Abstract
Background
Eosinophilic esophagitis (EE) involves marked accumulation of eosinophils in the esophageal mucosa that responds to swallowed fluticasone propionate (FP) in a subset of patients.
Objectives
We aimed to uncover the mechanism of action of swallowed FP in patients with EE by providing evidence for a topical effect in the esophagus by identifying a molecular signature for FP exposure in vivo.
Methods
Global microarray expression profiles, immunofluorescence microscopy, and cell signaling in esophageal tissue and cell lines were analyzed.
Results
Thirty-two transcripts exhibited altered expression in patients who responded to swallowed FP treatment. Esophageal FK506-binding protein 5 (FKBP51) mRNA levels were increased (P < .05) in FP responders compared with those seen in control subjects and patients with untreated active EE. After FP treatment of esophageal epithelial cells, FKBP51 mRNA and protein levels were increased in a dose- and time-dependent manner by FP treatment in vitro. FP-induced FKBP51 was steroid receptor dependent because RU486 completely inhibited gene and protein induction. The half-life of FKBP51 mRNA was 16 to 18 hours independent of FP treatment. FKBP51 overexpression reduced FP action as assessed by FP inhibition of IL-13–induced eotaxin-3 promoter activity.
Conclusions
Our results suggest that swallowed glucocorticoid treatment directly affects esophageal gene expression in patients with EE. In particular, increased FKBP51 transcript levels identify glucocorticoid exposure in vivo and distinguish FP responders from untreated patients with active EE and patients without EE. In addition, FKBP51 reduces glucocorticoid-mediated inhibition of IL-13 signaling in epithelial cells in vitro, suggesting that FKBP51 might influence FP responsiveness. We propose that esophageal FKBP51 levels have diagnostic and prognostic significance in patients with EE. (J Allergy Clin Immunol 2010;125:879-88.)
Keywords: Glucocorticoids, eosinophilic esophagitis, FKBP51, IL-13, esophageal epithelial cells
Eosinophilic esophagitis (EE) is characterized by an increased accumulation of eosinophils in the esophageal mucosa that is refractory to acid-suppressive therapy.1 Evidence suggests that the disease strongly associates with atopy and an antigen-driven, TH2-type immune response.2 EE predominantly affects male subjects in a ratio of approximately 3:1 (male/female).3,4 This chronic disease affects pediatric populations because age of onset can be as early as during the first year of life.3 Treatment strategies for EE include dietary restriction of food allergens or systemic glucocorticoid therapy. In addition, a subset of patients respond to swallowed glucocorticoid therapy delivered by means of asthma inhalation medications. The mechanism of action of swallowed glucocorticoids has not been examined, but the observation that patients with EE receiving inhaled corticosteroid therapy benefit from swallowing the same medication provides evidence for a local effect, although other reasons for the improvement exist (eg, increased systemic levels or delivery to the oral or gastric mucosa). Although swallowed or systemic glucocorticoids represent an effective therapy, their action is not sustained on discontinuation of the medicine.5 Additionally, a recent controlled clinical trial has reported that only 50% of patients with EE treated with swallowed fluticasone propionate (FP) respond to treatment compared with 9% in the placebo group.6 Determining the mechanism of action of swallowed corticosteroid therapy is likely to help improve efficacy, develop better drug formulation strategies, and identify mechanisms of steroid resistance, a common problem seen in patients with EE, as well as other allergic inflammatory disorders.7
Glucocorticoid treatment induces an anti-inflammatory effect by targeting various cell types involved in both the innate and adaptive immune responses. In the respiratory tract the action of glucocorticoids is mediated through effects on epithelial cells, at least in part.8,9 Several lines of evidence suggest that esophageal epithelial cells (keratinocytes) comprise a key cellular component involved in the pathogenesis of EE. Notably, on exposure to IL-13, a cytokine overproduced in patients with EE, esophageal epithelial cells express a transcriptome that markedly overlaps the EE transcriptome,10 a set of genes differentially expressed between biopsy specimens from control subjects and patients with EE.11 The gene exhibiting the highest increase in patients with EE and in IL-13–treated esophageal epithelial cells is eotaxin-3, an eosinophil chemoattractant and activator. It has therefore been proposed that IL-13 signaling in esophageal epithelial cells contributes to the pathogenesis of EE, particularly through induction of eotaxin-3. Global transcript profiling shows that 98% of the EE transcriptome is reversed to normal levels in patients who respond to FP.10 Given the importance of esophageal epithelial cells in patients with EE, these cells can serve as relevant targets of glucocorticoid treatment, particularly swallowed FP.
Glucocorticoids exert their action through a variety of mechanisms, including transcriptional inhibition of specific promoter response elements, destabilization of cytokine mRNA, and direct induction of cellular apoptosis. The action of glucocorticoids varies among cell types, and few studies exist12–15 that have examined the action of glucocorticoids on the esophagus in vivo or on esophageal cell lines. Accordingly, in this study we aimed to identify targets of swallowed FP treatment in the esophagus of patients with EE to further understand the molecular mechanisms underlying patient response to this drug. Using global expression profile analysis of esophageal tissue, we identified a panel of 32 transcripts altered by FP treatment in vivo in responders compared with untreated control patients and patients with active EE. Of these transcripts, only 1 gene exhibited levels that were differentially regulated in both responders and nonresponders; the remainder exhibited altered regulation only in responders and not in nonresponders. This suggested that nonresponders might exhibit a fundamental dysfunction in the glucocorticoid signaling pathway in their esophagus. Notably, levels of mRNA for FKBP51, a known steroid-induced gene in respiratory epithelial cells and lymphocytes,16–19 were increased in FP responders. FKBP51 was directly induced by glucocorticoids in esophageal epithelial cells, and we developed evidence that it acts as a negative regulator of FP action. We propose that swallowed FP therapy acts topically and mediates its effects by directly regulating gene expression in esophageal epithelial cells. Furthermore, we suggest that levels of esophageal transcripts, in particular FKBP51, serve as in vivo signatures for steroid exposure.
