Abstract
Neuroserpin is a member of the serpin superfamily. Point mutations in the neuroserpin gene underlie the autosomal dominant dementia, familial encephalopathy with neuroserpin inclusion bodies. This is characterized by the retention of ordered polymers of neuroserpin within the endoplasmic reticulum of neurons. pH has been shown to affect the propensity of several serpins to form polymers. In particular, low pH favors the formation of polymers of both α1-antitrypsin and antithrombin. We report here opposite effects in neuroserpin, with a striking resistance to polymer formation at acidic pH. Mutation of specific histidine residues showed that this effect is not attributable to the shutter domain histidine as would be predicted by analogy with other serpins. Indeed, mutation of the shutter domain His338 decreased neuroserpin stability but had no effect on the pH dependence of polymerization when compared with the wild-type protein. In contrast, mutation of His119 or His138 reduced the polymerization of neuroserpin at both acidic and neutral pH. These residues are at the lower pole of neuroserpin and provide a novel mechanism to control the opening of β-sheet A and hence polymerization. This mechanism is likely to have evolved to protect neuroserpin from the acidic environment of the secretory granules.
Keywords: neuroserpin, polymerization, familial encephalopathy with neuroserpin inclusion bodies, serpins
Introduction
Neuroserpin is a member of the serine proteinase inhibitor or serpin superfamily.1–4 It is expressed during the late stage of development in neurons of the central and peripheral nervous system and in the adult brain.5,6 The target proteinase of neuroserpin is tissue plasminogen activator (tPA),7 and thus, it is likely to be important in the control of synaptic plasticity8 and in learning and memory.9 Neuroserpin may also be important in the regulation of permeability between the vascular and nervous system compartments during normal brain function,9 and there is growing evidence that it can act as a neuroprotectant. Indeed, neuroserpin protects against cortical damage in mouse models of cerebral infarction.10–12 It can also limit neuronal excitotoxicity and reduce seizures in mouse models of epilepsy13,14 and affect the toxicity of the Aβ1-42 peptide that is central to the pathogenesis of Alzheimer's disease.15,16 Point mutations in the neuroserpin gene underlie the autosomal dominant dementia, familial encephalopathy with neuroserpin inclusion bodies (FENIB).17,18 This is characterized by the retention of ordered polymers of neuroserpin within the endoplasmic reticulum of neurons.17–21 Our previous work has shown that polymers of neuroserpin result from the sequential linkage between the reactive center loop of one molecule and β-sheet A of another.7,22 However, it has recently been proposed that such polymers are formed by a β-hairpin linkage between not only the reactive center loop but also s5A inserting into β-sheet A of another molecule.23 It is these polymers of neuroserpin that aggregate to form the cortical and subcortical periodic acid-Schiff-positive Collin's bodies that are the hallmark of the disease.17,18 Seven families with FENIB have now been described, which are caused by five different mutations within the neuroserpin gene.17,18,24–26 There is a clear correlation between genotype and phenotype that can be explained on the rate of polymerization.7,18–20,22 Those mutants that polymerize most rapidly are associated with the greatest number of cerebral inclusions and the earliest age of onset of disease. Neuroserpin can also be inactivated by intramolecular loop insertion to form a latent conformer that has also been isolated from the brains of individuals with FENIB.27
The polymerization reported for mutants of neuroserpin at physiological pH is similar to that reported for mutants of other members of the serpin superfamily.28,29 Such polymer formation is strikingly dependent on pH.30–33 For example, lowering the pH to 6.0 or less leads to the ready formation of polymers of antithrombin.32 This polymerization was markedly reduced at pH values above 6.0. The formation of polymers in plasminogen activator inhibitor (PAI-1) is similarly dependent on low pH.31 α1-Antitrypsin is most resistant to polymerization at pH 6.0–8.0, with an increase in polymerization rate at pH values above or below this range.30 The pH dependence of polymer formation has been attributed to protonation/deprotonation of His334 in antithrombin and α1-antitrypsin. This histidine forms a hydrogen bond network in the shutter domain that controls the opening and closing of β-sheet A.32 The effect of pH on the stability of neuroserpin is particularly important as this protein is stored in the acidic environment of the dense-core vesicles before secretion from the neurone.20,34–36 We report here that neuroserpin is resistant to polymer formation at acidic pH. This effect is not mediated by the shutter domain histidine but by other histidines, in particular His119 and His138.
Results
Low pH abrogates the polymerization of wild-type neuroserpin
The histidine in the shutter region is believed to play an important role in the hydrogen bond network that controls the patency of β-sheet A in both α1-antitrypsin and antithrombin.32 Neuroserpin contains a homologous histidine at position 338. We evaluated the role of pH on polymer formation by assessing the polymerization of wild-type neuroserpin over a range of pH values. No polymers of wild-type neuroserpin formed at or below pH 6.0 [Fig. 1(A)], but the rate of polymerization increased between pH 6.0 and 8.0 [Fig. 1(B,C)]. This was most marked between pH 6.0 and 7.0 [Fig. 1(D)]. The low pH stabilized wild-type neuroserpin in its active conformation as it was still able to form an SDS-stable complex with tPA [Fig. 1(E)]. This retention of activity excludes transition to inactive conformers such as the latent species. Circular dichroism (CD) measurements showed no change in the overall structure of wild-type neuroserpin over the range of pH tested [Fig. 2(A)]. Assessment of the thermal transition at 216 nm confirmed the increase in the stability of wild-type neuroserpin at lower pH values [Fig. 2(B)]. For example, the transition temperature of wild-type neuroserpin was 63.6°C at pH 6.0 but decreased to 58.9°C at pH 7.0 and to 57.1°C at pH 9.0, allowing us to calculate a mid-transition point at pH 6.6 (±0.1).
