Abstract
Successive activation of Wnt4 and Notch2 generates nephrons from the metanephric mesenchyme. Mesenchymal-to-epithelial transition requires Wnt4, and normal development of the proximal nephron (epithelia of glomeruli and proximal tubules) requires Notch2. It is unknown, however, whether Notch2 dictates the fate of the proximal nephron directly. Here, we generated a mutant strain of mice with activated Notch2 in Six2-containing nephron progenitor cells of the metanephric mesenchyme. Notch2 activation did not skew the cell fate toward the proximal nephron but resulted in severe kidney dysgenesis and depletion of Six2-positive progenitors. We observed ectopic expression of Wnt4 and premature tubule formation, similar to the phenotype of Six2-deficient mice. Activation of Notch2 in the progenitor cells suppressed Pax2, an upstream regulator of Six2, possibly through Hesr genes. Taken together, these data suggest that a positive feedback loop exists between Notch2 and Wnt4, and that Notch2 stabilizes, rather than dictates, nephron fate by shutting down the maintenance of undifferentiated progenitor cells, thereby depleting this population.
The mammalian kidney, the metanephros, is formed by reciprocally inductive interactions between two precursor tissues; namely, the metanephric mesenchyme and the ureteric bud. Glial cell line-derived neurotrophic factor (Gdnf) is secreted by the mesenchyme and attracts the ureteric bud toward the mesenchyme.1–3 The attracted ureteric bud in turn secretes Wnt9b and induces the mesenchyme.4 Upon this induction by the ureteric bud, mesenchymal cells secrete Wnt4, which plays an essential role in the mesenchymal-to-epithelial transition.5,6 The mesenchyme sequentially forms condensates, renal vesicles, C- and S-shaped bodies, and terminal epithelia of the glomeruli and renal tubules.
A previous report retrospectively suggested the presence of multipotent cells in embryonic kidneys, after demonstrating that cells in several portions of the nephron were derived from single stem cells by using LacZ gene transduction with a retrovirus into single mesenchymal cells.7 We prospectively proved this concept by establishing a novel colony assay. When we plated dissociated mesenchymal cells at a low density onto feeder cells stably expressing Wnt4, single cells formed colonies consisting of several types of epithelial cells that are found in glomeruli and renal tubules.8 We further found that only cells strongly expressing Sall1, a zinc finger nuclear factor expressed in the metanephric mesenchyme and essential for kidney development, formed colonies.8,9 Therefore, multipotent progenitors of nephron epithelial cells do exist and reside in the Sall1-high population of the mesenchyme.10
Nephron progenitors have also been shown to express another transcription factor, Six2. Kobayashi et al.11 marked Six2-positive cells using Six2Cre, a mouse strain expressing Cre recombinase under the control of the Six2 promoter, and they demonstrated that Six2-positive mesenchyme does indeed give rise to epithelia of the glomeruli and renal tubules of the nephron in vivo. Because Six2 is expressed in Sall1-high mesenchymal cells, the Sall1-high and Six2-positive mesenchyme may represent a nephron progenitor population in the embryonic kidney.12 Six2 is expressed in the undifferentiated mesenchyme that caps the ureteric bud (dorsal side of the ureteric bud), and its expression pattern is reciprocal to that of Wnt4, which is expressed near the ureteric stalk (ventral side). Wnt4 is upregulated in the dorsal portion of the mesenchyme in Six2-deficient mice, and the mice exhibit ectopic and premature tubulogenesis,13 suggesting that Six2 is required to maintain the undifferentiated progenitor population by opposing Wnt4-mediated epithelialization.
Six2 expression is regulated by the Pax2/Eya1/Hox11 complex, and the binding sites for this complex are present in the Six2 proximal promoter, which is essential for Six2 expression in the mesenchyme in vivo.14 Six2 expression is reduced in mice lacking each of the complex components, whereas crossing of these mutant mice revealed physiologic significance for the interactions among the complex components.14,15 Therefore, the Pax2/Eya1/Hox11 complex appears to be a bona fide regulator of Six2 in vivo. This complex also regulates the expression of Gdnf, which is essential for ureteric bud attraction.14
Six2-positive nephron progenitors differentiate into epithelia upon Wnt4 stimulation, but a fate decision is required for further differentiation toward glomerular podocytes or proximal and distal renal tubules. Notch2 is required for the differentiation of proximal nephron structures (podocytes and proximal tubules), because mesenchyme-specific Notch2 deletion in mice leads to impaired formation of these proximal structures.16–18 Although proximodistal polarity is still initiated in the absence of Notch2, Notch2 is essential for the final establishment of the proximal nephron fates. In humans, Notch2 haploinsufficiency causes Alagille syndrome, which is associated with renal abnormalities.19 Interestingly, endogenous Notch1 cannot compensate for Notch2 deficiency in mice, although Notch1 activity is present in the kidney, and increases in its activity enhance the formation of proximal tubules at the expense of podocytes and distal nephrons.18
To gain insights into Notch2 functions, we generated mice with activated Notch2 in Six2-positive nephron progenitors. Interestingly, Notch2 activation did not specify the proximal nephron fate but instead caused progenitor depletion by shutting down the Six2-mediated program for progenitor maintenance.
