Abstract
While a variety of methods exist to reconstruct ancestral sequences, all of them assume that a single phylogeny underlies all the positions in the alignment and therefore that recombination has not taken place. Using computer simulations we show that recombination can severely bias ancestral sequence reconstruction (ASR), and quantify this effect. If recombination is ignored, the ancestral sequences recovered can be quite distinct from the grand most recent common ancestor (GMRCA) of the sample and better resemble the concatenate of partial most recent common ancestors (MRCAs) at each recombination fragment. When independent phylogenetic trees are assumed for the different recombinant segments, the estimation of the fragment MRCAs improves significantly. Importantly, we show that recombination can change the biological predictions derived from ASRs carried out with real data. Given that recombination is widespread on nuclear genes and in particular in RNA viruses and some bacteria, the reconstruction of ancestral sequences in these cases should consider the potential impact of recombination and ideally be carried out using approaches that accommodate recombination.
ANCESTRAL sequence reconstruction (ASR) is one of the most popular uses of phylogenies, allowing us to test hypotheses about the evolution of ancestral genes and genomes (Liberles 2007). Moreover, inferred ancestral sequences can be synthesized in the laboratory, so their function can be studied in vitro Chang et al. 2002). There are many applications of ASR, including the reconstruction of ancestral biochemical pathways (Gabaldon et al. 2006) and paleoenvironments (Boussau et al. 2008; Gaucher et al. 2008), vaccine design (Gaschen et al. 2002), or the resurrection of ancestral viruses (Dewannieux et al. 2006).
A number of methods to reconstruct ancestral DNA and protein sequences have been developed during the last decades, in parallel with the development of methods for inferring phylogenies like maximum parsimony (MP), maximum likelihood (ML), and Bayesian approaches (Cunningham et al. 1998; Ronquist 2004; Liberles 2007). Several studies have shown that ASR works reasonably well (e.g., Koshi and Goldstein 1996; Zhang and Nei 1997; Cai et al. 2004; Hall 2006; Williams et al. 2006). Importantly, a common assumption of ASR methods is that all the positions in the alignment have evolved under the same phylogeny, and therefore that there is a unique, single most recent common ancestor (MRCA) for all the sequences in the sample. However, if recombination has occurred along the history of the sample, different parts of the alignment can have distinct evolutionary relationships (see Posada et al. 2002). In this case, each recombinant fragment will correspond to a particular genealogy or tree and will have its own MRCA (Figure 1). Indeed, all recombinant fragments finally coalesce into a single ancestor, which in the coalescent jargon is often referred to as the grand most recent common ancestor (GMRCA) (Griffiths and Marjoram 1997) (Figure 1). In the absence of recombination, the GMRCA and the fragment MRCA sequences are necessarily identical, as they refer to the same node. However, this is not necessarily true if there is recombination, because in this case different regions of the alignment will have their own MRCA at different times. Importantly, the reconstruction of the GMRCA can be very difficult, as changes in the GMRCA will be fixed at the fragment MRCAs (Figure 2). Moreover, it is known that ignoring recombination can bias phylogenetic estimation (Posada and Crandall 2002; Beiko et al. 2008) and derived inferences (Schierup and Hein 2000a,b). Given all these complexities, we expect recombination to bias ASR. Therefore, the consequences can be important, as recombination is widespread in nuclear, viral, and bacterial genomes (Posada et al. 2002; Awadalla 2003; Fraser et al. 2007; Gaut et al. 2007; Duret and Arndt 2008). Here we used computer simulations to assess and quantify the effect of recombination on ASR.
