without completely ablating potentially critical components that participate in multiple signaling networks.
The phenotypes of self-renewal and differentiation depend upon specific, coordinated and only partially understood patterns of gene expression, which are defined by both cell type and functionality. The ultimate goal of chemical genomics to enhance regenerative medicine is to understand the mechanism of reagents/drugs in terms of the transcriptional and regulatory machinery, which they affect. Hence molecular targets, whether on the cell membrane or in cytoplasm or nucleus, become “validated targets” once they can be correlated with the complex pattern of gene expression associated with the phenotypic outcome of treatment with the compound. The complexity of signal transduction pathways and the ubiquitous crosstalk between pathways however can substantially complicate this correlation. One has to also be always cognizant of the fact that even an extremely specific perturbation of a particular pathway, will undoubtedly cause compensatory responses in the system (be it cell or organism). Furthermore, it is doubtful that in vitro studies alone will suffice to adequately address these complex questions, as the cellular pathways that control self renewal and differentiation are features of living systems that need to be addressed within the context of these intact systems (i.e. combination of cell intrinsic and cell extrinsic events or alternatively stated the interactions between a stem cell and its niche).
Methodology
Understanding the cellular machinery that regulates self-renewal and differentiation requires techniques that perturb specific molecular interactions (or targets), while not directly affecting other molecular interactions in a living cell. Knockouts or siRNA knockdowns, by removal of a molecular target, essentially block all of the interactions of the specific gene/protein that is being targeted. Small molecule pharmacologic tools/drugs, by virtue of their generally small surface area or contact/binding surface, relative to their protein molecular target, have the potential to specifically block a subset of protein/protein interactions, while leaving others unaffected. This is especially important when considering large scaffolding proteins, which generally have multiple interactions under differing sets of conditions and are often responsible for the integration of a multiple signal transduction pathways.
The choice or design of the chemical libraries used for chemical genomic screening however is critical. Although not biased by restriction to known molecular targets, in order to achieve success, the compounds in the library may need to interact with large biomolecules that constitute the potential molecular targets. Although via synthetic organic chemistry, one can potentially synthesize an extraordinarily large array of stable molecules, only a small portion of this random chemical space is probably relevant to interaction with “bio-chemical” space. Nature utilizes a very simple set of building blocks, i.e. essentially 20 amino acids to synthesize all of the proteins, which serve as components of the cellular machinery that are sufficient to orchestrate the network specificity and fidelity required for life. Nature also effectively utilizes “optimized protein-structural motifs/domains” (i.e. PDZ, SH2 etc.) time and time again; therefore many macromolecules can be grouped into related “superfamilies” based upon similar 3-D architectures. Therefore targeting “superfamilies” with chemical libraries that contain priviledged scaffolds or motifs that recognize Nature’s proprietary set of architectures, would seem to represent a rational basis for chemical genomic approaches74–77.
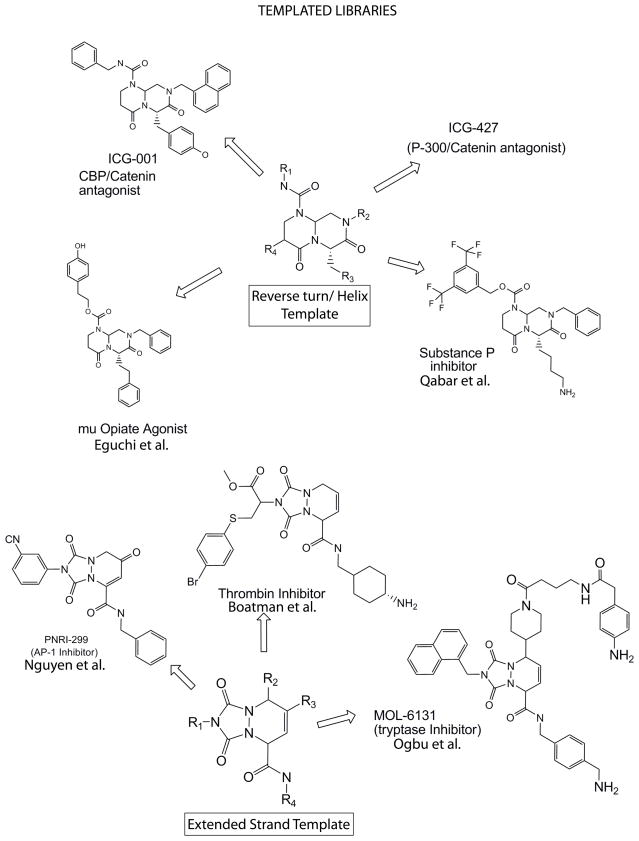
Following this line of thought a bit further; one can utilize “templated” chemical libraries, where diversity elements are presented around a basic frame or template that is known to interact with relevant “bio-chemical space”(Figure 2)78–81. This allows for specifically identifying target molecules within relevant “biochemical space” with distinct diversity elements (hence specificity). This approach also provides fundamental Structure Activity Relationship (SAR) information from high throughput screens, as each member of the library is related to all the other members of the library through the basic core template. By changing the diversity elements on the same core template, one can readily screen and identify “hits” for a range of structurally related members of a superfamily. Additionally, by changing the template slightly, yet maintaining the same diversity elements, one can potentially optimize interactions. Furthermore, high throughput combinatorial synthesis of templated libraries allows for relatively facile optimization of identified leads.
