Abstract
Motivation: Programs that evaluate the quality of a protein structural model are important both for validating the structure determination procedure and for guiding the model-building process. Such programs are based on properties of native structures that are generally not expected for faulty models. One such property, which is rarely used for automatic structure quality assessment, is the tendency for conserved residues to be located at the structural core and for variable residues to be located at the surface.
Results: We present ConQuass, a novel quality assessment program based on the consistency between the model structure and the protein's conservation pattern. We show that it can identify problematic structural models, and that the scores it assigns to the server models in CASP8 correlate with the similarity of the models to the native structure. We also show that when the conservation information is reliable, the method's performance is comparable and complementary to that of the other single-structure quality assessment methods that participated in CASP8 and that do not use additional structural information from homologs.
Availability: A perl implementation of the method, as well as the various perl and R scripts used for the analysis are available at http://bental.tau.ac.il/ConQuass/.
Contact: nirb@tauex.tau.ac.il
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The function of a protein is largely determined by its 3D structure. Therefore, the determination of a protein's structure is an important step in understanding how the protein achieves its function, and it can also aid in predicting protein function or designing experiments. However, experimental structure determination can be a long and difficult procedure, and naturally errors may occur (Kleywegt, 2009). This was recently demonstrated when several protein structures published in the Protein Data Bank (PDB) were discovered to be erroneous (Chang et al., 2006). Even when the structure determination process is correct, the determined structure may adopt a non-physiological fold, for example, due to non-physiological constraints imposed by the crystal in the case of X-ray crystallography. Such errors can cause confusion and mislead further research, so it is important to be able to spot them before the structures are published. Errors are even more frequent in computationally derived structures, which are built either by extrapolating from a homologous protein whose structure is already solved (Fiser and Sali, 2003; Ginalski, 2006) or by computer simulation (Das and Baker, 2008; Zhang and Skolnick, 2004). In the latter case, many alternative conformations might be generated during the simulation, and differentiating between erroneous conformations and structures that are more likely to be correct could help guide the simulation and limit the search space. Programs that try to numerically assess the correctness of a given structural model for a protein are called Model Quality Assessment Programs (MQAPs). The need for such programs is widely recognized by the structural biology community, as evidenced by the inclusion of a category for assessing MQAP performance in the biennial Critical Assessment of Techniques for Protein Structure Prediction (CASP) experiment, starting from its seventh round (CASP7; Cozzetto et al., 2007).
The two pioneering MQAPs, still widely used today, are Verify3D (Eisenberg et al., 1997) and ProSa (Wiederstein and Sippl, 2007). Both methods check the compatibility between the protein's structure and its sequence. Verify3D, for example, classifies each residue in the protein into one of the 18 classes according to the residue's structural environment in the input model. The propensity of each amino acid to exist in each such structural environment class is calculated according to statistics collected from structures in the PDB, and the final score given to the protein structure is the sum of propensities of the individual residues.
Newer MQAPs were recently assessed in the blind experiments of CASP7 (Cozzetto et al., 2007) and CASP8 (Cozzetto et al., 2009). The models given as input to the MQAPs were the ‘server models’, which are generated by the various servers participating in CASP shortly after the round starts, and long before the native structures are published. Many of the participating MQAPs, including QMEAN (Benkert et al., 2009) and MULTICOM-REFINE (Cheng et al., 2009), functioned similarly to Verify3D and ProSa, receiving only one structure as input and assigning it a quality score based on the compatibility of various features computed from the sequence with the predicted 3D structure. However, the most successful MQAPs in CASP8 were the consensus-based methods, such as Pcons (Larsson et al., 2009) and ModFOLDclust (McGuffin, 2009), which used as input the entire decoy set instead of just coordinates of a given model and took a consensus approach to rate each model according to how similar it was to the other structures in the set. This approach, while clearly advantageous in the setting of the CASP experiment, is not applicable in many scenarios in which few structures (possibly only one) are available, or when the decoy set is not likely to contain many correct models (Cozzetto et al., 2007; Wallner and Elofsson, 2008). The single-structure MQAPs that performed best in CASP8 were LEE and LEE-server. The group produced their own structural model for each target and ranked the decoys according to how similar they were to their model (Cozzetto et al., 2007). The models produced by the LEE group were homology based, so at least part of the success of the method could be attributed to the fact that it used additional constraints from structural models of homologous proteins. Two other methods from CASP8, SAM-T08-MQAU and SAM-T08-MQAO (Archie et al., 2009), also used such constraints taken from structural homologs, and they also performed significantly better than the rest of the single-structure methods. While homology-based approaches have proven very promising, they are only usable when reliable structural homologs exist. Therefore, there is still a need for devising methods that do not use additional structural information, neither from structural homologs nor from the other decoys. We term such methods ‘pure single-structure MQAPs’.
An alternative strategy to that of most single-structure MQAPs is to check the compatibility of the suggested 3D structure with the evolutionary conservation pattern of the sequence. There are various ways to calculate the conservation level (or evolutionary rate) of an amino acid position (Glaser et al., 2003; Mihalek et al., 2004). A residue that is conserved throughout evolution has undergone strong purifying selection; this suggests that the conserved residue is important for the protein's normal function (Brändén and Tooze, 1999). This observation has been used in many applications, such as identifying the active site of a protein, which is usually composed of a patch of clustered residues on the protein's surface (Nimrod et al., 2008). An interesting observation is that for most proteins, the structural core is composed mainly of such conserved residues (see for example, Fig. 1A). These residues are usually not involved directly in the mechanism of the protein's function. Rather, they are conserved because a mutation in such a buried residue would tend to perturb the architecture. The protein surface, in contrast, is mostly variable. If the association between residue accessibility and conservation level is strong enough, it might be used to differentiate between correct and incorrect model structures, as incorrect structures are unlikely to feature this pattern by chance.
