Abstract
Mutations in the COL4A5 gene cause X-linked Alport syndrome (XLAS). Understanding the correlation between clinical manifestations and the underlying mutations adds prognostic value to genetic testing, which is increasingly available. Our aim was to determine the association between genotype and phenotype in 681 affected male participants with XLAS from 175 US families. Hearing loss and ocular changes were present in 67 and 30% of participants, respectively. Average age of participants at onset of ESRD was 37 years for those with missense mutations, 28 years for those with splice-site mutations, and 25 years for those with truncating mutations (P < 0.0001). We demonstrated a strong relationship between mutation position and age at onset of ESRD, with younger age at onset of ESRD associated with mutations at the 5` end of the gene (hazard ratio 0.766 [95% confidence interval 0.694 to 0.846] per 1000 bp toward the 3` end; P < 0.0001). Affected participants with splice mutations or truncating mutations each had two-fold greater odds of developing eye problems than those with missense mutations; development of hearing impairment showed a similar trend. Hearing loss and ocular changes associated with mutations located closer to the 5` end of the gene. These strong genotype–phenotype correlations could potentially help in the evaluation and counseling of US families with XLAS.
Alport syndrome (AS) is a relatively frequent monogenic inherited kidney disorder characterized by gradual renal failure progressing to ESRD, sensorineural hearing loss, and ocular abnormalities.1–3 AS is caused by defects in type IV collagen, a major structural component of the basement membranes in the kidney, ear, and eye. Six genetically distinct type IV collagen α-chains have been identified. Defects in the COL4A5 gene, encoding collagen α-5 (IV) chain, located at Xq22, cause X-linked AS (XLAS), which accounts for 80% of AS.4,5 COL4A5 is a large gene comprising 51 exons.6,7 More than 440 mutations have been described to date in COL4A5.8,9 These mutations are spread throughout the gene without any identified mutational hot spot.
XLAS is clinically and genetically heterogeneous. Allelic heterogeneity is evidenced by the high number of mutations in the COL4A5 gene and the associated phenotypic variability.10,11 Clinically, the natural history of the nephropathy in XLAS is quite variable. Age at ESRD differs between families and in males ranges between the second and third decades; however, in milder cases, ESRD may be delayed until the fifth or sixth decade. Also, deafness occurs at variable ages.12–15 A wide variety of ocular changes, including anterior lenticonus, cataract, and maculopathy, have been reported among patients.12,16 Other extrarenal manifestations, such as diffuse esophageal leiomyomatosis, may be present in different families.16 Macrothrombocytopenia, previously considered a manifestation of AS, is now known to be a feature of the myosin heavy chain 9, nonmuscle (MYH9) family of disorders.17
Genetic testing has potential clinical value, because it is both noninvasive and accurate.18,19 More efficient screening methods are now available for mutation identification in patients with XLAS, and genetic testing also has the potential benefit of providing prognostic information20,21; however, the clinical and genetic heterogeneity associated with the disease have complicated study of the correlation between the phenotype and the underlying mutation, because this requires analysis of large numbers of families with XLAS. Although genotype–phenotype correlation has been studied in a European cohort,22,23 it has never been assessed in a large US population.
The purpose of this investigation was to study one of the most relevant clinical benefits of molecular studies, namely the correlation between the phenotype and the underlying genetic mutation. This study was undertaken in the largest cohort of US patients to date. Collection of genetic and clinical information on 681 participants from 175 families allowed performance of an accurate correlation of genotype–phenotype in XLAS.
Results
Mutation data were available for 681 male participants with XLAS from 175 families. Family size ranged from one to 53 participants. The patient characteristics are shown in Table 1. Phenotypic details of families and mutations are presented in Supplemental Table S1.
Table 1.