FIGURE 2.
templated library approach (reverse turn /helix template and extended strand template) and published examples therof
Target identification, the key bottleneck in chemical genomics, represents more than simple physical demonstration of a binding interaction between a small molecule and a potential “target”. For example, almost all small molecules will ‘bind” to serum albumin to some extent, yet this does not mean that serum albumin is the molecular target responsible for the relevant biological activity of the compound. Experimental methods in molecular biology (i.e. gain of function/loss of function type experiments) allow for the demonstration of the target-dependent mode of action of a small molecule and hence the “validation” of the drugs molecular target. Hence, the concepts of “target identification/validation” and “specificity of action” rest on the cumulative consistency of all the experimental evidence within the proposed model.
WNT Signaling in Stem Cell and Cancer Stem Cells: A complex cascade dissected via chemical genomics
The Wnt signaling cascade is an extremely complex signal transduction system involving 19 extracellular mammalian glycoprotein Wnt ligands, 10 Fzd family 7-TM spanning receptors and the co-receptors LRP 5, 6 as well as additional non-classical Wnt receptors (e.g. Ryk, Ror). Wnt ligands trigger a variety of intracellular responses, broadly associated with canonical (increase in nuclear β-catenin) and non-canonical (planar cell polarity, Ca++/PKC activation). In a gross oversimplification, the former is often associated with proliferation and lack of differentiation (for example as a hallmark of dysregulated Wnt signaling in cancer), whereas the latter is often associated with cell and organismal differentiation. Beyond classical Wnt activated translocation of β-catenin to the nucleus, a number of other factors (growth factors, prostaglandins, etc.) can induce the nuclear localization of β-catenin. β-catenin plays a key role in both aspects of the Wnt signaling cascade (canonical and non-canonical) through both its nuclear functions and cytoskeletal/cytoplasmic membrane interactions. Nuclear β-catenin drives the expression of a Wnt/catenin regulated-cassette of genes, whereas outside of the nucleus, β-catenin plays a critical role in cell-cell interactions and cellular polarization. Rather than being thought of as distinct signal transduction systems, we believe that the balance and coordination between nuclear/transcriptionally active β-catenin and cytoplasmic/cytoskeletal β-catenin, couples canonical with non-canonical Wnt signaling.
Wnt signaling plays important roles throughout development82. Although most would agree that Wnt signaling is important in stem cell biology, there is no consensus as to whether Wnt signaling is important for proliferation and maintenance of potency (pluri- or multipotency) (for e.g. see 48,46,83) or differentiation of stem/progenitor cells50,84. Wnt/β-catenin signaling has been demonstrated to maintain pluripotency in ES cells48 and is critical for the expansion of neural progenitors thereby increasing brain size85. However, Wnt/β-catenin signaling is also required for neural differentiation of ES cells86, fate decision in neural crest stem cells87 and Wnt3a has been reported to promote differentiation into the neural and astrocytic lineages by inhibiting neural stem cell maintenance88. Clearly, Wnt/β-catenin signaling also plays a critical role in lineage decision/commitment. These dramatically different outcomes upon activation of the Wnt signaling cascade has fueled enormous controversy concerning the role of Wnt signaling in maintenance of potency and induction of differentiation.
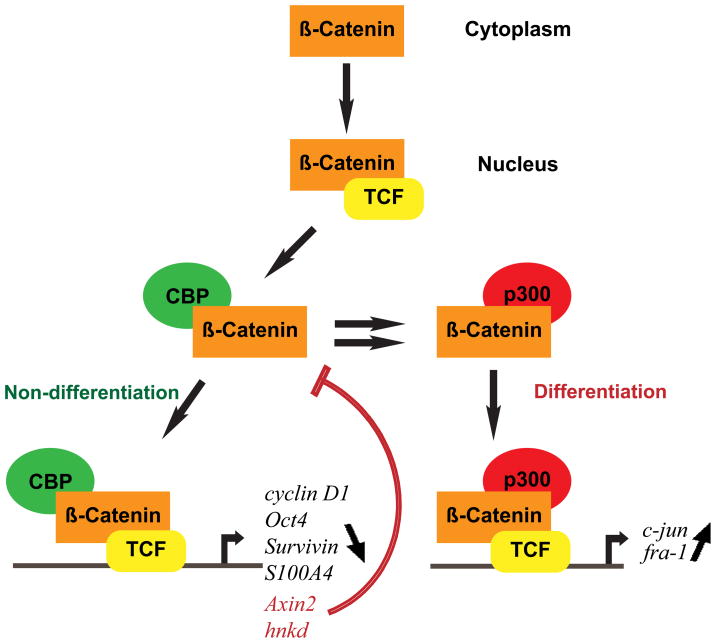
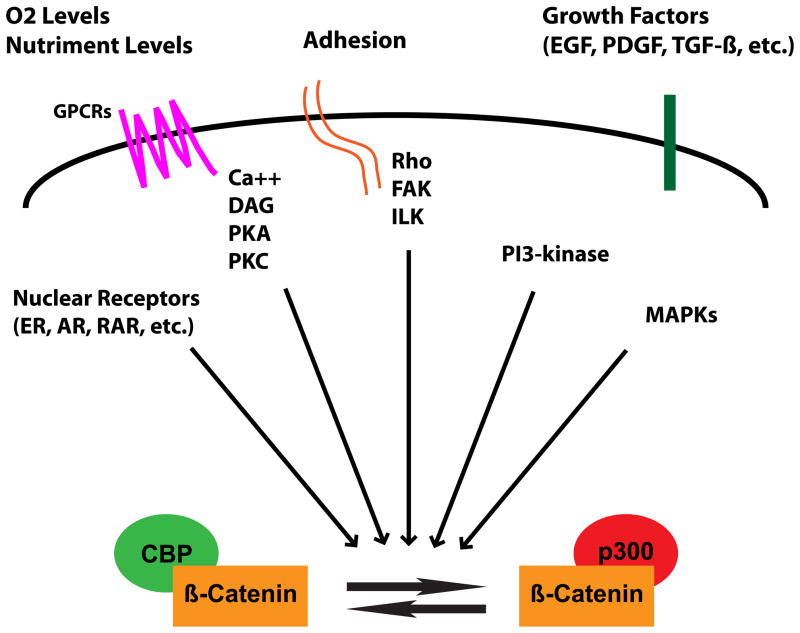
