Abstract
Contemporary genomic tools now allow the fast and reliable genotyping of hundreds of thousands of variants and permit an unbiased interrogation of the common variability across the human genome. These technical advances have been the basis of numerous recent investigations of genes underlying complex genetic traits, and the results for blood pressure and hypertension have been of particular interest. The pathophysiology of the complex genetic trait blood pressure and hypertension is unclear. The heritability of essential hypertension is high and insights can be gained by finding associated genes. Current genome-wide association studies (GWAS) have identified 10 to 20 loci in or near genes that generally were not expected to be associated with blood pressure or essential hypertension; more significant variants will be discovered when even larger and more refined studies become available. This article gives a short introduction to GWAS and summarizes the current findings for blood pressure and hypertension.
Keywords: Blood pressure, Hypertension, Genome-wide association study, Genomics
Introduction
For many years, hypertension genetics has been dominated by the stark contrast between the high heritability of blood pressure and hypertensive traits and the frustrating reality that no clearly reproducible genetic variant for essential hypertension could be found. Estimates for heritability in family studies range from 31% or 34% (single-measure systolic blood pressure [SBP] and diastolic blood pressure [DBP], averaged over three studies [1–3]) to 56% or 57% (long-term average SBP and DBP phenotype [1]), to 63% or 68% (24-hour profile of SBP and DBP [4]). Studies of rare familial forms of hypertension, on the other hand, have been extremely successful in identifying causal genes, illuminating the regulatory pathways of blood pressure [5], but these variants appear unrelated to essential hypertension in the general population.
Among the reasons put forward to explain the difficulty in identifying genes for blood pressure and hypertension are theoretical considerations suggesting that the power of linkage studies is low for variants with modest effects [6]. A large number of candidate genes have been tested for association with blood pressure and hypertension without convincing results; the candidate gene approach suffers for the very same reason why these investigations are carried out: the nature of essential hypertension has remained elusive since it was first described in 1877 [7].
Hypertension and blood pressure have been considered complex (or polygenic) genetic traits since the classic work of Pickering and colleagues [8] and have been prime examples of complex inheritance [9]. Successful identification of blood pressure genes therefore would not only explain some part of the nature of essential hypertension—elusive so far—but also would provide insight into the architecture of complex genetic traits in general.
Technologic advances now permit the genotyping of hundreds of thousands to more than a million single nucleotide polymorphisms (SNPs) on a single microarray at a reasonable cost [10, 11]. These genomic tools permit the interrogation of a large proportion of the common human genetic variation throughout the genome, a task that previously was not feasible. Association testing of every single SNP against hypertensive and blood pressure traits (a genome-wide association study, GWAS) opens the way for an unbiased investigation of genetic causes of these traits, which can be considered one of the first direct applications of the Human Genome Project [12] and the HapMap Project [13••].
This article gives a short introduction to GWAS and how to read GWAS, summarizes the results from studies on blood pressure and hypertension published so far, and highlights some of the general conclusions and limitations that have become apparent.
Architecture of Complex Genetic Disease
Complex genetic disease is determined by genes and the environment. The environmental factors of this equation are not discussed in this article, nor are interactions of genes and environment, which are likely to be important but currently are difficult to quantify. The impact of genes on complex genetic disease is thought to be largely determined by three basic characteristics of the disease-associated allelic variant: the frequency of the variant, the effect size of the variant on the phenotype, and the number of genetic variants acting on the phenotype.