Characteristics of 681 participants from 175 families with mutation classification
| Characteristic | N with Data | n (%) with Trait |
|---|---|---|
| Hypertension | 369 | 201 (54.47) |
| Proteinuria | 377 | 322 (85.41) |
| Gross hematuria | 345 | 168 (48.70) |
| Hematuria diagnosed by a physician | 405 | 367 (90.62) |
| ESRD | 609 | 364 (59.77) |
| Transplant | 610 | 255 (41.80) |
| Ocular change | 360 | |
| none | 251 (69.72) | |
| cataract | 42 (11.67) | |
| any combination | 66 (18.33) | |
| macular hole | 1 (0.28) | |
| Hearing loss | 401 | 268 (66.83) |
| Positive by audiometry | 356 | 317 (89.04) |
| Mutation type in patients | 681 | |
| large deletion | 31 (4.55) | |
| missense | 438 (64.32) | |
| small deletion | 64 (9.40) | |
| splice site | 78 (11.45) | |
| truncating | 70 (10.28) | |
| Gly-X-Y | 104 (15.27) | |
| Mutation type distribution in families | 175 families | |
| large deletion | 14 (8.00) | |
| missense | 89 (50.86) | |
| small deletion | 23 (13.14) | |
| splice site | 24 (13.71) | |
| truncating | 25 (14.29) | |
| Gly-X-Y | 50 (28.57) |
Renal Disease
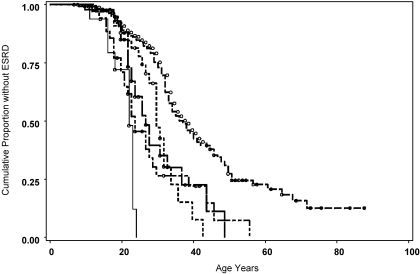
Age at onset of ESRD differed significantly among patients with different mutation types. The median time (95% confidence intervals [CIs]) from birth to ESRD and were as follows: Missense 37 years (34 to 40 years), splice site 28 years (26 to 32 years), truncating 25 years (21 to 31 years), large deletion 22 years (16 to 23 years), and small deletion 22 years (19 to 27 years; P < 0.0001; Figure 1, Table 2). There was no difference in time to onset of ESRD among patients with splice donor (28 years [25 to 32 years]) and splice acceptor mutations (30 years [18 to 38 years]; P = 0.87). Cox regression taking into account the possible correlation among members of the same family demonstrated a significant difference in survival time between mutation types with missense mutation as the reference: Truncating (hazard ratio [HR] 3.33; P < 0.0001), large deletion (HR 8.33; P < 0.0001), small deletion (HR 3.61; P < 0.0001), and splice site (HR 3.016; P < 0.0001). To verify that results were not biased on the basis of inclusion of multiple members from certain families, we performed a family analysis using a calculated average age at onset of ESRD or age without ESRD for each family. Mean age at onset of ESRD differed significantly among mutation categories: Missense 37.5 years (95% CI 33.5 to 44.0 years), splice site 29.0 years (95% CI 23.5 to 30.2 years), truncating 24.0 years (95% CI 20.0 to 28.5 years), large deletion 22.5 years (95% CI 16.0 to 24.0 years), and small deletion 26.0 (95% CI 18.5 to 54.0 years; P < 0.0001). Similarly, in Cox regression, the HRs were almost identical on the basis of individual or family analyses with missense mutation as the reference: Truncating (HR 3.85; P < 0.0001), large deletion (HR 6.71; P < 0.0001), small deletion (HR 2.25; P < 0.01), and splice site (HR 2.65; P < 0.002).
Figure 1.
Time to onset of ESRD associates with mutation type. From the left of figure, light weight solid line, large deletion; dots, splice site mutation; dash-dot, small deletion; heavy solid line, truncating mutation; dash, missense mutation; ○, censored data. The number of patients with ESRD or age without ESRD is indicated in Table 1.
Table 2.
Frequency of clinical characteristics in those with available clinical data
| Characteristic | Large Deletion | Missense | Small Deletion | Splice Site | Truncating | P |
|---|---|---|---|---|---|---|
| Hypertension | 17/26 (65.38) | 120/232 (51.72) | 21/34 (61.76) | 22/43 (51.76) | 21/34 (61.76) | 0.46 |
| Proteinuria | 22/34 (91.67) | 210/244 (86.07) | 25/33 (75.76) | 34/42 (80.95) | 31/34 (91.18) | 0.30 |
| Gross hematuria | 14/23 (47.01) | 110/234 (47.01) | 14/29 (48.28) | 20/34 (58.82) | 10/25 (40.00) | 0.44 |
| Hematuria diagnosed by a physician | 23/23 (100.00) | 229/252 (90.87) | 36/42 (85.71) | 47/49 (95.95) | 32/39 (82.05) | 0.07 |
| ESRD | 18/27 (66.67) | 211/385 (54.81) | 45/60 (75.00) | 50/70 (71.43) | 40/67 (59.70) | 0.007 |
| Age of onset of ESRD (years; median [95% CI]) | 22 (16 to 23) | 37 (34 to 40) | 22 (19 to 27) | 28 (26 to 32) | 25 (21 to 31) | <0.0001 |
| Transplant | 18/29 (62.07) | 126/385 (32.73) | 32/59 (54.24) | 42/70 (60.00) | 37/67 (55.22) | <0.0001 |
| Ocular changes | 13/26 (50.00) | 45/222 (20.27) | 12/33 (36.36) | 22/45 (48.89) | 17/34 (50.00) | <0.0001 |
| Hearing loss | 21/26 (80.77) | 121/238 (50.84) | 41/43 (95.35) | 43/50 (86.00) | 42/44 (95.45) | <0.0001 |
| Hearing loss by audiometry | 24/25 (96.00) | 187/222 (84.23) | 30/32 (93.75) | 42/43 (97.67) | 34/34 (100.00) | <0.006 |
Data are n/N (%) except where indicated.
