Abstract
Motivation: A major challenge in utilizing microarray technologies to measure nucleic acid abundances is ‘normalization’, the goal of which is to separate biologically meaningful signal from other confounding sources of signal, often due to unavoidable technical factors. It is intuitively clear that true biological signal and confounding factors need to be simultaneously considered when performing normalization. However, the most popular normalization approaches do not utilize what is known about the study, both in terms of the biological variables of interest and the known technical factors in the study, such as batch or array processing date.
Results: We show here that failing to include all study-specific biological and technical variables when performing normalization leads to biased downstream analyses. We propose a general normalization framework that fits a study-specific model employing every known variable that is relevant to the expression study. The proposed method is generally applicable to the full range of existing probe designs, as well as to both single-channel and dual-channel arrays. We show through real and simulated examples that the method has favorable operating characteristics in comparison to some of the most highly used normalization methods.
Availability: An R package called snm implementing the methodology will be made available from Bioconductor (http://bioconductor.org).
Contact: jstorey@princeton.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
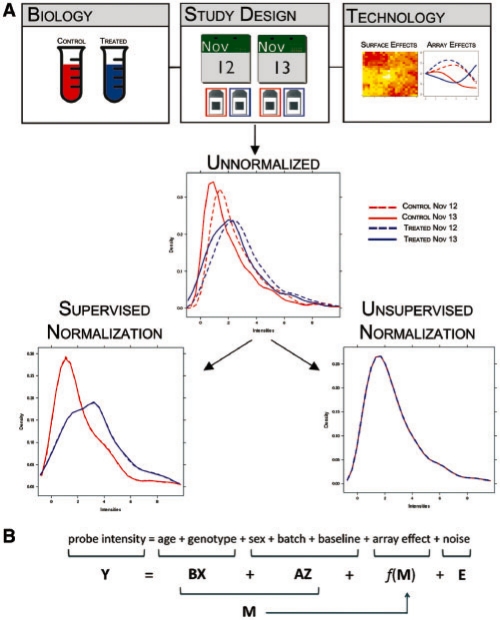
The microarray technology measures nucleic acid abundances, such as from mRNA, in biological samples by producing fluorescence intensities that reflect hybridization events between nucleotide sequences. Attached to the microarray are single-stranded DNA probes, which represent a short segment of complementary DNA within a gene. Differences among current microarray technologies are typically in terms of probe design and the number of fluorescent dyes applied to any given array. Regardless of the technology, statistical analysis of these data usually attempts to infer the relationship between nucleic acid abundance variation and biological phenotype. One of the main challenges of this inference process is normalization, which addresses the fact that factors not of biological relevance can influence the observed intensities (Bolstad et al., 2003; Dudoit et al., 2002; Tseng et al., 2001; Wu et al., 2004), complicating approaches used to infer relationships from these data (Dabney and Storey, 2007; Rattray et al., 2006; Wu and Irrizary, 2007). The major contribution of this paper is to provide a general ‘supervised’ framework for microarray normalization (Fig. 1). This approach is applicable to all major microarray technologies and performs the normalization in a manner supervised by all known variables relevant to the study.
Fig. 1.
A demonstration of the main ideas behind supervised normalization of microarrays. (A) A hypothetical example to demonstrate the differences between supervised and unsupervised normalization strategies. The three boxes arranged across the top display different types of potential effects. Each of these potentially influences the unnormalized observed intensities, which are presented as densities in the middle panel. The blue and red lines describe the different biological conditions, while the dashed and dotted lines describe the different dates. The differences among the four arise either from the biology or study design. After normalization with a supervised approach that takes all three effects into account when normalizing the data, the differences between the blue and red lines are still present, while the differences between the dashed and dotted lines have been removed. However, for unsupervised approaches, such as quantile normalization, the resulting data have been transformed so that all arrays have the same distribution, a result that clearly violates the biological relationship of interest. (B) An example of the model we fit to the probe-level data from a microarray study. The model has probe-specific terms, intensity-dependent terms and may include other terms such as probe composition effects or surface level spatial effects.
Some of the most highly used microarray normalization methods are what we call ‘unsupervised’ methods. These are normalization procedures that do not utilize the variables describing the study, specifically the biological variables of interest (Fig. 1). For example, suppose the goal of a microarray study is to identify genes differentially expressed with respect to an experimental treatment. Also, suppose that the arrays were processed in two separate batches. Unsupervised methods ignore the treatment and batch variables when performing the normalization. However, if the goal of normalization is to separate biologically meaningful signal from technical confounders, then it seems infeasible to do so without taking into account the signal explained by the study-specific variables, such as treatment and batch in this example.
