Abstract
MicroRNAs are endogenous, noncoding, small RNAs that regulate expression and function of genes, but little is known about regulation of microRNA in the kidneys under normal or pathologic states. Here, we generated a mouse model in which the proximal tubular cells lack Dicer, a key enzyme for microRNA production. These mice had normal renal function and histology under control conditions despite a global downregulation of microRNAs in the renal cortex; however, these animals were remarkably resistant to renal ischemia-reperfusion injury (IRI), showing significantly better renal function, less tissue damage, lower tubular apoptosis, and improved survival compared with their wild-type littermates. Microarray analysis showed altered expression of specific microRNAs during renal IRI. Taken together, these results demonstrate evidence for a pathogenic role of Dicer and associated microRNAs in renal IRI.
Ischemia-reperfusion injury (IRI) in organs and tissues leads to the development of ischemic diseases, including myocardial infarction and stroke in the brain.1 In kidneys, IRI is a major cause of acute renal failure or acute kidney injury (AKI), a disease associated with unacceptably high mortality rates.2–5 The pathogenesis of IRI is very complex and likely involves an interplay between the cells within the organs and a marked inflammatory response. Despite decades of research, the cellular and molecular basis of IRI remains to be fully understood, and effective therapies are currently lacking.
MicroRNAs are a family of endogenous noncoding RNAs of 19 to 25 nucleotides, which have emerged as important regulators of various physiologic and pathologic processes in eukaryotes.6–11 MicroRNA genes are transcribed to produce the primary microRNAs, which are cleaved by Drosha to generate the precursor microRNAs of approximately 70 nucleotides. Precursor microRNAs are then transported to the cytosol, where they are cleaved by Dicer to produce mature and functional microRNAs. MicroRNAs often bind to the 3′-untranslated region of target mRNAs to repress protein translation and regulate gene expression.6–11
MicroRNAs are expressed in kidney cells and tissues; however, very little is known about the role and regulation of microRNAs in renal pathophysiology.12,13 In this regard, Natarajan and colleagues14,15 have suggested the involvement of specific microRNAs in TGF-β signaling during diabetic nephropathy. Recent studies by Harvey, Shi, and Ho and their colleagues16–18 further demonstrated that conditional knockout of Dicer from podocytes leads to glomerular defects and rapid progression to ESRD in mice, suggesting an important role for microRNAs in podocyte function in glomeruli of the kidneys. Despite these phenomenal findings, the involvement of microRNAs in the regulation of other renal cell types and related kidney diseases is unknown. In this study, we established a mouse model with targeted Dicer deletion from renal proximal tubules. Using this model, we demonstrated an important role for Dicer and associated microRNAs in the pathogenesis of renal IRI.
We used the Cre-loxP system to establish the renal proximal tubule–specific Dicer-null (PT-Dicer−/−) mouse model. To this end, Dicer(flox/flox) mice with two loxP sites flanking the exon of the second RNaseIII domain of Dicer19 were crossed with PEPCK-CRE mice that had X-linked Cre gene under the control of a modified rat cytosolic-PEPCK promoter.20 The modified PEPCK promoter drives Cre expression predominantly in renal proximal tubular cells and marginally in hepatocytes.20 As shown in Figure 1A, after two rounds of breeding, we obtained Dicer(flox/flox)XCREY mice that were expected to have Dicer deletion in renal proximal tubules (PT-Dicer−/−). We also obtained offspring of all other possible genotypes in Mendelian frequency (data not shown). Because of X-linked Cre expression, we used only male mice in this study to ensure the correct genotype.
Figure 1.

Mouse model of targeted deletion of Dicer from renal proximal tubules. (A) Breeding protocol for the production of proximal tubule–specific Dicer knockout (PT-Dicer−/−) mice. This study uses only male littermate mice with confirmed genotypes for experiment. (B) PCR-based genotyping to confirm Dicer deletion from renal cortical tissues. Genomic DNA is isolated from renal cortical tissues of PT-Dicer−/− and PT-Dicer+/+ mice. The DNA samples (50 ng per lane) are PCR-amplified using the primer pair DicerF1 and DicerDel (sequences shown in the Concise Methods section). Actin is also amplified as control. (C) Immunoblot analysis of Dicer expression in renal cortical tissues. Renal cortical tissues are dissected from kidneys of PT-Dicer−/− and PT-Dicer+/+ mice for homogenization to collect whole-tissue lysates for immunoblotting of Dicer and β-actin.