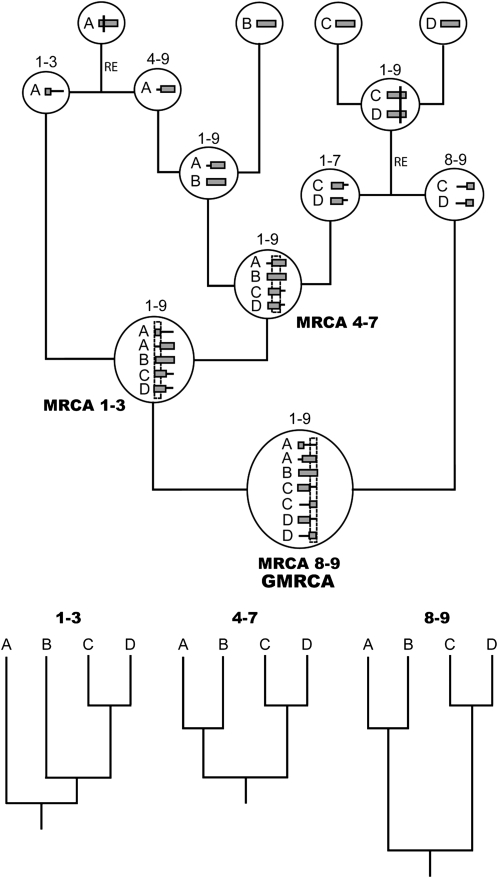
Figure 1.—
Example of an ancestral recombination graph. Inside each node (circles) there are recombinant segments with ancestral (shaded horizontal blocks) and nonancestral material (horizontal lines). RE indicates recombination events. Vertical lines across the segments indicate recombination breakpoints. Numbers above nodes indicate the nucleotide interval of ancestral material included. Note that each independent recombinant fragment (1–3, 4–7, and 8–9) has its own most recent common ancestor (MRCA), all of which finally coalesce into a grand most recent common ancestor (GMRCA). At the bottom, we can see the individual trees corresponding to each recombinant fragment.
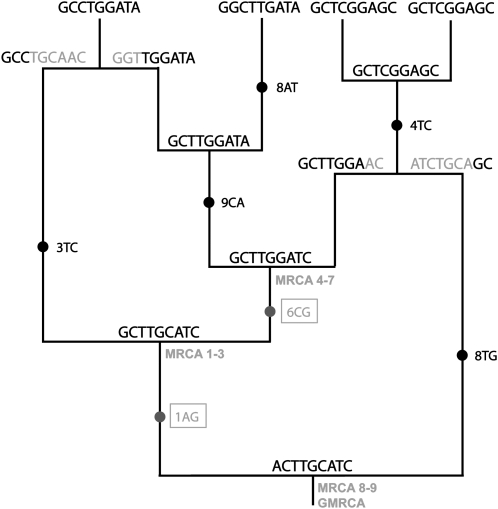
Figure 2.—
Example of the evolution of nucleotides on the ancestral recombination graph in Figure 1. Substitutions along branches are marked with solid circles, followed by the position and the states involved. The two changes that occur between the GMRCA and the MRCAs (enclosed in a box and shaded) are fixed in the sampled sequences, so the corresponding ancestral states in the GMRCA (1A and 6C) cannot be recovered from this sample.
MATERIALS AND METHODS
Simulation of recombinant sequences:
We simulated alignments of coding and noncoding nucleotide sequences under different scenarios, allowing for both intercodon and intracodon recombination, and where both GMRCA sequence and the MRCA fragments were known (Arenas and Posada 2010). In all cases, we used the same number of sequences (n = 11; 10 ingroup sequences + 1 outgroup sequence), alignment length (l = 999 nucleotides/333 codons) and effective population size (N = 1000). We explored three different values of the population mutation parameter (θ = 4Nμl = 50, 100, and 200) and six population recombination rates (ρ = 4Nrl = 0, 1, 4, 16, 64, and 128), where μ and r are the substitution and recombination rates per site per generation, respectively. Note that these rates encompass many different organisms, as they range from zero to extreme values like those observed in rapidly evolving pathogens (McVean et al. 2002; Stumpf and McVean 2003; Carvajal-Rodriguez et al. 2006). Noncoding nucleotide sequences were evolved under the JC69 substitution model (Jukes and Cantor 1969) while protein-coding sequences were evolved under the GY94 model (Goldman and Yang 1994), with a transition/transversion ratio of 0.5 and equal nonsynonymous (dN) and synonymous (dS) rates per nonsynonymous and synonymous site, respectively (ω = dN/dS = 1.0). We used simple substitution models to focus on the role of recombination. For every combination of parameters, we simulated 200 alignments. The latter resulted in ∼6, 11, and 19% average pairwise nucleotide distances, respectively for θ = 50, 100, and 200. Amino acid alignments were obtained by translation of the simulated coding sequences, assuming the universal genetic code; allowing us to include intracodon recombination at the protein level.