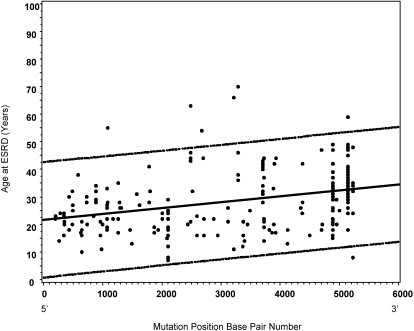
Cox regression with a robust estimator demonstrated a strong relationship between mutation position and age of onset of ESRD (HR 0.766 [95% CI 0.694 to 0.846] per 1000-bp increment toward the 3′ end of the gene; P < 0.0001) with younger onset of ESRD associated with mutation position at the 5′ end of the gene. Figure 2 shows a scatter plot of age at ESRD with mutation position for all patients who reached ESRD regardless of mutation type. Mutations located toward the 5′ end of the gene were associated with earlier age at ESRD onset.
Figure 2.
Age at onset of ESRD associates with mutation position (n = 364 with ESRD). Dotted lines represent upper and lower 95% confidence limits. R = 0.33, P < 0.0001 with mutations positioned at 5′ end of the gene associated with earlier age at ESRD onset.
Ocular Changes
The distribution of ocular changes differed significantly according to mutation type, with fewer ocular changes among patients with missense and small deletion mutations than among those with large deletions or splice site or truncating mutations (Table 2). Participants with deletions or splice or truncating mutations had two- to four-fold greater odds of developing ocular changes than those with missense mutations (Table 3). General estimating equation (GEE), accounting for repeated measures among families, also showed a significant increase in the odds of developing ocular changes among those with splice and truncating compared with missense mutations (Table 4). There was no difference in the incidence of ocular changes between patients with splice donor (52.2%) and splice acceptor (45.5%) mutations (P = 0.65). Logistic regression demonstrated a strong relationship between mutation position and development of ocular changes (odds ratio [OR] 0.841 [95% CI 0.738 to 0.958] per 1000-bp increment toward the 3′ end of the gene; P = 0.01), with ocular disorders associated with mutation position at the 5′ end of the gene. Those with ocular changes had earlier age at ESRD onset (median 24 years; 95% CI 22 to 27 years) than those without (median 33 years; 95% CI 32 to 42 years; P < 0.0001).
Table 3.
Probability of ocular changes and hearing loss by mutation type
| Mutation Type | Ocular Change |
Hearing Loss |
||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | Pa | |
| Large deletion | 3.93 (1.71 to 9.07) | 0.0013 | 4.02 (1.48 to 11.13) | 0.0064 |
| Small deletion | 2.25 (1.03 to 4.91) | 0.0421 | 19.82 (4.69 to 83.82) | <0.0001 |
| Splice site | 3.76 (1.93 to 7.35) | 0.0001 | 5.94 (2.57 to 13.74) | <0.0001 |
| Truncating | 3.93 (1.86 to 8.31) | 0.0003 | 20.31 (4.81 to 85.80) | <0.0001 |
aCompared with missense mutation.
Table 4.
Probability of ocular changes or hearing loss by mutation type using GEE with missense as reference
| Mutation Type | Ocular Changes |
Hearing Loss |
||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | Pa | |
| Large deletion | 2.09 (1.64 to 2.44) | 0.055 | 2.22 (1.80 to 2.50) | 0.009 |
| Small deletion | 1.94 (1.58 to 2.27) | 0.115 | 2.59 (2.12 to 2.70) | 0.002 |
| Splice site | 2.14 (1.86 to 2.37) | 0.0007 | 2.35 (2.00 to 2.55) | 0.0003 |
| Truncating | 2.15 (1.83 to 2.39) | 0.002 | 2.59 (2.29 to 2.68) | <0.0001 |
aCompared with missense mutation.
Hearing Loss
There was a significant difference in the distribution of self-reported hearing loss, with fewer hearing problems among participants with missense mutations than among those with small deletions, large deletions, or splice site or truncating mutations (Table 2). In logistic regression, not adjusting for repeated measures among families, those with large or small deletions or splice site or truncating mutations had 4- to 20-fold greater odds of developing hearing loss than those with missense mutations (Table 3). GEE accounting for repeated measures among families also showed a significant decrease in odds of hearing loss among those with missense mutations compared with patients with small or large deletions or splice site and truncating mutations (Table 4). In logistic regression, fewer people with missense mutations had hearing loss positive by audiometry compared with those with other mutation categories (Table 2). There was no difference in the incidence of hearing loss between patients with splice donor (81.5%) and splice acceptor (91.3%) mutations (P = 0.32). We also analyzed traits by family, designating presence or absence of hearing loss as a single count per family. There was a significant difference in the distribution of hearing loss between families according to mutation type (Table 5). Logistic regression demonstrated a strong relationship between mutation position and development of hearing loss (OR 0.826 [95% CI 0.726 to 0.937] per 1000-bp increment; P = 0.003), with hearing loss associated with mutation position at the 5′ end of the gene.