Figure 1.
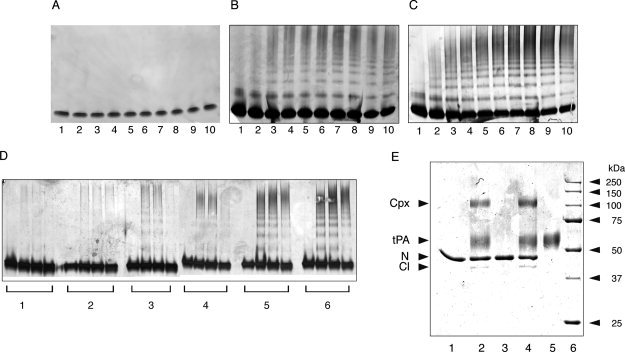
Neuroserpin was incubated at 45°C and 0.4 mg/mL and aliquots taken over time were analyzed by 3-12% w/v gradient nondenaturing PAGE. All lanes contain 2 μg of protein. A: Polymerization of wild-type neuroserpin at pH 6.0. B: Polymerization of wild-type neuroserpin at pH 7.0. C: Polymerization of wild-type neuroserpin at pH 8.0. Lanes 1-10 correspond to 0, 5, 10, 15, 20, 30, 45, 60, 120, and 180 min. D: Polymerization of neuroserpin at pH 6.0–7.0. Lanes 1-6 correspond to pH 6.0, 6.2, 6.4, 6.6, 6.8, and 7.0 at incubation times of 0, 2, 6, and 24 h. E: Complex formation between neuroserpin and tPA at different pH analyzed by 10% w/v SDS-PAGE. Lane 1, neuroserpin; lane 2, neuroserpin with two fold molar excess of tPA at pH 7.0; lane 3, neuroserpin incubated for 5 h at pH 5.0 and 45°C; lane 4, neuroserpin incubated for 5 h at pH 5.0 and 45°C, and then the pH was adjusted to 7.0 followed by the addition of two fold molar excess of tPA for 10 min; lane 5, tPA; lane 6, molecular mass markers. N, intact neuroserpin; Cl, reactive loop-cleaved neuroserpin; Cpx, the complex between neuroserpin and tPA. All lanes contain 1–2 μg of protein.
Figure 2.
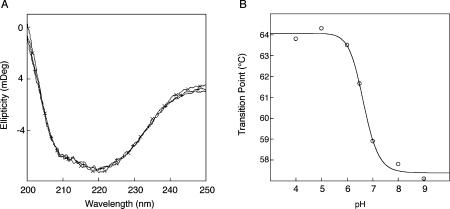
A: Far-UV spectra of wild-type neuroserpin at different pH. pH 6.0 (+ line), pH 7.0 (solid line), and pH 8.0 (× line). B: Transition temperature of neuroserpin with increasing pH as measured by circular dichroism. The transition temperature for the different pH values was calculated as described in the Materials and Methods section.
We also assessed the propensity of the Ser49Pro mutant of neuroserpin, responsible for the dementia FENIB, to polymerize at different pH. As with wild-type neuroserpin, there was no formation of polymers below pH 6.0, and an increase in polymerization between pH 6.0 and 8.0. Once again, CD measurements did not show any change in the overall structure of Ser49Pro neuroserpin at different pH (data not shown).
Other histidines are involved in neuroserpin polymerization
The absence of polymerization of wild-type neuroserpin at low pH suggests that His338 is not the only residue involved in the pH-dependent polymerization of neuroserpin. Our recent crystal structure of neuroserpin37 allowed us to identify His119 and His138 as other likely candidates (Fig. 3). We therefore mutated these residues along with His338 to glutamine. We also prepared and expressed His374Gln neuroserpin but were unable to purify the recombinant protein to homogeneity. There was no difference in CD spectra profiles among wild-type, His119Gln, and His138Gln neuroserpin at pH 7.0 [Fig. 4(A)]. However, replacing His338 by a glutamine resulted in a shift at 216 and 222 nm similar to that described previously for Ser49Pro neuroserpin.7 In the later, this difference was not due to a major conformational transition, but to an intermediate conformation in which the reactive loop was partly inserted in β-sheet A.7 There was no difference in the far UV CD spectra of His119Gln, His138Gln, or His338Gln neuroserpin between pH 5.0 and 8.0 (data not shown). Stability of the different mutants was assessed by CD profiles at 216 nm with increasing temperature. The His119Gln mutant was less stable than wild-type neuroserpin at pH values below 7.0, with a transition point that was 2–3°C lower between pH 5.0 and 6.5. However, it was more stable at pH values greater than 7.0, with a difference in transition temperature of more than 3°C [Fig. 4(B)]. By contrast, the His138Gln mutant showed a pattern that was similar to wild-type neuroserpin. Remarkably, His338Gln was the least stable mutant with a transition point that was 5–6°C lower than that of wild-type, His119Gln, and His138Gln neuroserpin at any of the measured pH. Despite these differences in thermostability, all the mutants were active as inhibitors of tPA at pH 7.4 [Fig. 4(C)].
