Introductory paragraph
Evidence suggests that testicular germ cell tumors (TGCT) have a strong underlying genetic component. We performed a genome-wide scan among 277 TGCT cases and 919 controls. Seven markers at 12p22 within c-KIT ligand (KITLG) reached genome-wide significance (P < 5.0 × 10−8). In independent replication, TGCT risk was increased 3-fold per copy of the major allele at rs3782179 and rs4474514 (OR=3.08, 95% CI 2.29, 4.13; OR=3.07, 95% CI 2.29, 4.13, respectively). We also replicated associations with rs4324715 and rs6897876 at 5q31.3 near sprouty 4 (SPRY4; P < 5.0 × 10−6 in discovery). Risk of TGCT was increased nearly 40% per copy of the major allele (OR=1.37, 95% CI 1.14, 1.64; OR=1.39, 95% CI 1.16, 1.66, respectively). All of the genotypes were associated with both seminoma and non-seminoma TGCT subtypes. These results demonstrate that common genetic variants affect TGCT risk and implicate KITLG and SPRY4 as TGCT susceptibility genes.
In the United States, testicular germ cell tumors (TGCT) are the most common cancers in young men, with a peak incidence among those aged 25 to 34 years1. The age-adjusted incidence in white men has doubled since 1975 and is now 6.6 per 100,000. The incidence in white non-Hispanic men is nearly five-fold higher than among black men1. The reasons for the increasing incidence and racial disparity in TGCT rates are unknown.
While environmental exposures have been postulated to play a role in the increasing incidence of TGCT, there also is evidence for a substantial genetic contribution to TGCT susceptibility. Brothers of TGCT patients have an eight- to 12-fold increased risk of disease, with the risk to monozygotic and dizygotic twins 75- and 35-fold increased, respectively, and fathers of patients have a four-fold increased risk2,3. Consistent with the high familial risks compared to most other cancer types and ethnic differences in TGCT risk, the proportion of TGCT susceptibility accounted for by genetic effects is estimated at 25%, and TGCT has the third highest heritability among all cancers4.
Results from linkage studies and candidate gene approaches, however, have produced limited insight into TGCT susceptibility factors. An initial report of linkage on Xq27 was not replicated nor have other loci been identified with significant effects, which suggests that multiple loci, potentially of weak to moderate effect, contribute to disease susceptibility5,6. The gr/gr deletion on the Y chromosome, studied as a candidate region, increases TGCT risk two- to three-fold, but carriage frequency of this variant is low (2–3%) suggesting it likely accounts for only a small component of risk7. Thus, despite the multiple lines of evidence suggesting a genetic etiology of TGCT, no genetic risk factor has been identified that can explain an appreciable proportion of TGCT cases.
To identify genes associated with TGCT development, we performed a genome-wide association study. Cases were 277 white, non-Hispanic men with pathologically defined TGCT seen at the University of Pennsylvania Health System (UPHS) or Fox Chase Cancer Center (FCCC) in Philadelphia, PA. We genotyped DNA extracted from venous blood using the Affymetrix® Genome-Wide Human SNP Array 6.0. We compared the frequency of observed genotypes among TGCT cases to those available from 919 white, non-Hispanic males from the Philadelphia region genotyped on the same Affymetrix platform (Table 1). Supplemental Figure 1 shows the quantile-quantile plot of χ2 values for observed versus expected allele frequencies based on Fisher's exact test for the 611,254 markers meeting quality control criteria, indicating little evidence of population stratification and evidence of excess disease associations8. The calculated genomic control inflation (λ) factor was 0.944, and hence we report unadjusted test statistics9.
Table 1.