Until recently, a rationale for the dichotomous behavior of Wnt/β-catenin signaling in controlling both proliferation and differentiation has been unclear. Recently, using a selective antagonist of the CBP/β-catenin interaction ICG-00189 (Figure 1, structure 22) that we identified utilizing a chemical genomic approach81, we have developed a model to explain the divergent activities of Wnt/β-catenin signaling (Figure 3). Our model highlights the distinct roles of the coactivators CBP and p300 in the Wnt/β-catenin signaling cascade90. The critical feature of the model is that CBP/β-catenin mediated transcription is critical for stem cell/progenitor cell maintenance and proliferation, whereas a switch to p300/β-catenin mediated transcription is the initial critical step to initiate differentiation and a decrease in cellular potency. Again, this represents an oversimplification. In reality, although a subset of the gene expression cassette that is regulated by the CBP/β-catenin arm is critical for the maintenance of potency and proliferation (e.g. Oct4, survivin, etc.), other genes that are regulated in this manner (e.g. hNkd and axin2) are in fact negative regulators of the CBP/β-catenin arm of the cascade (Figure 4)91,92. This inherently makes perfect sense. Assuming potency and activation of the CBP/β-catenin arm is the default pathway, at some point, in order for development to proceed, one must stop proliferation, exit cell cycle and initiate the process of differentiation (Figure 4).
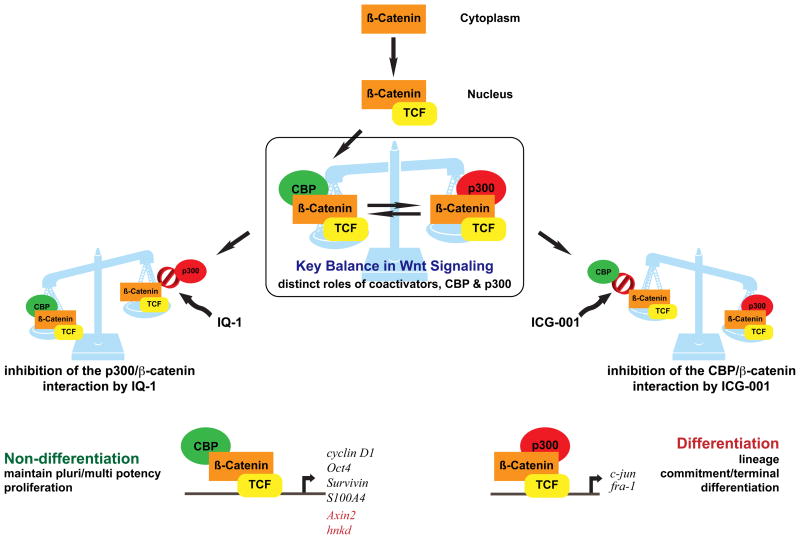
FIGURE 3.
Wnt signaling is a complex pathway, believed to be involved in the regulation of divergent processes, including the maintenance of pluripotency and commitment to differentiation. To resolve these divergent responses to activation of Wnt/β-catenin signaling, we have developed a model in which β-catenin/CBP-mediated transcription is critical for the maintenance of potency, whereas a switch to β-catenin/p300-mediated transcription is the first critical step to initiate differentiation. Hence the balance between CBP and p300-mediated β-catenin transcription regulates the balance between maintenance of potency and the initiation of commitment to differentiate. The interaction of β-catenin with either CBP or p300 can be modulated by two small molecules, respectively ICG-001 and IQ-1, identified in our laboratory. As a consequence, these small molecules regulate Wnt signaling pathway outcomes.
FIGURE 4.
a negative feedback normally turns off the CBP/β-catenin arm of the pathway and initiates differentiation via the p300/β-catenin arm.
Identification of the small molecule IQ-1 (Figure 1, structure 23, Figure 4) that allows for the Wnt/β-Catenin-driven long-term expansion of mouse ES cells by blocking the p300/β-catenin interaction, and thereby prevents spontaneous differentiation in the absence of LIF, is consistent and lends further credence to our model93. This study demonstrated that 23, in combination with Wnt3a to stimulate nuclear translocation of β-catenin, is sufficient to sustain pluripotency by decreasing the interaction of β-catenin with p300 and consequently increasing its interaction with CBP. Removal of 23 from the mES cultures rapidly leads to loss of pluripotency, even in the presence of Wnt3a. The switch to p300/β-catenin mediated transcription, whether induced pharmacologically (Figure 4), by removal of 23, or addition of 22, or naturally (e.g. LIF withdrawal), is critical to initiate a differentiative program with a more limited proliferative capacity 90,93. Importantly, the β-catenin coactivator switching mechanism is also critical in the maintenance of hES cell pluripotency and both mouse and human SSC populations94 (and M. Kahn et al. unpublished results).These studies represents additional “proof-of-principle” that complex phenotypes can be controlled by a single small molecule, isolated for its desirable and specific pharmacologic activity.