METHODS
Study design
Patients included in the study include those recruited from either Cincinnati Children’s Hospital Medical Center (50 patients) or Children’s Hospital, San Diego (1 patient). Patients ranged in age from 1.5 to 22.4 years (mean age, 8.9 ± 4.8 years). The population included 38 male and 13 female subjects. Samples were divided into groups according to the following characteristics: control patients (n = 14)—no history of EE, 0 eosinophils per high-powered field (hpf) in the esophagus at the time of biopsy, and no concomitant swallowed glucocorticoid treatment; patients with EE (n = 14)—24 or more eosinophils/hpf at the time of biopsy and no concomitant swallowed glucocorticoid treatment); patients with EE who responded to FP treatment (n = 13)—history of EE, 0 to 1 eosinophils/hpf at the time of biopsy, and concomitant swallowed fluticasone treatment; and patients with EE who did not respond fully to FP treatment (n = 8)—history of EE, 6 or more eosinophils/hpf at the time of biopsy, and concomitant swallowed FP treatment (see Table E1 in this article’s Online Repository at www.jacionline.org). It was noted that no significant difference existed in the average eosinophil count of untreated biopsy samples between FP responders and nonresponders (see Figure E1 and Table E2 in this article’s Online Repository at www.jacionline.org).
Patients taking swallowed FP were generally instructed to use an inhaler without a spacer, spray the medication into the pharynx, and not eat, drink, or rinse for 30 minutes after administration. Doses of swallowed FP are indicated for individual patients; when available, the time of the last dose of FP before endoscopy is listed (see Table E3 in this article’s Online Repository at www.jacionline.org). Biopsy specimens were collected from the distal esophagus on routine endoscopy and submerged in RNAlater (for RNA isolation), modified F-media (for culture), or 4% paraformaldehyde (for immunofluorescence studies). This study was approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center.
Microarray analysis
RNA extraction and microarray analysis were performed as previously described.11 To identify glucocorticoid-regulated genes, those transcripts differentially regulated between control subjects (n = 14) and patients with EE (n = 14) were identified by using the t test (P < .01). Genes differentially regulated between control subjects and FP responders (n = 13) were then identified by using a 2-fold change filter. Subsequently, the genes differentially regulated between control subjects and patients with EE were subtracted from those differentially regulated between control subjects and FP responders. These transcripts were then subjected to ANOVA (P < .01). Although not involved in the initial analysis, transcript values were also included for FP nonresponders (n = 8).
Quantitative real-time PCR
Real-time PCR was performed with the IQ5 system (Bio-Rad Laboratories, Hercules, Calif). Reactions were carried out with SYBR green mix (Bio-Rad Laboratories). The value obtained for each primer set was normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) value for the corresponding sample. Primer sequences are listed in Table E4 (available in this article’s Online Repository at www.jacionline.org).
Primary epithelial cell culture
Primary esophageal epithelial cells were generally cultured as previously described.10 Detailed methods of culture and treatment can be found in the Methods section in this article’s Online Repository at www.jacionline.org. This study was approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center.
TE-7 cell culture
The human esophageal cell line TE-7 was kindly provided by Dr Hainault (IARC, Lyon, France). These cells were maintained in RPMI medium (Invitrogen, Carlsbad, Calif) supplemented with 5% FBS (Atlanta Biologicals, Lawrenceville, Ga) and 1% penicillin/streptomycin (Invitrogen). Details regarding cell treatment and transfection can be found in the Methods section in this article’s Online Repository.
RNA isolation and reverse transcription
RNA was isolated from biopsy specimens as previously described11 by using the RNeasy kit (Qiagen, Hilden, Germany) per the manufacturer’s instructions. RNA was isolated from cells with Trizol (Invitrogen) according to the manufacturer’s instructions, except in actinomycin experiments, in which RNA was isolated with the RNeasy kit (Qiagen). RNA samples (100–1000 ng) were subjected to reverse transcription with Superscript II Reverse Transcriptase (Invitrogen) per the manufacturer’s protocol.
Protein extracts and Western blot analysis
TE-7 or primary esophageal epithelial cells were washed with PBS and incubated with MPER lysis buffer (Thermo Fisher Scientific, Rockford, Ill) supplemented with aprotinin (10 µg/mL), leupeptin (10 µg/mL), pepstatin (10 µg/mL), and sodium orthovanadate (1 mmol/L). Total protein (5–10 mg) was loaded onto 4–12% NuPage Tris-bis gels (Invitrogen), electrophoresed for 1.5 hours at 150 V, and transferred to nitrocellulose membranes, followed by Western blot analysis. Primary antibodies were diluted in TBS/0.1% Tween 20 containing 5% milk: goat anti-FKBP51 (1:1,000; R&D Systems, Minneapolis, Minn) and murine anti–β-actin (1:5,000; Sigma-Aldrich, St Louis, Mo). Secondary antibodies were incubated with the membranes: anti-goat horseradish peroxidase (1:10,000; Santa Cruz Biotechnology, Inc, Santa Cruz, Calif) and anti-mouse horseradish peroxidase (1:10,000; Cell Signaling Technology, Inc, Danvers, Mass). Blots were developed with ECL Plus reagent (GE Healthcare, Piscataway, NJ). Densitometric measurements were performed with Multi Gauge V3.0 (Fujifilm, Tokyo, Japan).