Reconstruction of ancestral sequences:
For every simulated alignment, we built neighbor-joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) phylogenetic trees using PAUP* (Swofford 2000) and HYPHY (Kosakovsky Pond et al. 2005) and rooted them using the outgroup. We carried out ASR using different methods and implementations: joint and marginal ML ASR in PAUP*, joint ML ASR in HYPHY, and marginal empirical Bayes ASR (Huelsenbeck and Bollback 2001) in PAML (Yang 2007). For nucleotides and codons the model of substitution assumed in all the analyses was the same model used to generate the data, thereby avoiding the effect of model misspecification. In the case of amino acids, the ASR was carried out assuming the WAG model (Whelan and Goldman 2001).
ASR for recombinant fragments:
We also devised a procedure that considers recombination during ASR. First, recombination breakpoints were located with GARD (Kosakovsky Pond et al. 2006). The resulting alignment fragments and corresponding NJ trees were redirected to HYPHY/PAUP* for joint ML ASR for each fragment. In the case of PAUP*, recombinant fragments lacking one of the four bases were pooled with the largest neighboring fragment. Also, breakpoints detected inside codons were moved to the nearest intercodon position. In addition, we repeated the reconstructions using the simulated (true) trees for each fragment.
Error measure:
Error was measured as the percentage of differences (at the nucleotide, codon, or amino acid level) between the inferred and simulated ancestral sequence at the ingroup MRCA. In the absence of recombination, this comparison is straightforward because there is only one MRCA for the whole alignment (and it is the same as the GMRCA). However, when there is recombination, there are several MRCAs for the different recombination fragments and one GMRCA. In this case, we computed two different errors, the distance from the estimated ancestral sequence to the simulated GMRCA and the average distance from the inferred ancestral sequence to the fragment MRCAs.
Phylogenetic error:
We also calculated the phylogenetic error between the inferred tree/s (one if recombination is ignored; several if recombination is accounted for) and the true tree/s (one if recombination has not occurred; several if there is recombination) for each segment. This error was estimated using two different metrics, the Robinson–Foulds (RF) distance (Robinson and Foulds 1981), which only considers differences in topology, and the branch score (BS) distance (Kuhner and Felsenstein 1994), which also considers differences in branch lengths.
Analysis of real data:
We analyzed two different alignments of the env region of HIV-1. The first data set was downloaded from the HIV Sequence Database (http://www.hiv.lanl.gov) and included the HIV-1 group M reference alignment plus an outgroup (40 sequences, 2514 bp). The second data set included only subtype B viruses and an outgroup (38 sequences, 2557 bp) (Doria-Rose et al. 2005). Sequence U19647 was too short, and therefore we removed it from the latter data set. In both cases, we realigned the sequences using MAFFT (Katoh and Toh 2008) and removed ambiguous positions with Gblocks (Talavera and Castresana 2007). We selected best-fit models with jModelTest (Posada 2008) and inferred ML trees using Phyml (Guindon and Gascuel 2003). We inferred ancestral sequences ignoring/considering recombination using the methodology described above and estimated population recombination rates with omegaMap (Wilson and McVean 2006). Then, we scanned the resulting sequences for known HIV-1 and CTL epitopes using ELF (http://www.hiv.lanl.gov/content/sequence/ELF/epitope_analyzer.html) and MHCPred (Guan et al. 2003), respectively, and for N-linked glycosylation sites using NetNGlyc (Gupta et al. 2004).
