Abstract
We show that most globus pallidus neurons, but very few neocortical interneurons, are generated from the ventral medial ganglionic eminence and dorsal preoptic area based on fate mapping using an Shh-Cre allele. The Shh-expressing subpallial lineage produces parvalbumin+ GABAergic neurons, ChAT+ cholinergic neurons, and oligodendrocytes. Loss of Nkx2-1 function from the Shh-expressing domain eliminated most globus pallidus neurons, whereas most cortical and striatal interneurons continued to be generated, except for striatal cholinergic neurons. Finally, our analysis provided evidence for a novel cellular component (Nkx2-1−;Npas1+) of the globus pallidus.
Introduction
The embryonic mouse subpallium generates the projection neurons of the basal ganglia, as well as telencephalic GABAergic, cholinergic, and dopaminergic interneurons. The subpallium has four principal progenitor domains: lateral ganglionic eminence (LGE), medial ganglionic eminence (MGE), preoptic area (POA), and septum [the caudal ganglionic eminence (CGE) contains caudal parts of the LGE and MGE] (Flames et al., 2007); each of these is further subdivided. For instance, Flames et al. (2007) proposed five MGE progenitor domain subdivisions: pMGE1–5.
Distinct progenitor domains generate distinct neurons and glia based on loss-of-function mutations and fate-mapping studies. Nkx2-1−/− mutants lack an identifiable globus pallidus (GP) and ∼50% of cortical interneurons [Lhx6+, parvalbumin+ (PV+), and somatostatin+ (SS+)] (Sussel et al., 1999; Pleasure et al., 2000; Xu et al., 2004; Butt et al., 2008; Nóbrega-Pereira et al., 2008). The Nkx2-1−/− MGE lacks Lhx6 and Lhx7(8) expression and is transformed toward an LGE identity (Sussel et al., 1999; Corbin et al., 2003; Butt et al., 2008). Lhx6 mutants fail to generate parvalbumin and somatostatin cortical interneurons (Liodis et al., 2007; Zhao et al., 2008), while Lhx8 mutants have fewer subpallial cholinergic neurons (Zhao et al., 2003; Mori et al., 2004; Fragkouli et al., 2005).
Transplantation of dissected MGE subdivisions provided evidence that somatostatin+ interneurons are preferentially derived from dorsal MGE, while parvalbumin+ interneurons are preferentially derived from ventral MGE (Flames et al., 2007; Wonders et al., 2008). Cre-mediated recombination fate mapping, using Cre driven by Lhx6, Nkx2-1, and Nkx6.2 bacterial artificial chromosome (BAC) transgenes, is consistent with the loss-of-function and transplantation analysis (Fogarty et al., 2007; Xu et al., 2008). Furthermore, the Shh+ domain in the ventral MGE/dorsal POA appears to be a source for oligodendrocytes (Fogarty et al., 2007; Petryniak et al., 2007).
Derivatives of the POA progenitor zones are less well characterized. Fate mapping with Dbx1-Cre provided evidence that the POA contributes cells to the preoptic area and to the amygdala (Hirata et al., 2009), whereas Nkx5.1-Cre shows that the POA also generates a specific subtype of neuropeptide Y+ (NPY+) cortical interneuron (Gelman et al., 2009).
Here, we report on the fate of the dorsal POA and the ventral-most MGE by performing fate mapping with an Shh-Cre allele (Harfe et al., 2004). These results show that most of the globus pallidus is generated from this region, whereas it generates surprisingly few cortical interneurons.
Next, we investigated the function of Nkx2-1 in Shh-expressing telencephalic cells using Shh-Cre and a floxed-Nkx2-1 allele (Kusakabe et al., 2006). Loss of Nkx2-1 function from the Shh-expressing subcortical domain eliminated most globus pallidus neurons, whereas most cortical and striatal interneurons continued to be generated, except for striatal cholinergic neurons. Thus, this analysis demonstrated that the dorsal MGE and caudal parts of the LGE (CGE) generated most of the cortical GABA interneurons, whereas the ventral-most MGE and dorsal POA primarily generated most of the globus pallidus and striatal cholinergic neurons. Finally, our analysis provided evidence for an Nkx2-1- and Shh-independent lineage that is Npas1+ and which constitutes ∼15% of globus pallidus cells.
Materials and Methods
Animals and tissue preparation
Mouse colonies were maintained in accordance with the protocols approved by the Committee on Animal Research at University of California, San Francisco.
Nkx2-1 floxed (Nkx2-1f) alleles and constitutive null alleles were generated by S. Kimura and genotyped as described previously (Kimura et al., 1996; Kusakabe et al., 2006; Mastronardi et al., 2006). Shh-Cre mice were provided by C. Tabin and genotyped as described previously (Harfe et al., 2004). The ROSA26 LacZ reporter mouse (ROSA) was used for Cre fate mapping as described by Soriano (1999). The Lhx6 BAC-GFP and Dlx1 BAC-GFP mouse transgenic lines were generated by GENSAT (http://www.gensat.org/index.html) and are described by Cobos et al. (2006). For staging of embryos, midday of the vaginal plug was defined as embryonic day 0.5 (E0.5). Mouse colonies were maintained in accordance with the protocols approved by the Committee on Animal Research at University of California, San Francisco. Breeding was performed as follows: Nkx2-1+/−;ShhCre/+ males were crossed with Nkx2-1f/f;ROSAR26R/+ females. Nkx2-1f/−; Shh+/+ or Nkx2-1f/+;ShhCre/+ mice were use as controls [heterozygote (HET)] and Nkx2-1f/−;ShhCre/+ mice were used as mutants. Adults were deeply anesthetized in a CO2 chamber and killed by cervical dislocation. Embryos were removed by cesarean section. Embryonic and postnatal day 0 (P0) mice were anesthetized by cooling; the brain was removed and immersion fixed in 4% paraformaldehyde (PFA) in phosphate-buffered solution (PB 0.1 m, pH 7.4) for 4–12 h. Two-month-old mice were anesthetized with Avertin (0.2 cc/10 g of body weight) and perfused intracardially with 4% PFA in PB before their brains were removed. Samples were cryoprotected in a gradient of sucrose 30%, frozen in embedding medium (OCT; Tissue-Tek), and sectioned using a cryostat.
In situ RNA hybridization
Nonradioactive in situ RNA hybridization was performed using digoxigenin-labeled riboprobes on 20 μm frozen sections as described on the Rubenstein laboratory website (http://physio.ucsf.edu/rubenstein/protocols/index.asp; Cobos DIG ISH protocol). Probe sequences have been previously described (Zhao et al., 2008; Long et al., 2009a). The following cDNAs were used: CoupTF1, Dbx1, ER81, Gad1, Gbx1, Gsx2 (Gsh2), Golf, Ikaros, Islet1, Lhx6, Lhx7(8), Lmo3, Lmo4, Npas1, Olig2, Pou3f1 (Oct6), Shh, somatostatin, and Zic1. Nkx2-1 exon 2 (Nkx2-1-E), Nkx2-1 full length (Nkx2-1-FL), Nkx6.2, Dlx1, and Dlx5 were generated in the Rubenstein laboratory. For more complete information about these cDNAs, see the study by Long et al. (2009a).
Immunohistochemistry
Brain sections were cut using a cryostat at 20 μm thickness for embryonic ages and P0, or at 40 μm for 2-month-old mouse brains. Twenty-micrometer sections were mounted on glass slides (Superfrost Plus, VWR) and 40 μm sections were kept free floating in PB buffer. Immunostaining was performed according to Zhao et al. (2008). The following primary antibodies were used: rabbit anti-MAFb (1:1000) (Bethyl Laboratories), rabbit anti-ER81 (1:20,000) (Covance), mouse anti-Nkx2-1 (1:50) (Novocastra Laboratories), rabbit anti-Nkx2-1 (1:1000) (BIOPAT Immunotechnologies), rabbit anti-PV (1:4000) (Swant), chicken anti-green fluorescent protein (GFP) (1:1000) (Aves Laboratories), rabbit anti-Olig2 (1:20,000) (kindly provided by John Alberta, Dana-Farber Cancer Institute), goat anti-choline acetyltransferase (ChAT) (1:250) (Millipore Bioscience Research Reagents), rabbit anti-Npas1 (1:1000) (kindly provided by Steve McKnight, University of Texas Southwestern, Dallas, TX), rabbit anti-phosphohistone 3 (PH-3) (1:200) (Millipore), rabbit anti-NPY (1:2000) (Immunostar), rat anti-SS (1:250) (Millipore Bioscience Research Reagents), rabbit anti-calretinin (CR) (1:2000) (Millipore Bioscience Research Reagents), guinea pig anti-β-galactosidase (bGAL) (1:1000) (kindly provided by Thomas Finger, University of Colorado School of Medicine, Denver, CO), mouse anti-β-III-tubulin (TUBIII) (1:1000) (clone TUJ1; Covance), mouse anti-bromodeoxyuridine (BrdU) (1:500) (clone B44; Becton Dickinson), rabbit anti-caspase-3 (1:500) (BD Pharmingen), rabbit anti-calbindin (CB) (1:4000) (Swant); mouse anti-nestin (1:200) (Millipore Bioscience Research Reagents), rabbit anti-brain lipid binding protein (BLBP) (1:1000) (Millipore Bioscience Research Reagents), rabbit anti-DARPP32 (1:2000) (Cell Signaling Technologies), and rabbit anti-TrkA (1:1000) (kindly provided by Louis Reichardt, University of California, San Francisco, CA). Alexa 488 and Alexa 594 secondary antibodies (Invitrogen) were used accordingly to the primary antibody species. Sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted with Vectashield mounting medium (Vector Laboratories). Immunoperoxidase method was used for ChAT staining with a biotinylated anti-goat IgG secondary antibody and the ABC elite kit (Vector Laboratories).