Age, family history of TGCT, and tumor type in the discovery and replication samples
| Status Total | Discovery | Replication | |||||
|---|---|---|---|---|---|---|---|
| Case n=277 | Control n=919 | Case n=371 | Control n=860 | ||||
| # | % | # | # | % | # | % | |
| Age (median, [interquartile range]) | 31a [24,39] | 57 [52,62] | 34 [28, 38] | 35 [30, 39] | |||
|
| |||||||
| Family history of TGCT | |||||||
|
| |||||||
| No | 239 | 86.3 | - | 314 | 84.6 | 769 | 89.4 |
|
| |||||||
| Yes | 30b | 10.8 | - | 11c | 3.0 | 10c | 1.2 |
|
| |||||||
| Unknown | 8 | 2.9 | - | 46 | 12.4 | 81 | 9.4 |
|
| |||||||
| Personal history of cryptorchidism | |||||||
|
| |||||||
| No | 245 | 88.5 | 330 | 88.9 | 844 | 98.1 | |
|
| |||||||
| Yes | 26 | 9.4 | 38 | 10.2 | 16 | 1.9 | |
|
| |||||||
| Unknown | 6 | 2.2 | 3 | 0.8 | 0 | 0 | |
|
| |||||||
| Tumor type | |||||||
|
| |||||||
| Seminoma | 85 | 30.7 | - | 230 | 62.0 | - | - |
|
| |||||||
| Non-seminoma | 180 | 65.0 | - | 141 | 38.0 | - | - |
|
| |||||||
| Unknown | 12 | 4.3 | - | 0 | 0 | - | - |
Age of diagnosis missing for six TGCT cases.
Sixteen cases were selected based on family history of TGCT; among non-selected (n=261) cases, the proportion reporting any family history of TGCT was 5.4%.
Denotes reported family history of TGCT among first degree relatives only.
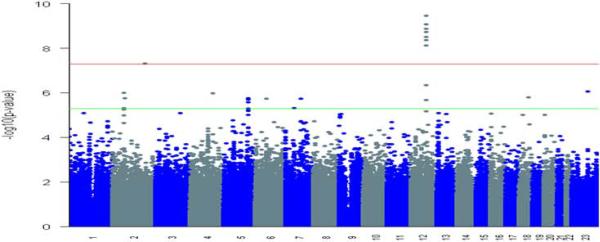
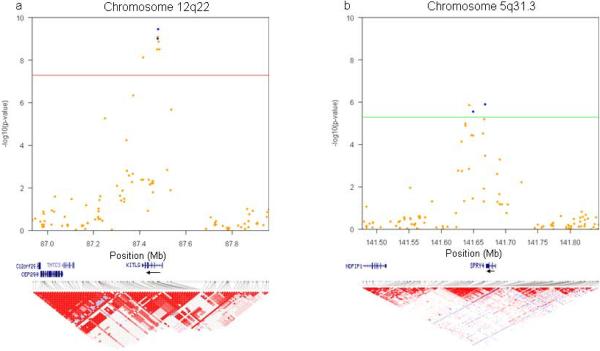
Eight markers reached statistical significance at a genome-wide threshold of P < 5.0 × 10−8 (Fig. 1, Supplementary Table 1). Seven of these (rs995030, rs1352947, rs1472899, rs3782179, rs3782181, rs4474514, rs11104952) including the most significant association (P = 3.54 × 10−10) at rs4474514 occurred within the KITLG (c-KIT ligand) gene region on 12q22 (Supplementary Fig. 2). These markers were in strong linkage disequilibrium with each other; pairwise D' and r2 measures were all > 0.99 (Fig. 2a). The eighth marker (rs3770112, P = 4.93 × 10−8) mapped to the integrin alpha 4 (ITGA4) gene on 2q31.3. Because no other markers in this genomic region (±10MB) reached statistical significance at P < 1.0 × 10−3, we suspected this association might have arisen purely by chance. We further investigated this possibility by imputing genotypes near rs3770112 based on publicly available HapMap genotypic data10. After imputation, the test of association at rs3770112 no longer surpassed the genome-wide threshold (P = 0.05); as well, all other markers in the region remained below the threshold for advancing to replication. The correlation between observed and imputed P values for the 23 markers that were in the same linkage disequilibrium block with rs3770112 was very high (r=0.96), and information content and maximum posterior call probability for rs3770112 were both > 0.998. Taken together, these results strongly suggested that the association observed in the discovery phase was a false positive (Supplementary Fig. 5). We selected two markers in KITLG (rs3782179, rs4474514) to bring forward into replication.