To verify the depletion of Dicer in proximal tubular cells, we examined the genomic DNA from kidney cortex, which consists mainly of proximal tubules. As shown in Figure 1B, PCR amplified a clear wild-type allele fragment from the wild-type (PT-Dicer+/+) tissues. In contrast, all PT-Dicer−/− tissues showed a strong Dicer deletion fragment, confirming Cre-mediated deletion of Dicer from proximal tubules. At the protein level, we examined Dicer expression in renal cortical tissues by immunoblot analysis. As shown in Figure 1C, wild-type tissues showed two Dicer isoform bands, both of which were markedly reduced in PT-Dicer−/− tissues. The immunoblot result further suggested Dicer knockout from proximal tubules in PT-Dicer−/− mice, because renal cortical tissues consist mainly of proximal tubules. Together, these results validated the PT-Dicer−/− conditional knockout model.
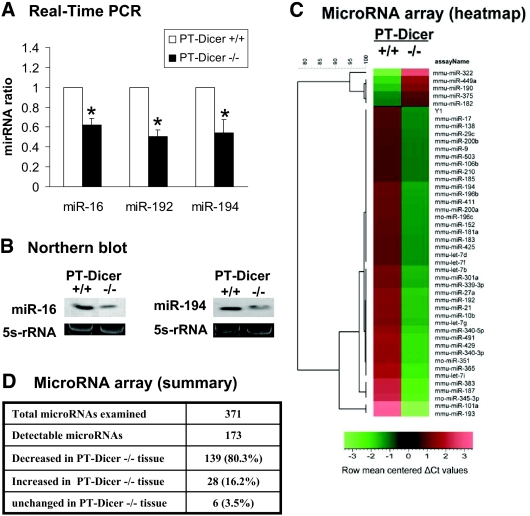
To confirm miRNA depletion in PT-Dicer−/− renal tissues, we initially examined miR-192 and miR-194, two miRNAs highly expressed in kidneys, and miR-16, a ubiquitously expressed miRNA.21,22 As shown in Figure 2A, real-time PCR detected significantly lower miR-192, miR-194, and miR-16 in renal cortical tissues from PT-Dicer−/− mice than those from PT-Dicer+/+ mice. Consistently, Northern blot analysis demonstrated that miR-16 and miR-194 expression was diminished in renal cortical tissues from PT-Dicer−/− mice (Figure 2B). We further conducted microarray analysis to compare miRNA expression in renal cortical tissues from PT-Dicer−/− and PT-Dicer+/+ mice. A representative heat map of the microarray is shown in Figure 2C. Among the 173 detectable miRNAs, the majority (139 [80.3%] miRNAs) were downregulated in PT-Dicer−/− tissues (Figure 2D). Of note, miRNA depletion was not complete in these tissues, which might be caused by (1) other cell types in kidney cortex that did not express Cre for Dicer ablation and (2) Dicer-independent maturation/production of specific miRNA species. Indeed, as reported previously,23,24 a small number of miRNAs were actually upregulated in PT-Dicer−/− tissues (Figure 2D), whereas the majority of miRNAs were depleted.
Figure 2.
Depletion of miRNAs from renal cortical tissues in PT-Dicer−/− mice. Total RNA is isolated from renal cortical tissues of PT-Dicer+/+ and PT-Dicer−/− mice. (A) Real-time PCR analysis of miR-16, miR-192, and miR-194. Real-time PCR is performed using the Taqman miRNA assay kit as described in the Concise Methods section. The value of each miRNA is normalized by the signal of snoRNU202, an internal control. The normalized values of PT-Dicer+/+ samples are arbitrarily set as 1; data of PT-Dicer−/−samples are expressed as mean ± SD (n = 3). *Significantly different from the PT-Dicer+/+ values. (B) Northern blot analysis of miR-16 and miR-194. Total RNA (10 μg per lane) is analyzed by Northern blotting as described in the Concise Methods section using a p32-labeled probe of miR-16 or miR-194. 5s-rRNA is shown as an RNA loading control. (C) Representative heat map of microRNA microarray analysis. Total RNA samples isolated from renal cortical tissues of PT-Dicer+/+ and PT-Dicer−/− mice are subjected to microRNA microarray analysis. The ΔCt values of all miRNAs are used to generate the heat map. (D) Summary of microRNA microarray results. The microarray included 371 miRNA species, 173 of which were detectable in the renal cortical tissues. The value of a specific miRNA from the PT-Dicer−/− sample is compared with that of PT-Dicer+/+ sample to show whether the miRNA is decreased, increased, or unchanged. The results are representative of duplicate analyses.