Microscopy and data collection
Bright-field images were taken for in situ RNA hybridization data by use of an SZX7 microscope (Olympus) and a DP70 camera (Olympus). Immunolabeling images were taken using a Nikon Eclipse 80i microscope (Nikon), a CoolSnap camera (Photometrics), and NIS-Elements BR 3.00 software (Nikon). Confocal images were taken using a confocal microscope (LSM510, Zeiss) (pinhole: 2 μm). The contrast and brightness of these images were adjusted for better visualization using Photoshop CS2 software (Adobe Systems).
Cell counting
Counting for Shh-Cre cell fate (bGAL+) and for transcription factor expression in the globus pallidus.
Confocal images were used for the quantification of data below (see Figs. 1, 3) (P0 and P60 analysis). The whole image area was used for counting. For counting data below [see Fig. 1A″ (E13.5), Fig. 4 (E18.5), and supplemental Fig. 2, available at www.jneurosci.org as supplemental material (E18.5)], counting was performed on a 10× magnification image. Boxes were drawn within the boundaries of the NKX2-1+ globus pallidus; the area of the boxes represented ∼1/4 of the globus pallidus area and had ∼100 DAPI+ cells for the E13.5 data and ∼150–200 DAPI+ cells for the E18.5 data. For both confocal and 10× data, images with red, green, and blue channels were created using Photoshop CS2 software. Only cells with clear DAPI staining were counted. Total numbers of red, green, and yellow cells were counted.
Figure 1.
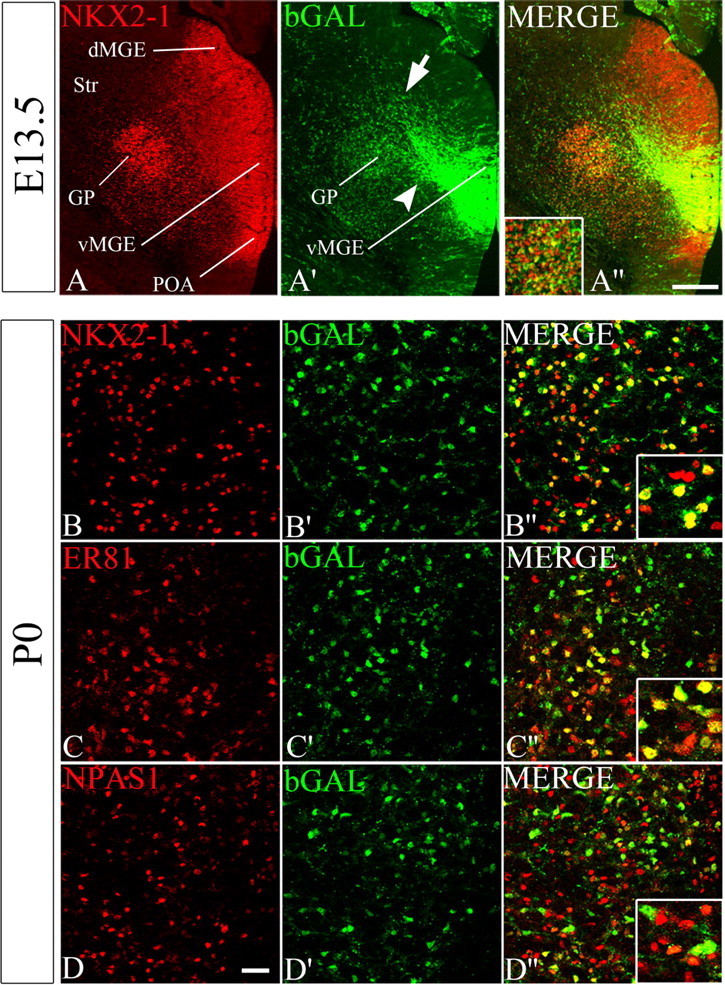
Cells derived from the Shh+ progenitor zone generate most of the globus pallidus neurons. A–A″, Coronal hemisection through the MGE and globus pallidus of an E13.5 ROSA;ShhCre/+ brain, costained with NKX2-1 and bGAL antibodies. Two streams of migrating bGAL+ cells are indicated in A′ by an arrow (SVZ migration) and arrowhead (migration to the GP). B–D″, Confocal images of coronal sections through the globus pallidus of a P0 ROSA;ShhCre/+ brain costained with bGAL and NKX2-1 antibodies (B–B″), bGAL and ER81 antibodies (C–C″), or bGAL and NPAS1 antibodies (D–D″). Higher-magnification images of globus pallidus cells are shown in A″–D″. Scale bars: (in A″) A–A″, 200 μm; (in D) B–D″, 100 μm. dMGE, Dorsal MGE; vMGE, ventral MGE; Str, striatum.
Figure 3.
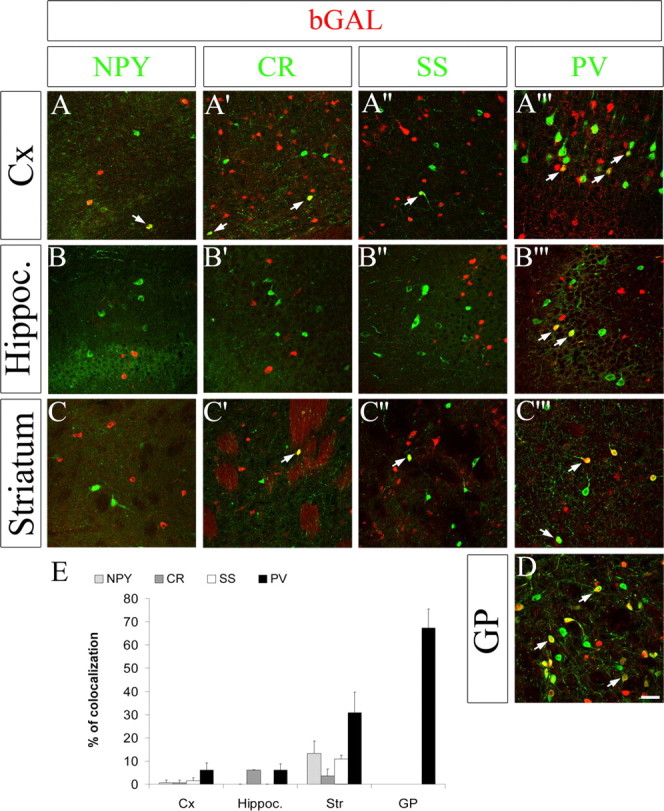
A–D, The Shh domain generates most neurons of the globus pallidus but few neocortical and hippocampal interneurons. Double immunofluorescence labeling for bGAL and markers for GABAergic neurons (NPY, CR, SS, and PV) defines which cell types are derived from the Shh lineage in the neocortex, hippocampus, striatum, and globus pallidus at P60 in ROSA;ShhCre/+ mice. White arrows indicate examples of bGAL+ cells that express GABAergic neuronal subtype markers. E, Histogram summarizes the percentage of bGAL+ cells that express the different markers for the indicated regions (average of 3 separate experiments ± SEM). Scale bar, 50 μm. Cx, Neocortex; Hippoc, hippocampus; Str, striatum.
Figure 4.
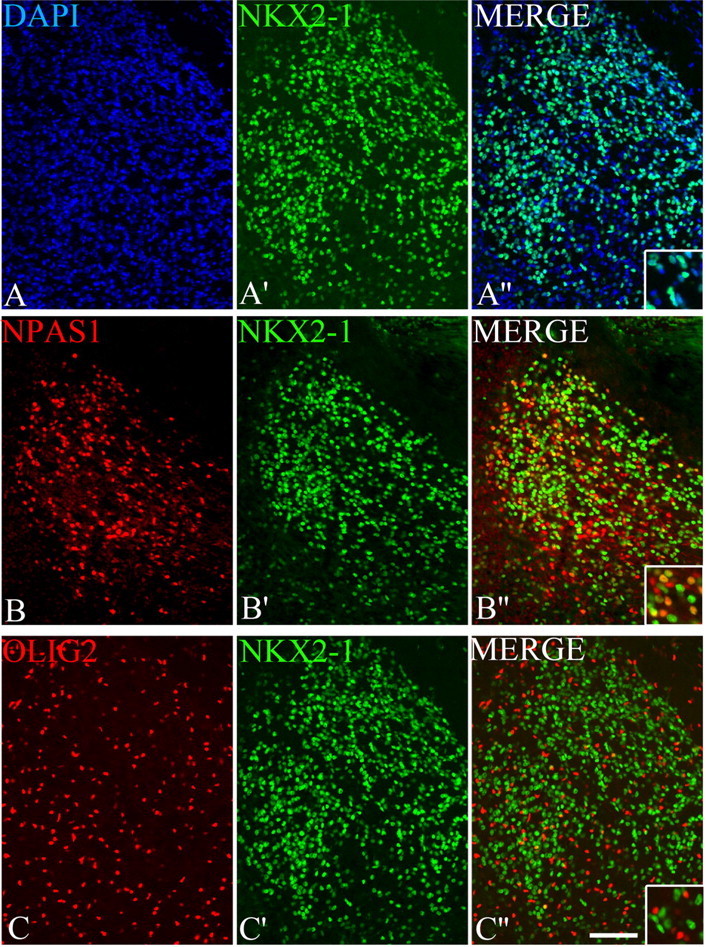
A–C, Expression of three transcription factors defines subsets of globus pallidus cells at E18.5. Double immunofluorescence and DAPI nuclear staining on wild-type coronal sections using NKX2-1, NPAS1, and OLIG2 antibodies. Higher magnifications of the merged images are shown in A′–C′ and A″–C″. Scale bar, 100 μm.