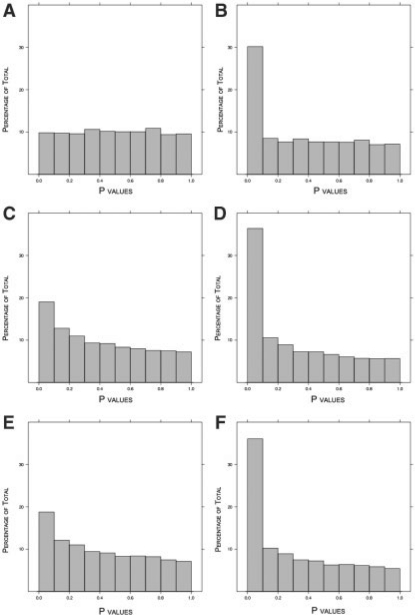
While unsupervised methods may show favorable operating characteristics in specialized settings—such as when biological variables contribute relatively negligible signal to the data—it has been shown they make assumptions about data that are commonly invalidated in practice (Dabney and Storey, 2007; Irizarry et al., 2006). As a simple motivating example meant to illustrate how easily these assumptions are violated, we simulated microarray data (extensive details are given in following sections) with signal due to a dichotomous biological variable and intensity-dependent array effects. We simulated 100 000 probes, 30% of which are differentially expressed. Figure 2 shows the P-value histograms corresponding to probes, which are not differentially expressed. Figure 2A is the method we propose in this work, where the P-values are correctly Uniform(0,1). Figure 2C and E show the P-values from the same probes when using invariant set normalization (ISN; Li and Wong, 2001) and quantile normalization (QN; Bolstad et al., 2003), respectively. It can be seen that both sets of P-values are anti-conservatively biased.
Fig. 2.
Results from simulated data with differential expression and array effects. The true proportion of null probes is π0 = 0.70. (A) P-value histogram of null probes after SNM normalization. (B) P-value histogram of all probes after SNM normalization. (C) P-value histogram of null probes after QN. (D) P-value histogram of all probes after QN. (E) P-value histogram of null probes after ISN. (F) P-value histogram of all probes after ISN.
We show that the main reason for the anti-conservative null P-value distribution from the two unsupervised normalization methods is that the known biological variable was not taken into account. We also show that unsupervised normalization methods may become more problematic and unpredictable as more study-specific variables are ignored. The basic reason for this is that when performing normalization, one cannot unbiasedly separate true biological signal from technical and other study-specific confounders unless both sets of variables are taken into account during the normalization process. On the other hand, supervised normalization methods (Baird et al., 2004; Dabney and Storey, 2007; Wolfinger et al., 2001; Wu and Irrizary, 2007; Wu et al., 2004) use all measured variables as a basis for normalization. Several supervised methods have been developed for the analysis of dual-channel microarrays (Baird et al., 2004; Dabney and Storey, 2007; Wolfinger et al., 2001), and recent work has demonstrated that results from supervised techniques offer a clear improvement over results obtained from unsupervised approaches (Dabney and Storey, 2007). However, these methods require highly structured experimental designs and are not applicable to single-channel microarrays or microarrays with several probes per gene.
We develop a framework for supervised normalization of microarray data, which is applicable to a large class of experimental designs and technologies, including single-channel arrays, dual-channel arrays and different probe designs (e.g. one probe per gene, probe sets and exon arrays). We focus the examples on common sources of confounding variation and demonstrate that the proposed supervised method accounts for their effects on the data without perturbing the resulting inference. We provide examples that show how the framework presented here can be used to analyze several types of microarray data, thus unifying the problems of data normalization for these alternative microarray technologies.
2 APPROACH
In any given study, there are a set of study-specific variables known to the researcher that are also capable of being included in the model used to perform inference. These study-specific variables fall into one of the two categories: biological variables or adjustment variables. We define biological variables to be those whose relationships with nucleic acid variation are the target of the statistical analysis. The other variables, which are utilized as covariates to account for other sources of variation, are what we call adjustment variables. Essentially, adjustment variables are all of the study-specific variables not of inferential interest in the study but may correctly explain variation in the data. These include technical variables such as dye, surface and probe-composition effects. They include study design variables, such as batch, technical replicates, or the dates on which the arrays were processed. Adjustment variables may also include those that are biological in nature, but not of interest in the study. For example, perhaps the goal is to characterize the relationship between an experimental treatment and gene expression, but the study is performed on both males and females. If the relationship between sex and expression is not of interest, then this variable would be included among the adjustment variables.
The basic idea of the proposed approach and how it differs from unsupervised methods is displayed in Figure 1. Figure 1A shows that the proposed normalization approach utilizes all that is known about the study (biology, study design and technology) in order to perform the normalization. This allows for the signal due to biology to be left intact in the normalized data. Figure 1B displays an example of the model we fit to the probe-level data from a microarray study. The model has probe-specific terms, intensity-dependent terms and may include other terms such as probe composition effects or array surface spatial effects. The probe-specific terms are partitioned between the biological variables and the adjustment variables. The intensity-dependent terms are written as smooth, random functions of these probe-specific terms. We jointly fit this model to all probes simultaneously, allowing all relevant variables to be properly included in the model. Our model fitting procedure is iterative in order to minimize confounding between the biological variation of interest and the other sources of variation.
3 METHODS
3.1 A general model
We first consider the situation where the complete data from a microarray experiment consists of three terms: the observed probe intensities, biological variables and adjustment variables. The intensities are usually presented as an m × n matrix, Y, where m and n describe the number of probes and arrays in the entire study, respectively. Define yij as the observed intensity for probe i = 1,…, m on array j =1,.., n, and yi = (yi1, yi2,…, yin) as the observed intensities for probe i across the n arrays. The k-th biological variable for array j, xkj, describes factors of interest such as disease status, experimental treatment, or time point. All d covariates for an individual sample j are denoted by the vector xj, and we group all n such vectors into a d × n matrix X = (x1,…, xn).