Figure 3.
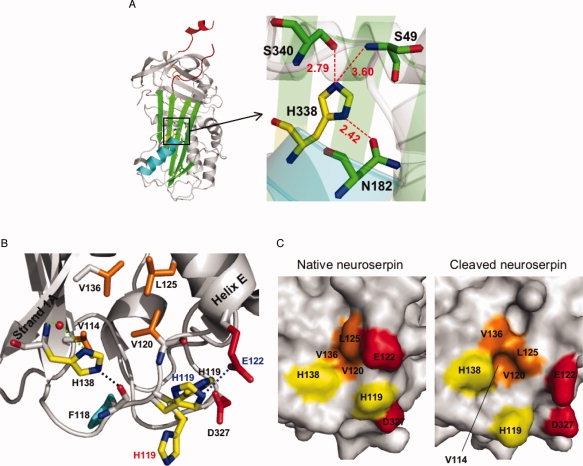
A: Schematic view of native neuroserpin (3FGQ).37 The reactive center loop is shown in red at the upper pole of the protein. The distances (in Å) between His338 and nearby residues in the shutter domain are shown (represented by the box on the left panel). B: Schematic view showing the interactions between strand 1A and helix E at the lower pole of native neuroserpin (3FGQ). The side chains of histidines (His138 and His119: yellow), the hydrophobic residues (Val114, Val120, Leu125, and Val136: orange), Asp327 (red), and Phe118 (cyan) are highlighted. His138 interacts with a water molecule (red sphere) by two hydrogen bonds (green dotted line), and the distance between NE2-His138 and O=C—Phe118 (black dotted line) is 2.50 Å. The distance between ND1-His119 and OD2-Asp327 is 2.64 Å. There is a π–π stacking interaction between the imidazole ring of His119 and O=C—Val120. For comparison, reactive loop-cleaved neuroserpin (3F02)42 and native neuroserpin obtained from different crystallizing conditions (3F5N)42 are superimposed, and His119 and Glu122 are shown with red (3F02) or blue letters (3F5N). The distance between NE2-His119 and OE1-Glu122 in 3F5N (blue dotted line) is 3.09 Å. C: Molecular surface of native neuroserpin (3FGQ: left) and reactive loop-cleaved neuroserpin (3F02: right). The side chains of His138, His119, Glu122, Asp327 and the hydrophobic residues (Val114, Val120, Leu125, and Val136) are highlighted with the same color as in panel B. Val114 is fully buried in native neuroserpin. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 4.
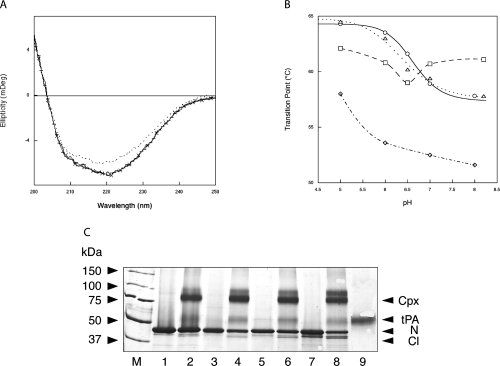
A: Far UV spectra of wild-type neuroserpin, His119Gln, His138Gln, and His338Gln at pH 7.0. Wild-type neuroserpin (solid line), His119Gln (crossed line), His138Gln (dashed line), and His338Gln (dotted line). B: Transition temperature of wild-type neuroserpin, His119Gln, His138Gln, and His338Gln at different pH. Wild-type neuroserpin (O), His119Gln (□), His138Gln (Δ), and His338Gln (◊). The transition temperature for the different pH values was calculated as described in the Results section. N = 3 for all experiments. C: Complex formation between wild-type and mutant neuroserpin and tPA at pH 7.4 analyzed by 10% w/v SDS-PAGE. Lane M, molecular mass markers; Lane 1, wild-type neuroserpin; lane 2, wild-type neuroserpin with tPA at pH 7.4; lane 3, His119Gln neuroserpin; lane 4, His119Gln neuroserpin with tPA at pH 7.4; lane 5, His138Gln neuroserpin; lane 6, His138Gln neuroserpin with tPA at pH 7.4; lane 7, His338Gln neuroserpin; lane 8, His338Gln with tPA at pH 7.4; lane 9, tPA. N, intact neuroserpin; Cl, reactive loop-cleaved neuroserpin; Cpx, the complex between neuroserpin and tPA. All lanes contain 1-2 μg of protein.
The polymerization of these mutants was then assessed over a range of pH values by nondenaturing PAGE. None of the mutants formed polymers at pH 6.5 or below. However, there was a marked difference between the neuroserpin mutants at pH values greater than 7.0. His338Gln neuroserpin did not polymerize faster than the wild-type protein at pH 7.0 or 8.0 [Figs. 1(A) and 5(A)], despite being less stable when assessed by thermal stability. Conversely, His119Gln neuroserpin, which is more stable than wild-type protein at pH 7.0 and 8.0, polymerizes far less readily, with no polymers appearing before 45 min at pH 7.0 and before 15 min at pH 8.0 [Fig. 5(B)]. Most striking was the His138Gln neuroserpin mutant, with no polymerization at pH 7.0, and the appearance of polymers at pH 8.0 but at a rate that was far slower than that of wild-type neuroserpin [Figs. 1(A) and 5(C)].