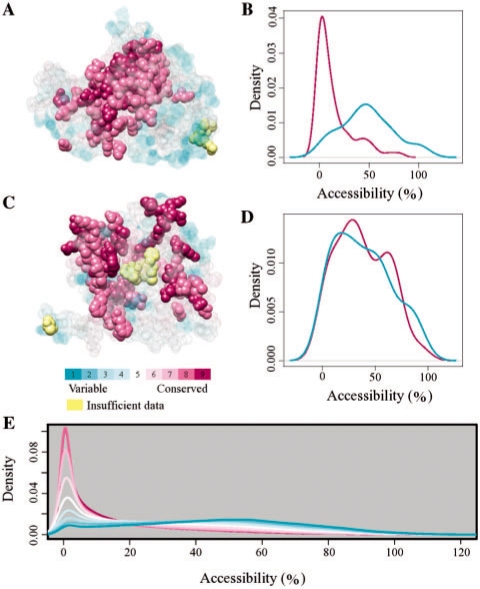
Fig. 1.
Tendency of conserved residues to be buried in correct structures. (A and C) The native structure for the CASP7 target T0289 (Aspartoacylase, PDB 2gu2A) in (A), and a poor model for the same target (model FPSOLVER-SERVER_TS1, GDT-TS = 7.9) in (C), colored by the ConSurf color map using the ConSurf-DB database. ConSurf colors 1–7 are semitransparent to show the cluster of conserved residues buried at the structural core in the native structure. (B and D) Distribution of relative residue accessibilities as calculated by Naccess for variable residues (cyan; ConSurf classes 1, 2, 3) and for conserved residues (purple; ConSurf classes 8, 9). The distributions are shown for the native structure in (B) and for the poor model in (D). (E) Distribution of relative residue accessibilities for all residues of all structures in the dataset, classified by their ConSurf conservation grades. The different grades are colored according to the ConSurf color scheme. There is a consistent shift to the right, with the most variable residues (ConSurf class 1) being most accessible. Molecular graphic images were generated using UCSF Chimera (Pettersen et al., 2004).
This conservation pattern has been used for computational modeling of proteins, both manually for checking the validity of a built model (Landau et al., 2007) and automatically for generating a Cα model of transmembrane proteins starting from a low-resolution cryo-EM map (Fleishman et al., 2004a, b, 2006). Conservation information has also been used for quality assessment in several studies.
The first conservation-based approach is to use the observation that conserved residues tend to be clustered in the native structure (Mihalek et al., 2003; Muppirala and Li, 2006; Schueler-Furman and Baker, 2003). This clustering is expected both for structurally conserved residues, as they form the structural core, and for functionally conserved residues, which are usually localized on the surface, at the functional site of the protein. Mihalek et al. (2003) used the evolutionary trace method to collect a set of conserved residues and quantified the set's tendency to cluster using a measure they termed the selection clustering weight (SCW). They applied this method to the Decoys ‘R’ Us decoy set (Samudrala and Levitt, 2000) and showed that indeed 78.1% of the decoys in the set were assigned a lower (less favorable) SCW score compared with the native structure. However, the assessment of a method by its ability to rank a native structure higher than decoys has been shown to be problematic (Handl et al., 2009). Schuler-Furman and Baker (Schueler-Furman and Baker, 2003) took a similar approach, adopting a simpler strategy for selecting the set of conserved residues based on entropy, as well as a different measure for quantifying the clustering. However, they validated their method in a more relevant scenario, showing that when the method is used to select decoys generated by ROSETTA (Das and Baker, 2008), there is a statistically significant enrichment in correct models.
The second approach to exploit conservation data is to use it initially to make contact predictions and subsequently use the predictions for quality assessment. Olmea et al. (1999) provided a set of contact predictions, using the observation that in pairs of conserved residues, as well in pairs of residues whose mutations are correlated, the members of the pair tend to be spatially close to each other in the 3D structure. They have shown that such predictions are usually more precise for native structures than for deliberately misfolded ones. They also used this information as a post-processing step in a threading method and showed that it improved the method's results. More recently, Miller and Eisenberg (Miller and Eisenberg, 2008) built an MQAP based on the agreement between such contact prediction information and the set of contacts in the proposed model. They checked their method on several of the CASP7 targets and proved that it performed significantly better than random.
While previous studies have suggested that evolutionary information can be used for quality assessment, the performance of these methods was never compared with that of other MQAPs. Furthermore, these methods only used the tendency of conserved residues to be spatially close to one other, which captures only partially the information that is present in the conservation–accessibility relation. In this study, we present a new very simple MQAP called ConQuass (conservation-based quality assessment), which is based on the correlation between each residue's degree of evolutionary conservation and its accessibility in the structure. We check the performance of ConQuass on the CASP8 dataset, and show our method to be comparable to the other pure single-structure MQAPs that participated in CASP8. We also show that ConQuass is complementary to existing methods and could potentially be integrated with them to improve their performance.
2 METHODS
2.1 Collecting a training set of known structures
The PISCES server (Wang and Dunbrack, 2003) was used to collect a non-redundant set of X-ray, full-atom protein structures from the PDB that have resolution better than 3.0 Å, R-factor better than 0.3 and sequence identity <25%. This resulted in a set of 6132 protein chains. Of those, we used only the 5648 proteins for which evolutionary conservation information was available in the ConSurf-DB database (Goldenberg et al., 2009).