Results
Generation of a Mouse Strain Allowing Notch2 Activation in a Cre Recombinase-Dependent Manner
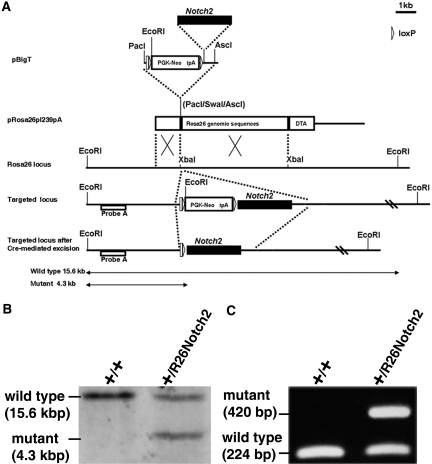
By using homologous recombination in embryonic stem (ES) cells, we generated R26Notch2 mice containing a controllable Notch2 cassette in the Rosa26 locus that confers ubiquitous expression.20 The Flag-tagged intracellular domain of Notch2 was introduced into the pBigT vector,21 which contains PGK-neo and three SV40 terminator sequences flanked by two loxP sites (Figure 1A). The pBigT vector containing FlagNotch2 was further inserted into a modified pRosa26-1 vector, which contains Rosa26 genomic sequences.21 Finally, the entire fragment was introduced into murine ES cells by electroporation. Southern blotting analysis and genomic PCR confirmed the correct homologous recombination, and the resultant clones were used to generate germline chimeras and heterozygous mice (Figure 1, B and C). All of the heterozygous mice were apparently normal and fertile. These mice did not express FlagNotch2 (Supplementary Figure 1), suggesting that there was no leaked expression of Notch2 in the absence of Cre recombinase activity.
Figure 1.
R26Notch2 mice are generated. (A) Targeting strategy of Flag-Notch2 in the Rosa26 locus. A Flag-Notch2 fragment is inserted into the pBigT vector containing an adenovirus splice acceptor sequence followed by a PGK-neo cassette and a tpA stop sequence flanked by two loxP sites. The resultant plasmid is inserted into pRosa26pl239pA (a modified pRosa26-1 vector) and subsequently integrated into the Rosa26 locus in ES cells by homologous recombination. (B) Targeted ES clones are confirmed by Southern blot analysis. Genomic DNA is digested with EcoRI and hybridized with probe A. (C) PCR amplification of mouse tail DNA.
Overexpression of Notch2 in Embryonic Nephron Progenitors Leads to Severe Dysgenesis
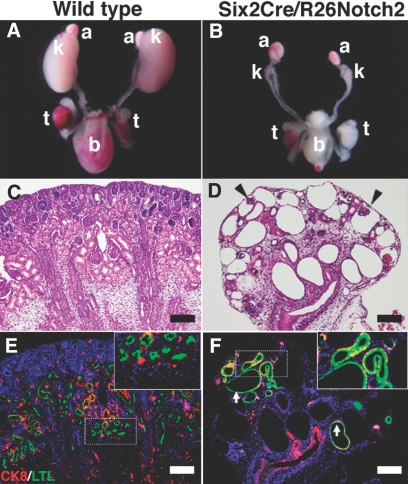
To gain insights into the roles of Notch2 in renal progenitors, we crossed R26Notch2 mice with Six2Cre mice specifically expressing Cre recombinase in renal progenitor populations in the metanephric mesenchyme (kindly provided by Dr. A.P. McMahon, Harvard University).11 The resulting Six2Cre/R26Notch2 mice were born at Mendelian frequency, but all of them died shortly after birth and displayed severe reductions in their kidney sizes (Figure 2, A and B). Histologic examination revealed that the mutant kidneys contained multiple glomerular cysts, dilated renal tubules, and thin cortexes (Figure 2, C and D). Some of the dilated tubules were positive for a proximal tubule marker, although it was not overproduced as observed for Notch1 activation when the same Cre line was used to ectopically express the intracellular domain of Notch1 in metanephric mesenchyme.18 These results suggest that, unlike Notch1 activation, Notch2 activation does not drive the cell fate toward the proximal nephron but leads to severe dysgenesis.
Figure 2.
Overexpression of Notch2 in embryonic nephron progenitors leads to severe kidney dysgenesis. (A) Kidneys in a newborn wild-type mouse (P0). k, kidney; t, testis; b, bladder; a, adrenal gland. (B) Kidney size is reduced in a newborn Six2Cre/R26Notch2 mouse (P0). (C and D) HE staining of newborn kidneys. Severe dysgenesis is observed in the Six2Cre/R26Notch2 kidney. Arrowheads show glomerular cysts. (E and F) Immunostaining for Lotus tetragonobulus lectin (LTL; green; proximal tubule marker) and cytokeratin 8 (CK8; red; ureteric bud marker). The upper right panels show higher-magnification images of the dotted square regions. They are ×2 of the original panels. Proximal tubules are not overproduced under Notch2 activation. Arrows, dilated proximal tubules. Scale bar = 100 μm.