Figure 1. Genome wide association results plotted for 277 TGCT patients and 919 controls.
The threshold for genome wide significance was P < 5.0 × 10−8 (top red line) based on Fisher's exact test. Markers that reached significance at P < 5.0 × 10−6 (bottom green line) based on Fisher's exact test also were considered.
Figure 2. Regional association plots and linkage disequilibrium structure.
(a, b) The −log10 of the P-value for the association of each discovery phase marker and TGCT status for segments of chromosomes (a) 12q22 [red line shows P < 5.0 × 10−] and (b) 5q31.3 [green line shows P < 5.0 χ 10−]. NCBI Build 36 was used for map locations. From each of these regions, two markers were taken into replication and are indicated in blue. Linkage disequilibrium structures for the GWAS data and based on r2 are shown.
Sixteen additional markers reached statistical significance at the P < 5.0 × 10−6 level (Supplementary Table S1). Of these, three (rs12521013, rs4324715, rs6897876) mapped 2.4 kb downstream of the SPRY4 (sprouty homolog 4) coding region on 5q31.3 (Fig. 2b, Supplementary Fig. 3), and two (rs17031166, rs1549383) mapped to a gene free region on 2p14 that is 500kb centromeric of SPRED2 (sprouty-related, EVH1 domain containing 2) (Supplementary Fig. 4). As both SPRY4 and SPRED2 have been implicated in the KIT/KITLG signaling pathway11,12, and as these two regions were the only ones that contained more than one marker surpassing threshold significance, we also chose two markers at each of these loci (SPRY4: rs4324715, rs6897876; 2p14: rs17031166, rs1549383) to bring forward for replication.
The replication set consisted of a population-based set of 371 TGCT cases and 860 controls, all white non-Hispanic, recruited from residents of the metropolitan Seattle-Puget Sound region, and parents of 204 of the cases. We observed associations with rs3782179 (Ptrend= 5.88 × 10−15) and rs4474514 (Ptrend= 5.88 × 10−15) in KITLG and with rs4324715 (Ptrend= 6.77 × 10−4) and rs6897876 (Ptrend= 3.67 × 10−4) proximal to SPRY4 (Table 2), but not with rs17031166 (Ptrend= 0.90) or rs1549383 (Ptrend= 0.88) near SPRED2. TGCT risk was increased three-fold per copy of the major A-allele in KITLG rs3782179 and rs4474514 (odds ratio (OR) = 3.08, 95% CI 2.29, 4.13; and OR=3.07, 95% CI 2.29, 4.13, respectively). Homozygous carriage of the major A-allele at these loci was associated with over a four-fold increased risk of TGCT (OR=4.56, 95% CI 1.78, 11.7; and OR=4.56, 95% CI 1.77, 11.7, respectively) compared with homozygous carriage of the minor G-allele. We noted weaker associations for the two markers close to SPRY4. TGCT risk was increased nearly 40% per copy of the major T-allele in rs4324715 (OR=1.37, 95% CI 1.14, 1.64) and major C-allele in rs6897876 (OR=1.39, 95% CI 1.16, 1.66); and risk was increased 65–80% with homozygous carriage of the major alleles (OR=1.81, 95% CI 1.26, 2.58; and OR=1.68, 95% CI 1.17, 2.42, respectively) compared with homozygous carriage of their corresponding minor alleles. In addition to the case-control analysis, we performed a case-parent triad analysis, which also showed that carriage of the risk allele for the markers in KITLG and proximal to SPRY4 are associated with TGCT. The per allele relative risks (RR) for rs3782179 and rs4474514 (KITLG) were 2.5 (95% CI 1.6, .9) and 2.6 (95% CI 1.6, 4.0), respectively, and for rs4324715 and rs6897876 (proximal to SPRY4) 1.5 (95% CI 1.2, 2.1) and 1.5 (95% CI 1.1, 2.0), respectively. These family-based estimates provide additional evidence that population stratification did not bias results in the replication phase.