Similarly, the ℓth adjustment variable for array j, zℓj, parameterizes variables to be fit in a probe−specific manner. Let rc denote the number of probe-specific adjustment variables. When the baseline value of yij is not of interest, this probe-specific intercept term is included among the adjustment variables. Define the vector zj and matrix Z in a fashion analogous to the one described for xj and X, respectfully. Finally, we assume that there are rf intensity-dependent effects, which we denote by ftj for effect t = 1,…, rf and array j.
Note that we construct X and Z such that bi = 0 represents the case where the biological variables of interest show no association with probe i. (This is straightforward to construct even for time course studies; Storey et al. 2005). A concrete example of X and Z can be found in the Supplementary Material.
We model yij as linear combinations of xj, zj, and intensity-dependent effects. The model for each probe intensity measurement is written as
| (1) |
where mij = ∑k=1d bikxkj + ∑ℓ=1rc aiℓzℓj. The coefficients bik and aiℓ describe the influence of the k-th biological and ℓ-th adjustment variable on probe i's intensity. We assume that the t-th intensity-dependent function ftj is a random smooth function such that E[ftj(m)|m] = 0 for all m. These are parameterized as Normal distributed coefficients applied to a B-spline basis (Supplementary Material). The zero expectation can be made without loss of generality because we allow for probe-specific offset terms (i.e. intercepts), which absorb any systematic deviation from these intensity−dependent functions. Finally, eij is the unexplained random error for probe i on array j with E[eij] = 0.
We can write model (1) for probe i data across all n arrays, yi, as
| (2) |
where bi and ai are 1 × d and 1 × rc vectors of the bik and aiℓ terms in (1), and ft(bi X + aiZ) = (ft1(mi1),…, ftn(min)). The model for the entire dataset Y can be written as
| (3) |
where B and A are m × d and m × rc matrices of coefficients, the i-th row corresponding to bi and ai, respectively. Also, ft(BX + AZ) is an m × n matrix with the i-th row equal to ft(biX + aiZ).
3.2 The goal of normalization
The statistical goal of microarray analysis is to infer relationships between biological variables and individual probes or sets of probes. Different microarray platforms employ different designs. Regardless of the platform, relevant biological variation can be identified using individual probes, or groups of probes that share certain biological characteristics; for example, probes mapping to the same gene, chromosomal position or pathway. We use the term ‘probe set’ for any such grouping of probes. We first establish a definition of a correct normalization when inference is performed on individual probes (e.g. when there is one probe per gene).
One property that a normalization should satisfy is that 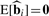 when bi = 0 and
when bi = 0 and 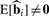 when bi ≠ 0. Stated more directly, the normalization procedure should preserve the biological relationships of interest in the expected values of the estimated coefficients corresponding to the biological variables. A second, less obvious property is that a normalization method should remove all latent structure shared across probes due to adjustment variables. Specifically, any remaining component of the aiZ + ∑t=1rfft(biX + aiZ) term will induce dependence in the error terms across probes, and unmodeled dependence has been shown to cause large-scale study−specific biases (Leek and Storey, 2007, 2008). An imperfect normalization procedure can bias the
when bi ≠ 0. Stated more directly, the normalization procedure should preserve the biological relationships of interest in the expected values of the estimated coefficients corresponding to the biological variables. A second, less obvious property is that a normalization method should remove all latent structure shared across probes due to adjustment variables. Specifically, any remaining component of the aiZ + ∑t=1rfft(biX + aiZ) term will induce dependence in the error terms across probes, and unmodeled dependence has been shown to cause large-scale study−specific biases (Leek and Storey, 2007, 2008). An imperfect normalization procedure can bias the 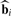 or induce new sources of latent structure shared across probes (Dabney and Storey, 2007).
or induce new sources of latent structure shared across probes (Dabney and Storey, 2007).
Violation of either of these properties can be seen in the joint distribution of P-values obtained from testing H0i : bi = 0 versus H1i : bi ≠ 0. Therefore, we propose the following criteria as the ideal properties for a normalization model to achieve.
Definition 1. —
Microarray Normalization Criteria.
The set of ‘null P-values’, obtained from testing H0i : bi = 0 versus H1i : bi ≠ 0 for all probes i where bi = 0, have a joint distribution following the Uniform(0,1) distribution.
The set of ‘alternative P-values’ obtained from the above hypothesis tests for all probes i where bi ≠ 0 have a joint distribution stochastically smaller than the Uniform(0, 1) distribution.
Correctly normalizing a microarray study is, therefore, a procedure that preserves the biological relationships of interest and the expected operating characteristics of the model fits at the probe level. The framework we introduce in this paper is designed to accomplish this goal, and numerical evidence we give indicates it does so when all relevant variables have been measured and included in the model.
We now explain how this definition relates to probe sets. The central challenge for inference on probe sets is that technical sources of variation are very much probe specific, while biological sources influence the entire set of probes. For example, intensity-specific effects, probe nucleotide composition and spatial effects are all most appropriately dealt with at the probe level, while differential expression in response to a treatment is best measured in terms of its effect across the entire set of probes measuring a given gene.
In our framework, a probe set summarization would combine all bi for all probes i corresponding to a given probe set, where the bi coefficients parameterize the biological relationships for each probe. A general probe set summarization for probe set k that takes this into account is
where the αi have been chosen to represent relative reliability of probe i, the set 𝒮k is the set of all probe indices in probe set k, and ∑i∈𝒮k αi = 1. Note that if bi = 0 for all i ∈ 𝒮k then ∑i∈𝒮k αibi = 0. In other words, preserving the proper operating characteristics of 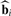 for all i also preserves them for
for all i also preserves them for 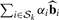 (notwithstanding issues with P-value calculations of multiple variables, which is unrelated to the normalization problem).