We generated three structures for each protein chain. The first contained only the given chain in isolation, without the other chains in the PDB structure. In addition, two versions of the chain in the context of its biological unit were generated using either the protein quaternary structure (PQS) server (Henrick and Thornton, 1998) or the progressive iterative signature algorithm (PISA) server (Krissinel and Henrick, 2007). Non-protein chains were removed. Finally, we removed each protein whose complex contained >26 protein chains and eliminated protein structures that were too big to run in Naccess (Hubbard and Thornton, 1993), leaving a total of 5543 proteins in the final set.
2.2 Features collected for each structure
2.2.1 Conservation
We collected the conservation level of each residue from the ConSurf-DB database (Goldenberg et al., 2009), which provides precalculated conservation profiles for every structure in the PDB. These profiles assign each residue to one of nine conservation levels, with 9 being the most conserved and 1 being the most variable. For some residues, the information in the multiple sequence alignment is not enough to compute the conservation level (for example, if that position consists mostly of gaps). In these cases, ConSurf-DB assigns the residue value of ‘insufficient data’.
2.2.2 Accessibility
We used the program Naccess to calculate the total relative accessibility for each residue (Hubbard and Thornton, 1993). We further normalized these accessibility values by transforming them into quantiles, so that the most buried residue in a given protein would get the value 0 and the most exposed would get the value 1. This normalization was done in light of the observation that some protein structures might overall be more accessible than others owing to their geometric properties, but within a single structure conserved residues still tend to be more buried compared with other residues in the same structure. Each residue was then classified into one of ten evenly distributed accessibility classes.
2.2.3 Structure quality features
For each structure, we extracted the resolution and the R- and free R-factors from the PDB as measures of the general structure quality. This was done in order to validate our assumption that the correlation between the level of burial and evolutionary conservation of the amino acids would increase with the structure quality (see Section 3.1.1).
2.2.4 Alignment quality features
We collected the following measures for each structure: (i) Nseq, the number of homologs in ConSurf-DB's alignment; (ii) Nseq20, the number of homologs in the alignment whose identity is >20% [the level of identity for each homolog is extracted from the PSI-BLAST output (Altschul et al., 1997), taken from ConSurf-DB]; (iii) Resnum, the number of residues with significant conservation information; (iv) %insig, the fraction of the protein residues whose conservation level is assigned the value ‘insufficient data’. These features were chosen to reflect the general quality of the alignment and evolutionary rates generated by ConSurf-DB for each protein.
2.2.5 Finding the optimal filtering cutoffs
The four measures of alignment quality that we collected could each help predict in advance whether a given protein would be well-suited for use with our method. To find the optimal way to integrate these features, we solved the following optimization problem: given a ratio X (called the filtering degree), find the optimal quadruple of cutoffs such that when filtering the dataset according to these cutoffs, X of the proteins in the dataset pass the filter, and their average ConQuass score (as defined in Section 2.3) is maximal. This problem was solved for each X in 0.01, 0.02,…, 1 using an exhaustive enumeration, enumerating for each cutoff over 50 discrete values distributed evenly across the dataset. In what follows we refer to proteins that passed the filter corresponding to a given filtering degree X as having a ‘high-quality alignment, according to the X-filter’, where a higher filtering degree corresponds to a more stringent requirement.
2.3 The ConQuass score
Similarly to Verify3D (Bowie et al., 1991), we built a 10 × 9 propensity matrix, where each cell gives the compatibility score for assigning a residue with conservation class c an accessibility class a, as given by the information value (Fano, 1961):
where P(c|a) is the probability of finding a residue of conservation class c in the accessibility class a, and P(c) is the overall probability of finding a residue in conservation class c. These probabilities are estimated using the conservation and accessibility levels of the residues in the dataset of known protein structures. The accessibilities were calculated using the biological unit given by PQS. We also tried using the PISA biological unit or the isolated chain, but the propensity matrices generated were very similar (data not shown). The final propensity matrix is shown in Supplementary Table S1.
ConQuass assigns each structure the average score of its residues:
where C and A are vectors of the same length L (number of amino acids in the protein), giving, respectively, the conservation and accessibility classes of the residues.
2.4 Assessment on the CASP dataset
In the model quality assessment category of CASP, the participating groups were asked to rank the models built by the participating automatic servers. We downloaded these server models, as well as the predictions of the participating MQAPs, from the CASP web site (http://predictioncenter.org). We also downloaded for each server model its global distance test total score (GDT-TS) (Zemla, 2003), which is in the range (0, 100] and is the standard quality evaluation score given by CASP.
For each CASP target, we downloaded conservation information, if available, from the ConSurf-DB database entry for the native structure. The same conservation information was aligned to all full-atom models of the target, as there is sometimes a shift between the residue sequence numbers in the native structures and in the CASP models. To this end, we ranked each such alignment by giving each column a score of +1 if the residue identity in the native matched that of the model, −2 if the residues did not match, −1 for an insertion/deletion and 0 if the residue was missing in one of the structures. The optimal alignment was then found using the Smith–Waterman algorithm (Smith and Waterman, 1981). For each model, we computed the accessibility class of each residue, as was done for the structures in the training set (see Section 2.1). The conservation levels and accessibilities were used to calculate the MQAP score for each model (see Section 2.3). We did not score targets whose native structure had no ConSurf-DB information.