Translation to cancer and other diseases of somatic stem cells
The similarities between normal adult stem cells and cancer stem cells (CSC), suggest that the signaling pathways (e.g. Wnt, Hedgehog and Notch) involved in regulating SSC maintenance are also involved in the regulation of CSC. Aberrant regulation of these same pathways leads to neoplastic proliferation in the same tissues95,96. Interestingly, progression of chronic myelogenous leukemia from chronic phase to blast crisis and imatinib resistance was correlated with increased nuclear β-catenin levels, a hallmark of increased Wnt/TCF/β-catenin transcription97. Recent studies have revealed that multi-drug resistance genes, including MDR-1, ABCG2, ABCA3 and BRCP1 are also intrinsically expressed in stem/progenitor cells from multiple adult tissues and that they may contribute to the side population (SP) phenotype of malignant cells98–100. Wnt/β-catenin signaling appears to play an important role in ABCB1/MDR-1 transcription. This observation was initially based upon the increased expression of MDR-1 associated with intestinal crypt cells, which carry a defective APC tumor suppressor gene in both the Min mouse and FAP patients101,102. Putative TCF binding elements were also identified in the ABCB1 promoter (−1813 to −275 bp)101. Canonical Wnt signaling is believed to play an important role in the maintenance of hematopoietic progenitors and also in the lineage commitment of progenitors during hematopoiesis. Expression of survivin, which we have demonstrated is a Wnt/CBP/β-catenin regulated gene103, expression is important during hematopoeisis104 and it is prominently upregulated in CD34+ hematopoietic stem/progenitor cells upon growth factor treatment. Survivin-deficient hematopoietic progenitors show defects in erythroid and megakaryocytic formation105. Recently, continued expression of survivin upon differentiation has been associated with teratoma formation by hES cells106. However, it is worth noting that β-catenin-deficient107 and even β,γ-double deficient108 mice maintain apparently normal hematopoiesis through the Wnt signaling cascade109, pointing to yet uncharacterized catenin-like molecule(s) that can compensate for the loss of both β and γ catenin.
Given the fact that multiple mutations can lead to the aberrant activation of nuclear β-catenin signaling, there is a clear need for drugs that attenuate the nuclear functions of β-catenin. The small molecule antagonist that our laboratory developed, 22, by binding to the coactivator CBP and not to its highly homologous relative p300, specifically downregulates a subset of Wnt/β-catenin-driven genes including S100A4 and survivin the number 1 and 4 transcriptomes up-regulated in cancer89. 22 has proven to be an invaluable tool in helping us to dissect the complex signal transduction pathways and interactions involved in the regulation of “stemness”.
AN INTEGRATED VIEW OF “STEMNESS”
Even from the abbreviated review above, certain characteristics about “stemness” in a variety of systems, i.e. embryonic stem cells, iPS cells, SSCs and CSCs appear to be conserved. For example, although not identical in mES and hES (possibly because of the status of the ES cell in these two species (i.e. stem versus epistem cell110), there are a number of critical pathways that are involved in the maintenance or loss of potency in all of these cell types (i.e. Wnt, Notch, Hedeghog, TGFβ/BMP, JAK/Stat, FGF/MAPK/PI3K). Another conclusion that one can draw from the studies described above, which again only represent a small fraction of the work already published (please see reviews listed above in the text), is that there are many potential points of intersection along these critical signaling networks, where one can utilize small molecules to modulate “stemness’ and the maintenance or loss of potency (i.e. initiation of differentiation). Finally, we believe that the ultimate decision for a cell to retain potency or initiate differentiation, although enormously complex, taking in hundreds if not thousands of inputs (e.g. concentrations of different growth factors, cytokines and hormones and the subsequent activation of different signal transduction complexes and kinase cascades, ionic concentrations (e.g. Ca++), nutrient levels, oxygen levels, genetic mutations, adhesion to substratum, etc.), in the end must be integrated and funneled down into a simple decision point, i.e. a yes/no binary decision (Figure 5). That is, after reading all of this input information, the cell must decide to either maintain its level of potency (be that ES or somatic stem cell), or to go on to differentiate and lose a level of potency. More recently, it has become clear that this decision process/point is also reversible (iPS cells demonstrating this most clearly, as well as much earlier work on transformation/immortalization of cells)26. Based upon our own investigations, we propose that this simple decision point that all of this information is funneled down into involves a change in coactivators (CBP versus p300) interacting with β-catenin (or catenin-like molecules in the absence of β-catenin) and more generally the basal transcriptional apparatus111. A model incorporating and summarizing this concept is depicted in Figure 5. We have specifically not specified the transcription factor TCF as the binding partner for β-catenin in this model. Although in classical Wnt signaling (e.g. in the intestinal stem cells) partnering with the TCF family of transcription factors is critical. In reality, this may represent only a fraction of the roles played by nuclear β-catenin in transcription, as β-catenin is known to interact with a much broader array of potential transcription partners (including the orphan nuclear receptors LRH-1 and Nurr1, nuclear receptors ER, AR, RAR and VDR, Sox and Smad family members and Oct4 itself to list but a few)112. We believe that this is an extremely fundamental switching mechanism that is present in mouse and human ES cells and that is conserved throughout essentially all stem/progenitor cell lineages. For example, we have found that the earliest and