Counting for cell numbers in sections from the Nkx2-1f/−;ShhCre/+ mutants and controls.
The quantification of cortical interneurons on E18.5 heterozygote and the mutant were performed on 10× images (see Fig. 10). The cortex and striatum were used as anatomical landmarks to match the rostrocaudal levels. We drew boxes (500 μm wide) from the ventricular zone to the pial surface. For the ventral pallium region, we used Gad1 staining on the heterozygote to define the size of the boxes (3.6 mm2 surface area). The surface areas used in the box counting were similar between heterozygote and mutant.
Figure 10.
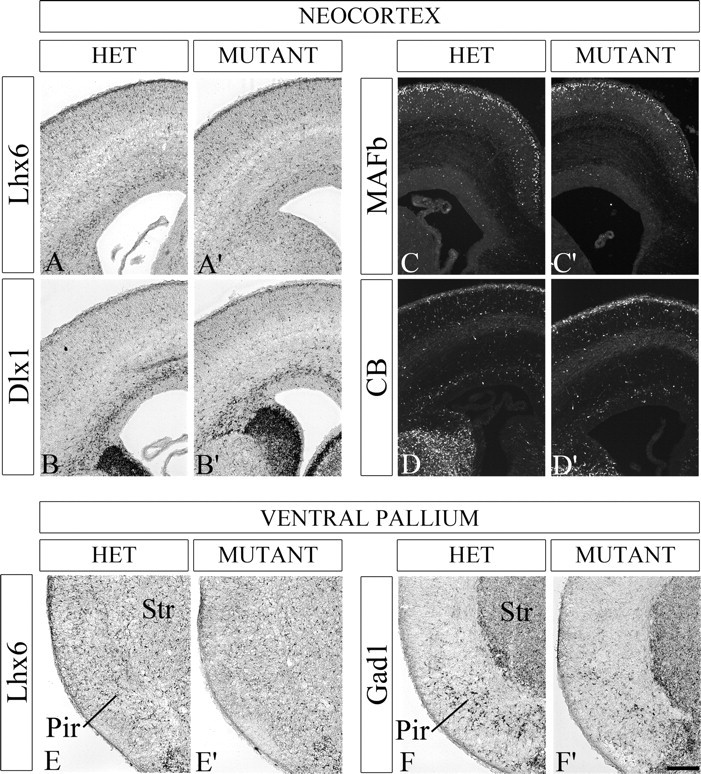
Most neocortical interneurons are present in the E18.5 Nkx2-1f/−;ShhCre/+ mutant. A–D′, Coronal hemisections showing the neocortex for HET and mutant labeled by in situ RNA hybridization using Lhx6 (A, A′) and Dlx1 (B, B′) riboprobes and immunostaining using MAFb (C, C′) and CB (D, D′) antibodies. Lhx6+ and Dlx1+ neocortical interneurons are reduced in the mutant by 10%; MAFb+ and CB+ interneurons are reduced 35 and 22%, respectively. E–F′, In situ RNA hybridization in the paleocortex using Lhx6 (E, E′) and Gad1(F, F′) riboprobes suggests interneuron reductions (25 and 40%, respectively). Scale bar, 200 μm. Pir, Piriform cortex; Str, striatum.
Results
The Shh-expressing progenitor domain of the dorsal POA and ventral MGE generated ∼70% PV+ neurons of the globus pallidus and ∼30% of PV+ striatal interneurons but only ∼10% of PV+ cortical and hippocampal interneurons
Nkx2-1 is expressed in the progenitor cells of the entire MGE and POA (Lazzaro et al., 1991; Shimamura et al., 1995; Sussel et al., 1999; Puelles et al., 2000; Flames et al., 2007; García-López et al., 2008). Its expression persists in subsets of postmitotic neurons, including those of the GP (pallidum) and striatal interneurons (Figs. 1A,B, 2) (Sussel et al., 1999; Marin et al., 2000). Furthermore, fate mapping using Nkx2-1-Cre confirms that the Nkx2-1-expressing cells give rise to these neurons (Xu et al., 2008). However, it is unclear what parts of the subpallium contribute to the GP and striatal interneurons.
Figure 2.
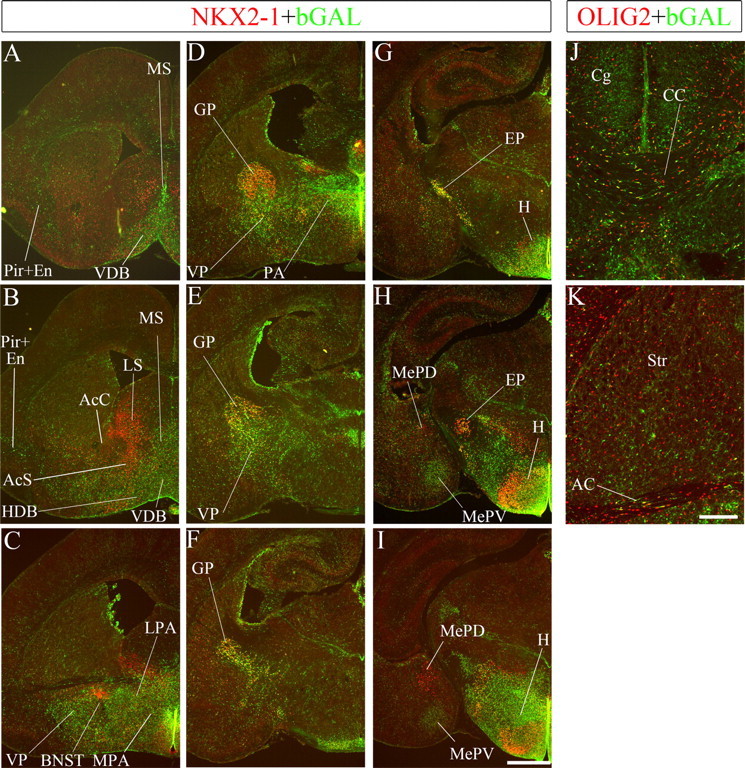
A–K, Fate-mapping analysis of cells in the Shh lineage in the P0 forebrain. NKX2-1 (red) or OLIG2 (red) and bGAL (green) protein expression in coronal sections of a ROSA;ShhCre/+ forebrain. Scale bars: (in I) A–I, 500 μm; (in K) J, K, 200 μm. AC, Anterior commissure; AcS and AcC, nucleus accumbens, shell and core; CC, corpus callosum; Cg, cingulate cortex; En, endopiriform nucleus; EP, endopeduncular nucleus; LS, lateral septum; H, hypothalamus; HDB, horizontal limb of the diagonal band; LPA, lateral preoptic area; MePD, posterodorsal part of the medial amygdala; MePV, posteroventral part of the medial amygdala; MPA, medial preoptic area; MS, medial septum; PA, preoptic area; Pir, piriform nucleus; Str, striatum; VDB, vertical limb of the diagonal band; VP, ventral pallidum.
Expression of ER81 at E10.5 and E11.5 suggests that ventral regions of the basal telencephalon (ventral MGE and dorsal POA) contribute to the GP (Flames et al., 2007). Here, we used Shh-GFP-Cre (Harfe et al., 2004) to determine the contribution of the POA and ventral MGE to the globus pallidus and interneuron populations. Cre expression from the Shh locus provides an approach to follow the fate of these cells following recombination of a reporter gene, such as LacZ, which encodes a bGAL (Soriano, 1999).
Analysis of the Shh-Cre-mediated recombination pattern at E11.5 and E13.5 showed a high density of bGAL+ ventricular zone (VZ) cells in the dorsal preoptic area and the ventral-most MGE [pPOA1 and pMGE5 (Flames et al., 2007)] and was very similar to the expression of Shh RNA; there was scattered recombination in the dorsal MGE regions and there were also occasional bGAL+ cells in the LGE, CGE, and cortex (Fig. 1A′; supplemental Fig. 1, available at www.jneurosci.org as supplemental material, and data not shown). In addition, there was migration of bGAL+ cells that appeared to be emanating from the ventral MGE and populating the deep and superficial MGE mantle zone, as well as cells migrating through the LGE and into the cortex (Fig. 1A′; supplemental Fig. 1A″–D″, available at www.jneurosci.org as supplemental material, and data not shown). At E11.5, GFP expression from the Shh-GFP-Cre allele closely matched the pattern of recombination except in two regions: (1) the region just dorsal to the zone of Shh expression in the VZ expressed more bGAL+ than GFP, suggesting that at an earlier developmental stage Shh was expressed in these progenitors and subsequently was downregulated (supplemental Fig. 1B‴), available at www.jneurosci.org as supplemental material); and (2) in a superficial part of the MGE mantle zone, the cells were GFP+ but bGAL− (i.e., had not undergone recombination) (supplemental Fig. 1A‴–C‴, available at www.jneurosci.org as supplemental material).
Analysis of the Shh-Cre recombination pattern at E11.5, E13.5, and P0 showed that extensive regions of the subcortical telencephalon had large numbers of cells derived from Shh-expressing cells, many of which continued to coexpress NKX2-1 protein (Figs. 1A″,B″, 2; supplemental Fig. 1A″–D″, available at www.jneurosci.org as supplemental material, and data not shown). This included substantial parts of the septum, diagonal band, ventral pallidum, preoptic area, and globus pallidus and small domains of the bed nucleus stria terminalis and amygdala. Recombination within the hypothalamus will be discussed elsewhere. In most regions, there was a high concordance between NKX2-1 protein expression and Shh-Cre recombination (particularly in the globus pallidus; see below), whereas in other regions, such as the medial amygdala, they appeared discordant; the ventral medial nucleus was derived from the Shh-expressing cells, whereas the dorsal medial nucleus was NKX2-1+ and not derived from the Shh lineage (Fig. 2H). In addition, there were scattered cells in the striatum and pallium (discussed below).