When comparing ConQuass to the MQAPs that participated in CASP7, we considered only the 16 MQAPs that had ranked at least 15 000 models. For MQAPs that participated in CASP8, we considered only the 22 pure single-structure methods that had ranked at least 20 000 models. We restricted each analysis to models that had been ranked by all considered methods (including ConQuass), and from this set we eliminated targets for which fewer than 100 ranked models were available. For each MQAP and each target, we calculated the Pearson correlation between the quality scores given by the MQAP and the GDT-TS scores downloaded from the CASP web site.
2.5 Integration of ConQuass with other methods
To demonstrate that the conservation information used in ConQuass is complementary to that used by other methods, we built three new MQAPs, integrating the score given by ConQuass (Section 2.3) with the scores given by Circle-QA (Terashi et al., 2007), QMEANfamily (Benkert et al., 2009) and MULTICOM-REFINE (Cheng et al., 2009), respectively. We chose Circle-QA because it was the leading pure single-structure method in CASP7, and we chose QMEANfamily and MULTICOM-REFINE because they were the leading pure single-structure methods in CASP8. For each integrated MQAP, the score we assigned to each model was a simple linear combination of the ConQuass score and the score produced by the other method (the two scores were each assigned a weight of 0.5). The analysis described in Section 2.4 was repeated for these three MQAPs. We compared the first MQAP (integration with Circle-QA) to MQAPs that had participated in CASP7 and compared the other two (integration with QMEANfamily or MULTICOM-REFINE) to MQAPs that had participated in CASP8.
3 RESULTS AND DISCUSSION
3.1 Experimentally determined structures match their conservation pattern
3.1.1 Examining a dataset of high-quality structures
It is widely recognized that residues buried in the protein core tend to be evolutionarily conserved, whereas residues on the surface are usually variable (Brändén and Tooze, 1999; Lichtarge et al., 1996). This implies that the accessibilities of variable residues should be shifted toward higher values in comparison with those of conserved residues, as indeed seems to be the case for many experimentally solved protein structures we examined (e.g. Fig. 1A and B). This characteristic is expected for true protein structures, and we would generally not expect to see it in incorrect models. Figure 1C and D show the evolutionary profile of an extremely poor model structure (analysis of an intermediate quality model of the same protein is provided in Supplementary Fig. S1). We first set out to measure the magnitude of this trend in real protein structures. For that purpose, we collected a comprehensive dataset of high-quality experimentally determined structures, which we can reasonably assume to contain mostly ‘correct’ structures (Section 2.1). For this dataset, it is obvious that the more variable residues are consistently more accessible than the conserved residues (Fig. 1E).
The information in this dataset was used to calculate a propensity matrix, giving the compatibility of each conservation class with each accessibility class (Section 2.3, and Supplementary Table S1). The matrix confirmed our intuitive expectations, giving high propensity scores to accessible-variable residues and to buried conserved residues. Consequently, the matrix was used to calculate each protein structure's ConQuass score, which was the average of the propensity scores of the protein's residues. A score was calculated for each structure in the dataset (Fig. 2), using the biological unit complexes as given by PQS (Henrick and Thornton, 1998). Only 7.9% of the structures received a negative score, meaning that for most structures the residues' conservation levels tended to be compatible with their accessibility levels. However, when we determined scores for the individual chains in the dataset without the context of the biological unit, more structures were assigned a negative score (12.5%). This was due to monomers exposing conserved interface residues that are actually buried in the physiological complex. We also tried to determine scores for the biological unit complexes given by PISA (Krissinel and Henrick, 2007) and the results were very similar to those obtained for the PQS complexes (data not shown). The ConQuass scores also seemed to become progressively higher for structures of higher quality, as measured by various structure quality measures such as resolution, R-factor and free R-factor (Fig. 3A).
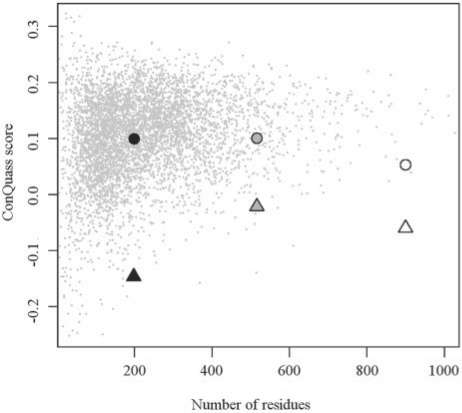
Fig. 2.
ConQuass scores assigned to experimental structures from the PDB and to a few erroneous models. The scatter plot shows the propensity score of the protein versus the number of residues with ConSurf information for all the structures in the dataset (in gray). Only structures that have ConSurf information for at least 40 residues were included. Also shown are pairs of incorrect (triangle) and correct (circle) structures for EmrE (black, 2f2m and 3b5d), Connexin (gray, 1txh and 2zw3) and MsbA (white, 1jsq and 3b5w). For each of these structures, the ConQuass score was calculated for the residues of all the chains in the biological unit. For models containing only the Cα-trace, the full-atom structures were rebuilt using MaxSprout and SCWRL4. The correct structure of MsbA (3b5w) was truncated to contain the same set of residues as the erroneous structure (1jsq).
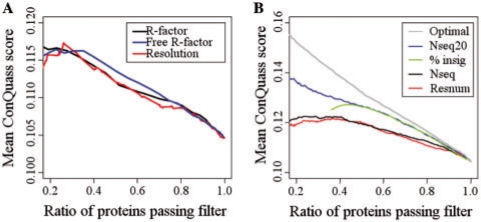
Fig. 3.