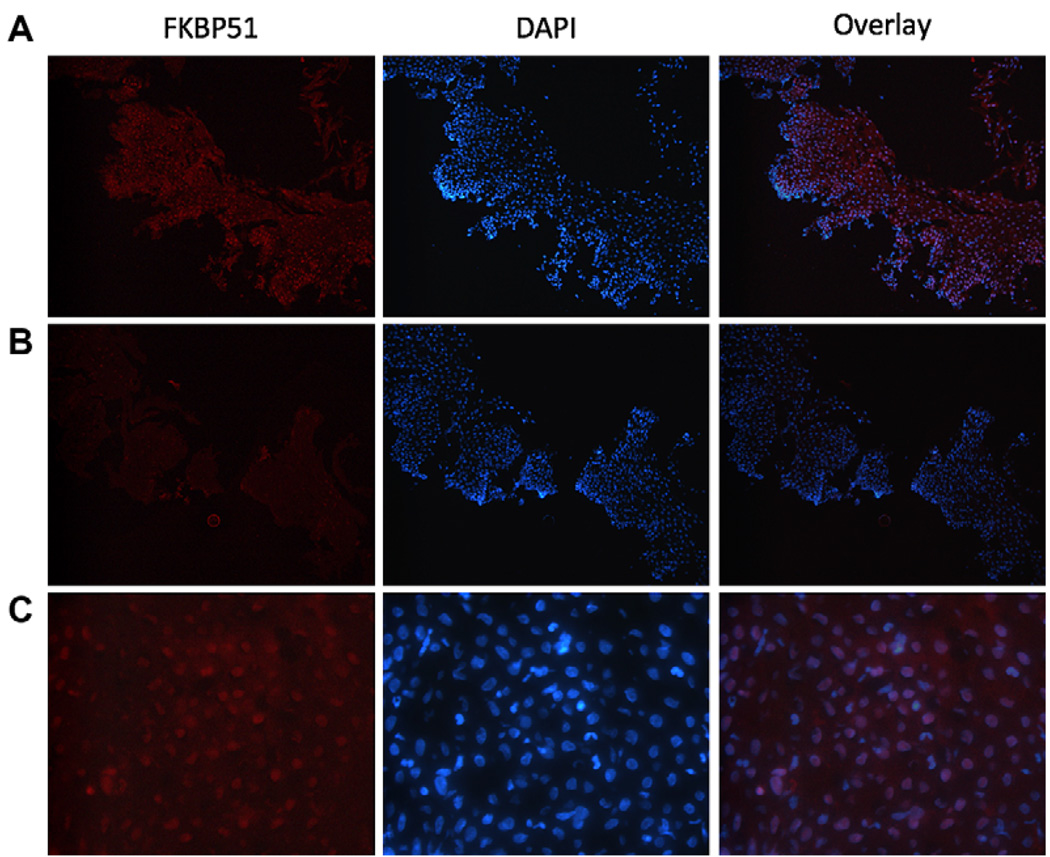
Immunofluorescence microscopy
Biopsy samples were collected as described above. After incubation in 4% paraformaldehyde, biopsy specimens were submerged in 30% sucrose and then embedded in OCT compound. Frozen sections cut from OCT compound–embedded esophageal biopsy samples were mounted on slides. Sections were fixed in ice-cold acetone for 10 minutes, incubated in blocking buffer (PBS, 1% saponin, and 3% FBS), and then incubated with either primary antibody or an equal concentration of control antibody in blocking buffer: anti-FKBP51, 10 µg/mL (R&D Systems, Inc); control, normal goat IgG (R&D Systems, Inc). Sections were incubated with Alexa 594–conjugated secondary antibody (1:250, Invitrogen). Sections were washed 3 times with PBS after each antibody incubation. Fluromount G containing 4′-6-diamidino-2-phenylindole, dihydrochloride was used for mounting. Sections were visualized with the BX51 microscope and MagnaFire imaging software (Olympus America, Inc, Center Valley, Pa).
Constructs
pHRL-TK, pGL3-Basic, and pCDNA3.1 were obtained from Promega (Madison, Wis). Construction of pEotaxin-3 has been described.20 The pEotaxin-3 3′ untranslated region (UTR) consists of coordinates 394 to 562 of NM_006072.4 (GenBank) in the XbaI/SalI sites of the pGL3-Promoter. pFKBP51 was constructed by inserting the FKBP51 open reading frame (coordinates 154 to 1535 ofNM_004117.2,GenBank) into the pCDNA3.1 polylinker.
Statistical methods
Data are expressed as means ± SEMs. Statistical significance was determined by using the Student t test (normal distribution, equal variance), 1-way ANOVA followed by the Tukey post test (>2 groups), or the Kruskal-Wallis test followed by a Dunn multiple comparison test (nonparametric test, >2 groups) with Prism 5.0 Software.
RESULTS
Identification of glucocorticoid-regulated genes in the esophagus
We performed genome-wide expression analysis of esophageal biopsy samples derived from 4 distinct patient populations, including control patients (n = 14), untreated patients with EE (n = 14), patients with EE who responded to FP treatment (n = 13), and patients with EE who did not respond fully to FP treatment (n = 8; see Table E1). We aimed to identify the subset of genes regulated exclusively by FP treatment and not by EE disease. More specifically, we sought to identify genes differentially regulated between untreated control subjects and FP responders, excluding genes that were dysregulated in untreated patients with EE. Accordingly, genes exhibiting altered expression between control subjects and untreated patients with EE were subtracted from the subset of genes that were differentially expressed between control subjects and FP responders (Fig 1, A). Thirty-two transcripts exhibited differential expression in FP responders compared with control subjects but were not included in the genes differentially expressed in untreated patients with EE at baseline (Fig 1, B, and see Table E5).
FIG 1.
Identification of glucocorticoid-regulated genes in patients with EE who respond to FP treatment.A, The average relative gene expression for each patient group.B,Average expression levels of transcripts identified in Fig 1, A. The scale is represented in Fig 1, A (right). C, Average expression of genes exhibiting increased transcript levels in FP responders. D, Average expression of genes showing decreased transcript levels in FP responders. EE, Untreated patients with EE; NL, control patients; NR, FP nonresponders; R, FP responders.
Eight transcripts represented 6 genes that exhibited increased expression in responders but not in nonresponders (Fig 1, C). Of the 17 transcripts (representing 15 genes) with decreased levels in responders, all showed levels in nonresponders that were similar to those seen in control subjects (Fig 1, D). Notably, 4 MHC class II genes were identified, as well as 3 genes of the collagen family. Six transcripts identified in the original analysis (Fig 1, B) were not included in either Fig 1, C or D, because they exhibited a change in expression in control subjects versus that seen in untreated patients with EE despite the analysis scheme devised to eliminate such transcripts. Of the subset of transcripts with increased levels in responders, 1 gene (F3, encoding for coagulation factor III or tissue factor) was identified that exhibited increased expression in both responders and nonresponders (Fig 1, C). Taken together, these results indicate that nonresponders have markedly blunted steroid-induced signaling responses in the esophagus because only 1 of 32 of the genes exhibited differential regulation in both responders and nonresponders.