Figure 5.
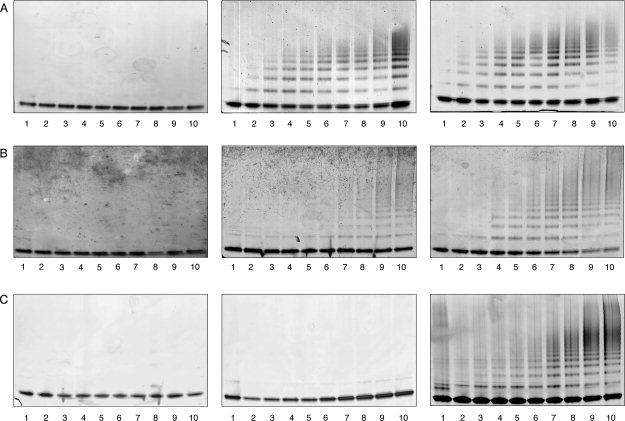
Neuroserpin mutants were incubated at 45°C and 0.4 mg/mL and aliquots taken over time were analyzed by 3-12% w/v gradient nondenaturing PAGE. All lanes contained 1.2 μg of protein. A: Left: Polymerization of His338Gln neuroserpin at pH 6.0. Middle: Polymerization of His338Gln neuroserpin at pH 7.0. Right: Polymerization of His338Gln neuroserpin at pH 8.0. B: Left: Polymerization of His119Gln neuroserpin at pH 6.0. Middle: Polymerization of His119Gln neuroserpin at pH 7.0. Right: Polymerization of His119Gln neuroserpin at pH 8.0. C: Left: Polymerization of His138Gln neuroserpin at pH 6.0. Middle: Polymerization of His138Gln neuroserpin at pH 7.0. Right: Polymerization of His138Gln neuroserpin at pH 8.0. Lanes 1-10 correspond to 0, 5, 10, 15, 20, 30, 45, 60, 120, and 180 min.
Discussion
pH has been shown to affect the propensity of several serpins to form polymers.30–33 Changes in pH had a striking effect on the stability of neuroserpin, as evidenced by the melting temperature but little effect on overall structure (Fig. 2). There was a marked effect on the rate of polymerization of wild-type neuroserpin (Fig. 1). These data contrast with the results of α1-antitrypsin30,38,39 and antithrombin,32 where a decrease in pH was accompanied by a reduction in stability. In the case of α1-antitrypsin and antithrombin, the results may be explained by the effect of pH on His334 in the shutter domain [Fig. 3(A)], as the expansion and opening of β-sheet A is dependent on a network of hydrogen bonds centered on this residue.32 Indeed, if His334 is replaced by an Ala or a Ser residue in α1-antitrypsin, there is increased polymerization and a decrease in Tm when compared with the wild-type protein.32
There is a homologous histidine residue at position 338 in neuroserpin [Fig. 3(A)]. However, our data show that protonation/deprotonation of His338 is not stabilizing neuroserpin at acidic pH as is the case with α1-antitrypsin. Indeed, the Tm values of His338Gln neuroserpin are lower than those of the wild-type protein at any pH between 5.0 and 8.0. The polymerization of His338Gln neuroserpin was similar to that of the wild-type protein at pH values between 6.0 and 8.0 [Figs. 1(A) and 5(A)], and like wild-type neuroserpin this mutant showed a higher Tm value at acidic than at neutral pH [Fig. 4(B)]. This difference between neuroserpin and other serpins can be explained by a change in hydrogen bonding in the shutter region. Our recent crystal structure of neuroserpin showed that the shutter region centered around His338 is different from that in α1-antitrypsin and antithrombin [Fig. 3(A)].37 Indeed, the crystal structures of both α1-antitrypsin (1QLP)40 and antithrombin (1E04)41 show that His334 forms two hydrogen bonds with Asn186 and Ser53 at neutral pH. However, at acidic pH the hydrogen bond between His334 and Ser53 is broken by protonation of His334, and so α1-antitrypsin and antithrombin become unstable.32,40 In contrast, in neuroserpin only one hydrogen bond is formed at neutral pH, between His338 and Asn182. The distance between His334 and Ser49 (3.60 Å) is too great to allow an additional hydrogen bond between these residues. At acidic pH, His338 is protonated and forms an extra hydrogen bond with Ser340. However, both His338 and Ser340 are in strand 5A, thus the hydrogen bond between them is less important for opening the shutter region than the hydrogen bond between His and Ser53 in antithrombin and α1-antitrypsin. Thus, neuroserpin is not destabilized at acidic pH. The stabilization of neuroserpin at acidic pH is therefore likely to be due to other His residues.