Compatibility of the structure with the evolutionary profile of the protein is higher for higher-quality structures or higher-quality multiple sequence alignments, as described by different quality measures. (A) The mean ConQuass score of the proteins in the dataset when filtering only for the top X proteins (x-axis), as measured by several crystallographic structure quality measures: the R-factor, free R-factor and the resolution. (B) As in (A), but when filtering by non-structural measures: the number of residues (red), the number of homologous sequences in the alignment (black), the ratio of residues with insignificant conservation information as measured by ConSurf (green) and the number of homologous sequences in the alignment with at least 20% identity with the query (blue). Also shown is the optimal ratio achieved by integrating these four measures (gray).
The conservation data was calculated according to the multiple sequence alignment generated automatically by ConSurf-DB, and it is possible that a high-quality structure would be assigned a low ConQuass score if an inadequate alignment was used. To discern these cases, we collected four measures that are indicative of the alignment quality or that could otherwise predict an incorrect ConQuass score for a protein model (see Section 2.2.4). As can be seen in Figure 3B, the ConQuass score becomes progressively higher as the dataset is filtered to leave only structures whose alignment is of higher quality according to any one of the four measures.
Obviously, a better indicator for how suitable a protein is for ranking with ConQuass can be achieved by integrating the different alignment quality measures. We used an exhaustive enumeration to find the optimal way to integrate these measures, each time filtering the database to leave only X% of the proteins such that the mean ConQuass score of the remaining proteins is maximal (Section 2.2.5). This procedure assumes that after filtering, a higher mean ConQuass score is achieved because the remaining proteins have a higher quality alignment. The integration achieves a much higher mean ConQuass score than does filtering by each measure separately (Fig. 3B; gray). The optimal cutoffs found for some selected filtering degrees are shown in Supplementary Table S2.
3.1.2 The conservation profile may reveal incorrect structures
To test whether the ConQuass score is capable of discriminating incorrect structures, we collected three examples of structural models that had been deposited in the PDB but were later found to be incorrect. All these structures also have corrected versions available, which we also scored using ConQuass (Fig. 2).
The first two examples are EmrE (Fig. 2; black) and MsbA (Fig. 2; white). Both structures were determined by Chang and coworkers using a faulty piece of in-house software, which caused the group to misinterpret the crystallization data and eventually yielded false models. Following the detection of the error in the software, the structures were retracted (Chang et al., 2006), and corrected versions have since been published (Chen et al., 2007; Ward et al., 2007). Calculating the ConQuass score for these structures is not straightforward, as they are all Cα-only models, with the exception of the erroneous EmrE structure. However, we were able to apply ConQuass after reconstructing the full-atom models using MaxSprout (Holm and Sander, 1991) and SCWRL4 (Krivov et al., 2009). Clearly, the correct structures are much more compatible with their conservation pattern than are the incorrect ones (Fig. 2).
The third example is the gap junction connexin channel (Fig. 2; gray), which was previously modeled by our group using low-resolution electron cryo-microscopy data (Fleishman et al., 2004b). The helix assignment of the model recently turned out to be wrong when an experimentally determined high-resolution structure of a homologous protein was reported (Maeda et al., 2009). For the purpose of comparing the two structures, we truncated the non-membrane residues from the experimental structure and also removed all non-Cα atoms. This procedure left us with two Cα-only models composed of the same set of residues. We then rebuilt the two full-atom models as above and scored them using ConQuass. While both the truncation of non-membrane residues and the full-atom reconstruction lowered the score for the crystallographic model (data not shown), it was still assigned a much higher score than the erroneous model (Fig. 2).
There are some cases in which a correct model seems not to match its conservation pattern, as denoted by a negative ConQuass score. However, a closer examination can usually provide an explanation for the low score. Some representative examples are discussed in Supplementary Section S1.1.
3.2 Ranking decoys in CASP
ConQuass may also assess how distant a given model is from the native structure. To show this, we checked how ConQuass scores models of varying quality for the same protein. A good source for such models is the biennial CASP experiment (Moult et al., 2009), where each round consists of several targets, corresponding to proteins whose structure have recently been solved (but not yet published), and each participant submits computational models in an attempt to predict the structure of each target. At the end of the round, the experimental structures are revealed, and the quality of each submitted model is measured by the similarity measure GDT-TS (Zemla, 2003), which is based on the superposition between the model and the native structure. The seventh and eighth CASP rounds included a quality assessment category (Cozzetto et al., 2007, 2009), in which different MQAPs participated and were consequently evaluated according to their performance. The models scored by the MQAPs were server models that were generated by the structure prediction servers participating in CASP and published shortly after the round began. The MQAPs were evaluated according to the correlation between the scores they gave the different models and the quality of those models as measured by GDT-TS. The scores given by the participating MQAPs are available for download from the CASP web site (http://predictioncenter.org), which allowed us to compare them with ConQuass. The results for the CASP8 set are presented below. The analysis of the CASP7 set showed a similar performance, and it is presented in Supplementary Section S1.2.
To be able to best compare ConQuass with the other pure single-structure methods, we have excluded from our analysis methods that use structural data from the other decoys or from homologs (a comparison of ConQuass with the latter methods is shown in Supplementary Fig. S2). For brevity, we will use the term MQAP in this section to refer only to pure single-structure methods.
3.2.1 Example of the performance on one CASP8 target
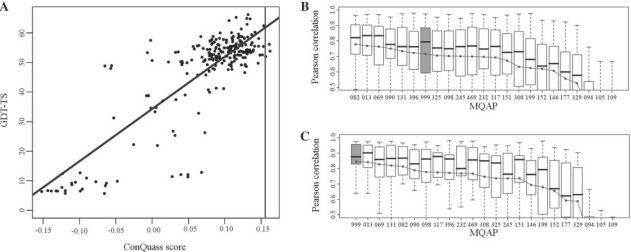
As an illustrative example, Figure 4A shows the GDT-TS values of the server models of CASP8 target T0449, plotted as a function of the assigned ConQuass scores. There is a striking correlation between the score and the structure quality, and the set of highly scored models was enriched with high-quality structures. The Pearson correlation in this case was 0.827, and the score of the native structure (Fig. 4A; vertical line) was higher than the scores of all the decoys except three.