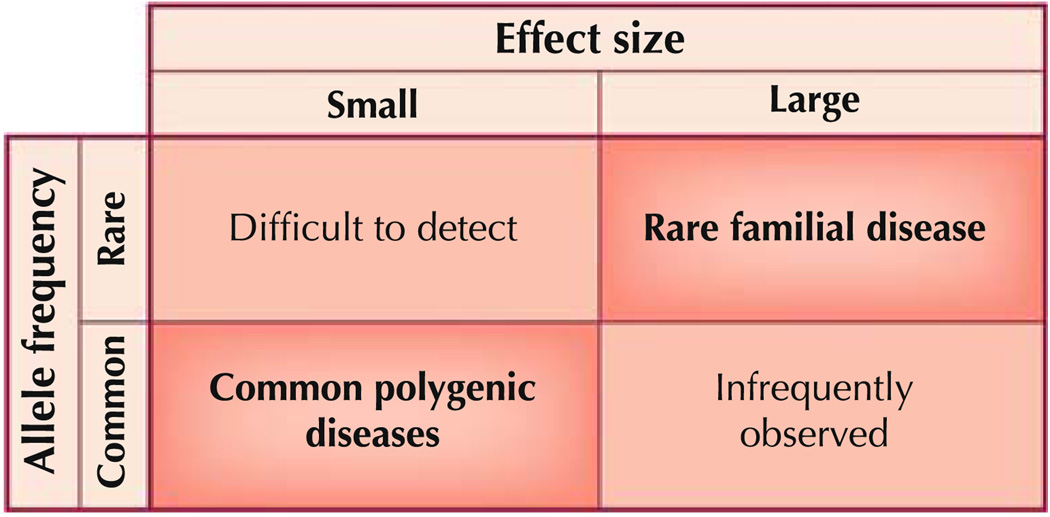
The relationship between frequency and effect size of a genetic variant is continuous and all possible combinations are thought to exist [14••]. Figure 1 depicts the classic types of combinations. The two combinations considered most often are a rare variant with a large effect size and a common variant with small effect size. The other two combinations are either very difficult to detect (rare variant with small effect size) or rarely observed (common variant with large effect size). Many monogenic hypertension syndromes occurring in rare families, such as those identified by Lifton and coworkers [5], are due to very rare variants with a large effect. These syndromes have distinctive biochemical and hormonal characteristics that allowed them to be classified as single-gene diseases before the identification of the actual genes affected. A different genetic principle must govern common traits such as hypertension. The frequency and the continuous nature of common traits are not compatible with rare variants harboring a large effect size. A model for the genetics of complex traits has been the “common disease-common variant” hypothesis [15, 16], which implies that common disease is due to allelic variants with a frequency greater than 5% in the general population and a small individual effect size. The current GWAS efforts address precisely this spectrum of genetic variation; genotyping is limited almost exclusively to allelic variants with a frequency of at least 5%, and the effect size of the variants identified so far is mostly small.
Fig. 1.
Spectrum of allele frequency and effect size in genetic disease
How much of the variability of the phenotype (or of the heritability) can be explained by multiple identified variants? How many disease-associated variants constitute the genetic component of blood pressure and hypertension? Precise numbers are yet unknown, but there are likely to be many more than the ones described in this review. An outlook can be gained from other traits: The Genetic Investigation of Anthropomorphic Traits (GIANT) Consortium has recently presented results of investigating the genetics of human height by a GWAS on 133,800 individuals. They found 318 independent genome-wide significant signals that explain about 14% of height variation [17]. It is interesting to see that even with such a large sample size, only a small fraction of the heritability of height (h2=0.8) could be explained. Another recent report, pertaining to schizophrenia, describes the development of a risk score including thousands of common alleles with very small effect (mostly nominally nonsignificant), which explains a sizable fraction of disease risk [18•]. Considering these results, it appears possible that common variants act on common disease at many loci (hundreds to thousands), explaining little individually but explaining a much larger share of the trait or disease collectively. Previous investigations of complex genetic disease by candidate gene studies or linkage analysis were not geared toward identification of variants with these features. The GWAS offers the first opportunity to test such hypotheses.
How to Read GWAS: A Short Introduction
The difficulty in understanding GWAS resides principally in an appreciation of the number of genetic variants tested for association with the phenotype, as hundreds of thousands to more than a million SNPs are investigated. For each SNP, an association test is performed yielding a P value and a regression analysis estimate of effect size (beta). Given the number of tests performed, the nominal P values need to be corrected for multiple testing, because highly significant results will arise by chance alone with many tests. The true significance threshold can be determined accurately by permutation testing [19]. Most current reports use an approximation: Significant associations have a P value smaller than 5 × 10−8, under the assumption that only 1 million independent tests are performed, even if a larger number of genetic variants is tested. The significance threshold 5 × 10−8, also termed “genome-wide significance,” is reached by dividing the usual alpha of 0.05 by 1 million (the effective number of tests performed). Such a Bonferroni correction is conservative, increasing the credibility of loci with a P value less than 5 × 10−8. A logical consequence of these requirements is the need for a large sample size; such highly significant results can be reached only by analyzing large samples (generally≥ 1000 participants), and this requirement is an important limitation of the method.