Table 5.
Distribution of clinical events among families
| Parameter | Large Deletion | Missense | Small Deletion | Splice Site | Truncating | P |
|---|---|---|---|---|---|---|
| Hypertension | 9/14 (64.29) | 62/80 (77.50) | 14/20 (70.00) | 15/21 (71.43) | 16/22 (72.73) | 0.84 |
| Proteinuria | 12/13 (92.31) | 77/80 (96.25) | 18/21 (85.71) | 19/20 (95.00) | 21/22 (95.45) | 0.47 |
| Gross hematuria | 8/11 (72.73) | 50/77 (64.94) | 12/17 (70.59) | 12/17 (70.59) | 7/19 (36.84) | 0.14 |
| Hematuria diagnosed by a physician | 12/12 (100.00) | 76/83 (91.57) | 17/20 (85.00) | 22/22 (100.00) | 20/24 (83.33) | 0.19 |
| ESRD | 10/14 (71.43) | 73/89 (82.02) | 20/23 (86.96) | 21/24 (87.50) | 23/25 (92.00) | 0.48 |
| Ocular changes | 6/14 (42.86) | 33/79 (41.77) | 11/21 (52.38) | 16/22 (72.73) | 12/22 (54.55) | 0.13 |
| Hearing loss | 13/14 (92.86) | 54/79 (68.35) | 21/21 (100.00) | 20/22 (90.91) | 23/23 (100.00) | 0.0002 |
Data are n/N (%). Presence of phenotype counted when one incidence within family.
Mutation Position on the Basis of Domain Structure of Collagen α-5(IV) Chain Protein
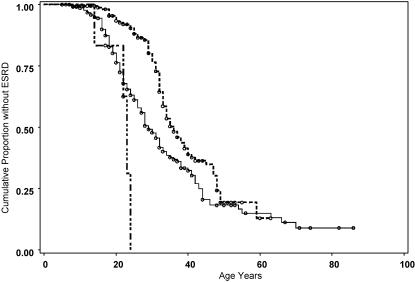
To dissect further the correlation between mutation position and phenotypical features of XLAS, we analyzed the mutation position on the basis of the affected structural domains of collagen α-5(IV) chain. Age at ESRD occurred earlier in patients with mutation affecting the signal peptide region (median 22 years; 95% CI 14 to 23 years), compared with the noncollagenous (NC1; median 36 years; 95% CI 33 to 40 years) or collagenous domain (median 29 years; 95% CI 27 to 32 years; P < 0.0001). There were insufficient observations within the NC2 region to permit comparative analysis of this domain (Figure 3). This raised the question of whether the mutation position on the gene regardless of affected structural domain had the greatest impact on phenotypic severity. To answer this question, we divided the collagenous domain into two parts: Mutation positions affecting base pairs 326 to 2448 and mutation positions affecting base pairs 2449 to 4570. When severity was related to disruption of the collagenous domain as opposed to a more 5′ position of mutation on the gene, no difference between the regions would be expected. Age at onset of ESRD was significantly earlier in the 326 to 2448 region (median 26 years; 95% CI 23 to 28 years) compared with 2449 to 4570 (median 38 years; 95% CI 31 to 42 years; P = 0.0023).
Figure 3.
Mutation position based on domain structure of collagen-5(IV) affects age at onset of ESRD. Long-short dash, signal peptide + NC2 (bp position 203 to 322); solid line, collagenous domain (bp position 323 to 4570); dashed line, NC1 (bp position 4583 to 5257). The individuals with mutations affecting the specific domains were as follows signal peptide + NC2, n = 6, with ESRD, n = 4; collagenous domain, n = 221, with ESRD, n = 130; NC1, n = 173, with ESRD, n = 81.
Glycine-X-Y Mutations
A total of 104 individuals had glycine-X-Y (Gly-X-Y) mutations affecting the collagenous domain. Among patients who had reached ESRD, age at ESRD was slightly earlier (median 33 years; 95% CI 29 to 41 years) for the 58 patients with Gly-X-Y mutations as compared with the 167 patients with missense mutations occurring in the same domain (median 38 years; 95% CI 34 to 44 years; P = 0.033). There was no relationship between mutation position and age at ESRD in those with Gly-X-Y mutations (HR 0.870 [95% CI 0.653 to 1.160] per 1000-bp increment toward the 3′ end of the gene; P = 0.343). There was no significant difference in the frequency of any other phenotype between Gly-X-Y mutations and non–Gly-X-Y mutations.