Most globus pallidus cells expressed NKX2-1 prenatally (E11.5, E13.5, and E15.5) and postnatally (P0) and were in the Shh lineage (Figs. 1A″,B″, 2D,E; also data not shown). At E13.5, ∼80% of the NKX2-1+ cells in the region of the developing globus pallidus were derived from the Shh-Cre lineage (Fig. 1A–A″). Analysis at P0 confirmed that the majority (∼70%) of NKX2-1+ globus pallidus cells were derived from Shh-Cre+ descendents (Fig. 2D,E). In the adult (P60), we found that ∼70% of PV+ globus pallidus neurons were derived from Shh-Cre descendents (Fig. 3D,E). PV+ projection neurons constitute a major fraction of globus pallidus neurons.
Other derivatives of the Shh-Cre lineage included cortical and striatal GABAergic interneurons (largely PV+) (Fig. 3), striatal TrkA+ cholinergic neurons (data not shown), and Olig2+ cells, presumably oligodendrocytes, populating several regions, but especially parts of the septum and telencephalic axon tracts (corpus callosum, hippocampal commissure, and anterior commissure) (Fig. 2J,K).
The Shh-expressing progenitor domain of the basal telencephalon generated most of the PV+ neurons of the globus pallidus (∼70%) (Fig. 3D) and a substantial fraction of PV+ striatal interneurons (∼30%) (Fig. 3C‴) but only a small fraction of PV+ cortical and hippocampal (∼5–10%) interneurons (Fig. 3A‴,B‴). Thus, this Shh domain is a rich source for subpallial, but not pallial, PV+ neurons. However, some neurons in the globus pallidus were not descended from Shh-expressing cells, suggesting heterogeneity of its developmental origins and perhaps neuronal subtypes. This hypothesis was confirmed by analysis of transcription factor expression (Fig. 4) and of the Nkx2-1 mutants described below.
Transcription factor expression in globus pallidus neurons
Our fate-mapping analysis suggested that not all globus pallidus neurons were derived from Shh-expressing cells, implying that there may be molecularly/developmentally distinct neuronal subtypes. To investigate this further, we compared the expression of the Dlx1, ER81, Lhx6, Nkx2-1, and Npas1 transcription factors in the globus pallidus at E15.5, E18.5, P0, and the adult ages. We did not have Dlx1 or Lhx6 antibodies and thus used mice expressing GFP from BAC transgenes to label these cells (Cobos et al., 2006).
At E18.5, ∼80% of globus pallidus cells (DAPI+) expressed NKX2-1 protein (Fig. 4A–A″). Double labeling of NKX2-1 with NPAS1 and OLIG2 showed that the globus pallidus had the following cell subtypes: 81% expressed NKX2-1 (54% were NKX2-1+;NPAS1−; 27% NKX2-1+;NPAS1+), 42% expressed NPAS1 (27% NPAS1+;NKX2-1+; 15% NPAS1+;NKX2-1−) (Fig. 4B–B″), and 13% expressed OLIG2 [probably oligodendrocytes (Petryniak et al., 2007)] (Fig. 4C–C″). While none of the NKX2-1+ cells coexpressed OLIG2, we do not know whether there were any NPAS1+;OLIG2+ cells because both antisera were rabbit, precluding double labeling.
Dlx1 (BAC-GFP), ER81, and Lhx6 (BAC-GFP) were coexpressed with ∼60–90% of NKX2-1+ cells (supplemental Fig. 2A″,C″,E″, available at www.jneurosci.org as supplemental material). Likewise, most of the cells expressing these genes also expressed NKX2-1 (supplemental Fig. 2A″,C″,E″, available at www.jneurosci.org as supplemental material). While ∼90% NKX2-1+ globus pallidus cells coexpressed Dlx1-GFP, only 65% of the NPAS1+ globus pallidus cells expressed Dlx1-GFP (supplemental Fig. 2D″,E″, available at www.jneurosci.org as supplemental material). Prenatally (E15.5), we observed similar results (data not shown).
Fate-mapping analysis (Shh-Cre) at P0 showed that the majority of NKX2-1+ (∼70%) and ER81+ (∼80%) GP cells were derived from Shh-Cre+ descendents (Fig. 1B″,C″). On the other hand, only ∼35% of globus pallidus cells expressing NPAS1 were derived from the Shh-Cre lineage (Fig. 1D″). Thus, while most globus pallidus cells expressed NKX2-1, Dlx1, ER81, and Lhx6 and were derived from Shh-Cre lineage, a large fraction of NPAS1+ globus pallidus cells represented a distinct population.
Loss of Nkx2-1 function in ventral MGE and POA progenitors disrupted migration to the globus pallidus
Mice constitutively lacking Nkx2-1 failed to generate a globus pallidus expressing ER81 and Lhx6 (Sussel et al., 1999). On the other hand, these mutants continued to express NPAS1 in the region of the globus pallidus (see Fig. 9L,L′), providing further evidence that a subset of these NPAS1+ neurons were independent of Nkx2-1 expression and function.
Figure 9.
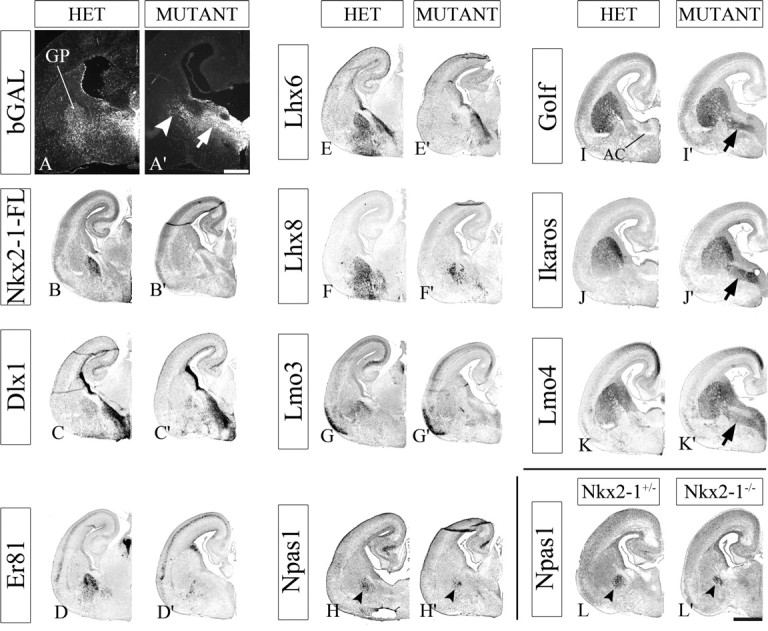
Nkx2-1 expression in the Shh domain is required to generate the E18.5 globus pallidus. A, A′, Coronal hemisections of HET (A) and Nkx2-1f/−;ShhCre/+ mutant (A′) telencephalons showing Cre-induced bGAL immunostaining; most mutant bGAL+ cells accumulate periventricularly in the vMGE/POA region in which the anterior commissure normally forms (arrow in A′; see supplemental Fig. 7, available at www.jneurosci.org as supplemental material, for additional sections). Some bGAL+ cells do migrate to the region of the globus pallidus (arrowhead, A′), many of which coexpress the striatal marker DARPP32 (see supplemental Fig. 7D,D′, available at www.jneurosci.org as supplemental material). B–H′, In situ RNA hybridization using Nkx2-1 full-length (Nkx2-1-FL; B, B′), Dlx1 (C, C′), Er81 (D, D′), Lhx6 (E, E′), Lhx8 (F, F′), Lmo3 (G, G′), and Npas1 (H, H′) riboprobes shows a loss of most of the globus pallidus in the mutant, except for some Npas1+ cells (arrowheads, H, H′) and Lhx8+ cells (many of these cells may be cholinergic neurons; see supplemental Fig. 8, available at www.jneurosci.org as supplemental material). I–K′, In situ RNA hybridization using Golf (I, I′), Ikaros (J, J′), and Lmo4 (K, K′) riboprobes reveals that cells with LGE/striatal properties accumulate in the region with most of the bGAL+ cells (arrows in I′, J′, K′). L, L′, Npas1 in the globus pallidus of Nkx2-1 constitutive null mutants (Nkx2-1−/−) at E18.5 (arrowheads in L, L′). Scale bars: (A′) A, A′, 500 μm; (in L′) B–L, B′–L′, 1 mm. AC, Anterior commissure.
To establish which basal telencephalic progenitor zones require Nkx2-1 function to generate the globus pallidus, we selectively removed Nkx2-1 function in cells expressing Shh-Cre using a floxed allele of Nkx2-1 (Kusakabe et al., 2006). We used the following cross: Nkx2-1f/f × Nkx2-1+/−; ShhCre/+ (with or without the ROSA LacZ Cre reporter) and compared the telencephalic phenotype of mutant (Nkx2-1f/−; ShhCre/+) and control (Nkx2-1+/−) offspring at E11.5, E13.5, E15.5, and E18.5 (they do not survive postnatally).
To establish the efficiency of Cre-mediated deletion, we studied the expression of Nkx2-1 RNA by in situ hybridization at E11.5, E13.5, E15.5, and E18.5. We used two different probes: one probe was complementary to the region that should be deleted by Cre-excision (exon 2 probe), and the other was complementary to the 5′ region of the transcript (full-length probe). Analysis with the exon 2 probe showed that Cre efficiently eliminated exon 2 expression in the progenitor domains of the ventral MGE (pMGE5) and the POA (pPOA1-2) throughout the rostrocaudal extent of the basal telencephalon (Figs. 5B,B′, 6B,B′, arrows; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). On the other hand, analysis with the full-length Nkx2-1 probe provided evidence that these progenitor domains continued to express Nkx2-1, albeit at lower levels, suggesting that they maintain some degree of their initial specification as MGE and POA progenitors (Figs. 5C,C′, 6C,C′; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material).