We next validated the expression pattern of 3 of the identified candidate genes using real-time PCR analysis. Genes were selected based on their known biology. FK506-binding protein 5 (FKBP51), which has a known role in glucocorticoid receptor biology; keratin 7 (KRT7), which promotes epithelial cell integrity and protects against physical damage; and H19, which is processed to a microRNA, showed increased transcript levels in FP responders compared with untreated patients by means of both microarray (Fig 2, A) and real-time PCR (Fig 2, B) analysis. On average, FKBP51 mRNA levels were 2-fold higher in FP responders compared with those seen in control subjects, whereas nonresponders exhibited levels of this transcript that were not significantly higher than those seen in control subjects (Fig 2, A).
FIG 2.
Verification of transcript levels of glucocorticoid-regulated genes by means of real-time PCR analysis. A, Average gene expression determined by means of microarray analysis is expressed as the fold change compared with that seen in control subjects. B, Transcript levels for the indicated gene were quantified by means of real-time PCR and normalized to GAPDH levels; samples used were collected from the same patients used for micro-array analysis. The graph displays the fold change compared with that seen in control subjects. EE, Untreated patients with EE; NL, control patients; NR, FP nonresponders; R, FP responders. *P < .05. **P < .01. ***P < .001.
FKBP51 expression patterns in esophageal tissue
We delineated the cell type or types responsible for FKBP51 expression in patients’ biopsy samples. Esophageal biopsy specimens were immunostained with antibody specific for FKBP51, and nuclei were stained with 4′-6-diamidino-2-phenylindole, dihydrochloride. In biopsy specimens obtained from untreated patients with EE, the basal layer of the esophageal tissue exhibited the strongest signal (Fig 3, A). The signal appeared most intense in the nuclei, although some cytoplasmic staining was apparent. Weaker cytoplasmic staining was also observed in the cells near the luminal side of the biopsy specimen (Fig 3, A). When compared with the hematoxylin and eosin–stained biopsy specimen (data not shown), the expression pattern of FKBP51 in patients’ biopsy specimens was consistent with its being present in epithelial cells. Staining with control antibody confirmed that the observed signal was specific for the FKBP51 antibody (Fig 3, B).
FIG 3.
Localization of FKBP51 expression in patients’ biopsy specimens. Esophageal biopsy sections were immunostained to visualize FKBP51 (red), and 4′-6-diamidino-2-phenylindole, dihydrochloride (DAPI) was used to visualize nuclei (blue). Magnification is ×200 for Fig 3, A and B, and ×800 for Fig 3, C. A, Esophageal biopsy specimen from an untreated patient with EE. B, Negative control (control antibody) for Fig 3, A. C, High-power magnification of biopsy specimen in Fig 3, A.
FKBP51 induction in esophageal epithelial cells
To determine whether FP directly induced FKBP51 in esophageal epithelial cells, we treated primary esophageal epithelial cells with FP and observed an increase (2.15 ± 0.4–fold) in FKBP51 protein expression after FP treatment (Fig 4, A). In addition, FP also induced FKBP51 mRNA and protein in the esophageal epithelial cell line TE-7 (Fig 4, B–D). Transcript levels of FKBP51 were increased by 4 hours after treatment and were further increased after 8 hours, after which a plateau was observed (Fig 4, C). Similarly, FKBP51 protein levels in TE-7 cells increased in a time-dependent manner, with maximum levels of protein observed by 24 hours after treatment (Fig 4, D). The induction of FKBP51 by FP was not specific for this steroid because FKBP51 mRNA (Fig 5, A) and protein (Fig 5, B and C) levels increased after treatment with dexamethasone. FKBP51 protein levels were similarly increased in primary esophageal epithelial cells after dexamethasone treatment (Fig 5, D).
FIG 4.
FKBP51 induction after glucocorticoid treatment of esophageal epithelial cells. For Fig 4, A, B, D, and E, protein extracts were subjected to SDS-PAGE and Western blot analysis for FKBP51 and actin; protein levels were quantified by means of densitometric analysis. The average fold change in the ratio of FKBP51 to actin for each sample compared with the untreated sample for 3 experiments is shown. For Fig 4, A through E, results are representative of 3 experiments. A, Primary esophageal epithelial cells were treated with 10−6 mol/L FP for 24 hours. B, TE-7 cells were treated with FP for 24 hours. C, TE-7 cells were treated with 10−7 mol/L FP. RNA was isolated, cDNA synthesis was performed, and real-time PCR analysis to detect FKBP51 and GAPDH transcripts was done. D, In a separate experiment TE-7 cells were treated for the indicated time with 10−7 mol/L FP. *P < .05. **P < .01. E, TE-7 cells were pretreated with 10−6 mol/L RU486 for 30 minutes. FP or dexamethasone (Dex) was then added for 24 hours. F, TE-7 cells were treated with 10−6 mol/L FP for 24 hours. Actinomycin D was then added (10 µg/mL). Cells were incubated for the indicated number of hours, at which time they were subjected to RNA isolation followed by cDNA synthesis. FKBP51 mRNA levels determined by means of real-time PCR were normalized to nanograms of reverse-transcribed RNA. The graph shows the ratio of FKBP51 per nanogram RNA as a percentage of the initial time point; each time point represents the mean ± SEM of 3 independent experiments. Half-life was calculated as the mean ± SEM of the half-life value for each of the 3 independent experiments. G, TE-7 cells were pretreated with 10 µg/mL CHX for 30 minutes. Subsequently, FP or dexamethasone was added for 24 hours. The graph shows FKBP51 mRNA expression normalized to GAPDH expression. The results are representative of 3 experiments. DMSO, Dimethyl sulfoxide.
FIG 5.