The PT-Dicer−/− mice developed normally and did not show obvious abnormalities in kidney size (data not shown), histology, or renal function (Figures 3 and 4A). These animals had blood urea nitrogen (BUN) levels of approximately 30 mg/dl and serum creatinine levels of approximately 0.4 mg/dl at the age of 8 to 10 weeks, which were comparable to the levels of wild-type PT-Dicer+/+ littermates. We monitored renal function, histology, and kidney size in PT-Dicer−/− mice for >6 months and did not find phenotypic alterations (data not shown).
Figure 3.
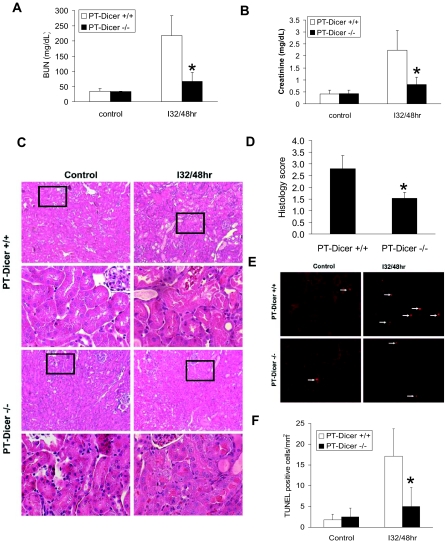
Resistance of PT-Dicer−/− mice to renal IRI. Male PT-Dicer+/+ and PT-Dicer−/− mice of 8 to 10 weeks of age are subjected to sham operation (Control) or 32 minutes of bilateral renal ischemia followed by 48 hours of reperfusion (I32/48h). (A and B) Blood samples are collected to measure BUN (A) and serum creatinine (B). Data are means ± SD. (C) Renal tissues are collected for hematoxylin and eosin staining to examine histology. Histologic images of both low and high magnifications. (D) Ischemia reperfusion–induced tubular damage in PT-Dicer+/+ and PT-Dicer−/− mice is evaluated and scored by histology. Data are means ± SD. (E) Renal cortical and outer medulla tissues are also collected for TUNEL assay of apoptosis. (F) Representative images of TUNEL assay. TUNEL-positive cells are quantified by cell counting in comparable regions of the tissues. Data are means ± SD. n ≥ 4. *Significantly different from PT-Dicer+/+ group.
Figure 4.
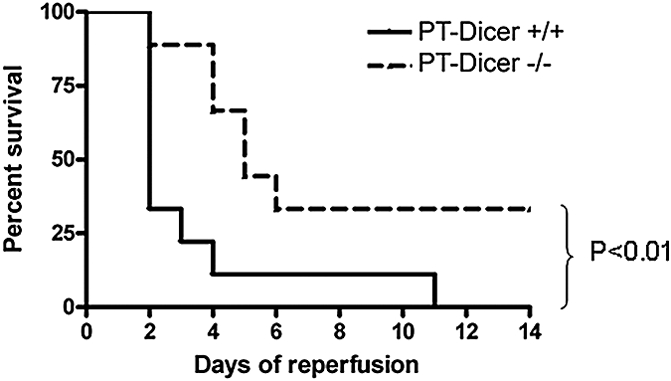
Survival of PT-Dicer−/− and PT-Dicer+/+ mice after severe renal IRI. PT-Dicer−/− mice (n = 9) and their wild-type PT-Dicer+/+ littermates (n = 10) we are subjected to 35 minutes of bilateral renal ischemia. Animal survival is monitored every day until 14 days of reperfusion. Statistical difference is determined by Kaplan-Meier survival analysis.