Figure 5.

Removing Nkx2-1 from the Shh domain alters cell migration, radial glia orientation, and proliferation in the E13.5 Nkx2-1f/−;ShhCre/+ mutant. A–H′, Immunofluorescence staining using bGAL (A, A′), nestin (E–F′), β-III tubulin (TUB-III; D, D′), and PH-3 (G–H′) antibodies. The mutant has fewer M-phase (PH-3+) SVZ cells [cell count: 107 for HET and 48 for mutant (45% reduction); G, G′, H, and H′]. B–C′, In situ RNA hybridization using Nkx2-1 exon 2 (Nkx2-1-E; B, B′) and full-length (Nkx2-1-FL; C, C′) probes demonstrating loss of Nkx2-1 exon 2 expression in the vMGE/POA (arrows) and globus pallidus, but maintenance of some Nkx2-1 transcripts in the VZ (C′). F, F′, H, H′, Higher-magnification pictures of regions illustrated by boxes in E, E′, G, and G′, respectively. Scale bars: (in A) A–D, A′–D′, G, G′, 250 μm; (in E′) E, E′, 150 μm); (in F′) F, F′, 75 μm; (in H′) H, H′, 125 μm. dMGE, Dorsal MGE; vMGE, ventral MGE.
Figure 6.
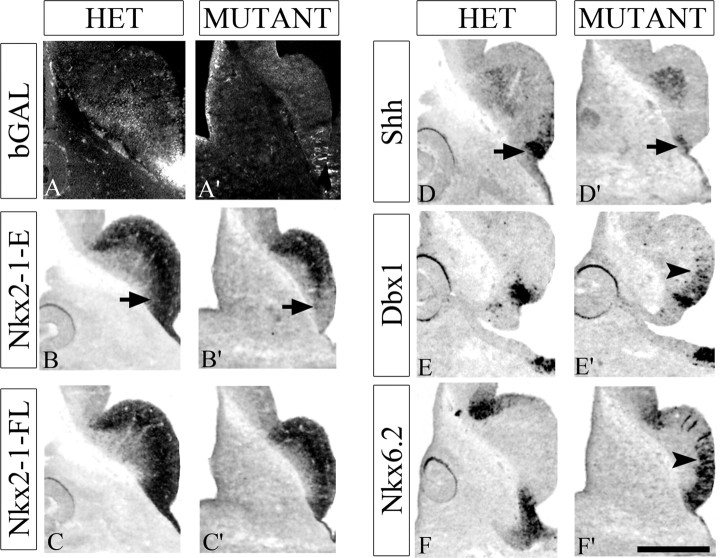
Nkx2-1 represses POA molecular features from being expressed in the MGE. Coronal hemisections of the E11.5 MGE from HET and Nkx2-1f/−;ShhCre/+ mutants. A, A′, bGAL immunohistochemistry shows a strong reduction in bGAL+ cells generated from the progenitor and mantle zones of the POA/vMGE. B–F′, In situ RNA hybridization using Nkx2-1 exon 2 (Nkx2-1-E; B, B′), Nkx2-1 full-length (Nkx2-1-FL; C, C′), Shh (D, D′), Dbx1 (E, E′), and Nkx6.2 (F, F′) riboprobes. Arrows indicates regions in which the mutant has reduced expression, and arrowheads point to ectopic expression. Scale bar, 500 μm.
Next, we followed the fate of the recombined cells by analyzing bGAL expression (the ROSA LacZ Cre reporter was included in the cross). At E11.5, bGAL expression in the VZ was greatly reduced, providing evidence that these cells failed to be generated or proliferate at the normal rate and/or survive; likewise, there was a large reduction of bGAL+ cells in the MGE mantle zone, suggesting that very few neurons in this lineage were produced by this age (Fig. 6A,A′).
At E13.5 and E18.5, bGAL fate-mapping analysis of the Nkx2-1f/−;ShhCre/+ mutant showed that Shh-expressing descendents failed to generate a globus pallidus and the tangentially migrating stream of subventricular zone (SVZ) cells of the MGE; rather, these cells accumulated in a deep mantle zone adjacent to the Shh+ progenitor zone (Figs. 5A,A′, 7A–C′; supplemental Fig. 7A′–C′, available at www.jneurosci.org as supplemental material).
Figure 7.
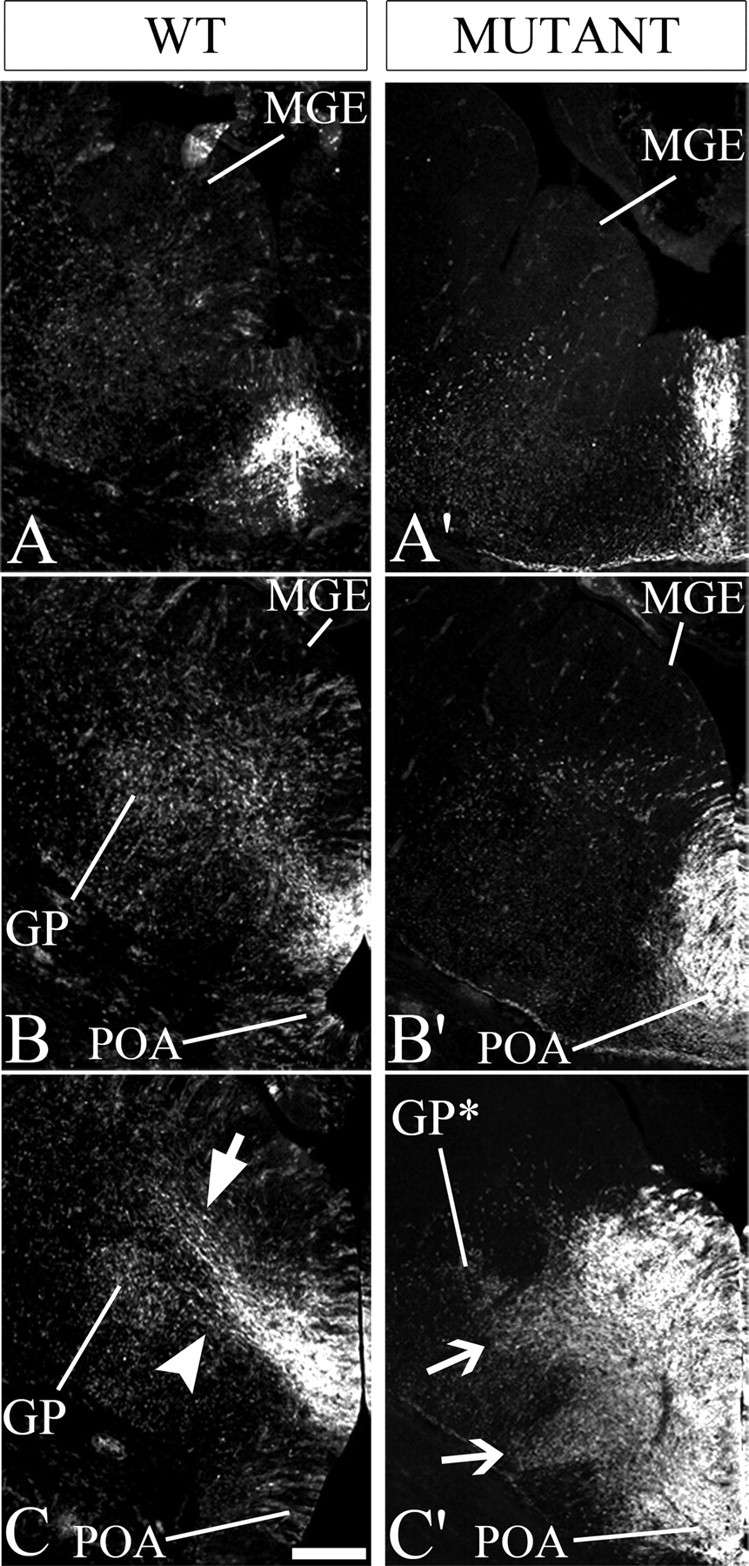
Nkx2-1 is required to produce cells that migrate to the globus pallidus and to restrict the size of the Shh+ progenitor domain at E13.5. Fates of Shh-Cre cells in wild-type (WT) and Nkx2-1f/−;ShhCre/+ (mutant) ventral telencephalon are detected by bGAL expression from the ROSA reporter. Immunofluorescence for bGAL on coronal hemisections (rostral to caudal; A, A′ to C, C′) shows expansion of the Shh progenitor domain in the mutant′s MGE and POA. Very few bGAL+ cells are detected in the region of the globus pallidus in the mutant (compare B′, C′ with B, C; also see supplemental Fig. 7B′,C′, available at www.jneurosci.org as supplemental material). In WT, there is a stream of bGAL+ cells migrating in the SVZ toward the LGE/striatum (arrow in C); some of this stream bifurcates toward the globus pallidus (arrowhead in C). In the mutant, most of the bGAL+ cells remain near the progenitor domains (B′, C′), although in a caudal region two migrations are seen (arrows in C′); one migration is toward to the region in which the globus pallidus normally forms (GP*) and one is in the POA region. Scale bar, 200 μm.