perhaps most important cellular decision point, i.e. at the embryonic 8 cell stage, is governed by differential coactivator usage by catenin (i.e. CBP/catenin is require for maintenance of inner cell mass and expression of Oct4, whereas a switch to p300/catenin initiates the formation of cdx2 positive trophectoderm113. Additionally, this switching mechanism appears to be important in a wide variety of somatic stem/progenitors (e.g. neural, cardiac, myogenic, hematopoietic etc.) as well as transformed somatic stem/progenitors (e.g. leukemic stem cells) (M. Kahn et al. unpublished results). It is also clear that there are numerous ways to perturb/modulate this coactivator decision process. For example, in a siRNA screen of the human kinome in HEK293 cells, we found that almost 25% of the kinases screened had significant effects on Wnt signaling (M. Kahn, H Ma and H. Nakanishi unpublished results). Therefore, we do not find it surprising that many small molecule kinase inhibitors modulate the “stemness’ or potency of ES and somatic stem cells. We are presently studying the effects of some of these kinase cascades on differential coactivator usage and have already demonstrated that activation of PKC93 and the MAPK cascade (M. Kahn, H Ma unpublished results) can differentially affect the interactions of β-catenin with CBP and p300. The ability to switch coactivators within the Wnt/catenin signaling cascade, thereby changes a gene expression cassette, including a change in miRNA expression (M. Kahn, J-L. Teo, P Manegold manuscript in preparation). We believe that this “regulation of expression cassettes” is critical to the proper control, maintenance and initiation of differentiation of both embryonic and somatic stem cells. Furthermore, we believe that the inability to properly initiate differentiation of somatic stem cells may underlie a wide range of diseases including fibrosis and Alzheimer’s disease90. Moreover, we believe that this is the underlying malfunction in essentially all cancers. Therefore, we would propose that cancer rather than being 10,000 different diseases (i.e. breast cancer is different than colon, is different than leukemia, etc.), is a disease in which 10,000 different mutations (some of which are tissue specific) (e.g. bcr/abl, K-Ras, Her2, etc.) can lead to aberrant regulation of the underlying equilibrium between catenin/CBP and catenin/p300; i.e. between proliferation and maintenance of potency and the initiation of differentiation (Figure 5) thereby aberrantly increasing the CBP/catenin interaction at the expense of the p300/catenin interaction. Finally, in regard to recent publications concerned with the increase in Wnt signaling with aging114,115 and the importance of stem cell homeostasis in aging and disease, it is interesting to speculate as to whether a progressive imbalance in this coactivator equilibrium is associated with the aging process more generally. This would also make sense since for the most part, cancer, fibrosis and neurodegeneration are diseases where risk increases substantially with aging.
FIGURE 5.
the ultimate decision for a cell to retain potency or initiate differentiation is dependent upon numerous inputs some of which are presented here: e.g. concentration of different growth factors, cytokines and hormones and the subsequent activation of different signal transduction complexes and kinase cascades, ionic concentrations (e.g. Ca++), nutrient levels, oxygen levels, genetic mutations, adhesion to substratum. In the end those multiple pathways must be integrated and funneled down into a simple decision point, i.e. a yes/no binary decision. We believe that Wnt signaling and the equilibrium between CBP-mediated and p300-mediated transcription play a central role in integrating these signals.
CONCLUDING REMARKS
As described above, there is a great expectation that small molecules will provide very useful tools for understanding the basic biology of stem cells as well as aiding the enablement of regenerative medicine. Concerns about specificity will always exist in regards to a small molecule and its molecular target, and the problem that a specific phenotype, or lack thereof, could be the result of an off-target effect not characterized in the initial elucidation of the mechanism(s) of action of the small molecule regulator. For example, targeting highly conserved motifs that are commonly utilized, for example the ATP binding motif, have demonstrated that the majority of the structures that target this motif fall within the pi-heterocycle category that mimic ATP81. This makes it quite difficult to definitively assign the mechanism of action of these compounds to one molecular target as there are often multiple members of the superfamily that are targeted with relatively similar efficiency. Yet, this should not deter us from seeking specific small molecules to use both as pharmacologic tools to better understand the complex networks that control maintenance and activation of stem cells or as therapeutic agents. We believe that the future of medicine clearly lies in achieving a better understanding about somatic stem cell homeostasis, as it is clear that the major limitations to increased life expectancy going forward (outside of threats from infectious agents) lie in being able to treat diseases of SSCs.
Finally, although we still have much to learn about the enormous complexities of this relatively simple yes/no decision process, i.e. to maintain the current level of potency or to initiate differentiation, an even more daunting question awaits us. That is attempting to accurately control lineage decision/commitment with small molecules. This decision process undoubtedly requires both the turning on, via combinatorial activation of a number of transcription factors and genes, as well as the simultaneous turning off in the same cell, a number of competing lineage programs and transcription factors. Specific small molecule pharmacologic agents unquestionably will play a critical role in helping us to unlock the keys to a more complete understanding of the complexities of “stemness” and lineage commitment.