Discussion
In this study, the natural history of XLAS and genotype–phenotype correlation were examined in the largest number of US patients to date. Microscopic hematuria was present in 91% of the participants, with 49% presenting with gross hematuria. Microscopic hematuria has been reported to be a presenting symptom in 80 to 100% of patients with AS in Europe and China.22,24,25 Signs of kidney impairment, including proteinuria, develop with disease progression. Proteinuria occurred in 85% of our cohort. Previously, the incidence has been reported to be as high as 95%.20 Over time, hypertension and ESRD occur; hypertension was diagnosed in 54% and ESRD in 60% of this group of participants. A mean age of ESRD between 16 and 35 years has been reported in XLAS and in this study was 37 years.
Sensorineural hearing loss is common in XLAS26 and has been detected in 80 to 85% of affected males22,27; our results demonstrated hearing loss in 67% of participants. The exclusion of other causes of hearing loss in this study may partially explain the lower incidence of hearing impairment. A limitation of this study is the lack of access to the audiometric reports for all participants. Thus, participant-reported hearing impairment may not be as sensitive as other reports in which hearing loss was based on audiometric testing.
Ocular changes, a characteristic clinical finding in XLAS, were found in 30% of participants examined. This included a 12% incidence of cataracts and 18% incidence combination of lens malformation or maculopathy. The frequency of specific ocular changes was previously reported in a limited number of studies. One study reported a 35% incidence of ocular changes,22 which parallels our results. Other studies reported incidence ranging from 30 to 72%.4,28,29 It is clear that the rate of ocular changes is high enough in XLAS that ophthalmologic examination should be considered for patients who present with hematuria.
The focus of this study was the evaluation of genotype–phenotype correlations in a large US population of male patients with XLAS. Missense was the most prevalent type of mutation detected in this study (51% of the families) followed by truncating (14%) and splice site (13.7%) mutations.
The association between severity of both renal and extrarenal manifestations of XLAS and the underlying mutation was reported previously.22,23,30–34 In a review of males in 195 European families, missense, splice site, and large deletion; nonsense; and frameshift mutations were correlated with less severe, moderately severe, and more severe phenotype presentations, respectively.
Our detailed study of genotype–phenotype correlation in XLAS has several unique characteristics. First, it is the only study in a large US population. Second, it is the only study to examine mutation position along the COL4A5 gene and among the different domains of the gene and associates results with the phenotype of XLAS. One other study examined mutation position; however, it was restricted to glycine mutations only.23 Third, it is the only study that has accounted for potential bias as a result of relatedness among the participants. Because some of the 175 families included multiple members, we statistically corrected for relatedness in our analyses to overcome this potential source of bias.
A correlation between the underlying mutation and phenotype was clearly shown in this study in several ways. Mutation type was associated with age at onset of ESRD. Missense mutations are associated with the best prognosis with an average age at onset of ESRD of 37 years (Figure 1). Mean age at onset of ESRD was 28 years for participants with splice site mutation, 25 years for those with truncating mutations, and 22 years for those with small deletions. Similar family-based analysis yielded an almost identical result, confirming that our statistical analysis of individuals appropriately corrected for potential bias as a result of inclusion of multiple members of some large families. This outcome supports the results of previous studies22,23 in a larger cohort of affected male participants. The number of participants with large deletions was too limited for the statistical significance to have any clinical relevance in this study. An influence of antihypertensive medication use on the rate of renal disease progression cannot be discounted in this study; however, the incidence of hypertension did not differ significantly between patients in the various mutation groups (Table 2).
Noteworthy is the correlation between mutation position and the age at ESRD. Mutations positioned at the 5′ end of the gene, regardless of type, were associated with younger age at onset of ESRD (Figure 2). The only previous study that analyzed the effect of mutation position on disease severity compared 98 glycine missense mutations located in exons 1 through 20 and 21 through 47 of COL4A5, respectively (n = 56 for ESRD), and found that exons 1 through 20 influenced ESRD in a less severe manner.23 In contrast, in this study, no relationship between position of Gly-X-Y mutation and the age of onset of ESRD (n = 58) was found. Study of larger cohorts may be necessary to detect a relationship between Gly-X-Y mutation position and age at onset of ESRD. It seems that deletions and truncating and splice site mutations, which change the structure of the protein, have the greatest effect on ESRD. One theory is that mutations resulting in protein truncation cause a more deleterious effect when they occur at the 5′ end of the gene as loss of the whole gene product ensues. In contrast, mutations located toward the 3′ end of the gene may result in production of a residual truncated protein product. Similar observation for missense mutations may indicate that protein stabilization requires the structural integrity of the NC2 and adjacent portion of the collagenous domain despite potential initiation of triple-helix formation at the NC1 domain.23
Results for hearing loss and ocular changes are similar to those for ESRD. Specifically, missense mutations were associated with a lower incidence of hearing impairment and ocular changes (Table 3). After accounting for the relatedness among the population, the difference remained significant (Table 4). The occurrence of both hearing loss and ocular changes was associated with mutations located at the 5′ end of the gene. Although the association of hearing loss and ocular changes with genotype has been previously reported,22,23,34 the association with mutation position is a novel finding in our study.