Fig. 4.
The ability of the ConQuass score to rank decoys in the CASP8 dataset. (A) Demonstration for target T0449. For each decoy, the GDT-TS (similar to the native structure) is plotted versus the ConQuass score. The vertical line is the ConQuass score assigned to the native structure. The Pearson correlation for this target was 0.827. (B) Box plots of the correlation values for the 22 MQAPs that ranked at least 20 000 models. The number signifying each MQAP (x-axis) is the number assigned in the original CASP8 experiment (see http://predictioncenter.org/casp8). Also shown is the box plot for the correlation values of ConQuass (999, gray). The correlations were calculated only for models ranked by all 23 MQAPs. The box plots were sorted by the mean correlation, indicated by the black dots. The figure is cut to show only the correlation range [0.5, 1] in order to make the differences between the methods more apparent (the uncut version is shown in Supplementary Fig. S3). (C) Same as (B), when looking only at targets with the highest quality alignment, using the 20% filter. Although ConQuass is ranked first here, the specific ordering of the top ranking methods is irrelevant, as the correlation values achieved by ConQuass are not significantly higher than those achieved by MULTICOM-REFINE (013).
3.2.2 Overall performance on all CASP8 targets
In our calculation on the CASP datasets (see Section 2.4), we used the conservation data recorded in the ConSurf-DB dataset. There are cases in which the alignment could have been manually improved in order to achieve a better performance, but we deliberately refrained from doing this to avoid biasing our results. Four CASP8 targets could not be ranked, because their native structures did not have any ConSurf-DB information. This usually happens when ConSurf-DB cannot find enough homologs to construct a meaningful alignment. Ten additional targets were cancelled by CASP8 or had no corresponding native structure listed in the CASP8 web site. A ConQuass score was given to each of the full-atom models of the remaining 114 targets.
CASP allowed each participating MQAP to choose to rank any subset of models, for any subset of targets. Indeed, many MQAPs are not applicable for all models. This makes performance comparison problematic. For example, it might be easier to assess the quality of full-atom models, and if so an MQAP (such as ConQuass) that ranks only such models would have an advantage over methods that also rank Cα models. To avoid this problem, we carried out all calculations on the set of 11 686 models and 75 targets that were ranked by all participating MQAPs. To avoid excluding too many models, only the 22 participating MQAPs that scored at least 20 000 models were used.
To evaluate the performance of each MQAP, we calculated for each CASP8 target the Pearson correlation between the scores determined by the MQAP and the GDT-TS values of all the models for that target. The sets of correlation values for each MQAP are plotted in Figure 4B. Our ranking of the methods is slightly different from the ranking published in the CASP8 proceedings (Cozzetto et al., 2009) due to differences in the ranking protocol (see a detailed explanation of the differences in Supplementary Section S1.3). However, as in the CASP8 results, the MQAPs that performed best according to our assessment were the different variants of QMEAN (Benkert et al., 2009) and MULTICOM (Cheng et al., 2009). The variants with the highest mean correlation were QMEANfamily (082) with a mean correlation of 0.778 and MULTICOM-REFINE (013) with a mean correlation of 0.768. Following the different variants of QMEAN and MULTICOM, the method with the next highest ranking, with a mean correlation of 0.722, was CIRCLE (396), which was the best performing single-structure MQAP in CASP7 (Cozzetto et al., 2007). ConQuass (999, Fig. 4B; gray) ranked next, with a mean correlation of 0.715.
As shown in Section 3.1.1, some structures were assigned a low ConQuass score because of a low-quality alignment rather than a low-quality model. Indeed, for some CASP8 targets, the native structure itself scored very low by ConQuass. Such targets were clearly not suitable for use with our method. Many of these cases could be discerned in advance by using the alignment quality measures presented in Section 2.2.4. To check how our method performs on more appropriate targets, we used the 20% filtering to select only the 17 targets with the highest quality alignment. The performance of the different methods for this subset of targets is shown in Figure 4C. With these targets, ConQuass performs significantly better, and it is ranked first with a mean correlation of 0.844. It is important to stress that the set of targets that are more suitable for use with ConQuass can be selected a priori, as all the features used for the filtering are based on the multiple sequence alignment alone.
The Pearson correlations we evaluated were calculated for each target independently, so scores produced by an MQAP that achieves a high correlation value can be used to select among alternative structural models for the same protein. However, in many scenarios one wants to evaluate the absolute quality of a single uncertain structural model without comparing it to other decoys. For these cases, it is informative to know the relation between the MQAP score and the structure quality, as measured by the GDT-TS. This relation, for all models of all CASP8 targets, is shown in Figure 5A. The overall correlation between the ConQuass score and GDT-TS is good (0.678), especially when filtering for targets with a high-quality alignment (0.843 if using the 20% filtering, Fig. 5A; red). The overall correlation was also compared with that of the other participating MQAPs (Supplementary Table S3).
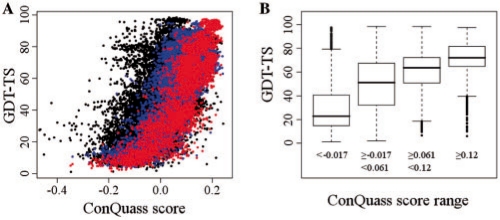
Fig. 5.