It’s not only what we have inherited from our father and mother that haunts us. It’s all sorts of dead ideas, and lifeless old beliefs, and so forth. They have no vitality, but they cling to us all the same, and we can’t get rid of them.
Henrik Ibsen
Acknowledgments
We would like to sincerely apologize to our colleagues whose work was omitted in this perspective due to space limitations. Work in our laboratory has been supported by grants from the US National Institutes of Health (RO1-HL386578), the California Institute for Regenerative Medicine, the Jeannik M. Littlefield-AACR Grants for Metastatic Colon Cancer, the Susan G. Komen Foundation and the V-Foundation. A. L. wishes to thank the Havner Family Foundation for fellowship support. We would like to thank Professor Nobuo Kato, Osaka University for his comments and in particular modifications to figures that we believe have significantly enhanced their clarity. Finally, M.K. wishes to express his sincere appreciation to Professors Gilbert Stork and Edwin Krebs for their insights over the years and inspiration for our studies.
Abbreviations
- iPS
induced Pluripotent Stem
- HSC
Hematopoietic Stem Cell
- ES
Embryonic Stem
- SSC
Somatic Stem Cell
- MSC
Mesenchymal Stem Cell/Multipotent Stromal Cell
- CSC
Cancer Stem Cell
- TIC
Tumor Initiating Cell
- LIF
Leukemia Inhibitory Factor
- NSC
Neural Stem Cell
- MEF
Mouse Embryonic Fibroblast
Biographies
Agnes Lukaszewicz obtained her masters of molecular and cell biology at l’Ecole Normal Supérieure de Lyon, France, followed by a PhD in Neuroscience on the role of cell cycle regulatory molecules in proliferation and differentiation of cortical neuroblasts. She then joined Dr. David J. Anderson’s laboratory at the California Institute of Technology, where her post-doctoral studies tackled the role of Cyclin Ds in neuronal specification and Neural Stem Cell maintenance. Following her interest on stem cell homeostasis, she recently joined Dr. Michael Kahn’s lab at the Center for Stem Cell and Regenerative Medicine, University of Southern California, where she is studying the role of Wnt signaling in the regulation of the mode of division of cortical precursors and its implications in neurogenesis.
Michael McMillan received his BS in chemistry from the University of Texas at Austin and his PhD in organic chemistry from Columbia University with Prof. Gilbert Stork. After graduation he went on an extended postdoctoral walkabout, including extended stays near the mouth of the Charles River in Massachusetts (MIT) and on the eastern shores of the San Francisco Bay in California (UC Berkeley). In 1994, he joined Molecumetics in Seattle, Washington to work on peptidomimetic chemistry. In 2007 he joined the Center for Stem Cell and Regenerative Medicine at USC.
Michael Kahn, is the Provost’s Professor of Medicine and Pharmacy at the University of Southern California with a joint appointment in the Department of Biochemistry and Molecular Biology, Keck School of Medicine and the Department of Pharmacology and Pharmaceutical Sciences in the School of Pharmacy. Dr. Kahn is also the co-director of the GI Oncology Program at the USC Norris Comprehensive Cancer Center. Dr. Kahn obtained his Ph.D. at Yale University with Professor Samuel Danishefsky and was an NIH Postdoctoral fellow at Columbia University with Professor Gilbert Stork. Dr. Kahn’s lab has been working extensively in the areas of chemical biology and particularly Wnt signaling in development, disease, stem cells and cancer stem cells for the past 9 years.
References
- 1.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113 doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 3.Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- 4.Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, Moore HD, Andrews PW. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–668. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- 5.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 6.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 7.Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 8.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 9.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 10.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 11.Byrne JA. Generation of isogenic pluripotent stem cells. Hum Mol Genet. 2008;17:R37–41. doi: 10.1093/hmg/ddn053. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–17. doi: 10.1016/j.cell.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 16.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS. Niwa, HPluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318 doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458 doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 26.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- 28.Warburton D, Perin L, Defilippo R, Bellusci S, Shi W, Driscoll B. Stem/progenitor cells in lung development, injury repair, and regeneration. Proc Am Thorac Soc. 2008;5:703–706. doi: 10.1513/pats.200801-012AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yacoub M, Suzuki K, Rosenthal N. The future of regenerative therapy in patients with chronic heart failure. Nat Clin Pract Cardiovasc Med. 2006;3(Suppl 1):S133–135. doi: 10.1038/ncpcardio0401. [DOI] [PubMed] [Google Scholar]
- 30.Taupin P. Stroke-induced neurogenesis: physiopathology and mechanisms. Curr Neurovasc Res. 2006;3:67–72. doi: 10.2174/156720206775541769. [DOI] [PubMed] [Google Scholar]
- 31.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 32.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 33.Miller RH, Bai L. The expanding influence of stem cells in neural repair. Ann Neurol. 2007;61:187–188. doi: 10.1002/ana.21106. [DOI] [PubMed] [Google Scholar]