(notwithstanding issues with P-value calculations of multiple variables, which is unrelated to the normalization problem).
Given that this general model for probe summarization lacks terms for adjustment variables, and that statistical inference will be carried out on these summarized values, the goal for normalization remains the same: namely, to remove the influence of adjustment variables on probe intensities in order to maintain their true relationships with the biological variables of interest. This is the motivation for formulating the above goal in terms of inference on probe-level data, making this goal relevant regardless of microarray platform, technology or biological question.
3.3 Supervised normalization of microarrays
Our proposed method, called ‘supervised normalization of microarrays’ (SNM), fits the model from Section 3.1 to all probes and arrays simultaneously. The big picture idea of the method is to fit a study-specific model based on that in (1)–(3) to yield estimates 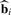 ,
, 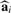 , and
, and 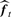 . (Note that there may be more terms in the model to account for other types of effects—see Supplementary Material.) The fitted model can then be used accordingly to perform subsequent analyses.
. (Note that there may be more terms in the model to account for other types of effects—see Supplementary Material.) The fitted model can then be used accordingly to perform subsequent analyses.
A key feature of SNM is that it involves simultaneously fitting the biological and study-specific adjustment variables. The fact that the intensity-dependent effects are functions of terms to be estimated by the model (i.e. BX + AZ) required us to develop a novel algorithm. The method we propose attempts to estimate a set of probes that are not associated to the biological variables (i.e. probes such that bi = 0). For ease of discussion, we call any probe i such that bi = 0 a ‘null probe’ and any probe i such that bi ≠ 0 an ‘alternative probe’. Once this set of null probes has been identified, we estimate the intensity-dependent effects using only these probes. Successfully doing so allows us to obtain estimates of the biological variables' coefficients satisfying the criteria from Section 3.2 . Thus, another key feature of SNM is that it specially handles the biological variables in order to obtain valid-fitted model coefficients of these variables.
The following are the main steps of the SNM algorithm; see Supplementary Material for more details, such as specifically how we fit smooth, random functions to model the intensity-dependent effects.

The reason we provide two different types of data adjustments in Step 6 is that exploratory analyses of the biological signal are best carried out on data where all signal not of interest has been modeled and removed. However, for statistical inference, it is necessary to account for the variation explained and degrees of freedom utilized by the adjustment variables, aiZ. It should be noted that the model fits we obtain at the final iteration of Steps 2–5 are exactly what would be obtained when fitting model (4) anyway.
4 RESULTS
Unsupervised normalization methods make data transformations based on graphical trends visible in diagnostic plots. QN (Bolstad et al., 2003) transforms the data so that a quantile–quantile plot comparing the ordered probe intensities between any two arrays in a study is equal to the line y = x (Fig. 1A); ISN (Li and Wong, 2001) transforms the data so that a plot comparing the intensities of a selected subset of probes between any two arrays is centered about the line y = x; and MA-normalization (Dudoit et al., 2002; Tseng et al., 2001; Yang et al., 2002) transforms the data so that the relationship between the difference between two groups and their average value is centered about the line y = 0. The implicit assumption behind each of these unsupervised methods is that transforming the data to satisfy these criteria removes all variation due to technical variables, but does not change the signal due to biological variables. Interestingly, an implicit assumption of QN is also that the probe intensity distributions among arrays is always exactly the same, regardless of biology or study design. This precludes different overall levels and variation of mRNA across biological conditions, as well as any overall asymmetric differential expression.
In this section, we demonstrate the operating characteristics of SNM through simulations and real data analysis, and make comparisons to the QN and ISN approaches. We also make comparisons to the eCADS method (Dabney and Storey, 2007), which is well suited for balanced two group comparisons on dual-channel arrays and has been shown to outperform the MA-normalization approach.
4.1 Simulations
While the set of biological and adjustment variables to be utilized may vary from study to study, we present simulations in the context of several variables most commonly used: array, batch and dye effects (Baird et al., 2004; Dabney and Storey, 2007; Wolfinger et al., 2001; Wu and Irrizary, 2007; Wu et al., 2004). For each type of variable we describe results from data simulations. In the first scenario, we demonstrate how to estimate array effects in the presence of biological signal. These simulations mimic the most commonly assumed models for single channel microarray data such as that provided by Affymetrix. Then, we extend this to a scenario where we also estimate batch effects, an adjustment variable that arises in many large microarray datasets, and, as shown below, can also be present in smaller studies. Finally, we apply SNM to a scenario based on two-color experiments where the signal is influenced by intensity-dependent dye and array effects.
A total of 100 simulated studies were carried out for each of the three scenarios, with the biological and technical effects simulated as follows. Data were simulated for a total of 100 000 probes and 12 arrays. The biological variable of interest is a dichotomous variable defining two groups (Groups 1 and 2) with six arrays sampled from each group. A randomly selected set of 30 000 probes were defined as differentially expressed between two groups, with the magnitude of the difference sampled from the Normal(1,1) distribution.