Renal ischemia-reperfusion is a major cause of AKI, a disease associated with high mortality. Although ischemic AKI involves multiple factors and may proceed in several phases, it is precipitated by sublethal and lethal damage to renal tubules.3–5 To gain insights into microRNA regulation in this kidney disease, we took advantage of the PT-Dicer−/− mouse model. Specifically, we examined and compared ischemic AKI in PT-Dicer+/+ and PT-Dicer−/− littermate mice. As shown in Figure 3A, 32 minutes of bilateral renal ischemia followed by 48 hours of reperfusion (I32/48h) led to an increase of BUN level from 32 to 217 mg/dl in PT-Dicer+/+ mice, which was markedly suppressed to 67 mg/dl in PT-Dicer−/− mice. Consistently, I32/48h resulted in serum creatinine increase to 2.2 mg/dl in PT-Dicer+/+ mice but only 0.8 mg/dl in PT-Dicer−/− littermates (Figure 3B). Thus, ischemia-induced loss of renal function was attenuated in PT-Dicer−/− mice.
Corroborating the functional analysis, renal histology revealed significantly less tissue damage in PT-Dicer−/− mice after ischemic injury. As shown in Figure 3C, I32/48h led to severe tubular damage or necrosis in renal cortex and outer stripe of outer medulla in PT-Dicer+/+ mice; especially, most of the tubules in the S3 segment seemed lysed. In sharp contrast, in PT-Dicer−/− mice, much fewer tubules were injured and the injury in individual tubules was also less severe, mainly showing tubular dilation and loss of brush boarder (Figure 3C). Quantification of injured tubules indicated that cortical and outer medulla tubular damage after renal ischemia-reperfusion was significantly ameliorated in PT-Dicer−/− mice (Figure 3D).
In addition to acute necrotic damage, tubular apoptosis contributes to the development of ischemic AKI.3–5 To determine whether Dicer and associated miRNAs are involved in tubular apoptosis, we examined renal tissues by terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (TUNEL) assay. Apoptotic cells were rare in control tissues of both PT-Dicer+/+ and PT-Dicer−/− mice (Figure 3, E and F). After I32/48h, the number of apoptotic cells increased to approximately 17/mm2 cortical tissue in PT-Dicer+/+ mice but only 5/mm2 cortical tissue in PT-Dicer−/− mice. The results of TUNEL assay were confirmed by immunofluorescence examination of caspase-3 activation in renal tissues (data not shown).
To examine the survival of PT-Dicer+/+ and PT-Dicer−/− mice after severe ischemic injury, we subjected the animals to 35 minutes of renal ischemia. As shown in Figure 4, a dramatic loss of animals was detected in PT-Dicer+/+ mice after 2 days of reperfusion, and all mice in this group died by day 11. In PT-Dicer−/− mice, there was a gradual loss of animals between days 2 and 6 of reperfusion; thereafter, all animals survived, and, as a result, 30% animals fully recovered during the observation period of 14 days. Kaplan-Meier analysis indicated that PT-Dicer−/− mice survived significantly (P < 0.01) better than PT-Dicer+/+ mice after renal ischemia-reperfusion.
Finally, we profiled microRNA expression during renal ischemia-reperfusion by microRNA microarrays. As summarized in Table 1, the expression of specific miRNAs showed significant changes, and, interestingly, whereas some were induced, others were downregulated. Notably, miRNA-132, -362, and -379 showed a continuous change during 12 to 48 hours of reperfusion, but other miRNAs changed at only one time point.
Table 1.
Microarray profiling of microRNA expression during renal ischemia-reperfusion
| Increased MicroRNAs |
Decreased MicroRNAs |
||
|---|---|---|---|
| 30 Minutes of Ischemia and 12 Hours of Reperfusion | 30 Minutes of Ischemia and 48 Hours of Reperfusion | 30 Minutes of Ischemia and 12 Hours of Reperfusion | 30 Minutes of Ischemia and 48 Hours of Reperfusion |
| mir-132 | mir-7 | mir-18a | mir-324-3p |
| mir-17–3p | mir-132 | mir-135b | mir-379 |
| mir-362 | mir-486 | mir-296 | mir-455-3p |
| mir-685 | mir-362 | mir-127 | |
| mir-687 | mir-467 | mir-322 | |
| mir-207 | mir-495 | mir-379 | |
| mir-489 | mir-668 | mir-487b | |
| mir-694 | mir-491 | ||
Total RNA was extracted from kidney cortex and outer medulla for TaqMan microRNA microarray analysis. Log2 fold changes over the sham operation group were determined for each microRNA in two microarray analyses. Listed are microRNAs that show an average log2 fold changes >2.