In wild-type mice, there was a robust cell migration parallel to radial glial processes expressing BLBP and nestin; these processes emanated from the Shh+ progenitor domain and extended through the SVZ of the MGE (Fig. 5E,F, and data not shown), and were continuous with cells tangentially migrating through the SVZ of the MGE, LGE and cortex (data not shown). Note that the radial processes of the Shh domain extended perpendicularly to the radial processes emanating from the adjacent MGE. On the other hand, in the Nkx2-1f/−;ShhCre/+ mutant, these radial glial processes appeared to have an altered trajectory from the Shh+ progenitor domain (Fig. 5E′,F′).
The reduced numbers of mantle zone cells in the Shh-Cre lineage was not accounted for by increased cell death, as we did not detect an increase in the number of apoptotic cells at E11.5 and E13.5 (based on caspase-3+ cells) (data not shown). There were defects in the progenitor zones of the caudoventral MGE and POA of the Nkx2-1f/−;ShhCre/+ mutant; the SVZ showed a ∼2-fold reduced proliferative index at E13.5 based on phosphohistone 3+ M-phase cells (Fig. 5H,H′). While we did not detect a phenotype in VZ proliferation (Fig. 5H,H′), many bGAL+ cells accumulated as radial clone-like clusters in the progenitor zone (Figs. 5A′, 7B′,C′). These cells were not ectopic immature neurons, as they did not express β-III-tubulin (Fig. 5D,D′, and data not shown). The morphology of the VZ in this region was altered; the bGAL+ progenitor domain was wider and extended further along the dorsoventral axis (Figs. 5A′, 7B′,C′). To explore whether these VZ and SVZ defects reflected an alteration in the regional identity of this progenitor zone, we performed in situ RNA hybridization.
Nkx2-1 expression in the POA and ventral MGE progenitor zones is required to repress ventral POA fate and to generate the globus pallidus
To assess whether selective deletion of Nkx2-1 from the Shh domain altered the molecular properties of the VZ, we used a panel of molecular markers that have expression boundaries within this region at E11.5 and E13.5 (Flames et al., 2007; Zhao et al., 2008). The exon 2 Nkx2-1 probe identified where recombination deleted Nkx2-1; this domain closely matched the VZ domain of Shh expression (Figs. 5A′,B′, 6A′,B′; supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Hybridization with the full-length Nkx2-1 probe showed that this region maintained Nkx2-1 transcripts (albeit at lower levels), suggesting that transcription and some aspects of fate were maintained (Figs. 5C′, 6C′, 8E′). Note that normal Nkx2-1 expression was maintained in the dorsal MGE along its rostrocaudal extent (supplemental Figs. 3, 4, available at www.jneurosci.org as supplemental material).
Figure 8.
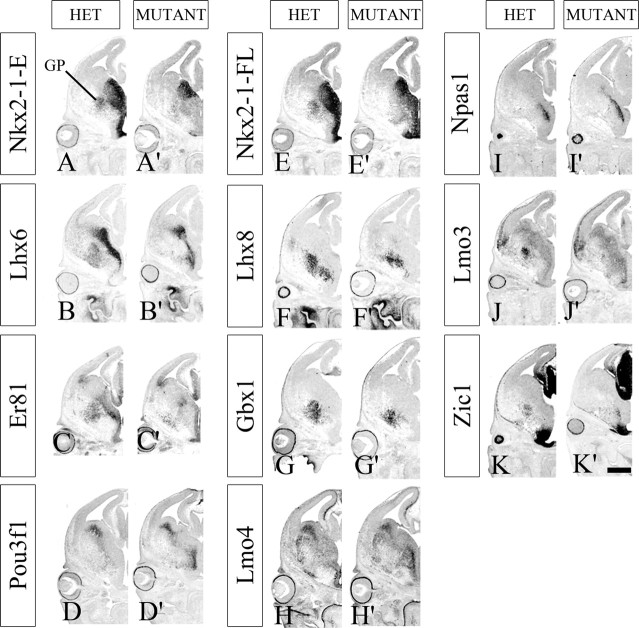
Nkx2-1 expression in the Shh domain is required to generate the E13.5 globus pallidus. A–K′, Coronal hemisections of HET and Nkx2-1f/−;ShhCre/+ mutant telencephalons studied by in situ RNA hybridization using Nkx2-1 exon 2 (Nkx2-1-E; A, A′), Lhx6 (B, B′), Er81 (C, C′), Pou3f1 (D, D′), Nkx2-1 full-length (Nkx2-1-FL; E, E′), Lhx8 (F, F′), Gbx1 (G, G′), Lmo4 (H, H′), Npas1 (I, I′), Lmo3 (J, J′), and Zic1 (K, K′) riboprobes. Scale bar, 500 μm.
Next, we studied the effect of removing Nkx2-1 from the Shh expression zone (ventral MGE and POA) at E11.5. Shh expression was reduced in most of the VZ (Fig. 6D,D′, arrows; supplemental Fig. 3, available at www.jneurosci.org as supplemental material), whereas Shh expression was maintained in the mantle zone (MZ) of the rostrodorsal MGE (Fig. 6D,D′). The region with preserved Shh MZ expression probably corresponds to the GFP+ Shh expression zone in which Cre recombination did not occur (supplemental Fig. 1A‴–D‴, available at www.jneurosci.org as supplemental material). Likewise, the Nkx2-1f/−;ShhCre/+ mutants showed reduced expression of ER81, Lhx6, and Lhx7(8) in the region in which Nkx2-1 was deleted (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). MGE expression of Lhx6 and Lhx7(8) was maintained both rostrodorsally and caudodorsally (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Deletion of Nkx2-1 by E11.5 did not show a major effect on the ventral POA, based on preserved expression of Dbx1, Nkx5.1 and Nkx6.2 at E11.5 and E13.5 (Fig. 6; supplemental Figs. 3, 4, available at www.jneurosci.org as supplemental material). On the other hand, the dorsal POA and ventral MGE showed ectopic expression of two of these ventral POA markers (Dbx1 and Nkx6.2) (Fig. 6E′,F′, arrowheads; supplemental Figs. 3, 4, available at www.jneurosci.org as supplemental material); in the ventral MGE, there were radial clusters of cells showing this ectopic expression. Despite, these changes, the expression of COUP-TF1, Nkx5.1, Olig2 and Zic1 in this region did not show major changes, although there were subtle changes in COUP-TF1 and Zic1 in the VZ of the POA (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Deletion of Nkx2-1 in the Shh domain led to a loss of most of the globus pallidus based on several criteria. In control brains at E13.5 and E15.5, we detected the globus pallidus based on its location and its expression of Dlx1, Gbx1, ER81, Lhx6, Lhx7(8), Lmo3, Nkx2-1, and Zic1 (Fig. 8; supplemental Figs. 4, 5, available at www.jneurosci.org as supplemental material). At E13.5, the mutant lacked expression of most of these markers in the region in which the globus pallidus was normally present (Fig. 8; supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Expression of NPAS1, a marker of a subtype of globus pallidus neurons (Fig. 4B″; supplemental Fig. 2D″, available at www.jneurosci.org as supplemental material), and cortical interneurons (Zhao et al., 2008) was not affected in the mutant at E13.5 (Fig. 8I,I′; supplemental Fig. 4, available at www.jneurosci.org as supplemental material). At this age, however, NPAS1 was not detected in the position of the globus pallidus but rather showed expression in a pattern that could reflect a tangential migration from a progenitor zone near pMGE5 or pPOA1.
At E18.5, molecular features of the globus pallidus continued to be deficient in the mutant, showing only scattered expression of ER81, Lmo3, and Nkx2-1 (full-length, exon 2) (Fig. 9; supplemental Fig. 6, available at www.jneurosci.org as supplemental material). On the other hand, the mutant had clusters of cells expressing Lhx6, Lhx7(8), Dlx1, and Npas1 in the general region in which the globus pallidus should form. The cells expressing Npas1 and Dlx1 (supplemental Fig. 6, available at www.jneurosci.org as supplemental material) could correspond to the Npas1+;Dlx1+;Nkx2-1− globus pallidus neurons observed in normal mice (Fig. 4; supplemental Fig. 2, available at www.jneurosci.org as supplemental material). While the Lhx6- and Lhx7(8)-expressing cells could represent some globus pallidus cells, perhaps produced by the dorsal MGE, they are found in the same region in which we detect cholinergic (ChAT+) neurons (Fig. 9; supplemental Figs. 6, 8, available at www.jneurosci.org as supplemental material).
Expression of Nkx2-1-regulated genes was not affected (or was only modestly altered) in regions in which Nkx2-1 was not deleted. This included the entire rostrocaudal extent of the dorsal MGE (supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). For instance, in rostral sections containing the immature anterior bed nucleus of the stria terminalis (BNST) and ventral pallidum, expression of Gbx1, ER81, Lhx6, Lhx7(8), Nkx2-1, and Zic1 appeared normal. In addition, in most regions, there was expression of these genes in MZ superficial to the GP. Many of the neurons that populate these layers are produced at early developmental stages. In the mutant, we found no major change in the numbers of early-born neurons in these regions generated by BrdU incorporation at E10.5 (data not shown), as also assessed by βIII-tubulin expression (Fig. 5D,D′). Thus, we conclude that most early neurogenesis (through ∼E10.5) in the basal telencephalon was not disrupted by deleting Nkx2-1 in the Shh-Cre lineage.
Surprisingly, Lhx6 expression in the mutant's BNST appeared partially preserved (Figs. 8B′, 9E′; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). We postulate that the remaining Lhx6+ BNST expression domain was generated by a ventral migration from the dorsal MGE.