The prevalence of ocular changes was 30% in this group of participants with XLAS. Time to onset of ESRD differed between those with any ocular change (24 years) and those without (33 years). Thus, ophthalmology referral is an important part of evaluation of patients with XLAS, not only as a valuable diagnostic tool but possibly as a predictive factor for the general prognosis of kidney function.
In summary, in the largest US population reported to date, a genotype–phenotype correlation for both renal and extrarenal manifestations of the XLAS was observed. Our results indicate that deduced premature termination of the collagen α-5(IV) chain associated with large and small deletions and truncating or splice mutations causes the most severe clinical phenotype. In contrast, missense mutations result in a less severe phenotype. Notably, mutations at the 5′ end of the gene have the greatest effect and the worst prognosis. The rate of progression of both renal and extrarenal manifestations of XLAS was associated with the type and location of the underlying mutation. Genetic analysis thus seems to be a better prognostic indicator than either renal or skin biopsy.
Concise Methods
Participants and Clinical Criteria
This study includes analyses of clinical and genetic information from 681 male participants with XLAS from 175 families with mutations in COL4A5, who entered the University of Utah Alport Syndrome study. Clinical diagnosis was based on either (1) family history of renal insufficiency or failure, coincident with hematuria, or hearing loss or anterior lenticonus or (2) one or more instances of kidney biopsies with histology morphologically consistent with AS. All participants gave informed consent, and the study was conducted according to a protocol approved by the University of Utah institutional review board.
Mutation Analysis
Mutations in COL4A5 were identified and confirmed by mapping of Eco R1, Taq1, and Pst 1 restriction sites,32 reverse transcriptase PCR and denaturing gradient gel electrophoresis,14 RNAse protection assays,14 allele-specific oligonucleotide probe tests,14 PCR and direct DNA sequencing,18 and multiplex genomic PCR-SSCP35 in different families.
Mutation Categories
Mutations were classified into (1) missense; (2) splice site (occurring at the donor or acceptor site); (3) truncating (when the substitution, deletion/insertion, or any other changes cause premature termination of translation of the collagen α-5 (IV) chain); (4) large deletion (when there is deletion >20 bases including whole exon deletions); or (5) small deletion (deletion of ≤20 bases).
All mutations were numbered on the basis of reference sequence Ensembl transcript ENST00000361603, Gene sequence NCBI Bld. 36, Chromosome X 107569210-107828031. Mutations are reported at the cDNA and protein levels (ENSP00000354505). Ensembl web site (http://www.ensembl.org) was used to locate the position of the mutations and based on that, a variable called mutation position was defined, which is then statistically associated with the clinical findings along with the mutation classes in two ways. First the mutations were classified on the basis of their bp location within COL4A5. Second, mutations were classified on the basis of the functional structural domain of the collagen α-5(IV) chain protein.6,7 These domains included signal peptide (c-DNA bp number 203 to 280), NC2 (c-DNA bp number 281 to 322), collagenous (c-DNA bp number 323 to 4570), and NC1 (c-DNA bp number 4583 to 5257). Gly-X-Y mutations located within the collagenous domain were also analyzed with regard to the mutation number on the basis of the mutation c-DNA number.
Phenotype Information
Questionnaires were designed to obtain participants' clinical history. Participants were contacted between 2005 and 2007, and demographic information and clinical data were collected through telephone interviews. The clinical data included information regarding ESRD, age at diagnosis of ESRD, diagnosis of hypertension, presence of proteinuria, occurrence of gross hematuria at any time, microscopic hematuria diagnosed by a physician, transplant history, ocular changes, and hearing problems. Ocular changes were classified as lens malformation, cataract, or maculopathy. Occurrence of hearing loss was per patient-described history, and data from participants with other possible causes of hearing impairment (e.g., significant noise exposure, ear infection or ruptured ear drum, trauma, surgery, medication) were excluded from the analysis.
Statistical Analysis
Descriptive statistics including frequency counts and means were used to describe the sample of participants with XLAS. χ2 test of independence or Fisher exact test was used to test the association of mutation position with categorical phenotypes. The Kaplan-Meier method and the log-rank statistic for time to ESRD onset were used to compare survival curves among mutation domains. Cox proportional hazards model was use, and potential correlation within members of the same family was accounted for and significance was tested using a robust variance estimator. Logistic regression and GEEs adjusting for the correlation within members of the same family were used to estimate the odds of developing clinical traits for each mutation type. Results were considered significant at P < 0.05.