FKBP51 transcript and protein levels are increased by dexamethasone (Dex) treatment of esophageal epithelial cells. A, RNA isolated from TE-7 cells treated with dexamethasone or FP for 24 hours was subjected to cDNA synthesis. FKBP51 transcript levels determined by means of real-time PCR analysis were normalized to GAPDH levels. B–D, Western blot analysis for FKBP51 and β-actin. Fig 5, B, TE-7 cells were treated with dexamethasone for 24 hours. Fig 5, C, TE-7 cells were treated with 10−6 mol/L dexamethasone. Fig 5, D, Primary esophageal epithelial cells were treated with 10–6 mol/L dexamethasone for 24 hours. In Fig 5, A–D, data are representative of 3 experiments per figure.
Glucocorticoid-induced FKBP51 is steroid receptor dependent
Cells were pretreated with the glucocorticoid receptor antagonist RU486 followed by FP or dexamethasone treatment. Western blot analysis showed that RU486 treatment abolished the FP- and dexamethasone-mediated increase in FKBP51 protein levels. Dexamethasone required an equimolar amount of RU486 for the inhibition, whereas FP-mediated induction of FKBP51 protein levels was inhibited by a 100-fold molar excess of RU486 (Fig 4, E). As such, FKBP51 induction is mediated through a glucocorticoid receptor ligand binding–dependent process.
TE-7 cells were treated with FP for 24 hours, followed by addition of actinomycin D to inhibit de novo transcription, to determine whether glucocorticoid treatment increased the stability of FKBP51 mRNA. At the indicated time points after initiation of actinomycin D treatment, FKBP51 mRNA levels were monitored. The half-life of FKBP51 mRNA in the absence and presence of FP treatment was similar (17.84 ± 2.03 hours and 16.44 ± 1.11 hours, respectively; P = .35; Fig 4, F). We next tested whether de novo protein synthesis was required for the increased transcript levels of FKBP51. TE-7 cells were pretreated with cycloheximide (CHX) at a dose (10 µg/mL) that inhibited protein synthesis in these cells more than 95%, as measured based on [35S]-methionine incorporation into protein (data not shown) followed by addition of vehicle or FP (10−6 mol/L) for 24 hours. Although an increase of FKBP51 transcripts was observed at baseline after CHX treatment, FKBP51 mRNA levels were even further increased in FP-treated cells in which protein synthesis was inhibited (Fig 4, G), suggesting that de novo protein synthesis was not absolutely required for subsequent FP-mediated induction of FKBP51 gene expression.
Effect of FKBP51 on glucocorticoid and IL-13 signaling in esophageal epithelial cells
IL-13 signaling in esophageal epithelial cells has been shown to have a critical role in EE pathogenesis by inducing the EE transcriptome, at least in part.10 When TE-7 cells were treated with IL-13, an increase in eotaxin-3 transcript levels was observed (Fig 6, A). When the cells were treated concomitantly with FP, the IL-13–mediated increase in eotaxin-3 mRNA and protein decreased significantly at all doses tested (Fig 6, A and B).
FIG 6.
IL-13–induced transcript and protein levels of eotaxin-3 are reversed by FP treatment of esophageal epithelial cells. A, RNA isolated from TE-7 cells treated with IL-13, FP, or both for 24 hours was subjected to cDNA synthesis. Transcript levels of eotaxin-3 were determined by means of real-time PCR analysis and normalized to GAPDH levels. B, TE-7 cells were treated with IL-13, FP, or both for 48 hours. ELISA was performed to detect eotaxin-3 in the supernatants. The dashed line indicates the detection limit of the assay. For Fig 6, A and B, data are representative of 3 experiments per figure. *P < .05. **P < .01.
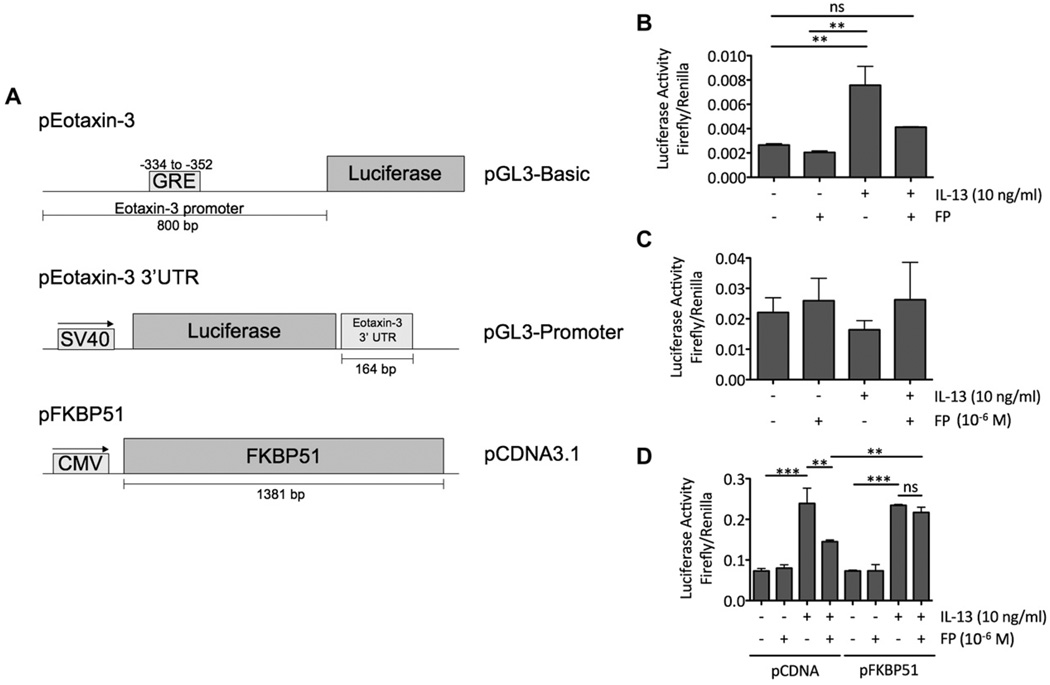
To understand the contribution of the promoter and 3′ UTR sequences of eotaxin-3 to the control of its gene expression in esophageal epithelial cells after IL-13 and FP treatment, we performed luciferase assays with TE-7 cells transiently transfected with constructs containing the eotaxin-3 5′ UTR upstream or the 3′ UTR downstream of a luciferase expression cassette. The cells were transfected with a luciferase reporter construct containing either 800 bp of the eotaxin-3 promoter 5′ of the firefly luciferase gene in the pGL3-Basic vector or a plasmid containing 164 bp of the eotaxin-3 3′ UTR downstream of the firefly luciferase gene in the pGL3-Promoter vector (Fig 7, A).10,11 Experiments with the promoter construct demonstrated a decrease in IL-13–induced eotaxin-3 promoter activity on concomitant FP treatment (Fig 7, B). Experiments with the 3′ UTR construct did not reveal significant modulation of mRNA stability on IL-13 or FP treatment (Fig 7, C). Taken together, these findings indicate that at least part of the observed decrease in IL-13–induced eotaxin-3 mRNA levels after FP treatment is mediated through modulation of the promoter activity through cis-acting sequences located within the immediate 5′ UTR of eotaxin-3.