Two types of P value plots have emerged as the standard presentation of GWAS results: −log10(P) genome-wide association plots (Manhattan plots) and quantile-quantile (QQ) plots.
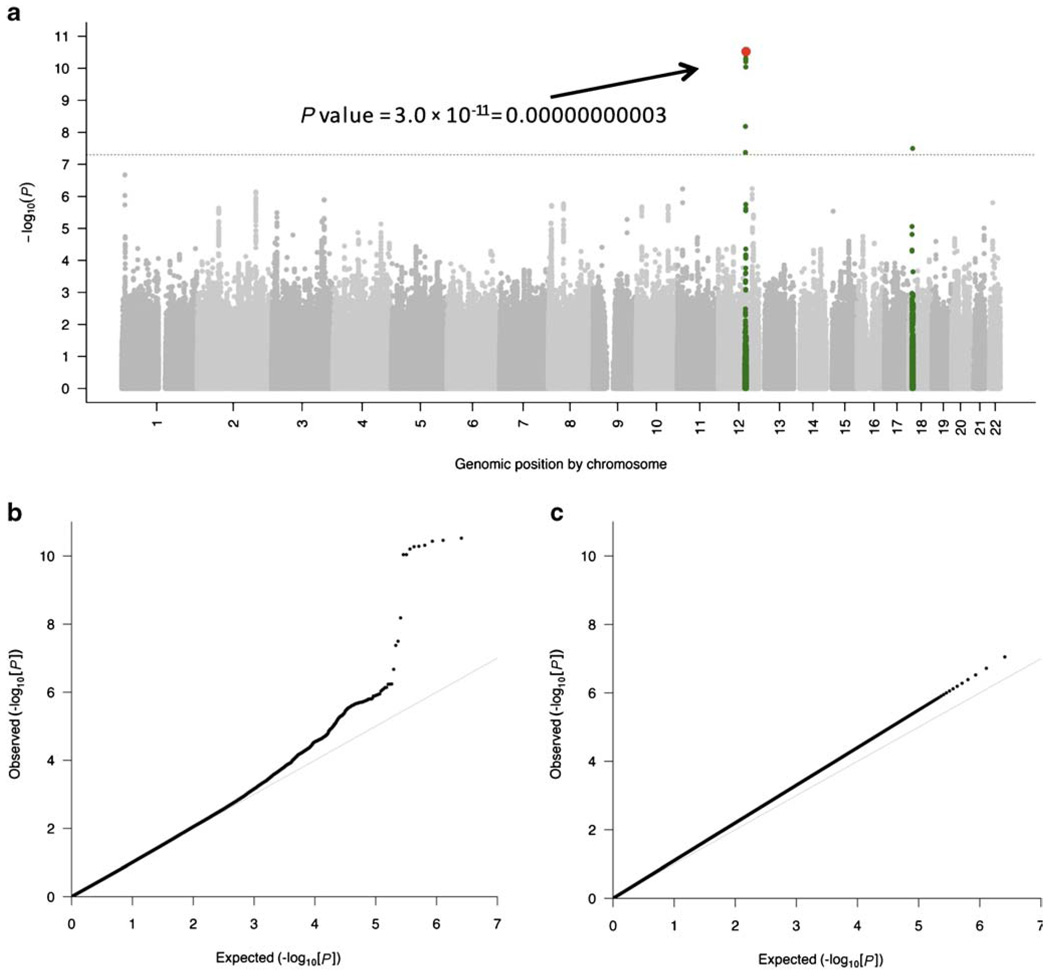
Manhattan plots represent the P values of the entire GWAS on a genomic scale (Fig. 2a). The P values are represented in genomic order by chromosome and position on the chromosome (x-axis). The value on the y-axis represents the −log10 of the P value (equivalent to the number of zeros after the decimal point plus one). For example, see the P value indicated in red on Fig. 2a. Because of local correlation of the genetic variants, arising from infrequent genetic recombination, groups of significant P values tend to rise up high on the Manhattan plot, making the graph look like a Manhattan skyline.