- 34.Bai L, Caplan A, Lennon D, Miller RH. Human mesenchymal stem cells signals regulate neural stem cell fate. Neurochem Res. 2007;32:353–362. doi: 10.1007/s11064-006-9212-x. [DOI] [PubMed] [Google Scholar]
- 35.Gerdoni E, Gallo B, Casazza S, Musio S, Bonanni I, Pedemonte E, Mantegazza R, Frassoni F, Mancardi G, Pedotti R, Uccelli A. Mesenchymal stem cells effectively modulate pathogenic immune response in experimental autoimmune encephalomyelitis. Ann Neurol. 2007;61:219–227. doi: 10.1002/ana.21076. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Rosen JM. Stem cells in the etiology and treatment of cancer. Curr Opin Genet Dev. 2006;16:60–64. doi: 10.1016/j.gde.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 38.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 39.Duval D, Malaise M, Reinhardt B, Kedinger C, Boeuf H. A p38 inhibitor allows to dissociate differentiation and apoptotic processes triggered upon LIF withdrawal in mouse embryonic stem cells. Cell Death Differ. 2004;11:331–341. doi: 10.1038/sj.cdd.4401337. [DOI] [PubMed] [Google Scholar]
- 40.Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 43.Chen S, Do JT, Zhang Q, Yao S, Yan F, Peters EC, Scholer HR, Schultz PG, Ding S. Self-renewal of embryonic stem cells by a small molecule. Proc Natl Acad Sci U S A. 2006;103:17266–17271. doi: 10.1073/pnas.0608156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vallier L, Pedersen RA. Human embryonic stem cells: an in vitro model to study mechanisms controlling pluripotency in early mammalian development. Stem Cell Rev. 2005;1:119–130. doi: 10.1385/SCR:1:2:119. [DOI] [PubMed] [Google Scholar]
- 45.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R7, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, Rao MS. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 47.Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 48.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 49.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 50.Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X, Cheng L. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- 51.Desbordes SC, Placantonakis DG, Ciro A, Socci ND, Lee G, Djaballah H, Studer L. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 53.Damoiseaux R, Sherman SP, Alva JA, Peterson C, Pyle AD. Integrated Chemical Genomics Reveals Modifiers of Survival in Human Embryonic Stem Cells. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duncan AW, Dorrell C, Grompe M. Stem Cells and Liver Regeneration. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bug G, Gul H, Schwarz K, Pfeifer H, Kampfmann M, Zheng X, Beissert T, Boehrer S, Hoelzer D, Ottmann OG, Ruthardt M. Valproic acid stimulates proliferation and self-renewal of hematopoietic stem cells. Cancer Res. 2005;65:2537–2541. doi: 10.1158/0008-5472.CAN-04-3011. [DOI] [PubMed] [Google Scholar]
- 57.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujino H, West KA, Regan JW. Phosphorylation of glycogen synthase kinase-3 and stimulation of T-cell factor signaling following activation of EP2 and EP4 prostanoid receptors by prostaglandin E2. J Biol Chem. 2002;277:2614–2619. doi: 10.1074/jbc.M109440200. [DOI] [PubMed] [Google Scholar]
- 59.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 64.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 65.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 66.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 67.Zhu S, Wurdak H, Wang J, Lyssiotis CA, Peters EC, Cho CY, Wu X, Schultz PG. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell. 2009;4:416–426. doi: 10.1016/j.stem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 68.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 69.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding S, Wu TY, Brinker A, Peters EC, Hur W, Gray NS, Schultz PG. Synthetic small molecules that control stem cell fate. Proc Natl Acad Sci U S A. 2003;100:7632–7637. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saxe JP, Wu H, Kelly TK, Phelps ME, Sun YE, Kornblum HI, Huang J. A phenotypic small-molecule screen identifies an orphan ligand-receptor pair that regulates neural stem cell differentiation. Chem Biol. 2007;14:1019–1030. doi: 10.1016/j.chembiol.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Warashina M, Min KH, Kuwabara T, Huynh A, Gage FH, Schultz PG, Ding S. A synthetic small molecule that induces neuronal differentiation of adult hippocampal neural progenitor cells. Angew Chem Int Ed Engl. 2006;45:591–593. doi: 10.1002/anie.200503089. [DOI] [PubMed] [Google Scholar]
- 73.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Che Y, Marshall GR. Privileged scaffolds targeting reverse-turn and helix recognition. Expert Opin Ther Targets. 2008;12:101–114. doi: 10.1517/14728222.12.1.101. [DOI] [PubMed] [Google Scholar]
- 75.Gianti E, Sartori L. Identification and selection of “privileged fragments” suitable for primary screening. J Chem Inf Model. 2008;48:2129–2139. doi: 10.1021/ci800219h. [DOI] [PubMed] [Google Scholar]
- 76.Muller G, Giera H. Protein secondary structure templates derived from bioactive natural products. Combinatorial chemistry meets structure-based design. J Comput Aided Mol Des. 1998;12:1–6. doi: 10.1023/a:1007954801605. [DOI] [PubMed] [Google Scholar]
- 77.Pettibone DJ, Freidinger RM. Discovery and development of non-peptide antagonists of peptide hormone receptors. Biochem Soc Trans. 1997;25:1051–1057. doi: 10.1042/bst0251051. [DOI] [PubMed] [Google Scholar]
- 78.Kahn M. Peptide Secondary Structure Mimetics: Recent Advances and Future Challenges. Synlett. 1993;11:821–826. [Google Scholar]