RESULTS
Impact of recombination on ASR:
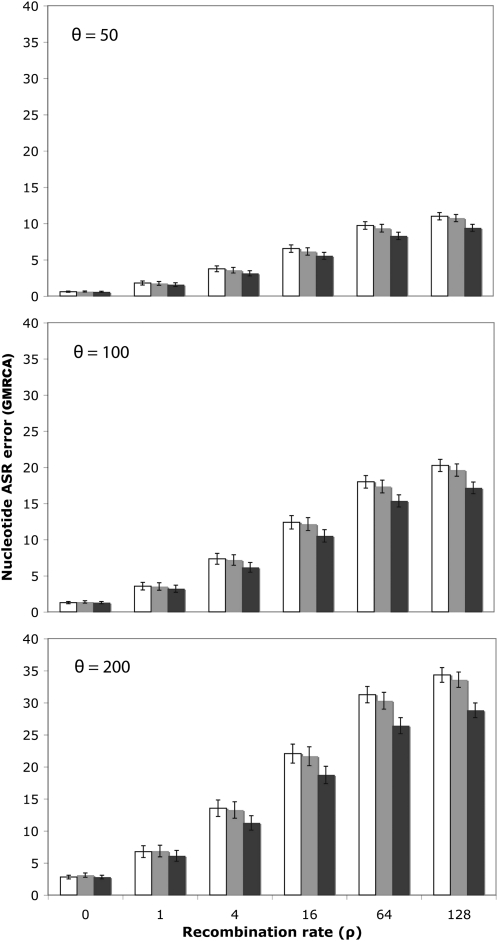
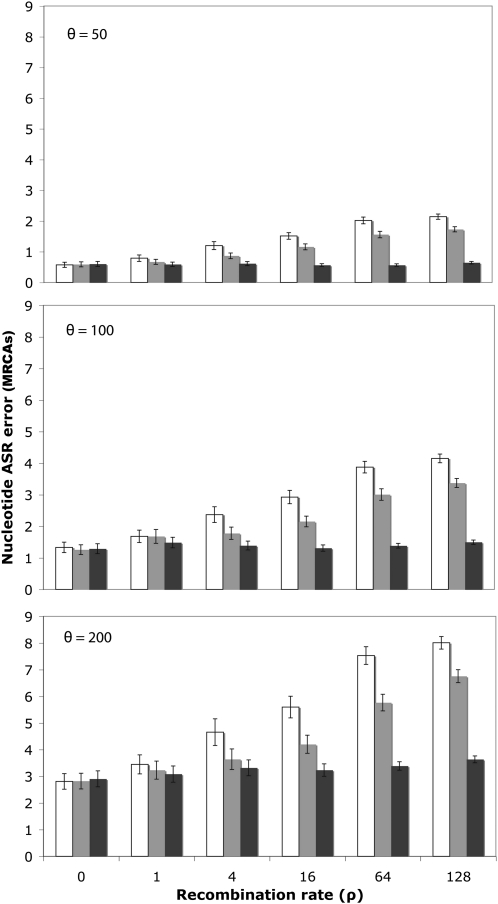
Recombination biased the reconstruction of the GRMCA sequence. For nucleotide sequences, the error reached up to 11, 20, and 36% for θ = 50, 100, and 200, respectively (Figure 3; open bars). For codons, the error was higher, up to 30, 50, and 72%, respectively (supporting information, Figure S1). For proteins, the error was also noticeable, up to 24, 41, and 62%, respectively (Figure S2; open bars). Note that if we just referred to the reconstruction at variable sites, which is where the ASR can make a difference, the error would increase notably, up to 39, 48, and 56% (nucleotides), up to 51, 63, and 76% (codons), and up to 51, 60, and 71% (proteins) for θ = 50, 100, and 200, respectively. In all cases, the error logically increased with the recombination rate and was larger for divergent sequences. Remarkably, this error was independent of the exact ASR algorithm (joint, marginal), phylogenetic framework (MP, NJ, ML) or software (PAUP*, PAML, HYPHY) (Figure S3). Recombination also confounded, although to a much less extent, the estimation of the MRCA for each individual recombinant fragment (Figure 4; Figure S4, Figure S5, and Figure S6).
Figure 3.—
Nucleotide ASR error as a function of the recombination rate. The percentage of nucleotide differences is shown between the inferred and the simulated GMRCA sequences, ignoring recombination (open), using the fragments and trees inferred by GARD (shaded), or using the simulated (true) fragments and trees (solid) for different levels of diversity (θ) and recombination (ρ). Error bars indicate 95% confidence intervals. In the example shown, ancestral nucleotide sequences were inferred using joint ML in HYPHY.
Figure 4.—
Error in the reconstruction of the fragment MRCAs as a function of the recombination rate. The percentage of nucleotide differences is shown between the inferred and the simulated fragment MRCAs sequences, ignoring recombination (open), using the fragments and trees inferred by GARD (shaded), or using the simulated (true) fragments and trees (solid) for different levels of divergence (θ) and recombination (ρ). Error bars indicate 95% confidence intervals. In this case ancestral nucleotide sequences were inferred using joint ML in HYPHY.