Persistent Nkx2-1 function is required to repress LGE fate in the ventral MGE
The MGE in constitutive Nkx2-1-null mice showed evidence of a fate transformation toward the LGE (Sussel et al., 1999), although further analysis showed that constitutive Nkx2-1 nulls maintain expression of Npas1 in the globus pallidus (Fig. 9L,L′). Here, we assessed whether deletion of Nkx2-1 in the ventral MGE/dorsal POA in the Nkx2-1f/−;ShhCre/+ mutant, ∼1 d after Nkx2-1 expression had been established, resulted in this fate switch. Above, we have shown that the ventral MGE and the dorsal POA lacked Nkx2-1 expression and failed to generate a GP. In addition to the dorsal expansion of POA markers (Dbx1 and Nkx6.2), the ventral MGE and the dorsal POA now expressed LGE/striatal markers in the SVZ [Lmo4 and Oct6 (Pou3f1)] and MZ (Golf, Ikaros, and Lmo4) at E13.5, E15.5, and E18.5 (Figs. 8, 9; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). Cre fate mapping at E18.5 showed that the same MGE region with ectopic Golf, Ikaros, and Lmo4 expression also contained derivatives of the Shh-Cre lineage (Fig. 9). Some of the cells derived from the Shh-Cre lineage expressed the striatal marker DARPP32 (supplemental Fig. 7D,D′, available at www.jneurosci.org as supplemental material).
BrdU birthdating analysis provided evidence that this region of ectopic striatal tissue was produced at ∼E13.5. BrdU administered at E13.5, and analyzed at E15.5, showed many BrdU-labeled cells ectopically collected in the deep MZ region of the Nkx2-1f/−;ShhCre/+ mutant caudoventral MGE (data not shown).
Gene expression analysis of striatal and pallial interneurons
Constitutive Nkx2-1 mutants failed to generate ∼50% of neocortical and hippocampal interneurons; paleocortical interneurons may have been even more reduced (Sussel et al., 1999; Pleasure et al., 2000; Xu et al., 2004). As discussed above, fate mapping with Shh-Cre showed that only a very small fraction of neocortical and hippocampal interneurons were derived from ventral MGE/dorsal POA (Fig. 3), providing evidence that most MGE-derived interneurons are produced by the dorsal MGE. To further assess this, we compared the numbers of neocortical interneurons at E13.5, E14.5, E15.5, and E18.5 in control and Nkx2-1f/−;ShhCre/+ mutants.
At E13.5 and E14.5, the mutants showed a reduction in neocortical interneurons expressing Dlx1+ (∼35 and ∼20% reduction) and Lhx6+ (∼45 and ∼48% reduction). More severe reductions were found in the paleocortex (data not shown).
By E18.5, however, the interneuron reductions found in the mutants were smaller compared with the earlier stages. Dlx1+ and Lhx6+ neocortical interneurons were reduced ∼10% (Fig. 10A,A′,B,B′). These decreases were consistent with the Shh-Cre fate-mapping data suggesting that <10% of adult cortical interneurons were derived from the Shh domain (Fig. 3). On the other hand, MAFb-, Npas1-, and somatostatin-expressing interneurons showed greater reductions (MAFb: ∼35%; calbindin: ∼22%; Npas1: ∼25%; somatostatin: ∼40%) (Fig. 10C,C′,D,D′; also data not shown), suggesting either that these molecular subtypes were preferentially affected or that alterations in Lhx6 expression levels affected gene expression in these cell types. The mutant ventral pallium (paleo/piriform cortex, claustrum, and endopiriform nucleus) also showed interneuron defects, with reductions of 25% of Lhx6+ and 40% of GAD1+ interneurons (Fig. 10E,E′,F,F′). Likewise, there was a large reduction of bGAL+ cells (Shh-Cre lineage) in the ventral pallium (supplemental Fig. 6A,A′, available at www.jneurosci.org as supplemental material).
Many striatal interneurons (including ∼30% of parvalbumin+) are derived from Shh-expressing cells based on Shh-Cre fate mapping (Fig. 3). At E18.5, Nkx2-1f/−;ShhCre/+ mutants showed striatal interneuron reductions. There was approximately a 10-fold reduction in the number of Lhx7(8)-expressing cells, and consistent with its known function, ChAT+ cholinergic striatal interneurons were reduced ∼4-fold (supplemental Fig. 8, available at www.jneurosci.org as supplemental material). Furthermore, Lhx6+ striatal interneurons were reduced, particularly in the ventral striatum/accumbens core (dorsal: ∼15% reduction; ventral: ∼50%). On the other hand, there was not an obvious reduction in the number of NPY+ and somatostatin+ striatal interneurons (data not shown); PV expression does not begin until postnatally and therefore could not be assessed. Medial septal expression of Lhx6 and Lhx7(8) were greatly reduced, consistent with Shh-Cre fate mapping (Fig. 2), whereas expression in the accumbens shell appeared preserved (supplemental Fig. 8, available at www.jneurosci.org as supplemental material).
Discussion
Shh-Cre-induced recombination in the telencephalon
Herein, we used the Shh-GFP-Cre allele, which makes a GFP-Cre fusion protein (Harfe et al., 2004), to perform fate mapping and to eliminate Nkx2-1 function in the ventral MGE and POA. Cre-mediated recombination in the ventricular zone was more extensive in the MGE and POA than either GFP or Shh RNA expression (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), suggesting that Shh expression was downregulated as development proceeds. Shh RNA expression wanes as development proceeds (Zhao et al., 2008), which explains why Flames et al., 2007, included Shh expression at E13.5 only in pPOA1.
Shh-Cre-mediated recombination was robust in most Shh-GFP+ cells. The only exception was in a superficial part of the MGE mantle zone, in which the cells were GFP+ but were bGAL− (i.e., no recombination) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). We do not understand why these cells fail to show Cre-mediated recombination. Consistent with this result, the Nkx2-1f/−;ShhCre/+ mutant continues to express Shh RNA in the MGE mantle zone (Fig. 6D,D′; supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Shh-expressing progenitor domains generate few neocortical and hippocampal interneurons, but substantial numbers of striatal interneurons
Previous studies suggest that the dorsal MGE is the predominant source for somatostatin+ cortical interneurons, whereas the ventral MGE is the predominant source for parvalbumin+ interneurons (Flames et al., 2007; Wonders et al., 2008). Fate mapping from the ventral MGE and the POA using Shh-Cre surprisingly showed that these regions produce very few neocortical and hippocampal interneurons, although there was a strong bias to generate the parvalbumin+ subtype (Fig. 3E). The morphological and gene expression evidence suggests that Shh-Cre-expressing cells induce robust recombination in pMGE5 and perhaps pMGE4, as well as pPOA2 and pPOA1 (domains defined by Flames et al., 2007). The results imply pMGE1-3 (perhaps pMGE4) generate most parvalbumin+ and somatostatin+ neocortical and hippocampal interneurons. The Shh-Cre domain may also produce interneurons that migrate to the ventral pallium (Fig. 2).
Furthermore, the ventral MGE and the dorsal POA produced a substantial fraction of striatal interneurons (GABAergic and cholinergic) (Fig. 3; supplemental Fig. 8, available at www.jneurosci.org as supplemental material). However, among the GABAergic subtypes, parvalbumin+ cells predominated. Thus, the Shh-Cre+ progenitors were biased toward generating parvalbumin+ neurons. This concept is further supported in the globus pallidus.
Shh-expressing progenitor domains generate neurons of the septal, pallidal, preoptic, and amygdaloid nuclei: evidence for cellular complexity of the globus pallidus
The Shh-Cre domain generates cells that contribute to the medial septum, the diagonal band, the pallidal complex (globus pallidus, and ventral pallidum), the preoptic complex (medial and lateral), the ventral medial amygdala, and ventral pallial structures (Fig. 2). We focused on the cellular and molecular complexity of the globus pallidus and found that NPAS1+;NKX2-1− cells are a novel cell type (Fig. 4).
Evidence supporting the existence of NPAS1+;NKX2-1− cells comes from analysis of Nkx2-1 constitutive and conditional mutants; both have NPAS1+ globus pallidus cells (Fig. 9H,H′,L,L′). Fate-mapping analysis at P0 showed that ∼70% of NKX2-1+ are derived from Shh-Cre+ descendents (Figs. 1, 2). On the other hand, only ∼35% of NPAS1+ globus pallidus cells were derived from the Shh-Cre lineage (Fig. 1). Thus, NKX2-1+ and NPAS1+;NKX2-1− cells represent distinct subtypes of globus pallidus neurons.
While the NKX2-1+ cells were largely derived from the Shh-Cre domain, the origin of the NPAS1+;NKX2-1− neurons is unclear. The ventral POA is probably not a source, based on fate mapping with Dbx1-Cre (Hirata et al., 2009). At E13.5, there were Npas1+ SVZ cells between the MGE and dorsal POA; this expression is maintained in the Nkx2-1f/−;ShhCre/+ mutant (Fig. 8I,I′). Perhaps these cells later migrate to the globus pallidus. We are uncertain about the location of the progenitors of these cells.
Finally, the fate mapping shows that ∼70% of globus pallidus neurons are parvalbumin+ (Fig. 3D,E). Thus, the majority of GABAergic neurons generated from the Shh expression domain, destined for the globus pallidus, striatum, and cortex, express parvalbumin.