- 79.Eguchi M, McMillan M, Nguyen C, Teo JL, Chi EY, Henderson WR, Jr, Kahn M. Chemogenomics with peptide secondary structure mimetics. Comb Chem High Throughput Screen. 2003;6:611–621. doi: 10.2174/138620703771981197. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen C, Teo JL, Matsuda A, Eguchi M, Chi EY, Henderson WR, Jr, Kahn M. Chemogenomic identification of Ref-1/AP-1 as a therapeutic target for asthma. Proc Natl Acad Sci U S A. 2003;100:1169–1173. doi: 10.1073/pnas.0437889100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McMillan M, Kahn M. Investigating Wnt signaling: a chemogenomic safari. Drug Discov Today. 2005;10:1467–1474. doi: 10.1016/S1359-6446(05)03613-5. [DOI] [PubMed] [Google Scholar]
- 82.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 84.Otero JJ, Fu W, Kan L, Cuadra AE, Kessler JA. Beta-catenin signaling is required for neural differentiation of embryonic stem cells. Development. 2004;131:3545–3557. doi: 10.1242/dev.01218. [DOI] [PubMed] [Google Scholar]
- 85.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 86.Zechner D, Fujita Y, Hulsken J, Muller T, Walther I, Taketo MM, Crenshaw EB, 3rd, Birchmeier W, Birchmeier C. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 87.Hari L, Brault V, Kleber M, Lee HY, Ille F, Leimeroth R, Paratore C, Suter U, Kemler R, Sommer L. Lineage-specific requirements of beta-catenin in neural crest development. J Cell Biol. 2002;159:867–880. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muroyama Y, Kondoh H, Takada S. Wnt proteins promote neuronal differentiation in neural stem cell culture. Biochem Biophys Res Commun. 2004;313:915–921. doi: 10.1016/j.bbrc.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 89.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Kim HY, Moon SH, Ha JR, Kahn M. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teo JL, Ma H, Nguyen C, Lam C, Kahn M. Specific inhibition of CBP/beta-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci U S A. 2005;102:12171–12176. doi: 10.1073/pnas.0504600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Creyghton MP, Roel G, Eichhorn PJ, Hijmans EM, Maurer I, Destree O, Bernards R. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 2005;19:376–386. doi: 10.1101/gad.328905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeng W, Wharton KA, Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 93.Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2007;104:5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hasegawa K, Teo J-T, Suemori H, Nakatsuji N, Pera M, Kahn M. ISSCR Annual Meeting; 2009. [Google Scholar]
- 95.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 96.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 97.Jamieson CH, Weissman IL, Passegue E. Chronic versus acute myelogenous leukemia: a question of self-renewal. Cancer Cell. 2004;6:531–533. doi: 10.1016/j.ccr.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 98.Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, Mori M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 99.Hirschmann-Jax C, Foster AE, Wulf GG, Goodell MA, Brenner MK. A distinct “side population” of cells in human tumor cells: implications for tumor biology and therapy. Cell Cycle. 2005;4:203–205. [PubMed] [Google Scholar]
- 100.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamada T, Takaoka AS, Naishiro Y, Hayashi R, Maruyama K, Maesawa C, Ochiai A, Hirohashi S. Transactivation of the multidrug resistance 1 gene by T-cell factor 4/beta-catenin complex in early colorectal carcinogenesis. Cancer Res. 2000;60:4761–4766. [PubMed] [Google Scholar]
- 102.Yamada T, Mori Y, Hayashi R, Takada M, Ino Y, Naishiro Y, Kondo T, Hirohashi S. Suppression of intestinal polyposis in Mdr1-deficient ApcMin/+ mice. Cancer Res. 2003;63:895–901. [PubMed] [Google Scholar]
- 103.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 104.Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: implication of survivin expression in normal hematopoiesis. Blood. 2001;98:2091–2100. doi: 10.1182/blood.v98.7.2091. [DOI] [PubMed] [Google Scholar]
- 105.Fukuda S, Foster RG, Porter SB, Pelus LM. The antiapoptosis protein survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood. 2002;100:2463–2471. doi: 10.1182/blood.V100.7.2463. [DOI] [PubMed] [Google Scholar]
- 106.Blum B, Bar-Nur O, Golan-Lev T, Benvenisty N. The anti-apoptotic gene survivin contributes to teratoma formation by human embryonic stem cells. Nat Biotechnol. 2009;27:281–287. doi: 10.1038/nbt.1527. [DOI] [PubMed] [Google Scholar]
- 107.Cobas M, Wilson A, Ernst B, Mancini SJ, MacDonald HR, Kemler R, Radtke F. Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. J Exp Med. 2004;199:221–229. doi: 10.1084/jem.20031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koch U, Wilson A, Cobas M, Kemler R, Macdonald HR, Radtke F. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008;111:160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- 109.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, Huelsken J, Held W. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 110.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 111.Deato MD, Tjian R. An Unexpected Role of TAFs and TRFs in Skeletal Muscle Differentiation: Switching Core Promoter Complexes. Cold Spring Harb Symp Quant Biol. 2008;73:217–225. doi: 10.1101/sqb.2008.73.028. [DOI] [PubMed] [Google Scholar]
- 112.Le NH, Franken P, Fodde R. Tumour-stroma interactions in colorectal cancer: converging on beta-catenin activation and cancer stemness. Br J Cancer. 2008;98:1886–1893. doi: 10.1038/sj.bjc.6604401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McKinnon T, Ma H, Hasegawa K, Kahn M. ISSCR Annual Meeting; Barcelona, Spain. 2009. [Google Scholar]
- 114.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 115.Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, Gutkind JS, Hwang PM, Finkel T. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]