Indeed, in the simulations, the average distance between the GMRCA sequence and the fragment MRCAs increased with the recombination and substitution rates (Table 1). Looking at the error figures for the GMRCA and fragment MRCAs (Figure 3; Figure S1 and Figure S2 vs. Figure 4; Figure S5 and Figure S6), it is clear that the ancestral sequences estimated were always much more similar to the fragment MRCAs than to the GMRCA, especially at high recombination and substitution rates. The ASR error relative to the GMRCA was several times larger regardless of the method or implementation used (data not shown).
TABLE 1.
Average nucleotide distance between the GMRCA sequence and the fragment MRCAs in the simulations, for different levels of diversity (θ) and recombination (ρ)
| θ | ρ | Distance (%) between GMRCA and fragment MRCAs |
|---|---|---|
| 50 | 0 | 0.00 ± 0.00 |
| 1 | 1.10 ± 0.25 | |
| 4 | 2.64 ± 0.35 | |
| 16 | 5.12 ± 0.47 | |
| 64 | 7.98 ± 0.52 | |
| 128 | 9.10 ± 0.49 | |
| 100 | 0 | 0.00 ± 0.00 |
| 1 | 2.14 ± 0.48 | |
| 4 | 5.15 ± 0.66 | |
| 16 | 9.74 ± 0.86 | |
| 64 | 14.91 ± 0.88 | |
| 128 | 16.92 ± 0.84 | |
| 200 | 0 | 0.00 ± 0.00 |
| 1 | 3.96 ± 0.86 | |
| 4 | 9.52 ± 1.15 | |
| 16 | 17.43 ± 1.39 | |
| 64 | 26.14 ± 1.35 | |
| 128 | 29.21 ± 1.26 |
Numbers after ± indicate 95% confidence intervals.
ASR considering recombination:
When ASR was carried out taking into account the recombination fragments delimited by GARD, the error in the reconstruction of the GMRCA always decreased, although not in a significant fashion (but note that in five out of five cases it is smaller, which itself is a nonsignificant result) (Figure 3; shaded bars). When the true trees and fragments were used in the analysis, the error decreased further, sometimes significantly (Figure 3; solid bars). The same trend was observed for amino acid sequences (Figure S2; shaded and solid bars).
In the case of the fragment MRCAs, the use of the segments identified by GARD reduced the ASR error significantly, ∼25%, for recombination rates higher than 1, independently of the substitution rate (Figure 4; shaded bars). When the true trees and fragments were used in the analysis, the error clearly decreased, although it was quite independent of the recombination rate (Figure 4; solid bars). The same trend was observed for amino acid sequences (Figure S6).
Relationship between phylogenetic error and ASR accuracy:
When recombination was ignored, the RF distance was significantly correlated across replicates with the ASR error [Pearson's correlation coefficient (corr.) = 0.75, P-value < 2.2e-16] and it was significantly larger for those sites that were assigned a wrong ancestral state (two-way ANOVA, P-value = 0.02). The BS distance was also correlated with the ASR error (corr. = 0.97, P-value < 2.2e-16), but it was independent of whether the assigned ancestral state was correct or wrong (P-value = 0.98). The same results were obtained when recombination was considered using GARD, although in this case the RF distances (but not the BS distances) were significantly smaller (two-way ANOVA, P-value < 2.2e-16).
Analysis of real data:
For the HIV-1 group M data set, the estimate of ρ was 8.5 and the number of recombination breakpoints detected by GARD was 4. For the HIV-1 subtype B data set the estimated ρ was 4.5 and the number of recombination breakpoints detected was 7. For HIV-1 M, the ancestral sequences inferred ignoring/considering recombination differed by 4.69% (118 nt) and 9.24% (70 aa), for nucleotide and amino acid ASR, respectively, while for HIV-1 B this difference was 3.56% (91 nt) and 4.82% (39 aa). When recombination was ignored, the number of epitopes identified in the inferred ancestral M sequence was 354, although when recombination was taken into account this number was 447. In the ancestral B sequence, 494 epitopes were detected in both cases, although these epitopes were not exactly the same. The number of CTL epitopes inferred was different depending on whether recombination was considered or not (Table S1), and the number of N-glycosylation sites inferred for the ancestral M sequence inferred ignoring/considering recombination was 14/17. For the HIV-1 B data, the number of detected N-glycosylation sites was 22 regardless of recombination, but the inference corresponded to different positions.