In summary, in this study, we established a conditional knockout mouse model with targeted Dicer deletion from renal proximal tubules. The conditional knockout mice did not show overt defects in kidney development, histology, and function. Interestingly, these animals are strikingly resistant to ischemic AKI. The results have therefore provided the first evidence of a role of Dicer and associated miRNA production in ischemic AKI.
In the conditional knockout mice, we detected the Dicer-deletion alleles in renal cortical tissues that mainly contain proximal tubules (Figure 1B). Moreover, immunoblot analysis showed a marked decrease in Dicer expression in renal cortical tissues, further indicating Dicer deletion from proximal tubules at the protein expression level (Figure 1C). The observation of a global depletion of microRNAs in renal cortical tissues provides functional evidence of Dicer deletion (Figure 2). Together, these results suggest that Dicer is mainly ablated from renal proximal tubules in the conditional knockout model, although the possibility of minor Dicer deletion in some other cell types, such as distal tubules and glomerular cells, cannot be completely excluded.
Conditional knockout of Dicer in specific cell types affects the development of various organs or tissues.19,25–30 In kidneys, deletion of Dicer from podocytes leads to glomerulopathy, early proteinuria, and rapid progression to ESRD;16–18; however, in our study, targeted Dicer deletion from renal proximal tubules did not show obvious phenotypes in the kidneys. During the observation period of 6 months, these mice had normal kidney size, renal histology, and renal function. The absence of phenotypes in this conditional Dicer knockout model is mostly likely due to the late turn-on feature of the PEPCK promoter that was used to drive Cre expression in proximal tubular cells in our study. In the kidneys, the cytosolic-PEPCK promoter is not activated in proximal tubular cells until 3 weeks after birth,31 a time point when kidney development has mostly completed in mice.32 Accordingly, in our study, Dicer deletion via PEPCK promoter-driven Cre expression likely occurred after the major events of kidney development. Thus, our results do not rule out a role of proximal tubular Dicer and associated miRNAs in kidney development.
The absence of renal developmental phenotypes in the PT-Dicer−/− mice supports the feasibility of using this model to investigate the possible involvement of tubular Dicer and miRNAs in renal pathophysiology in adult life. In this study, we demonstrated a remarkable resistance of the PT-Dicer−/− mice to ischemic AKI. After renal ischemia-reperfusion, these mice showed less tissue damage and fewer apoptotic cells, resulting in a significantly better renal function than their wild-type littermates. Notably, the PT-Dicer−/− mice survived significantly better after severe ischemic AKI. Our microarray analysis further revealed the changes of specific miRNAs during renal ischemia-reperfusion. Together, these observations support a critical role for Dicer and associated miRNA production in the pathogenesis of ischemic AKI. Further studies should identify the key miRNAs that contribute to ischemic AKI. In addition, investigation of the target genes of the miRNAs would gain insights into the pathogenesis of this disease.
Concise Methods
Mouse Strains
Dicer(flox/flox) mouse breeders were obtained from Dr. Michael McManus (University of California, San Francisco, CA).19 PEPCK-CRE mouse model was established in Dr. Volker Haase's laboratory as described previously.20 Dicer(flox/flox) mice were crossed to PEPCK-CRE mice to produce Dicer(flox/flox)XCREY as depicted in the breeding protocol in Figure 1A. Dicer(flox/flox)XCREY mice were Dicer-deleted in renal proximal tubules. These mice and wild-type littermates (male, 8 to 10 weeks old) were used for ischemic renal injury experiments. The mice were housed in Charlie Norwood VA Medical Center animal facility with a 12/12-hour light/dark cycle and food and water available ad libitum. All animal experiments were performed according to a protocol approved by the Institutional Animal Care and Use Committee of Charlie Norwood VA Medical Center at Augusta.