The relation between the ConQuass score assigned to the model and the model's quality. (A) Plot of the GDT-TS (similar to the native structure) versus the ConQuass score for the following models: all models in the CASP8 dataset (black, Pearson correlation 0.678); the models with the highest quality alignment by the 50% filter (blue, Pearson correlation 0.780); models that passed the less permissive 20% filter (red, Pearson correlation 0.843). (B) Box plots of the GDT-TS values for models in each ConQuass score quartile. For example, the median GDT-TS for the models scoring very low (below −0.017) is 22.8, and 50% of these models have GDT-TS values between 14.8 and 40.6. For the models scoring very high (>0.12), the median GDT-TS is 73.1, and 50% of these models have GDT-TS values between 65.0 and 81.7.
Figure 5B presents these results in a way that is more intuitive for interpreting the score given to a model by ConQuass. If a model is assigned a very low ConQuass score (below -0.017), it is expected to be of rather low-quality (median GDT-TS 22.8, most GDT-TS values in the range [14.8, 40.6]). However, if a model is assigned a very high ConQuass score (>0.12), it will very rarely be a low-quality structure (median GDT-TS 73.1, most GDT-TS values in the range [65.0, 81.7]).
3.2.3 Complementarity to other MQAPs
ConQuass uses the evolutionary conservation properties of the protein structure, a feature that is not directly used by any other contemporary MQAP. It therefore seems reasonable to suggest that ConQuass is complementary to the other prevalent methods. To support this claim, we scored the CASP8 models using two new MQAPs that were trivial integrations of ConQuass with, respectively, MULTICOM-REFINE and QMEANfamily, the two best performing single-structure MQAPs in CASP8 (see Section 2.5). The performance of these two integration methods was analyzed using the same procedure described above. The integration with ConQuass significantly improved the correlations achieved by both MULTICOM-REFINE (P-value ∼ 4.2e-14) and QMEANfamily (P-value ∼ 1.1e-08); see Supplementary Section S1.4.
4 CONCLUSION
Here we have presented ConQuass, a very simple MQAP based directly on the compatibility between the conservation and accessibility patterns of a given structural model. We studied the scores that ConQuass assigns to experimental structures, demonstrated its ability to discern erroneous models and checked the relation between the ConQuass scores given to different models and the models' resemblance to the native structure. We have also shown that ConQuass's performance is comparable to that of other pure single-structure MQAPs, despite being much simpler than most.
Our approach is different from previous MQAPs that used evolutionary conservation, which were based on the spatial clustering of the conserved residues. We feel our approach is more direct, since this clustering is mostly an effect of the conserved residues' tendency to be buried in the structural core (for a direct comparison with the method developed by Mihalek and coworkers; see Supplementary Section S1.5). ConQuass is also the first conservation-based approach to be rigorously compared with contemporary MQAPs. In addition, our score is based on summation of information that is local in the structure (the propensity of the conservation class of each residue for its accessibility class), so it should be adaptable to provide a local quality score for each residue of the structure, as is done by local quality assessment tools (Fasnacht et al., 2007). Preliminary tests for a local MQAP based on summing the propensities over a fixed-width window on the sequence have yielded promising results (data not shown).
In this study, we have clearly shown that evolutionary conservation is a powerful property for use in model quality assessment, so it would be advantageous for new MQAPs to integrate this property with other more commonly used properties. Evolutionary conservation is currently not used directly by any MQAP, although some methods, like QMEAN and MULTICOM, use it indirectly by comparing model surface accessibilities with the predicted accessibilities, which are associated, in part, with the evolutionary conservation. However, we have demonstrated that these methods do not use the conservation information to its full extent, as their results improve when their scores are integrated with those of ConQuass. In this work, we have followed a very naïve approach for such an integration, using a simple linear combination. Much better results would doubtless be obtained by a more intricate approach, for example by using the residue conservation as one of the features in a machine learning-based tool. In any case, such integration would have to take into account the quality of the alignment, as the evolutionary conservation property is more indicative for high-quality alignments. The same approach could also be used to integrate conservation in many other practices, such as finding the physiological complex of a crystal structure and scoring docking results. In addition, as the ConQuass score reflects the consistency between the alignment and the structure, its functionality could be reversed to check the quality of an alignment based on a high-quality structure.
While ConQuass is not the best performing of the examined MQAPs, many of which use a mixture of complex features including geometric and energetic properties of the structure, it has the advantage of being straightforward and easy to interpret. The conservation pattern of the protein is not used by most modeling and structural determination programs, so ConQuass gives independent support for a structural model, whether experimental or computational. If the model is assigned a low score, it is easy to visualize the discrepancy of the model with the conservation pattern by projecting it on the structure using ConSurf (Glaser et al., 2003), as we have done in the examples in Figure 1 and Supplementary Figure S4. This can either yield relevant insights regarding the mechanism associated with the structure (for example, hint that it may bind to another molecule; see Supplementary Section S1.1), lead to a rejection of the model (see Fig. 1C) or perhaps in some cases guide further refinements of the model.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Gilad Wainreb and Maya Schushan for helpful discussions.
Funding: Israel Science Foundation (grant 611/07); Edmond J. Safra Bioinformatics program at Tel-Aviv University (to M.K.).