DISCUSSION
Our simulations clearly show that recombination biases the reconstruction of ancestral nucleotide, codon, and amino acid sequences, regardless of the method and/or software used. The effect of recombination on ASR was stronger at higher recombination and substitution rates, but it was also considerable at low recombination rates. This trend was expected because tree height grows with the recombination rate due to an increment of incompatibilities in the data (Eyre-Walker et al. 1999; Schierup and Hein 2000a). The ASR error was largest at the codon level, suggesting that ASR with recombination is better accomplished at the nucleotide or amino acid level. The ASR error decreased when the true trees and fragments were used in the analysis, suggesting that it is due to the fact that in the presence of recombination the history of the whole alignment cannot be explained by a single phylogeny anymore, but by a set of distinct phylogenies for the different recombinant fragments. In this case, if we ignore recombination, the inferred tree can be an incorrect representation of the true underlying phylogenies (Posada and Crandall 2002), and the ASR would fail when trying to infer ancestral states at (wrong) nodes. In fact, in the presence of recombination the positions that were assigned a wrong ancestral state supported worse topologies but similar branch lengths than sites that were correctly reconstructed.
While the error in the reconstruction of the fragment MRCAs was independent of the recombination rate, the estimated GMRCA sequence became less accurate with increasing recombination rates. Indeed, we expect the GMRCA and the fragment MRCAs to be more different with increasing recombination rates, because in this case the height of the simulated tree and the number of recombinant fragments will be larger, and therefore the sum of branch lengths between the GRMCA and the MRCAs will be bigger. In most situations, the GMRCA will be much more difficult to estimate than the fragment MRCAs, because all the substitutions that occur between the GMRCA and the fragment MRCAs will be fixed, and the ancestral states at the sites involved will never show up in the sampled sequences. Therefore, the ancestral sequence reconstructed in the presence of recombination will always be closer to the fragment MRCAs than to the GMRCA sequence.
Detecting breakpoints and estimating independent trees for each recombinant fragment is of little help in obtaining a more accurate GMRCA sequence, but it can be very useful if we are interested in the fragment MRCAs. Although the fragment MRCAs will correspond to different nodes of the genealogy, their sequences could be used for a better depiction of the history of changes in the sample or to reconstruct specific protein domains. For example, in the case of HIV-1, the latter applies to the design of polyvalent vaccines, in which the interest is on particular epitopes spread across the entire sequence (Gao et al. 2003; Nickle et al. 2003; Doria-Rose et al. 2005). Still, a more robust method for the reconstruction of the ancestral (GMRCA) sequence in presence of recombination is clearly needed. A potential avenue could be the use of explicit phylogenetic networks (Huson and Bryant 2006), where one could do the ASR integrating over the different trees embedded in the network, as in Bayesian phylogenetics (Huelsenbeck and Bollback 2001). However, this might be quite challenging given the impact of recombination on the accuracy of phylogenetic networks (Woolley et al. 2008) and potential problems identifying the GMRCA (Castelloe and Templeton 1994). An alternative could be the reconstruction of rooted ancestral recombination graphs (Song and Hein 2005; Minichiello and Durbin 2006; Parida et al. 2008).
Nearly every published study incorporating ancestral sequence reconstruction assumes no recombination, despite the fact that recombination is widespread on the nuclear genome of many eukaryotes, and in particular in RNA viruses and some bacteria. In these cases, although the target is most often the GMRCA, what is being estimated in practice will be much closer to the fragment MRCAs. To what point the error induced by recombination has relevant implications should be discussed on a case-by-case basis and within the context of the intended use of the reconstructed sequences. Indeed, it is possible that just a few nucleotide changes in the inferred ancestral sequence can lead to different functional or evolutionary inferences, while in other cases a larger number of changes may have little impact on conclusions (Krishnan et al. 2004). In our example, recombination resulted in different predictions regarding the structure/activity of the inferred ancestral domains and epitopes. In any case, the impact of recombination on ASR should be kept in mind when making ancestral inferences from genes and proteins with a potential history of recombination.