Table 2.
Associations of TGCT with replicated SNP markers
| Gene | Markera | Risk allele | Genotype Countb | Phase | Per allele | OR (95% CI) Heterozygotec | Homozygoted | P-trende | |
|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | ||||||||
| KITLG | rs3782179 A/G | A | 597/276/44 | 229/45/3 | Discovery | 2.36 (1.73, 3.21) | 2.39 (0.71, 8.03) | 5.63 (1.73, 18.3) | 1.95 × 10−8 |
|
| |||||||||
| 515/285/38 | 309/49/5 | Replication | 3.08 (2.29, 4.13) | 1.31 (0.49, 3.48) | 4.56 (1.78, 11.7) | 5.88 × 10−15 | |||
|
| |||||||||
| rs4474514 A/G | A | 599/276/44 | 229/45/2 | Discovery | 2.45(1.79,3.35) | 3.59(0.84, 15.3) | 8.41 (2.02, 35.0) | 7.34 × 10−9 | |
|
| |||||||||
| 517/285/38 | 310/49/5 | Replication | 3.07 (2.29, 4.13) | 1.31 (0.49, 3.48) | 4.56 (1.77, 11.7) | 5.88 × 10−15 | |||
|
| |||||||||
| SPRY4 | rs4324715 T/C | T | 230/433/255 | 87/145/33 | Discovery | 1.59 (1.30, 1.93) | 2.59 (1.72, 3.89) | 2.92 (1.89, 4.53) | 3.57 × 10−6 |
|
| |||||||||
| 191/437/197 | 119/171/68 | Replication | 1.37 (1.14, 1.64) | 1.13 (0.82, 1.57) | 1.81 (1.26, 2.58) | 6.77 × 10−4 | |||
|
| |||||||||
| rs6897876 C/T | C | 282/429/207 | 114/135/28 | Discovery | 1.59 (1.31, 1.94) | 2.33 (1.50, 3.61) | 2.99 (1.90, 4.69) | 2.96 × 10−6 | |
|
| |||||||||
| 251/428/154 | 156/149/57 | Replication | 1.39 (1.16, 1.66) | 0.94 (0.66, 1.34) | 1.68 (1.17, 2.42) | 3.67 × 10−4 | |||
dbSNP rsnumber and major/minor alleles.
Number of individuals genotyped as homozygous for the risk allele/heterozygous for the risk allele/homozygous for the non-risk allele. Nomenclature for major/minor alleles based on calls from Affymetrix® Genome-Wide Human SNP Array 6.0. MAF for discovery phase markers given in Supplemental Table 1.
OR for heterozygous carriage of risk allele compared to homozygous carriage of non-risk allele.
OR for homozygous carriage of risk allele compared to homozygous carriage of non-risk allele.
Cochran-Armitage test for trend.
We did not observe an interaction between KITLG and SPRY4 marker genotypes. In the replication set, marker genotypes in KITLG and SPRY4 were associated with both seminoma and non-seminoma germ cell tumors without indication that genotype associations differed between the two subtypes (Table 3). In subgroup analyses among those without a family history of TGCT and among those without cryptorchidism, two strong and well-established risk factors for TGCT, the genotypic ORs associated with KITLG and SPRY4 markers were only negligibly attenuated (results not shown). These findings indicate that for the majority of cases, KITLG and SPRY4 do not exert their effect solely based on mechanisms involving these known risk factors. Because of limited numbers, it was not possible to examine the effect of KITLG and SPRY4 among those with positive family history or personal history of cryptorchidism.
Table 3.