Notch2 Overexpression Causes Nephron Progenitor Depletion
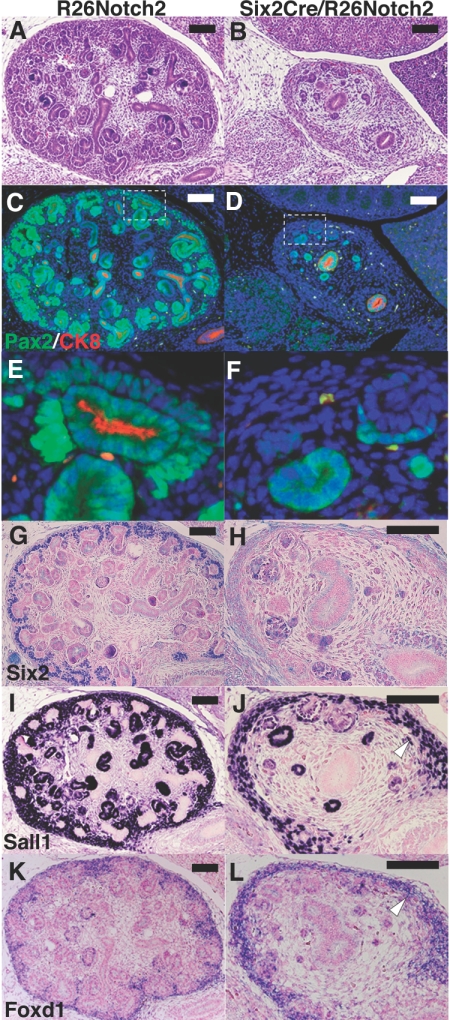
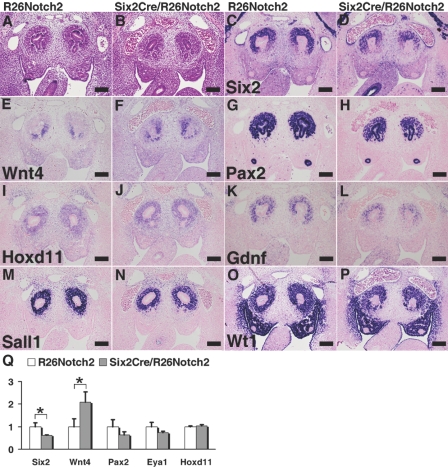
At embryonic day 14.5 (E14.5) during gestation, the mutant kidneys were already reduced in size and showed a thin cortical nephrogenic zone and poor nephron development (Figure 3, A and B). Immunostaining confirmed these findings. Although condensed metanephric mesenchyme (Pax2 positive/cytokeratin negative) surrounded the ureteric buds (Pax2 positive/cytokeratin positive) in the wild-type cortex accompanied by more differentiated Pax2-positive tubules, the mutant kidneys completely lacked the cortical mesenchyme and contained only small numbers of Pax2-positive scattered tubules (Figure 3, C–F). Ureteric branching was also impaired, and no ureteric bud-derived epithelia were observed in the mutant cortex (Figure 3, E and F). Consistent with these observations, the specific expression of Six2 in the nephron progenitors was significantly reduced in the mutant kidneys (Figure 3, G and H). Expression of Sall1, another marker for the progenitors, was also decreased in the mesenchyme, whereas its expression in the stroma was unaffected (Figure 3, I and J). Furthermore, Foxd1, a specific marker for the stroma, was also unaffected (Figure 3, K and L), reflecting the restricted activity of Cre recombinase in the mesenchyme but not in the stroma. These data suggest that Notch2 overexpression in the metanephric mesenchyme leads to nephron progenitor depletion.
Figure 3.
Notch2 overexpression causes nephron progenitor depletion. (A and B) HE staining at E14.5 during gestation. The mutant kidney is reduced in size and shows a thin cortical nephrogenic zone and poor nephron development, compared with the control kidney (R26Notch2). (C and D) Immunostaining for Pax2 (green) and cytokeratin 8 (CK8; red). (E and F) Higher-magnification images of the dotted square regions in panels C and D. They are ×6 of the panels C and D. Pax2-positive/cytokeratin-negative cells (arrowhead) that surround the ureteric bud-derived epithelia (arrow) are missing in the mutant cortex. Ureteric branching is also impaired. (G and H) Six2 immunostaining is significantly reduced in the mutant cortex. (I and J) Immunostaining of Sall1. Mesenchymal expression of Sall1 is reduced in the mutant kidney, whereas its stromal expression is retained (arrowhead). (K and L) In situ hybridization of Foxd1, a stromal marker. Foxd1 continues to be expressed in the mutant kidney (arrowhead). Scale bar = 100 μm.
