Abstract
Following earlier reports on modulation of poly(Q) toxicity in Drosophila by the developmentally active and stress-inducible noncoding hsrω gene, we investigated possible mediators of this modulation. RNAi-mediated downregulation of the large nuclear hsrω-n transcript, which organizes the nucleoplasmic omega speckles, suppressed the enhancement of poly(Q) toxicity brought about by reduced availability of the heterogeneous nuclear ribonucleoprotein (hnRNP) Hrb87F and of the transcriptional regulator, cAMP response element binding (CREB) binding protein (CBP). Levels of CBP RNA and protein were reciprocally affected by hsrω transcript levels in eye disc cells. Our data suggest that CBP and hnRNPs like Hrb57A and Hrb87F physically interact with each other. In addition, downregulation of hsrω transcripts partially rescued eye damage following compromised proteasome activity, while overexpression of hsrω and/or poly(Q) proteins disrupted the proteasomal activity. Rescue of poly(Q) toxicity by hsrω-RNAi required normal proteasomal function. We suggest that hsrω-RNAi suppresses poly(Q) toxicity by elevating cellular levels of CBP, by enhancing proteasome-mediated clearance of the pathogenic poly(Q) aggregates, and by inhibiting induced apoptosis. The direct and indirect interactions of the hsrω transcripts with a variety of regulatory proteins like hnRNPs, CBP, proteasome, Drosophila inhibitor of apoptosis protein 1 (DIAP1), etc., reinforce the view that the noncoding hsrω RNA functions as a “hub” in cellular networks to maintain homeostasis by coordinating the functional availability of crucial cellular regulatory proteins.
A number of human neurological disorders like Huntington's disease (HD), spinal and bulbar muscular atrophy (SBMA), dentatorubropallidoluysian atrophy (DRPLA), and several spinocerebellar ataxias (SCAs) are caused by expansion of polyglutamine stretches beyond the characteristic threshold range in different proteins (Gusella and MacDonald 2000; Everett and Wood 2004; Gatchel and Zoghbi 2005). The expanded polyglutamine domain confers novel toxicity to these proteins in neuronal cells such that these diseases are characterized by selective vulnerability of neurons despite a widespread expression of the diseased protein in brain and other tissues. A hallmark feature of most of these diseases is the formation of insoluble intracellular aggregates or inclusion bodies (IBs) in the affected human neurons as well as in cell culture and animal models due to abnormal folding of the mutant poly(Q) proteins (Davies et al. 1997; Klement et al. 1998; Chen et al. 2001, 2002). It is debated if the IBs are causal to or a consequence of disease pathogenesis or represent a cellular protective mechanism (DiFiglia et al. 1997; Saudou et al. 1998; Warrick et al. 1998; Yang et al. 2002; Yoo et al. 2003; Arrasate et al. 2004). Nevertheless, our earlier results (Mallik and Lakhotia 2009a) utilizing several fly models of poly(Q) disorders suggested a strong correlation between poly(Q) aggregation and neurodegeneration since an almost complete or significant inhibition of IB formation was seen following downregulation of the hsrω-n transcripts, which in turn was associated with reduction in Hsp70 levels and suppression of poly(Q)-induced eye degeneration.
Proteins with expanded poly(Q) tracts interact with and sequester many transcriptional regulators containing short poly(Q) repeats or proline-rich regions like cAMP response element binding (CREB) binding protein (CBP), TBP-associated factor 4 (TAFII130), Sp1 transcription factor (SP1), p53, etc., in the IBs and thus compromise their normal regulatory functions (Nucifora et al. 2001; Dunah et al. 2002; Li et al. 2002; Schaffar et al. 2004; Helmlinger et al. 2006). IBs also sequester molecular chaperones and components of the ubiquitin proteasome pathway (UPP), suggesting that misfolding, impaired degradation, and abnormal aggregation of proteins also contribute to the pathogenesis (Warrick et al. 1999; Chan and Bonini 2000; Cummings et al. 2001).
The hsrω gene produces independently regulated multiple noncoding transcripts (Lakhotia 2003). Their overexpression aggravates (Sengupta and Lakhotia 2006) and RNAi-based downregulation ameliorates (Mallik and Lakhotia 2009a) the poly(Q) toxicity. The large nucleus limited hsrω-n transcript organizes the nucleoplasmic omega speckles and is known to associate with a variety of proteins (Lakhotia et al. 1999; Prasanth et al. 2000; Jolly and Lakhotia 2006). We recently showed (Mallik and Lakhotia 2009a) that GMR-GAL4-driven coexpression of the hsrω-RNAi transgene and the pathogenic poly(Q) proteins does not affect the poly(Q) transgene transcription but prevents accumulation of the poly(Q) aggregates and consequently the associated pathogenesis (Mallik and Lakhotia 2009a). Thus the hsrω-RNAi-dependent recovery from poly(Q) pathogenesis must be effective after the mutant poly(Q) proteins are synthesized in the target cells. It was also seen that hsrω-RNAi does not suppress the pathological consequences of overexpression of wild-type or mutant tau protein and thus appears to be specific for poly(Q)-induced neurodegeneration (Mallik and Lakhotia 2009a).