Conflicts of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archie JG, et al. Applying undertaker to quality assessment. Proteins. 2009;77(Suppl. 9):191–195. doi: 10.1002/prot.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P, et al. Global and local model quality estimation at CASP8 using the scoring functions QMEAN and QMEANclust. Proteins. 2009;77(Suppl. 9):173–180. doi: 10.1002/prot.22532. [DOI] [PubMed] [Google Scholar]
- Bowie JU, et al. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Brändén C.-I, Tooze J. Introduction to Protein Structure. New York: Garland Pub.; 1999. [Google Scholar]
- Chang G, et al. Retraction. Science. 2006;314:1875. doi: 10.1126/science.314.5807.1875b. [DOI] [PubMed] [Google Scholar]
- Chen YJ, et al. X-ray structure of EmrE supports dual topology model. Proc. Natl Acad. Sci. USA. 2007;104:18999–19004. doi: 10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, et al. Prediction of global and local quality of CASP8 models by MULTICOM series. Proteins. 2009;77(Suppl. 9):181–184. doi: 10.1002/prot.22487. [DOI] [PubMed] [Google Scholar]
- Cozzetto D, et al. Assessment of predictions in the model quality assessment category. Proteins. 2007;69(Suppl. 8):175–183. doi: 10.1002/prot.21669. [DOI] [PubMed] [Google Scholar]
- Cozzetto D, et al. Evaluation of CASP8 model quality predictions. Proteins. 2009;77(Suppl. 9):157–166. doi: 10.1002/prot.22534. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, et al. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Fano RM. Transmission of Information; a Statistical Theory of Communications. New York: M.I.T. Press; 1961. [Google Scholar]
- Fasnacht M, et al. Local quality assessment in homology models using statistical potentials and support vector machines. Protein Sci. 2007;16:1557–1568. doi: 10.1110/ps.072856307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, et al. An automatic method for predicting transmembrane protein structures using cryo-EM and evolutionary data. Biophys. J. 2004a;87:3448–3459. doi: 10.1529/biophysj.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, et al. A C-alpha model for the transmembrane alpha helices of gap junction intercellular channels. Mol. Cell. 2004b;15:879–888. doi: 10.1016/j.molcel.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, et al. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- Ginalski K. Comparative modeling for protein structure prediction. Curr. Opin. Struct. Biol. 2006;16:172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Glaser F, et al. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Goldenberg O, et al. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handl J, et al. Artefacts and biases affecting the evaluation of scoring functions on decoy sets for protein structure prediction. Bioinformatics. 2009;25:1271–1279. doi: 10.1093/bioinformatics/btp150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J. Mol. Biol. 1991;218:183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. ‘NACCESS’, Computer Program. Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- Kleywegt GJ. On vital aid: the why, what and how of validation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:134–139. doi: 10.1107/S090744490900081X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krivov GG, et al. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, et al. Model structure of the Na+/H+ exchanger 1 (NHE1): functional and clinical implications. J. Biol. Chem. 2007;282:37854–37863. doi: 10.1074/jbc.M705460200. [DOI] [PubMed] [Google Scholar]
- Larsson P, et al. Assessment of global and local model quality in CASP8 using Pcons and ProQ. Proteins. 2009;77(Suppl. 9):167–172. doi: 10.1002/prot.22476. [DOI] [PubMed] [Google Scholar]
- Lichtarge O, et al. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Maeda S, et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ. Prediction of global and local model quality in CASP8 using the ModFOLD server. Proteins. 2009;77(Suppl. 9):185–190. doi: 10.1002/prot.22491. [DOI] [PubMed] [Google Scholar]
- Mihalek I, et al. Combining inference from evolution and geometric probability in protein structure evaluation. J. Mol. Biol. 2003;331:263–279. doi: 10.1016/s0022-2836(03)00663-6. [DOI] [PubMed] [Google Scholar]
- Mihalek I, et al. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J. Mol. Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- Miller CS, Eisenberg D. Using inferred residue contacts to distinguish between correct and incorrect protein models. Bioinformatics. 2008;24:1575–1582. doi: 10.1093/bioinformatics/btn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J, et al. Critical assessment of methods of protein structure prediction - Round VIII. Proteins. 2009;77(Suppl. 9):1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- Muppirala UK, Li Z. A simple approach for protein structure discrimination based on the network pattern of conserved hydrophobic residues. Protein Eng. Des. Sel. 2006;19:265–275. doi: 10.1093/protein/gzl009. [DOI] [PubMed] [Google Scholar]
- Nimrod G, et al. Detection of functionally important regions in “hypothetical proteins” of known structure. Structure. 2008;16:1755–1763. doi: 10.1016/j.str.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Olmea O, et al. Effective use of sequence correlation and conservation in fold recognition. J. Mol. Biol. 1999;293:1221–1239. doi: 10.1006/jmbi.1999.3208. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. Available at http://www.rbvi.ucsf.edu/chimera/(last accessed date April 5, 2010) [DOI] [PubMed] [Google Scholar]
- Samudrala R, Levitt M. Decoys ‘R’ Us: a database of incorrect conformations to improve protein structure prediction. Protein Sci. 2000;9:1399–1401. doi: 10.1110/ps.9.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler-Furman O, Baker D. Conserved residue clustering and protein structure prediction. Proteins. 2003;52:225–235. doi: 10.1002/prot.10365. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Terashi G, et al. Fams-ace: a combined method to select the best model after remodeling all server models. Proteins. 2007;69(Suppl. 8):98–107. doi: 10.1002/prot.21785. [DOI] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Quality assessment of protein models. In: Janusz MB, editor. Prediction of Protein Structures, Functions, and Interactions. Chichester: Wiley; 2008. pp. 143–157. [Google Scholar]
- Wang G, Dunbrack R.L., Jr. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- Ward A, et al. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl Acad. Sci. USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc. Natl Acad. Sci. USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.