Notch2 Overexpression Leads to Ectopic Wnt4 Expression and Premature Tubule Formation
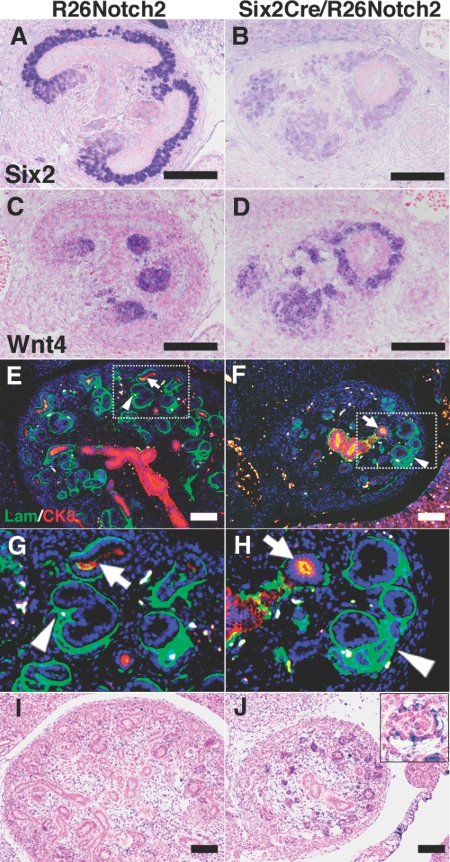
During normal development at E12.5, Six2 was expressed in the dorsal (outer) domain of the mesenchyme against the ureteric buds, whereas Wnt4 was expressed in the ventral (inner) domain to play an essential role in epithelial conversion of the mesenchyme (Figure 4, A and C). In the Notch2-activated mutant mice, Six2 expression was reduced, and Wnt4 was detected in the mesenchyme all around the ureteric buds (Figure 4, B and D), indicating that Wnt4 was ectopically expressed in the dorsal domain, which could accelerate the epithelial conversion of the otherwise undifferentiated nephron progenitors. Indeed, this notion was confirmed by double staining of laminin and cytokeratin at E14.5 (Figure 4, E–H). In the control mice, newly formed tubules of mesenchymal origin (laminin positive/cytokeratin negative) were located in close contact with and ventral to the cortical ureteric buds (laminin positive/cytokeratin positive). In contrast, the tubules in the mutant mice were observed dorsal to poorly branched ureteric buds that failed to develop into the cortex (Figure 4, G and H). Pax2 and cytokeratin staining further confirmed these results (Figure 2, C–F). As expected, all of the mesenchyme-derived epithelia in the mutants, including the renal tubules and glomerular podocytes but not the ureteric buds, overexpressed the intracellular domain of Notch2 (Figure 4, I and J, and Supplementary Figure 2, A and B). Even some of the dilated cystic epithelia continued to express the exogenous Notch2 (Supplementary Figure 2, C and D). These data indicate the occurrence of ectopic and premature epithelialization of the nephron progenitors, which could be the primary cause of the progenitor depletion. These phenotypes are very similar to those of Six2-deficient mice,13 further supporting that Notch2-mediated Six2 suppression is a key molecular event that leads to progenitor depletion.
Figure 4.
Notch2 overexpression leads to ectopic Wnt4 expression and premature tubule formation. (A) Immunostaining of Six2. Six2 is expressed dorsally to the ureteric buds in the control kidney (R26Notch2) at E12.5. (B) Six2 immunostaining is reduced in the Six2Cre/R26Notch2 kidney. (C) In situ hybridization of Wnt4. Wnt4 is expressed ventrally against the ureteric buds in the control kidney at E12.5. (D) Wnt4 expression is not confined to the ventral side of the ureteric buds in the Six2Cre/R26Notch2 kidney. (E) Immunostaining for laminin (Lam; green; epithelial marker) and cytokeratin 8 (CK8; red; ureteric bud marker). Mesenchyme-derived tubules (arrowhead) are formed ventrally to the cortical ureteric bud-derived epithelia (arrow) in the control kidney at E14.5. (F) Ectopic tubules (arrowhead) are observed dorsal to the poorly branched ureteric bud-derived epithelia (arrow). Scale bar = 100 μm. (G and H) Higher-magnification images of the dotted square regions in panels E and F. They are ×3 of the panels E and F (I and J) In situ hybridization of the intracellular domain of Notch2. Note that all of the mesenchyme-derived epithelia in the mutants express Notch2 (J). The upper right panel in J shows a higher-magnification image of glomerular podocytes. I and J are taken at ×100, and the inset is taken at ×400.
Pax2 Is Reduced and Hesr1/Hesr3 Are Increased in Notch2 Activation
Next, we investigated molecules that could be responsible for the Six2 suppression by Notch2. At E11.5, the mutant kidneys were histologically indistinguishable from the control kidneys (Figure 5, A and B). No significant differences in proliferation and apoptosis were observed in the mesenchyme at E11.5 (Supplementary Figure 3). However, Six2 expression was already reduced, and the Wnt4 domain had expanded dorsally (Figure 5, C–F). Pax2, Eya1, and Hox11 are known to bind to the Six2 promoter and directly regulate Six2 expression.14 Under Notch2 activation, the expression of Pax2 in the mesenchyme was reduced, whereas those of Eya1 and Hoxd11 remained unaffected (Figure 5, G–J, and data not shown). Gdnf, another direct target of the Pax2/Eya1/Hox11 complex, was also decreased in the mutant kidneys (Figure 5, K and L), which could explain the reduced branching of the ureteric buds. In contrast, Sall1 and Wt1 were minimally affected (Figure 5, M–P). Quantitative RT-PCR revealed similar tendencies (Figure 5Q). Six2 was reduced and Wnt4 was upregulated. A slight reduction in Pax2 was also observed, despite the fact that it was also expressed in the ureteric buds. There were no significant changes in Eya1 or Hoxd11, both of which were expressed in the mesenchyme. These data suggest that Notch2-dependent Six2 suppression could be mediated by inhibition of Pax2 expression. Other possibilities are described in the Discussion section.