To make our simulations as realistic as possible we based our sampling of baseline probe intensities on data from a commonly used spike-in study (Irizarry et al., 2003). Specifically, the baseline probe intensities were sampled from a distribution consisting of the mean raw probe intensities for each of the 201 800 probes used in this study.
The random variation terms eij were sampled from a Normal distribution with mean 0 and a probe-specific variance term that was sampled from the Uniform(0.2,0.4) distribution. This implies that each probe does not have the same variance. The array and dye functions were defined by randomly selecting coefficients for a two-dimensional B-spline basis function from a N(0,0.752), the probe-specific batch effects were sampled from a N(0,0.42).
For the first two scenarios, we compare the SNM normalized data to QN and ISN normalized data. For the third simulation, we compare the SNM results to those obtained from eCADS. For all three simulations we use Kolmogorov–Smirnov tests (KS-test) to assess the validity of each normalization method in terms of the criteria described in Section 3.2 defining the desired normalization operating characteristics. To assess Criterion I, we use a two-sided KS-test to compare the distribution of observed P-values from the null probes to a Uniform(0,1) distribution. To assess Criterion II, we use a one-sided KS-test to compare the distributions of P-values from the alternative probes to a Uniform(0,1) distribution. Both tests are performed at the 0.05 level, so we expect 5 or less significant tests out of 100 when the criteria are satisfied. The results of these simulations are summarized in Table 1.
Table 1.
Results from data simulations
Average 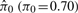 (π0 = 0.70) (π0 = 0.70) |
No. significant KS-tests |
|||||||
|---|---|---|---|---|---|---|---|---|
| Simulations | ISN | QN | SNM | eCADS | ISN | QN | SNM | eCADS |
| Array | 0.55 | 0.54 | 0.75 | NA | 100 | 100 | 1 | NA |
| Array + batch | 1.00 | 1.00 | 0.74 | NA | 100 | 100 | 4 | NA |
| Array + dye | NA | NA | 0.73 | 0.72 | NA | NA | 4 | 1 |
Shown are the average estimated π0 statistics and the KS-tests of Criterion I. The KS-tests were performed at the 0.05 level, so we expect 5 of 100 to be significant when Criterion I holds, and many >5 to be significant when the criterion does not hold. We do not include results of testing for Criterion II because all methods passed this criterion in these simulations.
4.1.1 Array effects
The data were simulated according to the above protocol where only the biological group differences and intensity-dependent array effects were included. In terms of the SNM model: Y is a 100 000 × 12 matrix of probe intensities; X is a 1 × 12 matrix indicating Group 2 membership; Z is a 1 × 12 matrix of 1's parameterizing the intercept term; and f(BX + AZ) is a 100 000 × 12 matrix that represents the intensity-dependent array effects. Note that under this construction, we achieve that bi = 0 is equivalent to no difference in mean probe intensities between Groups 1 and 2.
We normalized data from each of the 100 simulations using SNM, QN and ISN. Figure 2 presents representative histograms of P-values of null probes after normalization using SNM (Fig. 2A), QN (Fig. 2C) and ISN (Fig. 2E). In relation to Criterion I, we expect these P-values to be Uniform(0,1). While the SNM approach appears to meet this criterion, the null P-values from the unsupervised methods are clearly not Uniform(0,1). Of the 100 simulated studies, 1, 100 and 100 were significant for SNM, QN and ISN, respectively.
We examined the estimated π0 statistic for each simulation, which is the proportion of true null probes (Storey, 2002; Storey and Tibshirani, 2003). Recall that in this simulation the true π0 = 70%. The 95% confidence intervals of 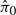 from the 100 simulations are 0.71 ± 0.04 for SNM, 0.54 ± 0.05 for QN and 0.55 ± 0.04 for ISN. The distributions of all P-values are presented in Figure 2B, D and F for SNM, QN and ISN, respectively. These results suggest that including the study-specific biological variables provides unbiased inference, while the inference derived from unsupervised methods is biased by the asymmetric biological signal.
from the 100 simulations are 0.71 ± 0.04 for SNM, 0.54 ± 0.05 for QN and 0.55 ± 0.04 for ISN. The distributions of all P-values are presented in Figure 2B, D and F for SNM, QN and ISN, respectively. These results suggest that including the study-specific biological variables provides unbiased inference, while the inference derived from unsupervised methods is biased by the asymmetric biological signal.
We also performed a KS-test of Criterion II, specifically whether the alternative P-values have a joint distribution stochastically smaller than the Uniform(0,1) distribution. Among the 100 simulated studies, 0 tests were significant for SNM, QN and ISN. These results suggest that SNM, QN and ISN satisify Criterion II.
Taken together, these results suggest that SNM satisifies Criteria I and II for a correctly normalized study in this scenario, while QN and ISN do not satisfy Criterion I. The bias of QN and ISN is anti-conservative, meaning that P-values have been spuriously made smaller than they should be.
4.1.2 Array effects plus batch effects
The simulated probe intensities for the second scenario included biological, array and batch effects as described above. Batch effects generally arise due to some need of the experimenter to manufacture or hybridize the arrays in groups; the real data example we consider below includes a batch effect. To apply SNM, we used the same model as in the previous simulation, except now Z is a 2 × 12 matrix parameterizing the probe-specific intercept terms in the first row and an indicator of batch in the second row.