Neuroserpin has two His residues (His119 and His138) at the lower pole of the molecule [Fig. 3(B)] where there is no histidine in α1-antitrypsin and antithrombin.37,40–42 Interestingly, the two His residues are very close to a hydrophobic pocket composed of Val114, Val120, Leu125, and Val136 between strand 1A and helix E [Fig. 3(B)]. This pocket is important in the control of opening of β-sheet A in PAI-1 and tengpin.43,44 Tengpin has two homologous residues in the hydrophobic pocket (tengpin/neuroserpin: Leu159/Leu125 and Ile170/Val136), and the N-terminal tail of tengpin makes contacts with the two residues and covers the hydrophobic patch. Exposure of this region by deletion of the N-terminal tail results in opening of β-sheet A, intramolecular insertion of the reactive loop, and ready conversion from the active monomer to the latent conformer of tengpin.4,44 The crystal structure of the native neuroserpin monomer shows that His119 can form a hydrogen bond with Asp327 when the imidazole ring is protonated at acidic pH.37 Another crystal structure of native neuroserpin reported by Ricagno et al.42 shows that His119 can also form a hydrogen bond with Glu122 [Fig. 3(B)]. H-NE2-Gln119 can form a hydrogen bond with Glu122 and/or Asp327 at any pH between 5.0 and 8.0, and thus His119Gln displays a constant Tm value at the pH range tested and forms fewer polymers at pH 7.0. The lower Tm value of His119Gln than wild-type neuroserpin at acidic pH might be explained by the loss of van der Waals interactions or the loss of a π–π stacking interaction between the imidazole ring of His119 and O=C—Val120. These interactions will contribute to reduce flexibility of His119 and helix E and may protect the hydrophobic pocket from exposure. The crystal structure of the cleaved form of neuroserpin simulates a situation in which the side chain of His119 moves away from the two acidic residues.42 In cleaved neuroserpin, His119 bends down, helix E is shifted, and the hydrophobic pocket is more open than in the native form [Fig. 3(C)]. Polymerization of His119Gln at pH 7.0 is likely to be due to a destabilization of His138 or the sum of effects from other His residues.
The data from the His138Gln neuroserpin mutant support this mechanism. His138Gln neuroserpin shows essentially the same thermostability as the wild-type protein at the pH range 5.0–8.0, indicating that this mutation does not affect the stability of the whole molecule. The crystal structure of native neuroserpin shows that one NH of the side chain of His138 forms a hydrogen bond with an oxygen atom of a water molecule [Fig. 3(B)]. When another N atom in the imidazole ring is protonated at acidic pH, a hydrogen bond can be formed with O=C—Phe118. This hydrogen bond will stabilize His138, thereby reducing the exposure of Val114 and Val136. Indeed, in cleaved neuroserpin, where His138 is shifted outside of the molecule as a result in the movement of strand 1A, Val114 and Val136 are more exposed than in native neuroserpin [Fig. 3(C)]. However, H-NE2-Gln138 can form a hydrogen bond with O=C—Phe118 at any pH between 5.0 and 8.0, thus Gln138 is not readily shifted and so prevents polymerization at pH 7.0. Polymerization of His338Gln at pH 8.0 is likely to be due to a destabilization of His138. Taken together, these data suggest the following mechanism: at pH 5.0–6.0 His119 and His138 are protonated, the hydrophobic pocket is less exposed than at pH 7.0-8.0, and thus neuroserpin is stabilized.
Neuroserpin also has His residues at positions 74 (helix C), 374 (loop between strands 1C and 4B), 396, and 405 (C-terminus). His74 has an equivalent position in α1-antitrypsin (His73) and is therefore unlikely to contribute to the pH effect observed with neuroserpin. None of the other histidines in neuroserpin is in locations likely to influence the movement of β-sheet A.37 The stabilization of neuroserpin at low pH may be physiologically relevant as neuroserpin is stored in the dense-core vesicles of cells that have a pH between 5.0 and 6.0, before its release in the extracellular environment.20,34–36 At neutral pH, the instability of neuroserpin might contribute to its faster clearance, either by forming unstable complexes with tPA7,45 or by internalization by the LDL receptor-related protein.46
In conclusion, this study showed the importance of the interaction between strand 1A and helix E in the polymerization of neuroserpin. The switching system for opening β-sheet A by changing the interactions at this site may be a common mechanism evolved by other serpins.43,44
Materials and Methods
Materials
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Hitchin, UK), and oligonucleotides were synthesized by MWG-Biotech AG (Ebersberg, Germany). The QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA). The expression vectors pQE81L and Ni-NTA agarose were from Qiagen (Crawley, UK), HiTrap Q HP was from GE Healthcare (Little Chalfont, UK), and the tPA substrate S-2288 (H-d-Ile-Pro-Arg-para-nitroanilide) was from Chromogenix (Quadratech, Epsom, UK). 1,5-Dansyl-Glu-Gly-Arg-chloromethylketone and tissue plasminogen activator (tPA) were from Calbiochem (Merck Biosciences, Nottingham, UK). Ampicillin, kanamycin, and isopropyl-ß-d-thiogalactopyranoside were from Melford Laboratories (Ipswich, UK).
Expression and purification of recombinant proteins
Histidines mutants were prepared by mutating His119, His138, His338, or His374 in the cDNA of wild-type neuroserpin to glutamine using the QuikChange site-directed mutagenesis kit. The genes were fully sequenced. Recombinant proteins were expressed and purified as described previously.7,22
Assays of complex formation with tPA
Wild-type or mutant neuroserpin was incubated at a 3:1 ratio with tPA (1.7 × 10−6 M) at 25°C as described previously.7,47 The proteins were separated by SDS-PAGE and visualized by Coomassie blue.