Figure 5.
Expression of Pax2 is impaired under Notch2 activation. (A) HE staining of a control kidney (R26Notch2) at E11.5. The ureteric bud invades into the mesenchyme. (B) HE staining of a Six2Cre/R26Notch2 kidney at E11.5. The mutant kidney is indistinguishable from the control kidney. (C and D) Six2 staining is reduced in the mutant mesenchyme. (E and F) In situ hybridization of Wnt4. The Wnt4 expression domain is expanded dorsally in the mutant kidney. (G and H) Pax2 staining in the mesenchyme is reduced in the mutant kidney, whereas its expression in the ureteric bud is unaffected. (I and J) In situ hybridization of Hoxd11. Hoxd11 expression is unaffected in the mutant kidney. (K and L) In situ hybridization of Gdnf. Gdnf expression is reduced in the mutant kidney. (M and N) Sall1 staining is minimally affected in the mutant kidney. (O and P) Wt1 staining is minimally affected in the mutant kidney. Scale bar = 100 μm. (Q) Quantitative RT-PCR of the E11.5 kidney. The columns represent means ± SD (n = 3). *P < 0.05, R26Notch2 mice versus Six2Cre/R26Notch2 mice.
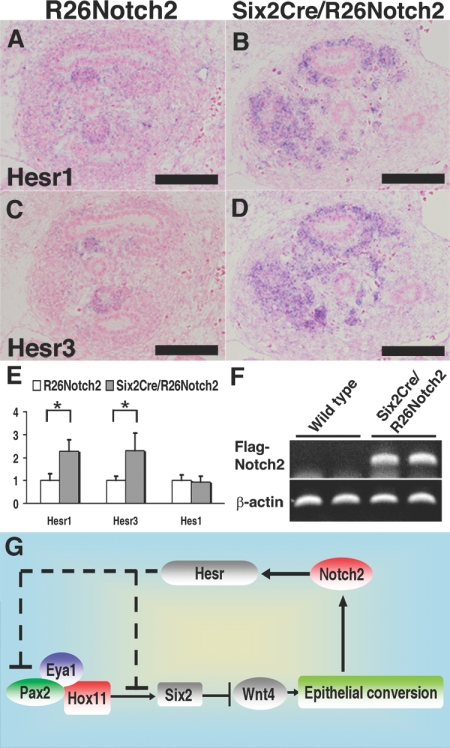
Canonical Notch signals are mediated by the Hes/Hesr gene family members, which suppress the target genes.22 Among the Hes/Hesr genes reported to be expressed in the metanephric mesenchyme,23,24 Hesr1 and Hesr3 were dramatically upregulated under Notch2 activation, as shown by in situ hybridization at E12.5 (Figure 6, A–D). Although not detected by in situ hybridization at E11.5, RT-PCR analyses revealed increases in Hesr1 and Hesr3 at this stage, as well as an increase in the exogenous intracellular domain of Notch2 (Figure 6, E and F), the latter of which is consistent with the activation timing of Six2Cre recombinase.11 Although the Hes1 and Hes5 promoters in the developing kidney are driven by endogenous Notch activity,25 we did not detect significant increases in these gene expressions under Notch2 activation (Figure 6E and data not shown). Therefore, we propose that Notch2 activates Hesr1/Hesr3, which in turn could suppress Pax2, thereby leading to Six2 reduction and ectopic Wnt4 expression (Figure 6G). Alternatively, Hesr proteins may directly inhibit Six2 expression.
Figure 6.
A Notch2-Wnt4-positive feedback loop stabilizes the differentiated state. (A and B) In situ hybridization of Hesr1. Hesr1 expression is significantly increased in the Six2Cre/R26Notch2 kidney at E12.5. (C and D) In situ hybridization of Hesr3. Hesr3 expression is significantly increased in the Six2Cre/R26Notch2 kidney at E12.5. Scale bar = 100 μm. (E) Quantitative RT-PCR of Hesr1 and Hesr3 in the E11.5 kidney. The columns represent means ± SD (n = 3). *P < 0.05, R26Notch2 mice versus Six2Cre/R26Notch2 mice. (F) RT-PCR of the exogenous intracellular domain of Notch2 in E11.5 kidneys from wild-type mice (n = 2) and Six2Cre/R26Notch2 mice (n = 2). (G) Proposed functions of Notch2 for the nephron progenitors. See the Discussion section for details.
Discussion
We have shown that Notch2 activation results in depletion of Six2-positive nephron progenitors, which could arise through premature differentiation toward renal tubules via ectopic Wnt4 activation. Six2 reduction should be a key molecular event in producing these phenotypes, because Six2Cre/R26Notch2 mice phenocopy Six2-deficient mice.13 Six2 expression is regulated by the Pax2/Eya1/Hox11 complex, and among these components, Pax2 was decreased under Notch2 activation, which could reasonably explain the Six2 reduction. Gdnf is another direct target of the Pax2/Eya1/Hox11 complex, and the observed reduction in Gdnf expression under Notch2 activation further supports the decreased activity of the complex.