Disclosures
None.
Supplementary Material
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1. Alport AC: Hereditary familial congenital haemorrhagic nephritis. Br Med 1: 504–506, 1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kashtan CE: Alport syndrome and thin glomerular basement membrane disease. J Am Soc Nephrol 9: 1736–1750, 1998. [DOI] [PubMed] [Google Scholar]
- 3. Kashtan CE: Michael AF Alport syndrome. Kidney Int 50: 1445–1463, 1996. [DOI] [PubMed] [Google Scholar]
- 4. Gregory MC, Shamshirsaz AA, Kamgar M, Bekherinia MR: Alport syndrome, Fabry disease, and nail patella syndrome. In: Diseases of the Kidney and Urinary Tract, 8th Ed., edited by Schrier RW. Boston, Little, Brown and Company, 2007, pp 540–569 [Google Scholar]
- 5. Kashtan CE: Familial hematuria due to type IV collagen mutations: Alport syndrome and thin basement membrane nephropathy. Curr Opin Pediatr 16: 177–181, 2004. [DOI] [PubMed] [Google Scholar]
- 6. Zhou J, Leinonen A, Tryggvason K: Structure of the human type IV collagen COL4A5 gene. J Biol Chem 269: 6608–6614, 1994. [PubMed] [Google Scholar]
- 7. Zhou J, Hertz JM, Leinonen A, Tryggvason K: Complete amino acid sequence of the human α5(IV) collagen chain and identification of a single base mutation in exon 23 converting glycine-521 in the collagenous domain to cystein in an Alport syndrome patient. J Biol Chem 267: 12475–12481, 1992. [PubMed] [Google Scholar]
- 8. The Human Gene Mutation Database at the Institute of Medical Genetics in Cardiff, 2008. Available at: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=COL4A5 Accessed April 3, 2009
- 9. Arup Laboratories: ARUP Online Scientific Resource: ALPORT database display page, 2009. Available at: http://www.arup.utah.edu/database/ALPORT/ALPORT_display.php Accessed June 30, 2009
- 10. Feingold J, Bois E, Chompret A, Broyer M, Gubler MC, Grünfeld JP: Genetic heterogeneity of Alport syndrome. Kidney Int 27: 672–677, 1985. [DOI] [PubMed] [Google Scholar]
- 11. Hasstedt SJ, Atkin CL, San Juan AC: Genetic heterogeneity among kindreds with Alport syndrome. Am J Hum Genet 38: 940–953, 1986. [PMC free article] [PubMed] [Google Scholar]
- 12. Kashtan CE: Alport syndromes: Phenotypic heterogeneity of progressive hereditary nephritis. Pediatr Nephrol 14: 502–512, 2000. [DOI] [PubMed] [Google Scholar]
- 13. Lemmink HH, Schröder CH, Monnens LA, Smeets HJ: The clinical spectrum of type IV collagen mutations. Hum Mutat 9: 477–499, 1997. [DOI] [PubMed] [Google Scholar]
- 14. Barker DF, Pruchno CJ, Xiang X, Atkin CL, Stone EM, Denison JC, Fain PR, Gregory MC: A mutation causing Alport syndrome with tardive hearing loss is common in the western United States. Am J Hum Genet 58: 1157–1165, 1996. [PMC free article] [PubMed] [Google Scholar]
- 15. Barker DF, Denison J, Atkin CL, Gregory MC: Common ancestry of three Ashkenazi-American families with Alport syndrome and COL4A5 R1677Q. Hum Genet 99: 681–684, 1997. [DOI] [PubMed] [Google Scholar]
- 16. Colville DJ, Savige J: Alport syndrome: A review of the ocular manifestations. Ophthalmic Genet 18: 161–173, 1997. [DOI] [PubMed] [Google Scholar]
- 17. Heath KE, Campos-Barros A, Toren A, Rozenfeld-Granot G, Carlsson LE, Savige J, Denison JC, Gregory MC, White JG, Barker DF, Greinacher A, Epstein CJ, Glucksman MJ, Martignetti JA: Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet 69: 1033–1045, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin P, Heiskari N, Zhou J, Leinonen A, Tumelius T, Hertz JM, Barker D, Gregory M, Atkin C, Styrkarsdottir U, Neumann H, Springate J, Shows T, Pettersson E, Tryggvason K: High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J Am Soc Nephrol 9: 2291–2301, 1998. [DOI] [PubMed] [Google Scholar]