Acknowledgments
We are grateful to Keith A. Crandall for his comments. We also thank the editor and the anonymous reviewers for their excellent suggestions. This work was supported by the Spanish Ministry of Science and Education (grant no. BIO2007-61411 to D.P. and a Formación de Personal Investigador fellowship BES-2005-9151 to M.A.).
Supporting Information is available online at http://www.genetics.org/cgi/content/full/genetics.109.113423/DC1.
References
- Arenas, M., and D. Posada, 2010. Coalescent simulation of intracodon recombination. Genetics 184 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla, P., 2003. The evolutionary genomics of pathogen recombination. Nat. Rev. Genet. 4 50–60. [DOI] [PubMed] [Google Scholar]
- Beiko, R. G., W. F. Doolittle and R. L. Charlebois, 2008. The impact of reticulate evolution on genome phylogeny. Syst. Biol. 57 844–856. [DOI] [PubMed] [Google Scholar]
- Boussau, B., S. Blanquart, A. Necsulea, N. Lartillot and M. Gouy, 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456 942–945. [DOI] [PubMed] [Google Scholar]
- Cai, W., J. Pei and N. V. Grishin, 2004. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 4 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Rodriguez, A., K. A. Crandall and D. Posada, 2006. Recombination estimation under complex evolutionary models with the coalescent composite-likelihood method. Mol. Biol. Evol. 23 817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelloe, J., and A. R. Templeton, 1994. Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol. Phylogenet. Evol. 3 102–113. [DOI] [PubMed] [Google Scholar]
- Chang, B. S., K. Jonsson, M. A. Kazmi, M. J. Donoghue and T. P. Sakmar, 2002. Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 19 1483–1489. [DOI] [PubMed] [Google Scholar]
- Cunningham, C. W., K. E. Omland and T. H. Oakley, 1998. Reconstructing ancestral character states: a critical reappraisal. Trends Ecol. Evol. 13 361–366. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet et al., 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16 1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose, N. A., G. H. Learn, A. G. Rodrigo, D. C. Nickle, F. Li et al., 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 79 11214–11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., and P. F. Arndt, 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4 e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker, A., N. H. Smith and J. Maynard Smith, 1999. How clonal are human mitochondria? Proc. R. Soc. Lond. B 266 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, C., W. P. Hanage and B. G. Spratt, 2007. Recombination and the nature of bacterial speciation. Science 315 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon, T., B. Snel, F. van Zimmeren, W. Hemrika, H. Tabak et al., 2006. Origin and evolution of the peroxisomal proteome. Biol. Direct. 1 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F., T. Bhattacharya, B. Gaschen, J. Taylor, J. P. Moore et al., 2003. Consensus and ancestral state HIV vaccines. Science 299 1515–1518. [DOI] [PubMed] [Google Scholar]
- Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao et al., 2002. Diversity considerations in HIV-1 vaccine selection. Science 296 2354–2360. [DOI] [PubMed] [Google Scholar]
- Gaucher, E. A., S. Govindarajan and O. K. Ganesh, 2008. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451 704–707. [DOI] [PubMed] [Google Scholar]
- Gaut, B. S., S. I. Wright, C. Rizzon, J. Dvorak and L. K. Anderson, 2007. Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8 77–84. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11 725–736. [DOI] [PubMed] [Google Scholar]
- Griffiths, R. C., and P. Marjoram, 1997. An ancestral recombination graph, pp. 257–270 in Progress in Population Genetics and Human Evolution, edited by P. Donelly and S. Tavaré. Springer-Verlag, Berlin.
- Guan, P., I. A. Doytchinova, C. Zygouri and D. R. Flower, 2003. MHCPred: a server for quantitative prediction of peptide-MHC binding. Nucleic Acids Res. 31 3621–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S., and O. Gascuel, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Gupta, R., E. Jung and S. Brubak, 2004. NetNGlyc: prediction of N-glycosylation sites in human proteins. http://www.cbs.dtu.dk/services/NetNGlyc/
- Hall, B. G., 2006. Simple and accurate estimation of ancestral protein sequences. Proc. Natl. Acad. Sci. USA 103 5431–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and J. P. Bollback, 2001. Empirical and hierarchical Bayesian estimation of ancestral states. Syst. Biol. 50 351–366. [PubMed] [Google Scholar]
- Huson, D. H., and D. Bryant, 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23 254–267. [DOI] [PubMed] [Google Scholar]
- Jukes, T. H., and C. R. Cantor, 1969. Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism, edited by H. M. Munro. Academic Press, New York.