Canonical Notch signals are mediated by the Hes/Hesr family members, which suppress the target genes.22 These family members either sequester positive basic helix-loop-helix transcription factors that bind to consensus sequences (E boxes) in the target promoters or actively repress the target genes by binding to different consensus sequences (N boxes or C sites) and recruiting the histone deacetylase (HDAC) complex. Among the family genes, Hesr1 and Hesr3 were upregulated under Notch2 activation. In addition, multiple potential N boxes and C sites exist in the proximal promoter of Pax2 but not in that of Hox11 (data not shown), and neither of these promoters contains E boxes. Therefore, we propose that Notch2 activates Hesr1/Hesr3, which could suppress Pax2 possibly by recruiting the HDAC complex, which in turn leads to Six2 reduction. Alternatively, Hesr proteins may directly inhibit Six2 expression, either by modulating the activity of the Pax2/Eya1/Hox11 complex or independently of the complex. It is also possible that the Notch2-mediated increase in Wnt4 secondarily affects the expression of Six2, although Hesr proteins are considered to be repressors, and Hesr-dependent Wnt4 activation has not been reported. To clarify these issues, it will be necessary to examine the binding sites of Hesr proteins and the possible interactions between Hesr proteins and the Pax2/Eya1/Hox11 complex, as well as to cross the activated Notch2 mice with Wnt4 or Hesr mutant mice.
Because Notch2 deficiency results in the absence of the proximal nephrons, endogenous Notch2 is critical for proximal nephron development, either by cell fate specification of the progenitors toward the proximal nephron or by stabilizing the proximal fate determined by other mechanisms. Although the former possibility is attractive, considering the lateral inhibition mechanisms of Notch in a variety of biologic processes, the latter is suggested because the initial proximodistal axis is still established under Notch2 deficiency, which is likely to be mediated by Lim1-dependent mechanisms.18,26 Our observations that Notch2 activation did not lead to proximal fate specification further support this hypothesis. In the mutant mice, Notch2 activation was observed from E11.5 and was detected in all of the mesenchyme-derived epithelia at E14.5. Nonetheless, proximal tubules were not overrepresented. In contrast, Notch1 deletion in the metanephric mesenchyme does not produce any specific phenotypes, whereas Notch1 activation causes overproduction of proximal tubules and reductions in kidney size.18 Cheng et al.18 suggested that Notch1 may be a weaker activator of the target promoters of Notch2, possibly owing to differences in their tertiary structures, and that the level of endogenous Notch1 could be below the threshold required to activate Notch2 targets. In their experiments, Notch1 was inserted into the Rosa26 locus, and the same Six2Cre mouse strain was used. Therefore, our data indicate that even in the overexpressed state, Notch1 and Notch2 may emit overlapping yet different signals in the nephron progenitors. However, side-by-side comparisons are required to exclude any influences of the genetic background.
Nephrons are generated from the metanephric mesenchyme by successive activation of Wnt4 and Notch2. Wnt4 is required for the mesenchymal-to-epithelial transition, whereas Notch2 plays a role in the development of the proximal nephron epithelia.5,27 Our observation that Notch2 activation leads to Wnt4 upregulation suggests the existence of a positive feedback loop (Figure 6G). Once Notch2 is activated, the feedback loop augments Wnt4 expression, which accelerates differentiation and inhibits reversion to the undifferentiated state. Therefore, we propose that Notch2 stabilizes, rather than specifies, the nephron fate by shutting down the undifferentiated progenitor program once directed toward the differentiation pathway. It is important to note that Notch functions are highly context dependent. Although our data support a molecular model of dysregulated Notch2 in nephron progenitors, different mechanisms may apply to endogenous Notch2 signaling in cells that are committed to an epithelial fate. Notch2 overexpression in these nascent epithelial cells would address this point.