Fig. 2.
a Manhattan plot (−log10[P] genome-wide association plot) of a genome-wide association study on systolic blood pressure in 29,136 individuals in Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE). The genome-wide significance level is set at 5 × 10−8 and plotted as the dotted line. Any single nucleotide polymorphism (SNP) within a region of 5 Mb containing a SNP reaching the genome-wide significance threshold is colored in green. The most significant SNP in this experiment is colored in red (rs2681492 in the ATP2B1 gene). The P value is indicated for demonstration. b Quantile-quantile (QQ) plot of the data shown in the Manhattan plot. c QQ plot of simulated data showing an early separation of the observed from the expected, suggesting population stratification. (a and b adapted from Levy et al. [22••], with permission.)
The QQ plot is a graphical representation of the deviation of the observed P values from the null hypothesis: the observed P values for each SNP are sorted from largest to smallest and plotted against expected values from a theoretical χ2-distribution. If the observed values correspond to the expected values, all points are on or near the middle line between the x-axis and the y-axis (null hypothesis: light gray line in Fig. 2b and c). If some observed P values are clearly more significant than expected under the null hypothesis, points will move towards the y-axis, as shown in Figure 2b. If there is an early separation of the expected from the observed (Fig. 2c), this means that many moderately significant P values are more significant than expected under the null hypothesis. This result is rarely due to thousands of true positives; more often, it is due to population stratification: systematic differences in allele frequencies between subpopulations of the collection of individuals investigated, so that a large number of P values are smaller than expected from chance alone.
GWAS on Blood Pressure and Hypertension
To date, 1 GWAS on blood pressure and hypertension have been published, including studies whose main objective was not blood pressure genetics. Table 1 summarizes these studies, their sample size, phenotype under investigation, and their key findings. Several different ethnicities have been examined, but most investigations have been centered on participants of European origin, partly because samples of European origin are more readily available, but also because the genetic analysis of African American individuals, for example, is more challenging as a result of incomplete accounting for admixture and because African genomes have undergone a higher number of recombinations than European genomes [20, 21].
Table 1.
Genome-wide association studies in blood pressure and hypertension: top findings
| Study (cohort) | Ethnicity | Phenotypes | N | Number of SNPs |
Top findings in GWAS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | rs Number | chr | Positiona | P valueb | Nearest genec | |||||
| Cho et al. [25] (KARE) |
A | SBP, DBP | 8842 | 2,156,535 | SBP | rs17249754 | 12 | 88,584,717 | 9.1×10−7 | ATPase, Ca++ transporting, plasma membrane 1 |
| DBP | rs17249754 | 12 | 88,584,717 | 1.2×10−6 | (ATP2B1) | |||||
| Yang et al. [29] | A | Young- onset HTN |
350 | 91,713 | HTN | rs9308945 | 2 | 34,138,356 | 7×10−4 | myeloid-associated differentiation marker-like (MYADML) |
| Kato et al. [30] | A | HTN | 940 | 80,795 | HTN | rs3755351 | 2 | 70,828,398 | 1.7×10−5d | adducin 2 (beta) (ADD2) |
| Adeyemo et al. [31•] (HUFS) |
AA | SBP, DBP, HTN |
1017 | 808,465 | SBP | rs5743185 | 2 | 190,446,083 | 2.1×10−11 | postmeiotic segregation increased 1 (PMS1) |
| SBP | rs16877320 | 6 | 16,031,005 | 3.4×10−9 | myosin regulatory light chain interacting protein (MYLIP) |
|||||
| SBP | rs17365948 | 8 | 102,026,053 | 1.6×10−8 | tyrosine 3-monooxygenase/tryptophan 5- monooxygenase activation protein, zeta polypeptide (YWHAZ) |
|||||
| SBP | rs12279202 | 11 | 9,388,666 | 4.8×10−8 | importin 7 (IP07) | |||||
| SBP | rs11160059 | 14 | 91,877,083 | 1.5×10−8 | solute carrier family 24 (sodium/potassium/ calcium exchanger), member 4 (SLC24A4) |
|||||
| Levy et al. [32] | E | SBP, DBP | 1260 | 70,987 | SBP | rs10493340 | 1 | 63,363,717 | 1.7×10−6 | forkhead box D3 (FOXD3) |
| (FHS) | DBP | rs1963982 | 8 | 73,269,470 | 3.3×10−6 | ankyrin repeat and MYND domain containing 1 (ANKMY1) |