Circular dichroism
CD experiments were performed using a JASCO J-810 spectropolarimeter in 20 mM sodium phosphate buffer at different pH. Thermal unfolding experiments were performed by monitoring the CD signal at 216 nm between 25 and 95°C using a heating rate of 1°C/min and a protein concentration of 0.4 mg/mL. The second derivative of the resulting data was used to calculate the inflection point of the transition and hence the Tm.30
Assessment of the effect of pH on the polymerization of wild-type and mutant neuroserpin
The polymerization of wild-type or mutant neuroserpin was assessed by nondenaturing PAGE in 20 mM sodium phosphate, 150 mM NaCl over a range of pH values. The proteins were incubated at 0.4 mg/mL and 45°C. Aliquots were taken over time, and 2 μg of protein was separated by 3–12% w/v gradient nondenaturing PAGE and visualized by silver staining.
Glossary
Abbreviations:
- CD
circular dichroism
- FENIB
familial encephalopathy with neuroserpin inclusion bodies
- PAGE
polyacrylamide gel electrophoresis
- PAI-1
plasminogen activator inhibitor 1
- tPA
tissue-plasminogen activator.
References
- 1.Silverman GA, Bird PI, Carrell RW, Coughlin PB, Gettins PG, Irving JI, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins: evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 2.Galliciotti G, Sonderegger P. Neuroserpin. Front Biosci. 2006;11:33–45. doi: 10.2741/1778. [DOI] [PubMed] [Google Scholar]
- 3.Miranda E, Lomas DA. Neuroserpa serpin to think about. Cell Mol Life Sci. 2006;63:709–722. doi: 10.1007/s00018-005-5077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huntington JA. Shape-shifting serpins—advantages of a mobile mechanism. Trends Biochem Sci. 2006;31:427–435. doi: 10.1016/j.tibs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Hastings GA, Coleman TA, Haudenschild CC, Stefansson S, Smith EP, Barthlow R, Cherry S, Sandkvist M, Lawrence DA. Neuroserpin, a brain-associated inhibitor of tissue plasminogen activator is localized primarily in neurons. Implications for the regulation of motor learning and neuronal survival. J Biol Chem. 1997;272:33062–33067. doi: 10.1074/jbc.272.52.33062. [DOI] [PubMed] [Google Scholar]
- 6.Krueger SR, Ghisu GP, Cinelli P, Gschwend TP, Osterwalder T, Wolfer DP, Sonderegger P. Expression of neuroserpin, an inhibitor of tissue plasminogen activator, in the developing and adult nervous system of the mouse. J Neurosci. 1997;17:8984–8996. doi: 10.1523/JNEUROSCI.17-23-08984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belorgey D, Crowther DC, Mahadeva R, Lomas DA. Mutant neuroserpin (S49P) that causes familial encephalopathy with neuroserpin inclusion bodies is a poor proteinase inhibitor and readily forms polymers in vitro. J Biol Chem. 2002;277:17367–17373. doi: 10.1074/jbc.M200680200. [DOI] [PubMed] [Google Scholar]
- 8.Berger P, Kozlov SV, Cinelli P, Kruger SR, Vogt L, Sonderegger P. Neuronal depolarization enhances the transcription of the neuronal serine protease inhibitor neuroserpin. Mol Cell Neurosci. 1999;14:455–467. doi: 10.1006/mcne.1999.0804. [DOI] [PubMed] [Google Scholar]
- 9.Yepes M, Lawrence DA. New functions for an old enzyme: nonhemostatic roles for tissue-type plasminogen activator in the central nervous system. Exp Biol Med. 2004;229:1097–1104. doi: 10.1177/153537020422901103. [DOI] [PubMed] [Google Scholar]
- 10.Docagne F, Nicole O, Marti HH, MacKenzie ET, Buisson A, Vivien D. Transforming growth factor-ß1 as a regulator of the serpins/t-PA axis in cerebral ischemia. FASEB J. 1999;13:1315–1324. doi: 10.1096/fasebj.13.11.1315. [DOI] [PubMed] [Google Scholar]
- 11.Yepes M, Sandkvist M, Wong MK, Coleman TA, Smith E, Cohan SL, Lawrence DA. Neuroserpin reduces cerebral infarct volume and protects neurons from ischemia-induced apoptosis. Blood. 2000;96:569–576. [PubMed] [Google Scholar]
- 12.Cinelli P, Madani R, Tsuzuki N, Vallet P, Arras M, Zhao CN, Osterwalder T, Rulicke T, Sonderegger P. Neuroserpin, a neuroprotective factor in focal ischemic stroke. Mol Cell Neurosci. 2001;18:443–457. doi: 10.1006/mcne.2001.1028. [DOI] [PubMed] [Google Scholar]