Genotyping
Genomic DNA was extracted from mouse tail biopsy for PCR-based genotyping. The Dicer-floxed allele was detected by PCR using primers DicerF1 (CCTGACAGTGACGGTCCAAAG) and DicerR1 (CATGACTCTTCAACTCAAACT) to produce a 420-bp floxed allele product and a 351-bp wild-type allele product.19 Dicer deletion in renal cortical tissues was confirmed by using primers DicerF1 and DicerDel (CCTGAGCAAGGCAAGTCATTC) for a 471-bp deletion allele product and a 1.3k-bp wild-type allele product.19 The presence of CRE gene was detected by PCR using the primer pair ACCTGAAGATGTTCGCGATTATCT and ACCGTCAGTACGTGAGATATCTT to amplify a 370-bp fragment.33
Real-Time PCR Analysis of miRNAs
Total RNA was extracted using the mirVana miRNA isolation kit (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's instruction. Forty nanograms of total RNA was converted to cDNA using the miRNA Reverse Transcription kit (Applied Biosystems). Real-Time PCR was carried out using the Taqman miRNA assay kit (Applied Biosystems), which included the sequence-specific primers for cDNA synthesis and Taqman probes for real-time PCR. Quantification was done using ΔCt values.
Northern Blot Analysis of miRNAs
Total RNA was extracted using the mirVana miRNA isolation kit. Ten micrograms of RNA was run on 15% acrylamide-bisacrylamide (19:1) gel containing 7 M urea in Tris-borate-EDTA buffer. The RNA was then transferred onto the Hybond-N+ membrane (Amersham, Piscataway, NJ) at 200 mA for 1 hour. After ultraviolet cross-linking, the membrane was dried at 80°C for 1 hour followed by prehybridization for 1 hour in Ultra-Hyb-Oligo hybridization buffer (Applied Biosystems/Ambion) at 37°C. The p32-labeled antisense probe of a specific miRNA was added to the prehybridization buffer and incubated overnight at 37°C. Then the membrane was washed in 2× SSC buffer (0.1% SDS) and exposed to x-ray film at −80°C.
MicroRNA Microarray
MicroRNA microarray was conducted by Genome-Explorations Inc. (Memphis, TN) with total RNA isolated from kidney cortical tissues.
Renal Ischemia-Reperfusion
Renal ischemia-reperfusion was induced in mice as described previously.34,35 Briefly, mice were anesthetized with 50 to 60 mg/kg (intraperitoneally) pentobarbital sodium and kept on a homoeothermic station to maintain body temperature at 37°C. Kidneys were exposed by bilateral flank incision, and the renal pedicles were clamped to induce ischemia. After ischemia, the arterial clamps were released for reperfusion. Sham control animals were subjected to identical operation but not renal pedicle clamping.
Renal Function, Histology, and Apoptosis
Renal analyses were conducted as described in our recent studies with minor modifications.34,35 Renal function was monitored by measuring BUN and serum creatinine using analytical kits from Biotron Diagnostics (Hemet, CA) and Stanbio Laboratory (Boerne, TX), respectively. Renal histology was examined by hematoxylin and eosin staining. Briefly, kidneys were collected and fixed with 4% paraformaldehyde at 4°C overnight. The kidneys were then paraffin-embedded and sectioned at 4 μm for hematoxylin and eosin staining. Histologic changes were evaluated by the percentage of injured/damaged renal tubules. Tissue damage was scored as follows: 0, no damage; 1, <25%; 2, 25 to 50%; 3, 50 to 75%; and 4, >75%. Pictures of representative fields were also recorded. Renal apoptosis was examined by TUNEL assay using the in situ Cell Death Detection kit from Roche Applied Science (Indianapolis, IN). Briefly, paraffin-embedded renal tissue sections were deparaffinized and permeabilized by 2 hours of incubation at 65°C in 0.1 M sodium citrate (pH 6.0). The sections were then exposed to a TUNEL reaction mixture containing TM red–labeled dUTP. TUNEL positive nuclei were identified by fluorescence microscopy.
Statistical Analysis
Kaplan-Meier analysis was conducted using the GraphPad Prism software. Statistical differences between two groups were determined by two-tailed unpaired t test (Microsoft Excel). P < 0.05 was considered statistically significant. Microarray data are representative of two separate analyses. Other qualitative data including PCR and Northern blots are representative of at least three experiments.