FIG 7.
Increased baseline FKBP51 levels affect glucocorticoid-mediated repression of IL-13–induced eotaxin-3 promoter activity. A, Constructs used in this study. CMV, Cytomegalovirus promoter. Cells were transfected with pHRL-TK and either pEotaxin-3 (B) or pEotaxin-3-3′UTR (C) or pHRL-TK, pEotaxin-3, and either pCDNA3.1 or pFKBP51 (D). Fig 7, B–D, After transfection, cells were treated with IL-13, FP, or both for 24 hours. Firefly and Renilla luciferase activities for each sample were then quantified. Eotaxin-3 promoter activity is expressed as the ratio of firefly to Renilla luciferase activity. For Fig 7, B–D, panels are representative of 3 experiments. ns, Nonsignificant. **P < .01. ***P < .001.
To test whether increased FKBP51 levels affected the FP-mediated decrease in IL-13–induced eotaxin-3 promoter activity, TE-7 cells were transfected with either empty vector (pCDNA3.1) or an expression construct that contained FKBP51 under the control of the CMV promoter (pFKBP51; Fig 7, A) to simulate high baseline levels of FKBP51. Cells transfected with the empty vector showed a 2-fold increase in eotaxin-3 promoter activity on IL-13 treatment, and this was reduced by 50% when the cells were concomitantly treated with FP (Fig 7, D). However, when cells were transfected with the FKBP51 expression vector, the FP-mediated decrease in IL-13–induced eotaxin-3 promoter activity was reduced (Fig 7, D).
DISCUSSION
In this study we identified a set of genes that exhibited differential expression in the esophagus of treated, FP-responsive patients with EE compared with untreated subjects. Our analysis uncovered several genes previously shown to be glucocorticoid responsive, including those encoding FKBP51, MHC class II, and collagen molecules. Glucocorticoids upregulate FKBP51, whereas they decrease expression of MHC class II and collagen genes in several cell types.16–19,21,22 The identification of such genes validates the analysis performed. Of the transcripts identified, all but 1 exhibited expression in nonresponders similar to that observed in untreated patients. This indicates that nonresponders might have a fundamental dysfunction in glucocorticoid signaling, at least in the esophagus. Tissue-specific glucocorticoid resistance has been reported in other inflammatory diseases, including rheumatoid arthritis, osteoarthritis, Crohn disease, ulcerative colitis, and asthma.23 Multiple mechanisms have been shown to account for glucocorticoid resistance, including aberrant interactions between transcription factors, coactivators, and corepressors; posttranslational modification of inflammatory mediators; and expression of glucocorticoid receptor isoforms.23 Interestingly, only 1 gene (F3) showed differential expression in both FP responder and FP non-responders compared with untreated subjects. This implies that F3 gene expression could be influenced by a nongenomic mechanism of glucocorticoid signaling. This gene might be especially useful to monitor patient compliance because it is expressed in all patients who take the drug, regardless of responsiveness.
The observed change in patients’ biopsy specimen gene expression in addition to previous literature reporting a low systemic bioavailability of oral FP24 supports the interpretation that swallowed FP treatment exerts a topical effect in the esophagus. The oral bioavailability of FP has been reported to be less than 1% at a dose greater than 20-fold higher than the dose for patients with EE in this current study24; this, coupled with the fact that FP undergoes extensive first-pass metabolism in the liver,25 makes it less likely that swallowed FP acts through a systemic route to alter gene expression in the esophagus. To our knowledge, this represents the first report suggesting direct evidence of a topical effect of glucocorticoids in the esophagus.
Here we report that FKBP51 was upregulated at the mRNA and protein levels in esophageal epithelial cells, including primary esophageal epithelial cells that were cultured from esophageal biopsy specimens. Based on the ability of RU486 to inhibit FKBP51 protein induction, the increased FKBP51 levels are likely glucocorticoid receptor dependent. We observed that increased transcript stability did not account for the upregulation of FKBP51 mRNA. Instead, increased FKBP51 mRNA levels are likely caused by increased transcription of the gene. Numerous putative glucocorticoid response elements (GREs) exist within the FKBP51 promoter and introns, and such sequences might mediate the increased transcription in cell types, including A549 cells.26 We observed that FKBP51 levels were increased at baseline on CHX treatment, suggesting that inhibition of protein synthesis up-regulates FKBP51. This could occur if a repressor of FKBP51 transcription required ongoing protein synthesis; alternatively, FKBP51 levels might be upregulated as part of a stress response. Despite this increased baseline expression, FKBP51 transcription still increased on glucocorticoid treatment in the presence of CHX, suggesting that at least a portion of the increased FKBP51 transcripts do not require de novo protein synthesis. These data collectively suggest that glucocorticoid signaling directly affects FKBP51 transcription in esophageal epithelial cells.
We observed that FP treatment inhibited IL-13–mediated eotaxin-3 promoter activity. This appears to be mediated through the 800 bp of promoter that contains 1 canonical GRE sequence. Previous reports have suggested that repression of IL-4–induced eotaxin-3 expression by FP occurs independently of this GRE sequence in lung epithelial cells.27 It remains to be tested whether this is the case in esophageal epithelial cells.