For regenerative medicine, it is desirable to be able to manipulate the cell fate specification of progenitors. However, our data suggest that premature Notch2 activation in the nephron progenitors does not serve this purpose. Instead, Notch2 signals could be used to stabilize the cell fate once specified. Therefore, the timing of Notch2 activation would be critical. Delayed activation of Notch2 by crossing R26Notch2 mice with Six2CreER or Wnt4Cre mice would provide information for solving this problem, as well as augmenting our understanding of kidney development.11 In addition, podocyte-specific Notch2 activation should be investigated, because Notch1 activation in this cell type leads to proteinuria and glomerulosclerosis.28,29
Concise Methods
Gene Targeting and Generation of Mutant Mice
The Notch2 fragment used for the experiment encompasses amino acids 1701 to 2470 and does not contain the transmembrane domain or S3 cleavage site but does contain the intact transactivation domain and C-terminal PEST sequences.30 The Flag-tagged intracellular domain of Notch2 was digested with SalI and NotI and inserted into the SalI-NotI sites of pBigT,21 a vector containing an adenovirus splice acceptor sequence followed by a PGK-neo cassette and a tpA stop sequence flanked by two loxP sites. The resulting plasmid was digested with PacI and AscI to release the entire floxed neo-tpA and Notch2 assembly, which was inserted into the PacI and AscI sites of a modified pRosa26-1 vector containing two homologous sequences to the Rosa26 locus flanking the inserted sequence.21 The resulting plasmid was linearized and used for electroporation. E14.1 ES cells (2 × 107) were electroporated with 50 μg of the targeting vector and allowed to grow on neomycin-resistant mouse embryonic fibroblasts in the presence of G418 (300 μg/ml). Successful targeting of 2 of 120 clones was confirmed by Southern blot analysis using genomic DNA digested with EcoRI. Primers (5′-AGG CGC CCG ATA GAA TAA AT-3′ and 5′-ACT CTT CCC CTC CCC CTA CT-3′) were used to generate a 607-bp 5′ probe (probe A; Figure 1A), which detected a 15.6-kb band for the wild-type sequence and a 4.3-kb band for the targeted sequence. Finally, the targeted ES clones were used to generate chimeric mice at the Center for Animal Resources and Development, Kumamoto University. All animal experiments were performed in accordance with institutional guidelines and ethics review committees.
Mouse Genotyping
Mice carrying the R26Notch2 allele were genotyped with primers for Neo (forward primer Neo F, 5′-AAG GGA CTG GCT GCT ATT GG-3′ and reverse primer Neo R, 5′-ATA TCA CGG GTA GCC AAC GC-3′) and Rosa26 (forward primer Rosa26 F, 5′-GAG TTC TCT GCT GCC TCC TG-3′ and reverse primer Rosa26 R, 5′-CCG ACA AAA CCG AAA ATC TG-3′). Mice carrying the Cre allele were genotyped with forward primer Cre 1 (5′-AGG TTC GTT CAC TCA TGG A-3′) and reverse primer Cre 2 (5′-TCG ACC AGT TTA GTT ACC C-3′). PCR amplifications were performed under identical conditions using GoTaq DNA polymerase (Promega) with denaturation at 95°C for 5 minutes, followed by 35 cycles of 95°C for 30 seconds, 58°C for 60 seconds, and 72°C for 30 seconds, and a final extension at 72°C for 7 minutes. The PCR products were analyzed by electrophoresis in a 1.2% agarose gel and visualized by ethidium bromide staining.
In situ Hybridization and Immunostaining
Histologic examinations were performed as described previously.9,31 Mice were fixed in 10% formalin, embedded in paraffin, and cut into 6-μm sections. In situ hybridization was performed using an automated Discovery System (Ventana) according to the manufacturer's protocols. The probes for Wnt4,5 Eya1,32 and Hesr1 and Hesr333 were described in the cited papers. Templates for other probes were generated by RT-PCR and sequenced. Immunostaining was carried out automatically using a BlueMap or DABmap kit and the automated Discovery System (Ventana) or manually for immunofluorescence staining. The following primary antibodies were used: anti-Pax2 (Covance); anti-Six2;34 anti-Sall135,36 (PPMX Perseus Proteomics); anti-cytokeratin (Sigma); anti-laminin (Sigma); and anti-DDDDK tag (Abcam).
Quantitative RT-PCR of the E11.5 Kidney
RNA was isolated from the dissected kidneys on both sides using an RNeasy Plus Micro Kit (Qiagen), and then reverse transcribed with random primers and Superscript III (Invitrogen). Quantitative PCR was carried out using a real-time PCR System (Applied Biosystems) and Thunderbird SYBR qPCR Mix (Toyobo). Two-step standard cycling conditions and sequence-specific primers were used (Supplementary Table 1). We analyzed the dissociation curves after each reaction to assess the specificity of the quantification. All of the samples were normalized by the β-actin expression using the relative standard curve method. We carried out three independent experiments, and representative data are shown.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank A.P. McMahon for the Six2Cre mice; K. Hozumi, R. Kageyama, P.X. Xu, Y. Kawakami, and F. Costantini for plasmids; and K. Kawakami for the anti-Six2 antibody. We also thank N. Takeda, J. Nakai, Y. Tsurumoto, and T. Ohmori for technical assistance, and S.S. Tanaka and M. Ikeya for helpful discussions. This study was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and by the Global COE Program (Cell Fate Regulation Research and Education Unit), MEXT, Japan.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
REFERENCES
- 1. Moore MW, Klein RD, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryan AM, Carver-Moore K, Rosenthal A: Renal and neuronal abnormalities in mice lacking GDNF. Nature 382: 76–79, 1996. [DOI] [PubMed] [Google Scholar]
- 2. Pichel JG, Shen L, Sheng HZ, Granholm AC, Drago J, Grinberg A, Lee EJ, Huang SP, Saarma M, Hoffer BJ, Sariola H, Westphal H: Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382: 73–76, 1996. [DOI] [PubMed] [Google Scholar]
- 3. Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M: Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382: 70–73, 1996. [DOI] [PubMed] [Google Scholar]