SNM was applied to normalize data from each of the 100 simulated studies. In the KS-test of Criterion I, 4 out of 100 were significant. The 95% confidence intervals for 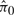 was 0.72 ± 0.04. In the KS-test of Criterion II, 0 out of 100 were significant. Thus, it appears SNM again provides a valid normalization in this simulation scenario.
was 0.72 ± 0.04. In the KS-test of Criterion II, 0 out of 100 were significant. Thus, it appears SNM again provides a valid normalization in this simulation scenario.
Given that there is an extra study-specific variable, we applied QN and ISN in two ways. First, we ignored the batch variable throughout the analysis. In other words, after normalization, batch was not utilized as an adjustment variable when testing the probes for differential expression between Groups 1 and 2. Of the 100 simulated studies, all 100 were significant for the KS-test of Criterion I for both QN and ISN. The 95% confidence intervals for the 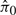 were 1 ± 0 for both QN and ISN.
were 1 ± 0 for both QN and ISN.
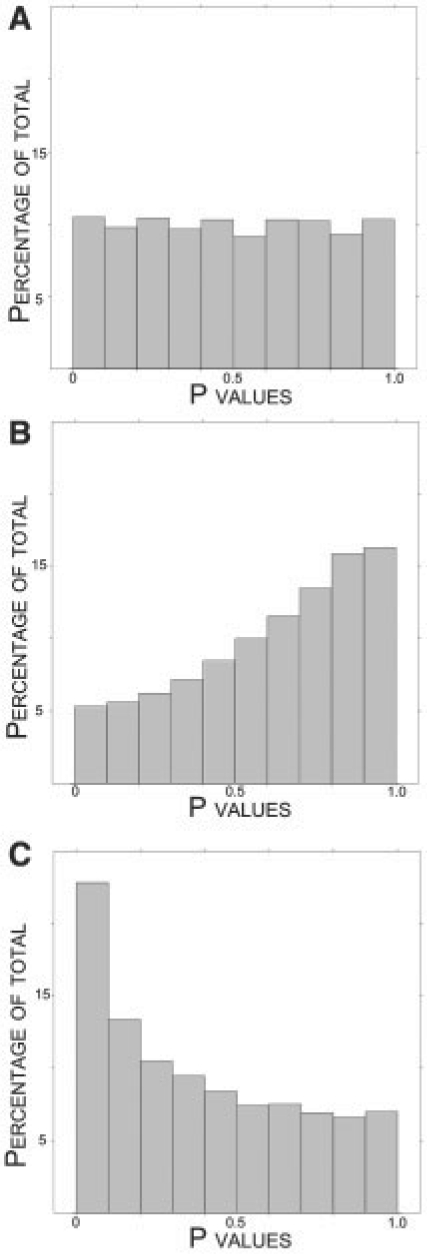
Figure 3B shows a plot of the null probe P-values when applying QN. (The analogous plot for ISN shows the same qualitative result and can be found in Supplementary Fig. 1E.) It can be seen that the null P-values have been biased in a conservative fashion, pushed towards one. This is due to the fact that ignoring the batch variable induced systematic variation in the probe intensities that was not taken into account.
Fig. 3.
Summary of null P-values from simulated data with differential expression, batch and array effects. The true proportion of null probes is π0 = 0.70. (A) P-value histogram of null probes after the SNM normalization. (B) P-value histogram of null probes after QN. (C) P-value histogram of null probes after QN using a model that includes a term for the batch effects.
We then applied QN and ISN in a second way, this time including batch as an adjustment variable in the test of differential expression after normalization. This would likely be the recommended way to include the batch variable when utilizing an unsupervised approach. Specifically, the unsupervised normalization is carried out ignoring the batch variable in addition to the biology variable, and then both are included in the analysis on the normalized data. Of the 100 simulated studies, all 100 were significant for the KS-test of Criterion I for both QN and ISN. The 95% confidence intervals for the 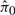 were 0.52 ± 0.03 for QN and 0.53 ± 0.04 for ISN. Among the 100 simulations, 0 were significant for the criterion II test for all three normalization methods.
were 0.52 ± 0.03 for QN and 0.53 ± 0.04 for ISN. Among the 100 simulations, 0 were significant for the criterion II test for all three normalization methods.
Figure 3C shows a plot of the null probe P-values when applying QN in this manner. (Again, the analogous plot for ISN shows the same qualitative result and can be found in Supplementary Fig. 1F.) Now, the null P-values are anti-conservatively biased; they are pushed toward zero as in the previous scenario. This follows because the systematic variation due to batch has now been modeled at the inference stage, and the anti-conservative bias due to array effects has returned. Additionally, since now both the biology and batch variables were not utilized in the unsupervised normalization, the null P-value bias and fluctuation from study to study have become more unpredictable.
4.1.3 Array effects plus dye effects
The probe intensities for the third simulation scenario included biological effects, array effects and intensity-dependent dye effects as described above. Dye effects arise in experiments that use two-color microarrays because of different incorporation rates of the Cy3 and Cy5 fluorescent dyes. To apply SNM, all terms in the model were defined exactly as in the first scenario, except we now defined f1(BX + AZ) to be a 100 000 × 12 matrix that represents the intensity-dependent array effects and f2(BX + AZ) to be a 100 000 × 12 matrix that represents the intensity-dependent dye effects. The study-specific model for this case is, therefore, Y = BX + AZ + f1(BX + AZ) + f2(BX + AZ) + E.