FKBP51 has been shown to act as a negative regulator of glucocorticoid signaling.28,29 In fact, baseline FKBP51 levels in airway epithelial cells negatively correlate with response to glucocorticoid treatment in asthmatic patients.30 Additionally, new world primates exhibit high levels of FKBP51, and this correlates with general glucocorticoid resistance in these animals. 17,31,32 Herein, we show that increasing baseline FKBP51 levels in the esophageal cell line TE-7 correlate with a decreased ability of glucocorticoid to repress IL-13–mediated eotaxin-3 promoter activity (Fig 8).
FIG 8.
Model to describe the regulation and potential function of FKBP51 in patients with EE. FP represses IL-13–induced eotaxin-3 expression while inducing FKBP51 gene expression through a mechanism likely dependent on the glucocorticoid receptor (GR). FKBP51 additionally inhibits glucocorticoid receptor (GR)–mediated signaling and thus dampens glucocorticoid-mediated repression of IL-13–induced eotaxin-3 promoter activity.
Biopsy specimens of patients who respond to swallowed FP treatment exhibit high levels of FKBP51, a negative regulator of glucocorticoid signaling.28,29,31,32 However, we note that the observed high FKBP51 transcript levels occurred after treatment. In contrast, we showed that high baseline FKBP51 levels before treatment negatively affect glucocorticoid signaling in vitro. Therefore we propose that although increased FKBP51 levels after FP treatment serve as a readout of functional glucocorticoid signaling, the actual biological function of FKBP51 in part involves negative regulation of glucocorticoid signaling to dampen the anti-inflammatory response so that homeostatic conditions can be restored after the glucocorticoid-mediated resolution of inflammation.
In summary, we have shown that (1) FP responders express a unique set of esophageal genes (including FKBP51), providing evidence that swallowed glucocorticoids mediate their role through a topical effect in the esophagus in vivo, and (2) FP nonresponders have a blunted expression of steroid-induced esophageal transcripts, suggesting impaired glucocorticoid signaling in these patients. Further proof for the ability of glucocorticoids to induce esophageal transcripts is derived by in vitro studies with esophageal epithelial cells, which have demonstrated that (1) glucocorticoids directly induce FKBP51 through a RU486-inhibited mechanism, indicating the direct involvement of the glucocorticoid receptor; (2) FKBP51 mRNA half-life is approximately 16 to 18 hours and importantly not regulated by glucocorticoids; and (3) FKPB51 overexpression inhibits glucocorticoid-mediated transcriptional repression of eotaxin-3. Taken together, our data provide molecular evidence that swallowed glucocorticoids mediate their action through a topical effect in the esophagus and that posttreatment FKBP51 esophageal levels serve as a measure of functional glucocorticoid signaling.
Clinical implications: Swallowed glucocorticoids induce disease remission by regulating local gene expression in the esophagus. Understanding the mechanism by which topical glucocorticoids induce remission from EE will provide insight into disease pathogenesis, steroid resistance, and additional molecular pathways to serve as targets for therapeutic intervention.
Supplementary Material
Acknowledgments
Supported in part by National Institutes of Health grant RO1 DK76893 and U19 AI070235, American Heart Association grant 09POST2180041, the Food Allergy Project, the Campaign Urging Research for Eosinophilic Disorders (CURED), and the Buckeye Foundation.
We thank Drs Michael Farrell, Michael Bates, Kathleen Campbell, and Mitchell Cohen for their referral of patients. We are grateful to Bridget Buckmeier, Emily Stucke, and Tommie Grotjan for research assistance, as well as to the Food Allergy Project, the Campaign Urging Research for Eosinophilic Disorders (CURED), and the Buckeye Foundation for their generous support.
Abbreviations used
- CHX
Cycloheximide
- EE
Eosinophilic esophagitis
- FKBP51
FK506-binding protein 5
- FP
Fluticasone propionate
- F3
Coagulation factor III gene
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase gene
- GRE
Glucocorticoid response element
- hpf
High-power field
- UTR
Untranslated region
Footnotes
Disclosure of potential conflict of interest: J. M. Caldwell has received a postdoctoral grant from the American Heart Association. C. Blanchard has received research support from the National Institutes of Health, the Digestive Health Center CCHMC, and the American Partnership for Eosinophilic Disorders. M. H. Collins was a subcontractor as a clinical study central review pathologist for GlaxoSmithKline and Ception Therapeutics; was a consultant as a clinical study central review pathologist for Meritage Pharma; and is a Member of the Medical Advisory Panel for the American Partnership for Eosinophilic Diseases. S. S. Aceves has intellectual property patent royalties in Meritage Pharma and is on the Medical Advisory Board for the American Partnership for Eosinophilic Disorders. M. E. Rothenberg is a speaker and consultant for Merck; is a consultant for Centocor, Ception Therapeutics, Nycomed, and Array Biopharmra; has received research support from the National Institutes of Health, the Food Allergy and Anaphylaxis Network, and the Dana Foundation; is on the Medical Advisory Board for the American Partnership for Eosinophilic Disorders; and is on the Executive Council for the International Eosinophil Society. The rest of the authors have declared that they have no conflict of interest.