- 4. Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP: Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell 9: 283–292, 2005. [DOI] [PubMed] [Google Scholar]
- 5. Stark K, Vainio S, Vassileva G, McMahon AP: Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 372: 679–683, 1994. [DOI] [PubMed] [Google Scholar]
- 6. Kispert A, Vainio S, McMahon AP: Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development 125: 4225–4234, 1998. [DOI] [PubMed] [Google Scholar]
- 7. Herzlinger D, Koseki C, Mikawa T, al-Awqati Q: Metanephric mesenchyme contains multipotent stem cells whose fate is restricted after induction. Development 114: 565–572, 1992. [DOI] [PubMed] [Google Scholar]
- 8. Osafune K, Takasato M, Kispert A, Asashima M, Nishinakamura R: Identification of multipotent progenitors in the embryonic mouse kidney by a novel colony-forming assay. Development 133: 151–161, 2006. [DOI] [PubMed] [Google Scholar]
- 9. Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, Katsuki M, Asashima M, Yokota T: Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128: 3105–3115, 2001. [DOI] [PubMed] [Google Scholar]
- 10. Nishinakamura R: Stem cells in the embryonic kidney. Kidney Int 73: 913–917, 2008. [DOI] [PubMed] [Google Scholar]
- 11. Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP: Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3: 169–181, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takasato M, Osafune K, Matsumoto Y, Kataoka Y, Yoshida N, Meguro H, Aburatani H, Asashima M, Nishinakamura R: Identification of kidney mesenchymal genes by a combination of microarray analysis and Sall1-GFP knockin mice. Mech Dev 121: 547–557, 2004. [DOI] [PubMed] [Google Scholar]
- 13. Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G: Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J 25: 5214–5228, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM: A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol 27: 7661–7668, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wellik DM, Hawkes PJ, Capecchi MR: Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev 16: 1423–1432, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T: Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128: 491–502, 2001. [DOI] [PubMed] [Google Scholar]
- 17. Cheng HT, Miner JH, Lin M, Tansey MG, Roth K, Kopan R: Gamma-secretase activity is dispensable for mesenchyme-to-epithelium transition but required for podocyte and proximal tubule formation in developing mouse kidney. Development 130: 5031–5042, 2003. [DOI] [PubMed] [Google Scholar]
- 18. Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R: Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development 134: 801–811, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB: NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 79: 169–173, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soriano P: Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71, 1999. [DOI] [PubMed] [Google Scholar]
- 21. Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F: Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kageyama R, Ohtsuka T, Kobayashi T: The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development 134: 1243–1251, 2007. [DOI] [PubMed] [Google Scholar]
- 23. Piscione TD, Wu MY, Quaggin SE: Expression of Hairy/Enhancer of Split genes, Hes1 and Hes5, during murine nephron morphogenesis. Gene Expr Patterns 4: 707–711, 2004. [DOI] [PubMed] [Google Scholar]
- 24. Chen L, Al-Awqati Q: Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol 288: F939–F952, 2005. [DOI] [PubMed] [Google Scholar]
- 25. Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R: Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem 281: 5106–5119, 2006. [DOI] [PubMed] [Google Scholar]
- 26. Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR: Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development 132: 2809–2823, 2005. [DOI] [PubMed] [Google Scholar]
- 27. Kopan R, Cheng HT, Surendran K: Molecular insights into segmentation along the proximal-distal axis of the nephron. J Am Soc Nephrol 18: 2014–2020, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Niranjan T, Bielesz B, Gruenwald A, Ponda MP, Kopp JB, Thomas DB, Susztak K: The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat Med 14: 290–298, 2008. [DOI] [PubMed] [Google Scholar]
- 29. Waters AM, Wu MY, Onay T, Scutaru J, Liu J, Lobe CG, Quaggin SE, Piscione TD: Ectopic notch activation in developing podocytes causes glomerulosclerosis. J Am Soc Nephrol 19: 1139–1157, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hozumi K, Abe N, Chiba S, Hirai H, Habu S: Active form of Notch members can enforce T lymphopoiesis on lymphoid progenitors in the monolayer culture specific for B cell development. J Immunol 170: 4973–4979, 2003. [DOI] [PubMed] [Google Scholar]
- 31. Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, Nishinakamura R: The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 133: 3005–3013, 2006. [DOI] [PubMed] [Google Scholar]
- 32. Xu PX, Zheng W, Laclef C, Maire P, Maas RL, Peters H, Xu X: Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development 129: 3033–3044, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R: The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem 278: 44808–44815, 2003. [DOI] [PubMed] [Google Scholar]
- 34. Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K: Tissue and developmental distribution of Six family gene products. Int J Dev Biol 42: 141–148, 1998. [PubMed] [Google Scholar]
- 35. Sato A, Kishida S, Tanaka T, Kikuchi A, Kodama T, Asashima M, Nishinakamura R: Sall1, a causative gene for Townes-Brocks syndrome, enhances the canonical Wnt signaling by localizing to heterochromatin. Biochem Biophys Res Commun 319: 103–113, 2004. [DOI] [PubMed] [Google Scholar]
- 36. Yuri S, Fujimura S, Nimura, Takeda N, Toyooka Y, Fujimura Y, Aburatani H, Ura K, Koseki H, Niwa H, Nishinakamura R: Sall4 is essential for stabilization, but not pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells 27: 796–805, 2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.