||||
| Org et al. [33] (KORA S3) |
E | SBP, DBP, HTN |
1977 | 395,912 | SBP | rs12153297 | 5 | 162,604,350 | 3.46×10−7 | cyclin Gl (CCNG1) |
| HTN | rs11646213 | 16 | 81,200,152 | 2.34×10−6 | Cadherin 13, H-cadherin (heart) (CDH13) | |||||
| WTCCC [34••] (BRIGHT) |
E | HTN | 5000 | 469,557 | HTN | rs7961152 | 12 | 24,872,878 | 7.39×10−6 | branched chain aminotransferase 1, cytosolic (BCAT1) |
| Sabatti et al. [35] (NFBC 1966) |
E | SBP, DBP | 4730 | 329,091 | “No significant finding” | |||||
| Saxena et al. [36] (DGI) |
E | SBP, DBP | 2931 | 386,731 | “No significant finding” | |||||
| Wang et al. [24] (AFDS) |
E | SBP, DBP | 542 | 79,447 | SBP | rs6749447 | 2 | 168,749,632 | 7.6×10−5 |
serine threonine kinase 39 (STE20/SPS1 homolog, yeast) (STK39) |
| Newton-Cheh et al. [23••] |
E | SBP, DBP | 34,433 | ~2,500,000 | SBP | rs17367504 | 1 | 11,785,365 | 1×10−5 |
5,10-methylenetetrahydrofolate reductase (NADPH) (MTHFR) |
| (GBPGen) | SBP | rs11191548 | 10 | 104,836,168 | 3×10−7 | 5'-nucleotidase, cytosolic II (NT5C2) | ||||
| SBP | rs12946454 | 17 | 40,563,647 | 4×10−6 | phospholipase C, delta 3 (PLCD3) | |||||
| DBP | rsl6998073 | 4 | 81,403,365 | 7×10−9 | fibroblast growth factor 5 (FGF5) | |||||
| DBP | rsl530440 | 10 | 63,194,597 | 3×10−6 |
chromosome 10 open reading frame 107 (C10orfl07) |
|||||
| DBP | rs653178 | 12 | 110,492,139 | 1×10−7 | ataxin 2 (ATXN2) | |||||
| DBP | rs1378942 | 15 | 72,865,396 | 6×10−8 | c-src tyrosine kinase (CSK) | |||||
| DBP | rs16948048 | 17 | 44,795,465 | 5×10−6 | zinc finger protein 652 (ZNF652) | |||||
| Levy et al. [22••] (CHARGE) |
E | SBP, DBP, HTN |
29,136 | ~2,500,000 | SBP | rsl004467 | 10 | 104,584,497 | 2×10−6 |
cytochrome P450, family 17, subfamily A, polypeptide 1 (CYP17A1) |
| SBP | rs381815 | 11 | 16,858,844 | 5.8×10−7 |
pleckstrin homology domain containing, family A member 7 (PLEKH7) |
|||||
|
SBP, DBP, HTN |
rs2681492, rs2681472 |
12 |
88,537,220, 88,533,090 |
3.0×10−11, 3.7×10−8, 1.7×10−8 |
ATPase, Ca++ transporting, plasma membrane 1 (ATP2B1) |
|||||
| SBP,DBP | rs3184504 | 12 | 110,368,991 | 5.7×10−7 | SH2B adaptor protein 3 (SH2B3) | |||||
| DBP | rs9815354 | 3 | 41,887,655 | 7.8×10−7 | unc-51-like kinase 4 (C. elegans) (ULK4) | |||||
| DBP | rsll014166 | 10 | 18,748,804 | 8.7×10−7 |
calcium channel, voltage-dependent, beta 2 subunit (CACNB2) |
|||||
| DBP | rs2384550 | 12 | 113,837,114 | 1.3×10−7 | T-box 3 (TBX3) | |||||
| DBP | rs6495122 | 15 | 72,912,698 | 8.1×10−7 | complexin 3 (CPLX3) | |||||
Shown in bold + italic if significant in initial scan (multiple-testing corrected) and replicated in another population; bold if significant (multiple-testing corrected) after adding additional data from another population
All coordinates are given on the March 2006 human reference sequence (NCBI Build 36.1)
Unless otherwise indicated, the P value of the initial GWAS or GWAS meta-analysis is indicated, not taking into account replication genotyping results
The physically closest RefSeq gene
For combined data
A Asian ancestry, AA Americans of African origin, AFDS Amish Family Diabetes Study, BRIGHT British Genetics of Hypertension, CHARGE Cohorts for Heart and Aging Research in Genomic Epidemiology, chr chromosome, DGI Diabetes Genetics Initiative, DBP diastolic blood pressure, E European ancestry, FHS Framingham Heart Study, GBPGEN Global BP Gen, GWAS genomewide association study, HTN hypertension, HUFS Howard University Family Study, KARE Korea Association Resource, KORA Kooperative Gesundheitsforschung in der Region Augsburg, NFBC Northern Finland Birth Cohort, SBP systolic blood pressure, SNP single nucleotide polymorphism, WTCCC Wellcome Trust Case Control Consortium
Only two of the published studies on blood pressure traits (CHARGE BP and Global BP Gen) have identified an association withstanding correction for multiple testing (“genome-wide significance”) within the study that could clearly be replicated in an independent study [22••, 23••]. The P values for the corresponding SNPs are highlighted in Table 1. All of these variants have been found in individuals of European origin. A thorough testing in other ethnic groups of the strongest associations is still outstanding. Both studies have very large sample sizes (near 30,000 participants) and are meta-analyses of several individual GWAS.