- Katoh, K., and H. Toh, 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics 9 286–298. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L., S. D. Frost and S. V. Muse, 2005. HYPHY: hypothesis testing using phylogenies. Bioinformatics 21 676–679. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L., D. Posada, M. B. Gravenor, C. H. Woelk and S. D. Frost, 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23 1891–1901. [DOI] [PubMed] [Google Scholar]
- Koshi, J. M., and R. A. Goldstein, 1996. Probabilistic reconstruction of ancestral protein sequences. J. Mol. Evol. 42 313–320. [DOI] [PubMed] [Google Scholar]
- Krishnan, N. M., H. Seligmann, C. B. Stewart, A. P. De Koning and D. D. Pollock, 2004. Ancestral sequence reconstruction in primate mitochondrial DNA: compositional bias and effect on functional inference. Mol. Biol. Evol. 21 1871–1883. [DOI] [PubMed] [Google Scholar]
- Kuhner, M. K., and J. Felsenstein, 1994. A simulation comparison of phylogeny algorithms under equal and unequal evolutionary rates. Mol. Biol. Evol. 11 459–468. [DOI] [PubMed] [Google Scholar]
- Liberles, D. A., 2007. Ancestral Sequence Reconstruction. Oxford University Press, Oxford.
- McVean, G., P. Awadalla and P. Fearnhead, 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minichiello, M. J., and R. Durbin, 2006. Mapping trait loci by use of inferred ancestral recombination graphs. Am. J. Hum. Genet. 79 910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle, D. C., M. A. Jensen, G. S. Gottlieb, D. Shriner, G. H. Learn et al., 2003. Consensus and ancestral state HIV vaccines. Science 299 1515–1518. [DOI] [PubMed] [Google Scholar]
- Parida, L., M. Mele, F. Calafell and J. Bertranpetit, 2008. Estimating the ancestral recombinations graph (ARG) as compatible networks of SNP patterns. J. Comput. Biol. 15 1133–1154. [DOI] [PubMed] [Google Scholar]
- Posada, D., 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25 1253–1256. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 2002. The effect of recombination on the accuracy of phylogeny estimation. J. Mol. Evol. 54 396–402. [DOI] [PubMed] [Google Scholar]
- Posada, D., K. A. Crandall and E. C. Holmes, 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36 75–97. [DOI] [PubMed] [Google Scholar]
- Robinson, D. F., and L. R. Foulds, 1981. Comparison of phylogenetic trees. Math. Biosci. 53 131–147. [Google Scholar]
- Ronquist, F., 2004. Bayesian inference of character evolution. Trends Ecol. Evol. 19 475–481. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., and J. Hein, 2000. a Consequences of recombination on traditional phylogenetic analysis. Genetics 156 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., and J. Hein, 2000. b Recombination and the molecular clock. Mol. Biol. Evol. 17 1578–1579. [DOI] [PubMed] [Google Scholar]
- Song, Y. S., and J. Hein, 2005. Constructing minimal ancestral recombination graphs. J. Comput. Biol. 12 147–169. [DOI] [PubMed] [Google Scholar]
- Stumpf, M. P., and G. A. McVean, 2003. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 4 959–968. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2000. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Talavera, G., and J. Castresana, 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56 564–577. [DOI] [PubMed] [Google Scholar]
- Whelan, S., and N. Goldman, 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18 691–699. [DOI] [PubMed] [Google Scholar]
- Williams, P. D., D. D. Pollock, B. P. Blackburne and R. A. Goldstein, 2006. Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput. Biol. 2 e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D. J., and G. McVean, 2006. Estimating diversifying selection and functional constraint in the presence of recombination. Genetics 172 1411–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley, S. M., D. Posada and K. A. Crandall, 2008. A comparison of phylogenetic network methods using computer simulation. PLoS ONE 3 e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang, J., and M. Nei, 1997. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J. Mol. Evol. 44 S139–S146. [DOI] [PubMed] [Google Scholar]