Associations of KITLG and SPRY4 SNP markers with seminoma and non-seminoma TGCTa
| Seminoma OR (95% CI) | Non-Seminoma OR (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Marker | Risk allele | Per allele | Heterozygoteb | Homozygotec | Per allele | Heterozygoteb | Homozygotec |
| KITLG | rs4474514 A/G | A | 2.97 (2.08, 4.23) | 1.42 (0.42, 4.87) | 4.70 (1.44, 15.4) | 3.27 (2.06, 5.18) | 1.13 (0.25, 5.1) | 4.34 (1.03, 18.23) |
|
| ||||||||
| KITLG | rs3782179 A/G | A | 2.95 (2.07, 4.21) | 1.42 (0.42, 4.87) | 4.67 (1.43, 15.3) | 3.30 (2.08, 5.24) | 1.13 (0.25, 5.1) | 4.39 (1.04, 18.45) |
|
| ||||||||
| SPRY4 | rs6897876 C/T | C | 1.38 (1.11, 1.72) | 1.11 (0.72, 1.73) | 1.78 (1.14, 2.79) | 1.39 (1.07, 1.81) | 0.72 (0.43, 1.2) | 1.55 (0.93, 2.56) |
|
| ||||||||
| SPRY4 | rs4324715 T/C | T | 1.40 (1.13, 1.73) | 1.39 (0.92, 2.09) | 1.98 (1.27, 3.09) | 1.32 (1.01, 1.71) | 0.83 (0.52, 1.32) | 1.60 (0.97, 2.62) |
Analyses reflect the replication set only
OR for heterozygous carriage of risk allele compared to homozygous carriage of non-risk allele.
OR for homozygous carriage of risk allele compared to homozygous carriage of non-risk allele
We have identified variation at 12q22 as a major risk locus for TGCT susceptibility. For rs3782179 and rs4474514, we observed a three-fold increased risk of disease per major allele and a 4.5-fold increased risk of disease for homozygous carriage of the major allele. The identified region contains KITLG, also known as stem cell factor, the ligand for the receptor tyrosine kinase, c-KIT. The KITLG/KIT signaling pathway plays an important role in gametogenesis, hematopoesis and melanogenesis13. In mouse models, Kitl (encoded at the steel [Sl] locus) is required for multiple aspects of primordial germ cell (PGC) development, including proliferation, migration and survival14,15. Kitl plays a crucial role in the migration of PGCs from the hindgut and subsequent targeting to the genital ridges, and down regulation of Kitl in the midline triggers localized apoptosis of PGCs15. Based on similarity in cellular ultrastructure, patterns of imprinting, and gene expression, multiple human studies have suggested that TGCT arise from PGCs16. Delayed differentiation of PGCs has been associated with development of testicular germ cell carcinoma in situ among patients with intersex conditions and abnormalities of chromosomal number17. These data support a role for KITLG in TGCT susceptibility.
Furthermore, loss of the transmembrane form of Kitl, which leads to decreased PGC number, has been identified as a TGCT susceptibility locus in the 129/Sv mouse18. In humans, activating mutations of KIT are the most common somatic point mutations in TGCT, present in 25% of seminomas, although rarely identified in non-seminomas19. Thus, both germline variation and somatic mutations in the KITLG/KIT signaling pathway are associated with TGCT. In addition, KITLG/KIT signaling plays an important role in male fertility20, and mutations in Kitl lead to decreased germ cell number. Our findings suggest that the reported epidemiological association between TGCT and male infertility21 may be due, in part, to a common genetic basis.
As KITLG plays a role in determining level of pigmentation22, we postulated that inherited variation at this locus could provide a genetic explanation for the observed differences in TGCT incidence in whites and blacks. KITLG has undergone strong positive selection in the European and East Asian populations, with an extended haplotype of 400kb23. Data from HapMap Phase 3 show significant differences (P = 4.3 × 10−20) in the frequency of the risk alleles of KITLG (rs3782179 and rs4474514) when comparing the CEU (major allele frequency = 0.80) and ASW (African ancestry in Southwest USA: major allele frequency = 0.25) populations10. This finding suggests that inherited variation in KITLG may explain, in part, the observed differences in TGCT incidence between whites and blacks.
We also observed an association between TGCT risk and variation at 5q31 just downstream of SPRY4. As with KITLG, the major allele was associated with increased risk. SPRY4 is one of a family of four genes (SPRY1–4) that have been implicated as negative regulators of the RAS-ERK-MAPK signaling pathway in response to growth factors24. Expression analyses and tumor studies have shown that SPRY4 is the most significantly downregulated gene when KIT signaling is inhibited by imatinib mesylate in gastrointestinal stromal tumors, supporting a functional relationship between the two proteins12.