Only one SNP reached genome-wide significance for hypertension in a primary GWAS meta-analysis, whereas in the same study, four SNPs reached this threshold for SBP and six reached it for DBP [22••]. It is possible that the differences in the number of significant findings are due to differences in power; continuous traits have greater power than discrete traits and therefore the chances of obtaining a significant result are higher with a continuous trait. It is important to emphasize that given the effect sizes observed and the number of tests performed, the power is low, even in GWAS with 30,000 participants.
In total, 14 independent loci have been identified so far for blood pressure traits that reached genome-wide significance, including replication in independent cohorts [22••, 23••, 24]. For three of these loci, two studies find SNPs that are close to each other physically and correlated (linkage disequilibrium r2, 0.4–1) [22••, 23••]. Therefore it is likely that they point to the same causal variant.
It is important to point out that the exact identity of the genes driving the association with blood pressure or hypertension cannot be determined on the basis of these data alone. All “nearest genes” indicated in Table 1 are necessarily only educated guesses about the gene containing the causal variant until the functional mechanism can be identified. Nevertheless, it is possible that the genes nearest to the variants identified are the genes through which the variant exercises its effect. The 14 loci are in or near genes encoding six enzymes (including three kinases and one cytochrome), two solute channels, two transcription factors, one growth factor, one cell signaling protein, one structural protein, and one hypothetical gene.
Assuming we have identified the genes correctly on the basis of proximity, it is striking that only two genes out of 14—CYP17A1 (cytochrome P450, family 17, subfamily A, polypeptide 1) and 5,10-methylenetetrahydrofolate reductase (MTHFR)—would have been identified as a blood pressure candidate gene before these studies. The protein encoded by CYP17A1 is a key enzyme in the steroidogenic pathway and has 17α-hydroxylase and 17,20-lyase activities. Loss of function of CYP17A1 can lead to an uncommon form of congenital adrenal hyperplasia with features that include hypertension. The protein encoded by MTHFR catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5methyltetrahydrofolate in the methionine metabolism.
All of the other 12 genes closest to the variants identified were previously largely unsuspected of involvement in blood pressure regulation. There are certainly very plausible biologic candidates on this list. For example, plasma membrane ATP2B1 (ATPase, Ca++ transporting, plasma membrane 1) is an ion-transport ATPase at the cell membrane and plays a critical role in intracellular calcium homeostasis. A variant near this gene has been found for SBP, DBP, and hypertension by one group [22••], and the variant is the only genome-wide significant variant identified by GWAS for hypertension so far. Furthermore, it is interesting that the top finding of another blood pressure GWAS (for both SBP and DBP) also identified a variant near this gene as the best finding [25]. For many of the remaining genes, a compelling biologic explanation can also be found, but it may be wise to wait until the biology of their action can be better explained, as proof by proximity may be a fallacy.