REFERENCES
- 1.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard C, Rothenberg ME. Basic pathogenesis of eosinophilic esophagitis. Gastrointest Endosc Clin N Am. 2008;18:133–143. doi: 10.1016/j.giec.2007.09.016. x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orenstein SR, Shalaby TM, Di Lorenzo C, Putnam PE, Sigurdsson L, Mousa H, et al. The spectrum of pediatric eosinophilic esophagitis beyond infancy: a clinical series of 30 children. Am J Gastroenterol. 2000;95:1422–1430. doi: 10.1111/j.1572-0241.2000.02073.x. [DOI] [PubMed] [Google Scholar]
- 4.Franciosi JP, Tam V, Liacouras CA, Spergel JM. A case-control study of sociodemographic and geographic characteristics of 335 children with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:415–419. doi: 10.1016/j.cgh.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Schaefer ET, Fitzgerald JF, Molleston JP, Croffie JM, Pfefferkorn MD, Corkins MR, et al. Comparison of oral prednisone and topical fluticasone in the treatment of eosinophilic esophagitis: a randomized trial in children. Clin Gastroenterol Hepatol. 2008;6:165–173. doi: 10.1016/j.cgh.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Konikoff MR, Noel RJ, Blanchard C, Kirby C, Jameson SC, Buckmeier BK, et al. A randomized, double-blind, placebo-controlled trial of fluticasone propionate for pediatric eosinophilic esophagitis. Gastroenterology. 2006;131:1381–1391. doi: 10.1053/j.gastro.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Leung DY, Bloom JW. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 2003;111:3–23. doi: 10.1067/mai.2003.97. [DOI] [PubMed] [Google Scholar]
- 8.Stellato C. Glucocorticoid actions on airway epithelial responses in immunity: functional outcomes and molecular targets. J Allergy Clin Immunol. 2007;120:1247–1265. doi: 10.1016/j.jaci.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 9.Schleimer RP. Glucocorticoids suppress inflammation but spare innate immune responses in airway epithelium. Proc Am Thorac Soc. 2004;1:222–230. doi: 10.1513/pats.200402-018MS. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunin AG, Nikolaev DV. Effect of acute and chronic glucocorticoid treatments on epithelial cell proliferation in the esophagus and small intestine of rats. J Gastroenterol. 1999;34:661–667. doi: 10.1007/s005350050316. [DOI] [PubMed] [Google Scholar]
- 13.Gololobova MT. [The duration of the periods of the mitotic cycle in mouse esophageal epithelium following hydrocortisone administration] Biull Eksp Biol Med. 1970;69:97–100. [PubMed] [Google Scholar]
- 14.Zboralske FF, Karasek MA. Growth characteristics of human esophageal epithelial cells in primary explant and serial culture. In Vitro. 1984;20:109–118. doi: 10.1007/BF02626651. [DOI] [PubMed] [Google Scholar]
- 15.Menard D, Arsenault P. Maturation of human fetal esophagus maintained in organ culture. Anat Rec. 1987;217:348–354. doi: 10.1002/ar.1092170405. [DOI] [PubMed] [Google Scholar]
- 16.Baughman G, Wiederrecht GJ, Chang F, Martin MM, Bourgeois S. Tissue distribution and abundance of human FKBP51, and FK506-binding protein that can mediate calcineurin inhibition. Biochem Biophys Res Commun. 1997;232:437–443. doi: 10.1006/bbrc.1997.6307. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds PD, Ruan Y, Smith DF, Scammell JG. Glucocorticoid resistance in the squirrel monkey is associated with overexpression of the immunophilin FKBP51. J Clin Endocrinol Metab. 1999;84:663–669. doi: 10.1210/jcem.84.2.5429. [DOI] [PubMed] [Google Scholar]
- 18.Wan Y, Nordeen SK. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol Endocrinol. 2002;16:1204–1214. doi: 10.1210/mend.16.6.0848. [DOI] [PubMed] [Google Scholar]
- 19.Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M. Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. J Clin Endocrinol Metab. 2003;88:277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- 20.Blanchard C, Durual S, Estienne M, Emami S, Vasseur S, Cuber JC. Eotaxin-3/CCL26 gene expression in intestinal epithelial cells is up-regulated by interleukin-4 and interleukin-13 via the signal transducer and activator of transcription 6. Int J Biochem Cell Biol. 2005;37:2559–2573. doi: 10.1016/j.biocel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Schwiebert LM, Schleimer RP, Radka SF, Ono SJ. Modulation of MHC class II expression in human cells by dexamethasone. Cell Immunol. 1995;165:12–19. doi: 10.1006/cimm.1995.1181. [DOI] [PubMed] [Google Scholar]
- 22.Shull S, Cutroneo KR. Glucocorticoids coordinately regulate procollagens type I and type III synthesis. J Biol Chem. 1983;258:3364–3369. [PubMed] [Google Scholar]
- 23.Kino T, Chrousos GP. Tissue-specific glucocorticoid resistance-hypersensitivity syndromes: multifactorial states of clinical importance. J Allergy Clin Immunol. 2002;109:609–613. doi: 10.1067/mai.2002.123708. [DOI] [PubMed] [Google Scholar]
- 24.Falcoz C, Oliver R, McDowall JE, Ventresca P, Bye A, Daley-Yates PT. Bioavailability of orally administered micronised fluticasone propionate. Clin Pharmacokinet. 2000;39 suppl 1:9–15. doi: 10.2165/00003088-200039001-00002. [DOI] [PubMed] [Google Scholar]
- 25.Harding SM. The human pharmacology of fluticasone propionate. Respir Med. 1990;84 suppl A:25–29. doi: 10.1016/s0954-6111(08)80004-2. [DOI] [PubMed] [Google Scholar]
- 26.Hubler TR, Scammell JG. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones. 2004;9:243–252. doi: 10.1379/CSC-32R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsukura S, Kokubu F, Kurokawa M, Kawaguchi M, Kuga H, Ieki K, et al. Molecular mechanisms of repression of eotaxin expression with fluticasone propionate in airway epithelial cells. Int Arch Allergy Immunol. 2004;134 suppl 1:12–20. doi: 10.1159/000077787. [DOI] [PubMed] [Google Scholar]
- 28.Wochnik GM, Ruegg J, Abel GA, Schmidt U, Holsboer F, Rein T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem. 2005;280:4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 29.Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 30.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scammell JG, Denny WB, Valentine DL, Smith DF. Overexpression of the FK506-binding immunophilin FKBP51 is the common cause of glucocorticoid resistance in three New World primates. Gen Comp Endocrinol. 2001;124:152–165. doi: 10.1006/gcen.2001.7696. [DOI] [PubMed] [Google Scholar]
- 32.Westberry JM, Sadosky PW, Hubler TR, Gross KL, Scammell JG. Glucocorticoid resistance in squirrel monkeys results from a combination of a transcriptionally incompetent glucocorticoid receptor and overexpression of the glucocorticoid receptor co-chaperone FKBP51. J Steroid Biochem Mol Biol. 2006;100:34–41. doi: 10.1016/j.jsbmb.2006.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.