In summary, our results demonstrate that common genetic variants at the 12p22 and 5q31 loci are associated with TGCT and strongly implicate KITLG as a susceptibility gene in the pathogenesis of TGCT. In addition, these observations may explain, in part, two important features of the disease: the increased incidence in whites and the epidemiological association with male infertility.
METHODS
Genome-wide association study
For the discovery phase, we initially selected 353 TGCT patients seen at UPHS (n=303) and FCCC (n=50); all cases were from the Philadelphia region. TGCT cases from UPHS were from an ongoing clinic-based case-control study of genetic susceptibility of TGCT for which study participants were asked to complete a self-administered questionnaire that elicited information on known and presumptive risk factors for TGCT. TGCT cases from FCCC were obtained from the Biosample Repository Facility, which collects and stores blood samples and obtains information on family history of cancer, risk factors and demographics from participating patients. We classified each TGCT patient according to the histological diagnosis of his tumor: seminoma or non-seminoma (including yolk sac, choriocarcinoma, embryonal, teratoma and mixed cell type TGCT) germ cell tumor. Only those with primary disease in the testis were included.
Male controls (n=932) were selected from PennCATH, a UPHS single-center, hospital-based study of angiographic coronary artery disease (CAD) in almost 4,000 subjects undergoing cardiac catheterization. This study investigates the association of biochemical and genetic factors for CAD and its risk factors25; information on personal history of cancer was not collected. All controls were from the Philadelphia region, 90% were 46 years or older and had already passed the peak age of TGCT development. Based on available age-specific TGCT rates, we estimated that only four TGCT cases would be expected to have arisen in this control group1. It is unlikely that this potential small misclassification of phenotype would have biased results appreciably.
Controls had been genotyped previously using the Affymetrix® Genome-Wide Human SNP Array 6.0 platform and had passed genotyping quality controls measures analogous to those used for TGCT cases (see below).
We used the Affymetrix® Genome-Wide Human SNP Array 6.0 to obtain genotypes for TGCT cases. We used the Birdseed algorithm to determine genotypes for the combined TGCT case and CAD control sample set26. Among the 353 case samples, 18 subsequently were excluded for not meeting case eligibility (two Leydig cell tumors, one female germ cell tumor erroneously coded as TGCT, 15 non-TGCT samples) and 11 replicate samples with lower genotyping call rates were excluded. Of the 324 unique samples from TGCT cases, 19 (5.9%) were excluded because of a low (< 95%) genotyping call rate, eight (2.5%) because of lower than expected genotypic heterozygosity across called markers (FST ≥ 0.06), and 20 (6.2%) because of Asian or African ancestry as determined by multidimensional scaling (MDS)27; no cases were excluded for cryptic relatedness (proportion of genotypes IBD for all cases was < 0.20). Among the 932 CAD controls, 13 were excluded because of female or ambiguous sex.
After excluding 224,705 (24.7%) markers with a minor allele frequency (in the total sample) < 0.05, 1,594 (0.2%) that deviated from Hardy-Weinberg equilibrium (HWE; P < 1 × 10−7), 71,978 (7.9%) with an individual genotype call rate < 0.95, and 233 (0.03%) invalid markers, 611,112 markers remained in the discovery phase.
To further investigate potential bias that could arise from our choice of control group, we compared the minor allele frequencies of markers brought into replication between those controls with verified coronary heart disease (n=700) and those without (n=219). We noted no statistically significant differences for the six markers, nor were there noted differences comparing these controls to population-based controls used in the replication phase (data not shown).
Replication study
To replicate findings of the discovery phase, we used 371 cases, 860 controls and 204 sets of mothers and fathers of cases from a population-based case-control study of TGCT in western Washington State. Methods for recruitment of TGCT cases and parents in this study previously have been published28. Briefly, all cases had first, primary TGCT diagnosed between 1999 and 2007 and were residents of three urban counties of western Washington aged 18 to 44 years at diagnosis. Control subjects did not have a personal history of TGCT and were frequency-matched on age and ascertained from the general population of the three counties using random digit telephone dialing. Family history of TGCT among first-degree relatives and personal history of cryptorchidism was ascertained through self-administered questionnaires. Only cases and controls who self-identified as white, non-Hispanic were included in the replication study.