- 13.Yepes M, Sandkvist M, Coleman TA, Moore E, Wu JY, Mitola D, Bugge TH, Lawrence DA. Regulation of seizure spreading by neuroserpin and tissue-type plasminogen activator is plasminogen-independent. J Clin Invest. 2002;109:1571–1578. doi: 10.1172/JCI14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebeurrier N, Liot G, Pascual Lopez-Atalaya J, Orset C, Fernandez-Monreal M, Sonderegger P, Ali C, Vivien D. The brain-specific tissue-type plasminogen activator inhibitor, neuroserpin, protects neurons against excitotoxicity both in vitro and in vivo. Mol Cell Neurosci. 2005;30:552–558. doi: 10.1016/j.mcn.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Kinghorn KJ, Crowther DC, Sharp LK, Nerelius C, Davis RL, Chang HT, Green C, Gubb DC, Johansson J, Lomas DA. Neuroserpin binds Aβ and is a neuroprotective component of amyloid plaques in Alzheimer disease. J Biol Chem. 2006;281:29268–29277. doi: 10.1074/jbc.M600690200. [DOI] [PubMed] [Google Scholar]
- 16.Fabbro S, Seeds NW. Plasminogen activator activity is inhibited while neuroserpin is upregulated in the Alzheimer disease brain. J Neurochem. 2009;109:303–315. doi: 10.1111/j.1471-4159.2009.05894.x. [DOI] [PubMed] [Google Scholar]
- 17.Davis RL, Shrimpton AE, Holohan PD, Bradshaw C, Feiglin D, Collins GH, Sonderegger P, Kinter J, Becker LM, Lacbawan F, Krasnewich D, Muenke M, Lawrence DA, Yerby MS, Shaw CM, Gooptu B, Elliott PR, Finch JT, Carrell RW, Lomas DA. Familial dementia caused by polymerization of mutant neuroserpin. Nature. 1999;401:376–379. doi: 10.1038/43894. [DOI] [PubMed] [Google Scholar]
- 18.Davis RL, Shrimpton AE, Carrell RW, Lomas DA, Gerhard L, Baumann B, Lawrence DA, Yepes M, Kim TS, Ghetti B, Piccardo P, Takao M, Lacbawan F, Muenke M, Sifers RN, Bradshaw CB, Kent PF, Collins GH, Larocca D, Holohan PD. Association between conformational mutations in neuroserpin and onset and severity of dementia (erratum 2002, 360, 1102) Lancet. 2002;359:2242–2247. doi: 10.1016/S0140-6736(02)09293-0. [DOI] [PubMed] [Google Scholar]
- 19.Miranda E, Römisch K, Lomas DA. Mutants of neuroserpin that cause dementia accumulate as polymers within the endoplasmic reticulum. J Biol Chem. 2004;279:28283–28291. doi: 10.1074/jbc.M313166200. [DOI] [PubMed] [Google Scholar]
- 20.Miranda E, MacLeod I, Davies MJ, Perez J, Römisch K, Crowther DC, Lomas DA. The intracellular accumulation of polymeric neuroserpin explains the severity of the dementia FENIB. Hum Mol Genet. 2008;17:1527–1539. doi: 10.1093/hmg/ddn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takasawa A, Kato I, Takasawa K, Ishii Y, Yoshida T, Shehata MH, Kawaguchi H, Mohafez OM, Sasahara M, Hiraga K. Mutation-, aging-, and gene dosage-dependent accumulation of neuroserpin (G392E) in endoplasmic reticula and lysosomes of neurons in transgenic mice. J Biol Chem. 2008;283:35606–35613. doi: 10.1074/jbc.M804125200. [DOI] [PubMed] [Google Scholar]
- 22.Belorgey D, Sharp LK, Crowther DC, Onda M, Johansson J, Lomas DA. Neuroserpin Portland (Ser52Arg) is trapped as an inactive intermediate that rapidly forms polymers: implications for the epilepsy seen in the dementia FENIB. Eur J Biochem. 2004;271:3360–3367. doi: 10.1111/j.1432-1033.2004.04270.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 24.Davis RL, Holohan PD, Shrimpton AE, Tatum AH, Daucher J, Collins GH, Todd R, Bradshaw C, Kent P, Feiglin D, Rosenbaum A, Yerby MS, Shaw CM, Lacbawan F, Lawrence DA. Familial encephalopathy with neuroserpin inclusion bodies. Am J Pathol. 1999;155:1901–1913. doi: 10.1016/S0002-9440(10)65510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gourfinkel-An I, Duyckaerts C, Camuzat A, Meyrignac C, Sonderegger P, Baulac M, Brice A. Clinical and neuropathologic study of a French family with a mutation in the neuroserpin gene. Neurology. 2007;69:79–83. doi: 10.1212/01.wnl.0000265052.99144.b5. [DOI] [PubMed] [Google Scholar]
- 26.Coutelier M, Andries S, Ghariani S, Dan B, Duyckaerts C, van Rijckevorsel K, Raftopoulos C, Deconinck N, Sonderegger P, Scaravilli F, Vikkula M, Godfraind C. Neuroserpin mutation causes electrical status epilepticus of slow-wave sleep. Neurology. 2008;71:64–66. doi: 10.1212/01.wnl.0000316306.08751.28. [DOI] [PubMed] [Google Scholar]
- 27.Onda M, Belorgey D, Sharp LK, Lomas DA. Latent S49P neuroserpin forms polymers in the dementia familial encephalopathy with neuroserpin inclusion bodies. J Biol Chem. 2005;280:13735–13741. doi: 10.1074/jbc.M413282200. [DOI] [PubMed] [Google Scholar]
- 28.Lomas DA, Mahadeva R. α1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest. 2002;110:1585–1590. doi: 10.1172/JCI16782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belorgey D, Hägglöf P, Karlsson-Li S, Lomas DA. Protein misfolding and the serpinopathies. Prion. 2007;1:15–20. doi: 10.4161/pri.1.1.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dafforn TR, Mahadeva R, Elliott PR, Sivasothy P, Lomas DA. A kinetic mechanism for the polymerization of α1-antitrypsin. J Biol Chem. 1999;274:9548–9555. doi: 10.1074/jbc.274.14.9548. [DOI] [PubMed] [Google Scholar]