An important consideration is the effect size of the variants identified. The genetic model in all studies with significant findings shown Table 1 is additive, and each copy of the risk allele is associated with an increase of about 1 mm Hg in SBP or about 0.5 mm Hg in DBP, on average. This increase is small, well below the measurement error, but nevertheless highly significant in large cohorts. It is also important to keep in mind clinical observational data indicating that prolonged increases of 5 mm Hg in the usual DBP are associated with a 34% increase in stroke and a 21% increase in coronary heart disease [26].
Based on these findings, it appears that common variants associated with blood pressure phenotypes have a very small effect size. There is no such thing as “the blood pressure gene.” It is likely that many genes act conjointly, and the individual contribution of each gene is very small. One group has described a risk score using the best 10 findings for each blood pressure phenotype, and the conjoint effect amounts to several millimeters mercury of blood pressure [22••]. Although interesting, such analyses are victim of the “winner’s curse” [27], as genome-wide significant findings often have large effect sizes in the studies identifying them, much larger than can be shown in replication studies; it remains to be seen how much lower the effect size estimate will be in an independent population. The variants significant so far explain only a very small fraction of the heritability of blood pressure traits, for many potential reasons [28••]. Most notably, further effector variants may be found at lower allele frequencies at the same genes in a scenario that would fall between the categories described in Fig. 1.
Conclusions
GWAS permit for the first time the investigation of most genetic variability due to common variants in the human genome. Application of this technology to blood pressure traits and hypertension has identified more than a dozen loci that are reproducibly associated with blood pressure traits in large cohorts. The genes closest to the variants identified are largely not suspected of involvement in blood pressure regulation; they may not be causal genes because they are chosen by proximity alone. The effect sizes of the variants identified are small and currently explain about 1% of the phenotypic variability (after correcting for major confounders such as sex, age, and body mass index).
Why is so little of the heritability explained? Dr. McKusick’s 1960 article on hypertension genetics [9] is followed by a quote from Oliver Wendell Holmes: “We are all tattooed in our cradle by the beliefs of our tribe.” Might it be that heritability estimates for blood pressure traits are overestimated and familial environment and habits determine more of blood pressure variability than predicted? Can rare variants explain more of the phenotypic variability? Are current experimental approaches missing an unknown, yet major, biologic phenomenon? It is also possible that gene-environment interactions play an important role, but it is currently not possible to quantify them. At this stage, it is important to emphasize for the clinician that the predictive power of the genetic variants identified for blood pressure and hypertension, even taken collectively, is so small that they are not clinically useful, though this limitation may change in the future. Conversely, the current studies have discovered new pathways regulating blood pressure and hypertension, and there is at least a possibility that they may be acted upon by small molecules.
Findings of GWAS are an encouraging step in blood pressure genetics, and they open the way for subsequent investigations. Studies with larger sample sizes are under way; they will have greater power and are likely to uncover additional blood pressure variants. Further GWAS on alternative phenotypes (eg, pulse pressure and mean arterial pressure) and on refined phenotypes (eg, long-term average blood pressure) are expected to appear soon. One important contributor will be the International Consortium on Blood Pressure (ICBP)-GWAS, formed by joining together the CHARGE BP [22••] and Global BP Gen [23••] consortia, with a total sample size of close to 70,000 individuals. Large-scale experiments to investigate blood pressure traits in nonwhite populations (eg, the CARE consortium) are also under way. These investigations will help in better understanding of the genetics of blood pressure and hypertension, with potential benefits for prediction, diagnosis, and treatment.
Acknowledgments
Dr. Ehret receives partial funding for his work on blood pressure and hypertension though the “FEHGAS Study” (National Heart, Lung, And Blood Institute grant 5R01HL086694-02; Dr. Ehret has additional affiliations with the Cardiology Center, Geneva University Hospital, Geneva, and the Institute of Social and Preventive Medicine, Lausanne University Hospital, Lausanne, both in Switzerland. Dr. A. Chakravarti, Principal Investigator) and through a Special Program University Medicine (SPUM) grant funded by the Swiss National Science Foundation (Dr. M. Bochud, Principal Investigator).