SNM and eCADS were used to normalize data from each of the 100 simulated studies. Of the 100 studies, 4 KS-tests were significant for Criterion I when applying SNM and 1 was significant when applying eCADS. The 95% confidence intervals for the 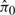 were 0.73 ± 0.03 for SNM and 0.72 ± 0.03 for eCADS. Among the 100 simulations for both SNM and QN, 0 were significant for the Criterion II test. These results show that SNM and eCADS behave similarly in this scenario, both providing acceptable normalizations. In fact, eCADS can be viewed as a special case of the more general SNM framework.
were 0.73 ± 0.03 for SNM and 0.72 ± 0.03 for eCADS. Among the 100 simulations for both SNM and QN, 0 were significant for the Criterion II test. These results show that SNM and eCADS behave similarly in this scenario, both providing acceptable normalizations. In fact, eCADS can be viewed as a special case of the more general SNM framework.
4.2 Vascular development study
A study was carried out designed to measure the association between gene expression levels and aging in the mouse aorta. Aorta were micro-dissected from mice aged 1, 7, 14 and 60 days, RNA was extracted, a single RNA pool was generated for each age, and each pool was hybridized to a single Affymetrix mouse430 version 2 microarray. This entire experiment was carried out twice, resulting in two technical batches.
We first verified that there are intensity-dependent array effects and effect due to the two batches. To this end, we regressed age on the unnormalized data by for each probe, ignoring batch and intensity-depdendent effects. On the residuals resulting from this regression, we investigated the relationship across arrays using hierarchical clustering (Supplementary Fig. 2A). From this clustering, it can be seen that there is a strong batch effect in this study. Finally, the relationship between overall intensity and the array-specific residuals were plotted for each array (Supplementary Fig. 2B). This result shows that there exist non-linear trends between intensity and the remaining error. Taken together, these results suggest that batch and intensity-dependent array effects contribute to the variation of probe level measurements.
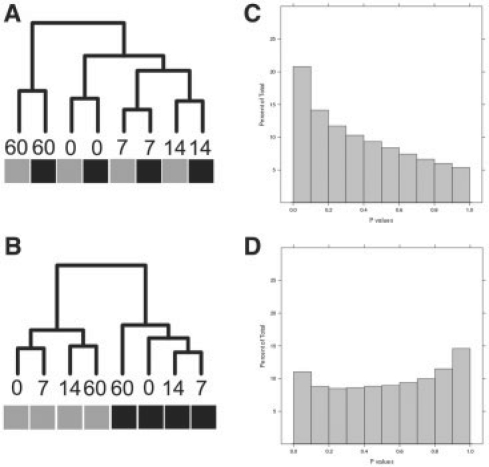
Next, we applied SNM to the study. In relation to model (3), Y is a 450 000 × 8 vector of observed intensities, X parameterizes the different ages and Z represents the parameterized probe-specific intercepts and batch effects. The results when applying SNM are shown in Figure 4. First, note that the histogram of P-values (Fig. 4C) suggests that age has a pronounced effect on differential expression (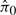 = 0.53). Many genes with known roles in vascular biology exhibited robust changes in expression across this time series, suggesting that the experiment-captured biological signal. For example, previous work identified a cluster of seven genes whose expression is activated soon after birth (List C Elastic Fiber Genes from McLean et al. 2005). Moreover, the relationship across samples, as described by a clustering dendrogram, correctly places the replicate arrays for each age on adjacent nodes (Fig. 4A).
= 0.53). Many genes with known roles in vascular biology exhibited robust changes in expression across this time series, suggesting that the experiment-captured biological signal. For example, previous work identified a cluster of seven genes whose expression is activated soon after birth (List C Elastic Fiber Genes from McLean et al. 2005). Moreover, the relationship across samples, as described by a clustering dendrogram, correctly places the replicate arrays for each age on adjacent nodes (Fig. 4A).
Fig. 4.
Results from Vascular Development Study obtained from QN and SNM. The relationship between samples after normalization are presented as a clustering dendogram. The labels for each node denote the corresponding age of the sample hybridized to that array, and the colored boxes indicate the batch. Note that the SNM results correctly position biological replicate samples on adjacent nodes (A), and predicts a robust effect of age on gene expression [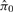 = 0.51 (C)]. Conversely, the first bifurcation in the QN data separates the data by the batch (B) and these data suggest there is no effect of age on gene expression [
= 0.51 (C)]. Conversely, the first bifurcation in the QN data separates the data by the batch (B) and these data suggest there is no effect of age on gene expression [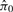 = 1 (D)].
= 1 (D)].
We also investigated the results when these data are normalized with the two unsupervised normalization procedures considered above, ISN and QN. The histogram of resulting P-values from QN is shown in Figure 4D. Results for the ISN method are nearly identical (as in the simulations) and are not shown here. Note that both approaches estimate 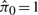 , suggesting that there are no genes differentially expressed between the four ages explored in this study. This result directly contradicts previously published data from a similar study (McLean et al., 2005) . Moreover, the clustering of the normalized data, shown in Figure 4B, suggests that batch exhibits a stronger influence on the variation of the QN normalized data than does the biology. This analysis provides a real data example that supports the results from the data simulations explored earlier, and the results suggest that the SNM approach outperforms the unsupervised approaches in this example.