Nkx2-1 is required for dorsoventral patterning within the ventral MGE and POA
Previous studies of the constitutive and conditional Nkx2-1-null mouse demonstrated that Nkx2-1 is essential for specifying MGE and repressing LGE (and dorsal CGE) identity (Sussel et al., 1999; Corbin et al., 2003; Butt et al., 2008). Herein, we deleted Nkx2-1 from the Shh-expressing progenitors in the ventral MGE and POA, beginning at ∼E9.5 [Shh expression in the MGE begins shortly after Nkx2-1 (Crossley et al., 2001; Shimamura et al., 1995]. Perdurance of Nkx2-1 transcripts in the ventral MGE and POA, showed that these regions were initially specified to express Nkx2-1 (Fig. 6C,C′, 8E,E′; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). Continued Nkx2-1 function in the ventral MGE was required to (1) repress dorsal spread of POA properties (Dbx1, Nkx6.2, Islet1) (Fig. 6E,E′,F,F′; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material), (2) maintain normal levels of Shh expression in the VZ (Fig. 6D,D′; supplemental Fig. 3, available at www.jneurosci.org as supplemental material), (3) repress induction of LGE/striatal properties (DARRP32, Dlx5, Golf, Ikaros, Islet1, Lmo4, Pou3f1) (Figs. 8D,D′,H,H′, 9I,I′,J,J′,K,K′; supplemental Figs. 5–7, available at www.jneurosci.org as supplemental material), and (4) repress induction of dCGE properties (COUP-TFI) (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Thus, persistent Nkx2-1 function represses expression of both dorsal (LGE/dCGE) and ventral (POA) properties in the ventral MGE.
The reduced expression of Shh (Fig. 6D,D′; supplemental Fig. 3, available at www.jneurosci.org as supplemental material) could cause non-cell-autonomous effects (Xu et al., 2005; Gulacsi and Anderson, 2006), which may extend the phenotype to adjacent regions of the MGE and POA. This could contribute to the decrease in proliferative index (Fig. 5H,H′).
Nkx2-1 expression in the Shh domain is more important in the generation of the globus pallidus and striatal interneurons than neocortical and hippocampal interneurons
Many facets of MGE and POA development were not derailed in the Nkx2-1f/−;ShhCre/+ mutant. For instance, the ventral POA continues to express Nkx5.1 (supplemental Figs. 3, 4, available at www.jneurosci.org as supplemental material). Furthermore, Nkx2-1 expression in the rostral and caudal MGE appeared normal, consistent with the localized domain of Shh-Cre expression in the middle of the basal telencephalon. The progenitor model of Flames et al. (2007) posited that dorsal MGE domains are present in the rostral (pMGE1-3) and caudal (pMGE3) MGE. We suggest that Shh-Cre expression is centered in MGE5 and POA1; MGE4 and POA2 may also be affected by the action of Shh-Cre. Expression of Nkx2-1-dependent genes continued in the dorsal MGE domains [(Gbx1, Lhx6, and Lhx7(8)], showing that these regions maintained their fate (Fig. 8B,B′,F,F′,G,G′; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). We propose that pMGE1-3 generate the lion's share of MGE-derived neocortical and hippocampal interneurons.
Loss of Nkx2-1 from the Shh domain reduced the number of striatal interneurons. Striatal Lhx7(8), ChAT, P75, and TrkA expression was almost eliminated (supplemental Fig. 8, available at www.jneurosci.org as supplemental material; also data not shown), consistent with the findings of Zhao et al. (2003), Mori et al. (2004), and Fragkouli et al. (2005). Lhx6 expression in striatal interneurons was also reduced (supplemental Fig. 8, available at www.jneurosci.org as supplemental material), particularly in the ventral striatum and the core of the nucleus accumbens, suggesting the ventral MGE/POA preferentially produces interneurons for ventral striatal structures. Likewise, we saw a trend for a greater reduction of ventral pallial (∼25%) than dorsal pallial (∼10%) interneurons (Fig. 10).
While the number of Lhx6+ and Dlx1+ neocortical interneurons was reduced only ∼10% at E18.5, their numbers were more severely reduced (∼50%) at E13.5 and E14.5 (data not shown). This suggests that the ventral MGE/POA has a prominent role in producing early-born interneurons, whereas the dorsal MGE is more important for late-born interneurons.
Nkx2-1-dependent and -independent cellular constituents of the globus pallidus
Most globus pallidus neurons were derived from Shh+ and Nkx2-1+ progenitors (Figs. 1B″, 3D,E) (Xu et al., 2008). Nkx2-1 mutants have reduced expression of transcription factors in the globus pallidus: Dlx1, ER81, Gbx1, Npas1, Lhx6, Lhx7(8), Lmo3, and Zic1 (Figs. 8, 9; supplemental Figs. 3–6, available at www.jneurosci.org as supplemental material). Thus, we propose Nkx2-1-positive regulation of ER81, Lhx6, Lhx7(8), Lmo3, and Gbx1 [consistent with Sussel et al. (1999) and Du et al. (2008)]. We also propose at least three Nkx2-1 independent pathways: (1) general pathways for subcortical GABAergic neuronal development requiring Dlx1/2/5/6, Gsx2 and Ascl1/Mash1 in Nkx2-1+ cells (Long et al., 2009a,b) (B. Wang, K. Campbell, J. Rubenstein, unpublished results); and (2) Npas1+ globus pallidus neurons that are not in the Nkx2-1-dependent and Shh-Cre lineage; and (3) Zic1, whose expression is not lost in the Nkx2-1f/−;ShhCre/+ mutant; Zic1+ migrating cells fail to populate the globus pallidus.
Loss of Nkx2-1 in the Shh+ progenitor domain alters radial glial orientation and cell migration
The Shh+ progenitor domain generates radial processes that are oriented tangentially to the radial glial in more dorsal parts of the MGE (Figs. 5E,F). This perpendicular orientation is probably related to the orthogonal orientation of the ventricular zones in these adjacent regions. We speculate that Shh+ progenitor domain's putatively tangential radial glial processes contribute to the migratory pathway of globus pallidus neurons. Radial glial processes that extend from the Nkx2-1f/−;ShhCre/+ mutant's Shh+ progenitor domain appeared to have an altered trajectory (Fig. 5E′,F′), and most of the bGAL+ cells generated from the Shh domain failed to migrate to their correct subcortical and cortical destinations (Figs. 5A,A′, 7A–C′, 9A,A′). Furthermore, Shh+ progenitor domain marked the location where the anterior commissure crosses the midline (García-López et al., 2008). The anterior commissure fails to form in the Nkx2-1f/−;ShhCre/+ mutants (Fig. 9I–J′; supplemental Figs. 5, 6, available at www.jneurosci.org as supplemental material); perhaps the tangential radial glia processes participate in guiding the commissural axons.
Footnotes
This work was supported by research grants to J.L.R.R. from Nina Ireland and the Weston Havens Foundation and National Institute of Mental Health R37 Grants MH049428 and R01 MH081880, and by funding to P.F. from Swiss National Science Foundation Fellowships PBGE33-112882 and PA0033-117463. We thank the following people for the cDNAs: CoupTF1 (Ming Tsai), Dbx1 (Sanwei Lu), ER81 (Tom Jessell), Gad1 (Brian Condie), Gbx1 (Mike Frohman), Gsx2 (Gsh2) (Steve Potter), Golf (Richard Axel), Ikaros (Katia Georgopoulos), Islet1 (Tom Jessell), Lhx6 and Lhx7(8) (Vassilis Pachnis), Lmo3 (T. H. Rabbitts), Lmo4 (Gordon Gill), Npas1 (Steve McKnight), Olig2 (David Anderson), Pou3f1 (Oct6) (M. G. Rosenfeld), Shh (Andy McMahon), somatostatin (Thomas Lufkin), and Zic1 (Jun Aruga).
References
- Butt SJ, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59:722–732. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Long J, Thwin M, Rubenstein JLR. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;1:82–88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Rutlin M, Gaiano N, Fishell G. Combinatorial function of the homeodomain proteins Nkx2.1 and Gsh2 in ventral telencephalic patterning. Development. 2003;130:4895–4906. doi: 10.1242/dev.00717. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Ohkubo Y, Rubenstein JL. Evidence that coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon define patterning centers for the telencephalic and optic vesicles. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135:1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marín O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur J Neurosci. 2005;21:2923–2938. doi: 10.1111/j.1460-9568.2005.04141.x. [DOI] [PubMed] [Google Scholar]
- García-López M, Abellán A, Legaz I, Rubenstein JL, Puelles L, Medina L. Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development. J Comp Neurol. 2008;506:46–74. doi: 10.1002/cne.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Martini FJ, Nóbrega-Pereira S, Pierani A, Kessaris N, Marín O. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J Neurosci. 2009;29:9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulacsi A, Anderson SA. Shh maintains Nkx2.1 in the MGE by a Gli3-independent mechanism. Cereb Cortex. 2006;16:i89–i95. doi: 10.1093/cercor/bhk018. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hirata T, Li P, Lanuza GM, Cocas LA, Huntsman MM, Corbin JG. Identification of distinct telencephalic progenitor pools for neuronal diversity in the amygdala. Nat Neurosci. 2009;12:141–149. doi: 10.1038/nn.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Kawaguchi A, Hoshi N, Kawaguchi R, Hoshi S, Kimura S. Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol Endocrinol. 2006;20:1796–1809. doi: 10.1210/me.2005-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009a;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 2009b;19:i96–i106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi C, Smiley GG, Raber J, Kusakabe T, Kawaguchi A, Matagne V, Dietzel A, Heger S, Mungenast AE, Cabrera R, Kimura S, Ojeda SR. Deletion of the Ttf1 gene in differentiated neurons disrupts female reproduction without impairing basal ganglia function. J Neurosci. 2006;20:13167–13179. doi: 10.1523/JNEUROSCI.4238-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, Kitanaka J, Takemura M, Misawa H, Ikawa M, Okabe M, Wanaka A. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur J Neurosci. 2004;19:3129–3141. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- Nóbrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marín O. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2-1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Taylor L, Welagen J, Mbata IC, Xiang JZ, Anderson SA. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314:127–136. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132:4987–4998. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2-1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Marín O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A. 2003;100:9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]