- 19. Inoue Y, Nishio H, Shirakawa T, Nakanishi K, Nakamura H, Sumino K, Nishiyama K, Iijima K, Yoshikawa N: Detection of mutations in the COL4A5 gene in over 90% of male patients with X-linked Alport's syndrome by RT-PCR and direct sequencing. Am J Kidney Dis 34: 854–862, 1999. [DOI] [PubMed] [Google Scholar]
- 20. Tazón-Vega B, Ars E, Burset M, Santín s, Ruíz P, Fernández-Llama P, Ballarín J, Torra R: Genetic testing for X-linked Alport syndrome by direct sequencing of COL4A5 cDNA from hair root RNA samples. Am J Kidney Dis 50: 257–269, 2007. [DOI] [PubMed] [Google Scholar]
- 21. Gubler MC: Diagnosis of Alport syndrome without biopsy? Pediatr Nephrol 22: 621–625, 2007. [DOI] [PubMed] [Google Scholar]
- 22. Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, Weber M, Gross O, Netzer KO, Flinter F, Pirson Y, Verellen C, Wieslander J, Perrson U, Tryggvason K, Martin P, Hertz JM, Schroder C, Sanak M, Krejkova S, Carvalho MF, Saus J, Antignac C, Smeets H, Gubler MC: X-linked Alport syndrome: Natural history in 195 families and genotype–phenotype correlations in males. J Am Soc Nephrol 11: 649–657, 2000. [DOI] [PubMed] [Google Scholar]
- 23. Gross O, Netzer KO, Lambrecht R, Seibold S, Weber M: Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: Impact on clinical counseling. Nephrol Dial Transplant 17: 1218–1227, 2002. [DOI] [PubMed] [Google Scholar]
- 24. Wang F, Ding J, Guo S, Yang J: Phenotypic and genotypic features of Alport syndrome in Chinese children. Pediatr Nephrol 17: 1013–1020, 2002. [DOI] [PubMed] [Google Scholar]
- 25. Wei G, Zhihong L, Huiping C, Caihong Z, Zhaohong C, Leishi L: Spectrum of clinical features and type IV collagen alpha-chain distribution in Chinese patients with Alport syndrome. Nephrol Dial Transplant 21: 3146–3154, 2006. [DOI] [PubMed] [Google Scholar]
- 26. Izzedine H, Tankere F, Launay-Vacher V, Deray G: Ear and kidney syndromes: Molecular versus clinical approach. Kidney Int 65: 369–385, 2004. [DOI] [PubMed] [Google Scholar]
- 27. Gubler M, Levy M, Broyer M, Naizot C, Gonzales G, Perrin D, Habib R: Alport's syndrome: A report of 58 cases and a review of the literature. Am J Med 70: 493–505, 1981. [DOI] [PubMed] [Google Scholar]
- 28. Gubler MC, Antignac C, Deschênes G, Knebelmann B, Hors-Cayla MC, Grünfeld JP, Broyer M, Habib R: Genetic, clinical and morphologic heterogeneity in Alport's syndrome. Adv Nephrol 22: 15–35, 1993. [PubMed] [Google Scholar]
- 29. Flinter FA, Cameron JS, Chantler C, Houston I, Bobrow M: Genetics of classic Alport's syndrome. Lancet 2: 1005–1007, 1988. [DOI] [PubMed] [Google Scholar]
- 30. Kashtan CE: Alport syndrome: An inherited disorder of renal, ocular, and cochlear basement membranes. Medicine (Baltimore) 78: 338–360, 1999. [DOI] [PubMed] [Google Scholar]
- 31. Tryggvason K, Zhou J, Hostikka SL, Shows TB: Molecular genetics of Alport syndrome. Kidney Int 43: 38, 1993. [DOI] [PubMed] [Google Scholar]
- 32. Barker DF, Hostikka SL, Zhou J, Chow LT, Oliphant AR, Gerken SC, Gregory MC, Skolnick MH, Atkin CL, Tryggvason K: Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248: 1224–1227, 1990. [DOI] [PubMed] [Google Scholar]
- 33. Antignac C, Knebelmann B, Drouot L, Gros F, Deschênes G, Hors-Cayla MC, Zhou J, Tryggvason K, Grünfeld JP, Broyer M: Deletions in the COL4A5 collagen gene in X-linked Alport syndrome: Characterization of the pathological transcripts in nonrenal cells—Correlation with disease expression. J Clin Invest 93: 1195–1207, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan R, Colville D, Wang YY, Rigby L, Savige J: Alport retinopathy results from “severe” COL4A5 mutations and predicts early renal failure Clin J Am Soc Nephrol 5: 34–38, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barker DF, Denison JC, Atkin CL, Gregory MC: Efficient detection of Alport syndrome COL4A5 mutations with multiplex genomic PCR-SSCP. Am J Med Genet 98: 148–160, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.