, suggesting that there are no genes differentially expressed between the four ages explored in this study. This result directly contradicts previously published data from a similar study (McLean et al., 2005) . Moreover, the clustering of the normalized data, shown in Figure 4B, suggests that batch exhibits a stronger influence on the variation of the QN normalized data than does the biology. This analysis provides a real data example that supports the results from the data simulations explored earlier, and the results suggest that the SNM approach outperforms the unsupervised approaches in this example.
5 DISCUSSION
In this article, we described a framework for the normalization of microarray data. The central premise of this work is that the study under investigation should inform the normalization process. The framework is intended to be general enough to address most microarray studies, regardless of platform or biological goal. The examples we explored were selected as they are commonly occurring in practice and clearly describe the effects on inference of ignoring relevant information from an analysis. We note that the study-specific models utilized in this paper will not satisfy all studies. Rather, we view this work as a general supervised normalization framework based on a well-defined goal, namely to remove the effects of adjustment variables without biasing inference, and the examples as a means to illustrate the benefit of this approach. A researcher would utilize this framework to build a study-specific model and then apply the proposed algorithm, which is quite general.
The results presented in this paper demonstrate some of the problems associated with unsupervised normalization methods. For example, the simulations show how such approaches can introduce signal in the presence of asymmetric biological variation. Similarly, the simulations and vascular development example make clear that unsupervised approaches ignore effects of study design on expression data. Finally, these approaches make the assumption that few genes are differentially expressed, an assumption that is clearly false in many settings. These and other limitations of unsupervised methods are the motivation for this work.
The framework is designed to handle a variety of study designs. We presented a modeling strategy that is designed to model relevant study-specific variables (e.g. probe-specific biological effects, probe-specific technical effects and intensity-dependent effects). The flexibility of our modeling strategy is demonstrated by the fact that it straightforwardly derives previously described supervised normalization models for more specialized experimental designs and technologies (Baird et al., 2004; Dabney and Storey, 2007; Wolfinger et al., 2001). Another benefit is that other terms described in the literature can be included. We describe such a process for probe sequence effects in the Supplementary Material. These and other benefits support the claim that we have presented a flexible set of models for the normalization and analysis of microarray data.
Model building is paramount to successful implementation of any supervised normalization, including ours. Properly defining the model for a given study requires that relevant biological and study-specific adjustment variables of interest are known to the analyst, a situation that is rarely present in the clinical setting. The vascular development example shows one of many approaches to diagnose a model from a given dataset. In this work, we focused less on the difficulties associated with this approach; however, future work will address diagnosing models from the existing data.
Given the potential difficulties associated with knowing the true model, we base our framework on a goal that can be used to understand the validity of a study-specific model. The idea to remove the effects of technical variables without biasing inference has its roots in ideas originally presented in Dabney and Storey (2007); Leek and Storey (2007, 2008). To our knowledge, we are the first to explicitly state a goal for microarray data normalization. Three positive contributions of this goal are: (i) it unifies results from any particular instance of a study-specific model; (ii) basing a normalization algorithm on this goal clarifies its purpose and expected operating characteristics; and (iii) attaining this goal suggests that the experimenter understands all systematic sources of variation present in the data. This last point is important as it allows a user to understand when their assumed model accounts for variation from all relevant study-specific variables.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Robert Mecham and Dr Sean McLean, Washington University School of Medicine, for providing the mouse vascular array data.
Funding: National Institutes of Health grants T32 HG00035 and T32 CA80416 (to B.H.M.); PNW Prostate SPORE CA097186 and R01 DK069690 (to P.S.N.); R01 HG002913 (J.D.S.).
Conflict of Interest: none declared.
REFERENCES
- Baird D, et al. Normalization of microarray data using a spatial mixed model analysis which includes splines. Bioinformatics. 2004;20:3196. doi: 10.1093/bioinformatics/bth384. [DOI] [PubMed] [Google Scholar]
- Bolstad B, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Dabney A, Storey J. Normalization of two-channel microarrays accounting for experimental design and intensity-dependent relationships. Genome Biol. 2007;8:R44. doi: 10.1186/gb-2007-8-3-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, et al. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin. 2002;12:111–140. [Google Scholar]
- Irizarry R, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R, et al. Feature-level exploration of a published Affymetrix GeneChip control dataset. Genome Biol. 2006;7:404. doi: 10.1186/gb-2006-7-8-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J, Storey J. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J, Storey J. A general framework for multiple testing dependence. Proc. Natl Acad. Sci. 2008;105:18718. doi: 10.1073/pnas.0808709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:0032–1. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean S, et al. Extracellular matrix gene expression in the developing mouse aorta. In: Miner Jeffrey H., editor. Extracellular Matrix in Development and Disease. Vol. 15. San Diego, CA: Elsevier; 2005. of Advances in Developmental Biology. [Google Scholar]
- Rattray M, et al. Propagating uncertainty in microarray data analysis. Brief. Bioinformatics. 2006;7:37–47. doi: 10.1093/bib/bbk003. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. B. 2002;64:479–498. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc. Natl Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, et al. Significance analysis of time course microarray experiments. Proc. Natl Acad. Sci. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng G, et al. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 2001;29:2549. doi: 10.1093/nar/29.12.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger R, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irrizary R. A statistical framework for the analysis of microarray probe-level data. Ann. Appl. Stat. 2007;1:333. [Google Scholar]
- Wu Z, et al. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- Yang Y, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.