- 31.Zhou A, Faint R, Charlton P, Dafforn TR, Carrell RW, Lomas DA. Polymerization of plasminogen activator inhibitor-1. J Biol Chem. 2001;276:9115–9122. doi: 10.1074/jbc.M010631200. [DOI] [PubMed] [Google Scholar]
- 32.Zhou A, Stein PE, Huntington JA, Carrell RW. Serpin polymerization is prevented by a hydrogen bond network that is centered on His-334 and stabilized by glycerol. J Biol Chem. 2003;278:15116–15122. doi: 10.1074/jbc.M211663200. [DOI] [PubMed] [Google Scholar]
- 33.Stanley P, Serpell LC, Stein PE. Polymerization of human angiotensinogen: insights into its structural mechanism and functional significance. Biochem J. 2006;400:169–178. doi: 10.1042/BJ20060444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill RM, Parmar PK, Coates LC, Mezey E, Pearson JF, Birch NP. Neuroserpin is expressed in the pituitary and adrenal glands and induces the extension of neurite-like processes in AtT-20 cells. Biochem J. 2000;345:595–601. [PMC free article] [PubMed] [Google Scholar]
- 35.Parmar PK, Coates LC, Pearson JF, Hill RM, Birch NP. Neuroserpin regulates neurite outgrowth in nerve growth factor-treated PC12 cells. J Neurochem. 2002;82:1406–1415. doi: 10.1046/j.1471-4159.2002.01100.x. [DOI] [PubMed] [Google Scholar]
- 36.Ishigami S, Sandkvist M, Tsui F, Moore E, Coleman TA, Lawrence DA. Identification of a novel targeting sequence for regulated secretion in the serine protease inhibitor neuroserpin. Biochem J. 2007;402:25–34. doi: 10.1042/BJ20061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takehara S, Onda M, Zhang J, Nishiyama M, Yang X, Mikami B, Lomas DA. The 2.1-Å crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability. J Mol Biol. 2009;388:11–20. doi: 10.1016/j.jmb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Devlin GL, Chow MK, Howlett GJ, Bottomley SP. Acid denaturation of α1-antitrypscharacterization of a novel mechanism of serpin polymerization. J Mol Biol. 2002;324:859–870. doi: 10.1016/s0022-2836(02)01088-4. [DOI] [PubMed] [Google Scholar]
- 39.Boudier C, Bousquet J-A, Schauinger S, Michels B, Bieth JG. Reversible inactivation of serpins at acidic pH. Arch Biochem Biophys. 2007;466:155–163. doi: 10.1016/j.abb.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Elliott PR, Pei XY, Dafforn TR, Lomas DA. Topography of a 2.0Å structure of α1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000;9:1274–1281. doi: 10.1110/ps.9.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson DJ, Langdown J, Li W, Luis SA, Baglin TP, Huntington JA. Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation. J Biol Chem. 2006;281:35478–35486. doi: 10.1074/jbc.M607204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricagno S, Caccia S, Sorrentino G, Antonini G, Bolognesi M. Human neuroserpstructure and time-dependent inhibition. J Mol Biol. 2009;388:109–121. doi: 10.1016/j.jmb.2009.02.056. [DOI] [PubMed] [Google Scholar]
- 43.Zhou A, Huntington JA, Pannu NS, Carrell RW, Read RJ. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat Struct Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q, Buckle AM, Law RH, Pearce MC, Cabrita LD, Lloyd GJ, Irving JA, Smith AI, Ruzyla K, Rossjohn J, Bottomley SP, Whisstock JC. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 2007;8:658–663. doi: 10.1038/sj.embor.7400986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barker-Carlson K, Lawrence DA, Schwartz BS. Acyl-enzyme complexes between tissue-type plasminogen activator and neuroserpin are short-lived in vitro. J Biol Chem. 2002;277:46852–46857. doi: 10.1074/jbc.M207740200. [DOI] [PubMed] [Google Scholar]
- 46.Makarova A, Mikhailenko I, Bugge TH, List K, Lawrence DA, Strickland DK. The low density lipoprotein receptor-related protein modulates protease activity in the brain by mediating the cellular internalization of both neuroserpin and neuroserpin-tissue-type plasminogen activator complexes. J Biol Chem. 2003;278:50250–50258. doi: 10.1074/jbc.M309150200. [DOI] [PubMed] [Google Scholar]
- 47.Renatus M, Engh RA, Stubbs MT, Huber R, Fischer S, Kohnert U, Bode W. Lysine 156 promotes the anomalous proenzyme activity of tPA: X-ray crystal structure of single-chain human tPA. EMBO J. 1997;16:4797–4805. doi: 10.1093/emboj/16.16.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]