Genotyping was accomplished using pre-designed TaqMan SNP Genotyping Assays according to manufacturer's specifications. Genotyping was run in duplicate for 1034 marker pairs (an average of 172 sample pairs per each of the six markers in replication). In total, six (0.58%) calls were discordant; the Spearman correlation coefficient was > 0.99. Genotyping calls were made without knowledge of case or duplicate status. We also re-genotyped the majority (94–99%) of TGCT cases from the discovery phase for markers in Table 2. Concordance between genotype calls obtained from the Affymetrix® chip and TaqMan assays for these four makers was 100%.
For both the genome-wide scan and replication study, all participants provided written informed consent approved by their local Institutional Review Boards.
Statistical analysis
For the discovery phase, we used PLINK software to adjust for missing genotypes and calculate rates of heterozygosity29. Population stratification was assessed using multidimensional scaling (MDS) methods and all markers were tested for HWE. PLINK also was used to determine genotypic associations among the 277 TGCT cases and 919 CAD controls. Statistical significance was assessed using Fisher's Exact test, and for top hits we determined ORs and 95% CIs for the per allele, heterozygous, and homozygous effects of the minor allele (Supplementary Table 1).
Imputation was conducted using a computationally efficient hidden Markov model based algorithm as implemented in software MACH30. MACH combines our genotyped data with phased chromosomes from the HapMap CEU samples and then infers the unknown genotypes in the study sample probabilistically by searching for similar stretches of flanking haplotype in the HapMap CEU reference sample. We only analyzed markers that passed the following imputation QC criteria: R2 > 0.3, and MAF > 0.05 in both cases and controls. To account for uncertainty involved in the imputation, we analyzed case-control associations for imputed SNP markers using software SNPTEST31.
For the replication phase, analyses were performed using SAS v9.1.3 (SAS Institute, Cary, NC). We used unconditional logistic regression to determine per allele associations and associations of homozygous and heterozygous carriage of risk alleles with case status (overall and among specified subgroups), and present unadjusted ORs because age was not a confounder in our data. We assessed trend across genotype categories by the Cochran-Armitage test for trend.
Models containing markers coded on an ordinal scale (additive model) and a cross-product term were made to test for marker-marker interaction. To estimate and compare the associations within TGCT subtypes, we used multinomial logit models to obtain simultaneously the OR and 95% CI for the association between markers and each level of outcome after adjusting for age.
Supplementary Material
Acknowledgments
We would like to thank both the men with and without TGCT, and parents of TGCT patients, who contributed to the study. We thank K. D'Andrea, B. Wubbenhorst, and S. Fish for their expert assistance with DNA extraction and genotyping. We thank D. Pucci, K. Robertson, E. Ellis Ohr, A. Mackley, M. Shellenberger, C. Panks, K. Leach, and D. Jacke for patient recruitment and database maintenance. The study was financially supported by the Abramson Cancer Center at the University of Pennsylvania, Lance Armstrong Foundation, and NIH grants R01CA114478 (P.A.K, K.L.N.); R01CA085914 (C.C, J.R.S, S.M.S.); and Fox Chase Cancer Center Support Grant P30 CA006927 (A.K.G.). Recruitment of the UPHS controls was supported by the Cardiovascular Institute of the University of Pennsylvania. Genotyping was supported by GlaxoSmithKline through an Alternate Drug Discovery Initiative research alliance award (to M.P.R. and D.J.R.) with the University of Pennsylvania School of Medicine.
Footnotes
URLs. PLINK, http://pngu.mgh.harvard.edu/purcell/plink/.