Disclosures
None.
Acknowledgments
The study was supported by grants from the National Institutes of Health, Veterans Administration, and the Intramural Grants Program of Medical College of Georgia. Z.D. is a Veterans Administration Research Career Scientist.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
REFERENCES
- 1. Semenza GL: Series introduction: Tissue ischemia—Pathophysiology and therapeutics. J Clin Invest 106: 613–614, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schrier RW, Wang W, Poole B, Mitra A: Acute renal failure: Definitions, diagnosis, pathogenesis, and therapy. J Clin Invest 114: 5–14, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonventre JV, Weinberg JM: Recent advances in the pathophysiology of ischemic acute renal failure. J Am Soc Nephrol 14: 2199–2210, 2003. [DOI] [PubMed] [Google Scholar]
- 4. Devarajan P: Update on mechanisms of ischemic acute kidney injury. J Am Soc Nephrol 17: 1503–1520, 2006. [DOI] [PubMed] [Google Scholar]
- 5. Molitoris BA: Transitioning to therapy in ischemic acute renal failure. J Am Soc Nephrol 14: 265–267, 2003. [DOI] [PubMed] [Google Scholar]
- 6. Ruvkun G: The perfect storm of tiny RNAs. Nat Med 14: 1041–1045, 2008. [DOI] [PubMed] [Google Scholar]
- 7. Carrington JC, Ambros V: Role of microRNAs in plant and animal development. Science 301: 336–338, 2003. [DOI] [PubMed] [Google Scholar]
- 8. Bushati N, Cohen SM: microRNA functions. Annu Rev Cell Dev Biol 23: 175–205, 2007. [DOI] [PubMed] [Google Scholar]
- 9. van Rooij E, Olson EN: MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest 117: 2369–2376, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winter J, Jung S, Keller S, Gregory RI, Diederichs S: Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234, 2009. [DOI] [PubMed] [Google Scholar]
- 11. Croce CM, Calin GA: miRNAs, cancer, and stem cell division. Cell 122: 6–7, 2005. [DOI] [PubMed] [Google Scholar]
- 12. Kato M, Arce L, Natarajan R: MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 4: 1255–1266, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saal S, Harvey SJ: MicroRNAs and the kidney: Coming of age. Curr Opin Nephrol Hypertens 18: 317–323, 2009. [DOI] [PubMed] [Google Scholar]
- 14. Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R: TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11: 881–889, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R: MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 104: 3432–3437, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH: Podocyte-specific deletion of Dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19: 2150–2158, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA: Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol 19: 2069–2075, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP: Podocyte-selective deletion of Dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 19: 2159–2169, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ: The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A 102: 10898–10903, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rankin EB, Tomaszewski JE, Haase VH: Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res 66: 2576–2583, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ: Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res 32: e188, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM: An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A 101: 9740–9744, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ: Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 105: 2111–2116, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P: Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med 204: 1553–1558, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT: Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A 105: 5614–5619, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X: Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A 103: 2208–2213, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD: Essential role for Dicer during skeletal muscle development. Dev Biol 311: 359–368, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE: The miRNA-processing enzyme Dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 16: 1041–1049, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS: MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 56: 2938–2945, 2007. [DOI] [PubMed] [Google Scholar]
- 30. Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS: Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci U S A 106: 10266–10271, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hanson RW, Reshef L: Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu Rev Biochem 66: 581–611, 1997. [DOI] [PubMed] [Google Scholar]
- 32. Matsusaka T, Miyazaki Y, Ichikawa I: The renin angiotensin system and kidney development. Annu Rev Physiol 64: 551–561, 2002. [DOI] [PubMed] [Google Scholar]
- 33. Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA: Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274: 305–315, 1999. [DOI] [PubMed] [Google Scholar]
- 34. Brooks C, Wei Q, Cho S, Dong Z: Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Invest 119: 1275–1285, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei Q, Yin XM, Wang MH, Dong Z: Bid deficiency ameliorates ischemic renal failure and delays animal death in C57BL/6 mice. Am J Physiol Renal Physiol 290: F35–F42, 2006. [DOI] [PubMed] [Google Scholar]