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance,
•• Of major importance
- 1.Levy D, DeStefano AL, Larson MG, et al. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham Heart Study. Hypertension. 2000;36(4):477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Pilia G, Chen WM, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genetics. 2006;2(8):e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rijn MJ, Schut AF, Aulchenko YS, et al. Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood-pressure-related genes. J Hypertens. 2007;25(3):565–570. doi: 10.1097/HJH.0b013e32801449fb. [DOI] [PubMed] [Google Scholar]
- 4.Tobin MD, Raleigh SM, Newhouse S, et al. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation. 2005;112(22):3423–3429. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 5.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104(4):545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 6.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 7.Mahomed FA. On the sphygmographic evidence of arteriocapillary fibrosis. Trans Path Soc. 1877;28:394–397. [Google Scholar]
- 8.Pickering GW, Keen H, Rose G, Smith A. The nature of essential hypertension. Lancet. 1959;274(7110):1027–1030. [PubMed] [Google Scholar]
- 9.McKusick VA. Genetics and the nature of essential hypertension. Circulation. 1960;22(5):857–863. [Google Scholar]
- 10.Fan JB, Chen X, Halushka MK, et al. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 2000;10(6):853–860. doi: 10.1101/gr.10.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. BioTechniques. 2002 Suppl:56–58. 60–61. [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13. Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. This is a report of phase 2 of the HapMap project, a large collaborative effort to identify common variation in the human genome.
- 14. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev. 2008;9(5):356–369. doi: 10.1038/nrg2344. This is a review of the genetic challenges and opportunities of GWAS.
- 15.Chakravarti A. Population genetics—making sense out of sequence. Nat Genet. 1999;21(1 Suppl):56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 16.Lander ES. The new genomics: global views of biology. Science. 1996;274(5287):536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 17.Lango Allen H, Lettre G, Estrada K, et al. The identification of over 135 loci involved in adult height variation provides important insights into the contribution of common variation to a model complex trait [abstract]. Presentation at the 59th Annual Meeting of the American Society of Human Genetics; October 20–24, 2009; Honolulu, HI. [Google Scholar]
- 18. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. This article describes a method to construct a risk score using thousands of nominally nonsignificant SNPs.
- 19.Arking DE, Pfeufer A, Post W, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38(6):644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 20.Ehret GB, Morrison AC, O’Connor AA, et al. Replication of the Wellcome Trust genome-wide association study of essential hypertension: the Family Blood Pressure Program. Eur J Hum Genet. 2008;16(12):1507–1511. doi: 10.1038/ejhg.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehret GB, O’Connor AA, Weder A, et al. Follow-up of a major linkage peak on chromosome 1 reveals suggestive QTLs associated with essential hypertension: GenNet study. Eur J Hum Genet. 2009;17(12):1650–1657. doi: 10.1038/ejhg.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41(6):677–687. doi: 10.1038/ng.384. This reference and reference [23••] are the largest published GWAS. Collectively, they identify more than a dozen blood pressure and hypertension genes.
- 23. Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41(6):666–676. doi: 10.1038/ng.361. This reference and reference [22••] are the largest published GWAS on blood pressure traits so far. Collectively, they identify more than a dozen blood pressure and hypertension genes.
- 24.Wang Y, O’Connell JR, McArdle PF, et al. Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009;106(1):226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 26.MacMahon S, Peto R, Cutler J, et al. Blood pressure, stroke, and coronary heart disease. Part 1, Prolonged differences in blood pressure: prospective observational studies corrected for the regression dilution bias. Lancet. 1990;335(8692):765–774. doi: 10.1016/0140-6736(90)90878-9. [DOI] [PubMed] [Google Scholar]
- 27.Goring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69(6):1357–1369. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. This meeting report on the “missing heritability” of complex genetic traits discusses in detail possible reasons why only little heritability can be explained so far.
- 29.Yang HC, Liang YJ, Wu YL, et al. Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. PloS One. 2009;4(5):e5459. doi: 10.1371/journal.pone.0005459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato N, Miyata T, Tabara Y, et al. High-density association study and nomination of susceptibility genes for hypertension in the Japanese National Project. Hum Mol Genet. 2008;17(4):617–627. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 31. Adeyemo A, Gerry N, Chen G, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5(7):e1000564. doi: 10.1371/journal.pgen.1000564. This is the only GWAS on hypertension in African American individuals published to date.
- 32.Levy D, Larson MG, Benjamin EJ, et al. Framingham Heart Study 100 K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8 Suppl 1:S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Org E, Eyheramendy S, Juhanson P, et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18(12):2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. This report describes the first large GWAS on seven complex genetic traits.
- 35.Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]