References
- 1.Ries LAG, et al. SEER Cancer Statistics Review, 1975–2005. National Cancer Institute; Bethesda, MD: 2008. [Google Scholar]
- 2.Swerdlow AJ, De Stavola BL, Swanwick MA, Maconochie NE. Risks of breast and testicular cancers in young adult twins in England and Wales: evidence on prenatal and genetic aetiology. Lancet. 1997;350:1723–8. doi: 10.1016/s0140-6736(97)05526-8. [DOI] [PubMed] [Google Scholar]
- 3.Hemminki K, Li X. Familial risk in testicular cancer as a clue to a heritable and environmental etiology. Br. J. Cancer. 2004;90:1765–1770. doi: 10.1038/sj.bjc.6601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int. J. Cancer. 2002;99:260–6. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 5.Rapley EA, et al. Localization to Xq27 of a susceptibility gene for testicular germ-cell tumours. Nat. Genet. 2000;24:197–200. doi: 10.1038/72877. [DOI] [PubMed] [Google Scholar]
- 6.Crockford GP, et al. Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum. Mol. Genet. 2006;15:443–51. doi: 10.1093/hmg/ddi459. [DOI] [PubMed] [Google Scholar]
- 7.Nathanson KL, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am. J. Hum. Genet. 2005;77:1034–43. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 9.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 10.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakioka T, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–51. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 12.Frolov A, et al. Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol. Cancer Ther. 2003;2:699–709. [PubMed] [Google Scholar]
- 13.Roskoski R., Jr. Signaling by Kit protein-tyrosine kinase--the stem cell factor receptor. Biochem. Biophys. Res. Commun. 2005;337:1–13. doi: 10.1016/j.bbrc.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 14.Mahakali Zama A, Hudson FP, 3rd, Bedell MA. Analysis of hypomorphic KitlSl mutants suggests different requirements for KITL in proliferation and migration of mouse primordial germ cells. Biol. Reprod. 2005;73:639–47. doi: 10.1095/biolreprod.105.042846. [DOI] [PubMed] [Google Scholar]
- 15.Runyan C, et al. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development. 2006;133:4861–9. doi: 10.1242/dev.02688. [DOI] [PubMed] [Google Scholar]
- 16.Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer. 2005;5:210–22. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 17.Rajpert-de Meyts E, Hoei-Hansen CE. From gonocytes to testicular cancer: the role of impaired gonadal development. Ann. N. Y. Acad. Sci. 2007;1120:168–80. doi: 10.1196/annals.1411.013. [DOI] [PubMed] [Google Scholar]
- 18.Heaney JD, Lam MY, Michelson MV, Nadeau JH. Loss of the transmembrane but not the soluble kit ligand isoform increases testicular germ cell tumor susceptibility in mice. Cancer Res. 2008;68:5193–7. doi: 10.1158/0008-5472.CAN-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes S, et al. COSMIC 2005. Br. J. Cancer. 2006;94:318–22. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blume-Jensen P, et al. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3'-kinase is essential for male fertility. Nat. Genet. 2000;24:157–62. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- 21.Richiardi L, Akre O. Fertility among brothers of patients with testicular cancer. Cancer Epidemiol. Biomarkers Prev. 2005;14:2557–62. doi: 10.1158/1055-9965.EPI-05-0409. [DOI] [PubMed] [Google Scholar]
- 22.Miller CT, et al. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–89. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki A, et al. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell. Biol. 2003;5:427–32. doi: 10.1038/ncb978. [DOI] [PubMed] [Google Scholar]
- 25.Lehrke M, et al. CXCL16 is a marker of inflammation, atherosclerosis, and acute coronary syndromes in humans. J. Am. Coll. Cardiol. 2007;49:442–9. doi: 10.1016/j.jacc.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 26.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 27.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr JR, et al. Risk of testicular germ cell cancer in relation to variation in maternal and offspring cytochrome p450 genes involved in catechol estrogen metabolism. Cancer Epidemiol. Biomarkers Prev. 2005;14:2183–90. doi: 10.1158/1055-9965.EPI-04-0749. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J Hum. Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am. J Hum. Genet. 2006;S79:2290. [Google Scholar]
- 31.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.