With a view to understand how depletion of these transcripts suppresses poly(Q) aggregation and thus the neurotoxicity, we investigated interaction(s) of the hsrω transcripts with heterogeneous nuclear ribonucleoproteins (hnRNPs), CBP, and components of the UPP, which are some of the reported modifiers of poly(Q) pathogenesis (Bonini and Fortini 2003; Nelson et al. 2005; Shao and Diamond 2007; Branco et al. 2008). We show for the first time that in developing eye disc cells hsrω transcript levels reciprocally affect cellular levels of CBP. Our results also suggest that CBP and hnRNPs like Hrb57A and Hrb87F physically interact in eye disc cells. Various hnRNPs and some other RNA-binding/processing proteins are known to be sequestered by the hsrω-n transcripts as part of cellular regulation (Lakhotia et al. 1999; Prasanth et al. 2000; Jolly and Lakhotia 2006) and downregulation of hsrω transcripts abolishes the omega speckles (Mallik and Lakhotia 2009a). Therefore, we believe that hsrω–RNAi augments the availability of hnRNPs, etc., in functional compartments. The enhanced availability of various hnRNPs and CBP following depletion of the hsrω transcripts thus seems to contribute to suppression of poly(Q) toxicity by not only reducing the formation of IBs but also compensating their functional depletion in poly(Q)-expressing cells. The present results also confirm our earlier suggestion (Mallik and Lakhotia 2009a) that the residual toxicity of the poly(Q) proteins is taken care of by the proteasomal clearance machinery since the almost complete suppression of the poly(Q) toxicity by hsrω-RNAi required functional proteasomal activity in eye disc cells.
We earlier reported (Mallik and Lakhotia 2009b) that RNAi-mediated depletion of the hsrω transcripts also suppresses induced apoptosis in Drosophila through stabilization of Drosophila inhibitor of apoptosis protein 1 (DIAP1) and through suppression of c-Jun N-terminal kinase (JNK) signaling. Since poly(Q)-induced neurodegeneration also involves enhanced apoptosis and hyperactivation of JNK signaling (Evert et al. 2000; Berke et al. 2004; Morfini et al. 2006; Scappini et al. 2007), these appear to be additional components through which depletion of hsrω transcripts can suppress the neurotoxicity. It is remarkable that the noncoding hsrω transcripts affect several components implicated in the poly(Q) pathogenesis.
MATERIALS AND METHODS
Fly stocks:
All flies were raised on standard agar–cornmeal medium at 24° ± 1°, and fly crosses were carried out at the same temperature unless otherwise stated. Oregon R+ was used as wild type. The UAS-127Q, UAS-MJDtr-Q78(S), UAS-httex1p Q93, UAS-hsrω-RNAi2, UAS-hsrω-RNAi3, EP93D, and EP3037 transgenic lines have been described previously (Mallik and Lakhotia 2009a). The GMR-GAL4 (Hay et al. 1994), P{Act5C-GAL4}25F01/CyO (Ekengren et al. 2001), and UAS-Pros261.B; UAS-Prosβ21 (Belote and Fortier 2002) stocks were obtained from the Bloomington Stock Center. UAS-CBP FL-AD (acetylase dead version, F2161A), UAS-CBP RNAi, UAS-CBP ΔNZK, UAS-CBP ΔQ, and UAS-CBP ΔBHQ, all homozygous viable stocks, were provided by J. P. Kumar (Kumar et al. 2004; Anderson et al. 2005). The UAS-DTS5-11 (Schweisguth 1999) transgenic line, obtained from N. M. Bonini, produces a GAL4-inducible dominant-negative form of the β-subunit of proteasome. UAS-UbG76V-GFP (Dantuma et al. 2000) was obtained from J. Paul Taylor and was used as a GFP reporter for proteasome function. The P{w+ tsr+}/P{w+ tsr+}, ry Df(3R)Hrb87F/ry Df(3R)Hrb87F (Zu et al. 1996) stock was provided by S. Haynes. The Df(3R)Hrb87F deletion removes the Hrb87F coding region together with ∼0.6 kb of the flanking testis-specific Tsr gene, resulting in recessive male sterility of Df(3R)Hrb87F homozygotes, although the Df(3R)Hrb87F/+ heterozygotes are normally fertile; to compensate for this, the stock carries a transgenic copy of Tsr on chromosome 2 (Zu et al. 1996).
Appropriate crosses were performed following standard protocols to obtain progenies of desired genotypes.
Examination of eye structure:
For recording images of external morphology of adult eyes, flies of the desired genotype were etherized and their eyes photographed using a Sony Digital Camera (DSC-75) attached to a Zeiss Stemi SV6 stereobinocular microscope. The surface architecture of adult eyes was examined by the nail polish imprint method (Arya and Lakhotia 2006).
The arrangement of photoreceptor rhabdomeres in adult eyes was examined by pseudopupil analysis using a 60× plan-apo oil immersion objective of a Nikon E800 microscope (Sengupta and Lakhotia 2006).
Pupal lethality assay:
The numbers of larvae of different genotypes that pupated in a given cross were counted and the fate of these pupae was monitored. Numbers of pupae that died before or after differentiation and of flies that emerged were counted separately.
RNA:RNA in situ hybridization:
A 445-bp PCR-amplified fragment corresponding to the Drosophila nejire (CBP) transcript was cloned into the pGEMR-T Easy (Promega, Madison, WI) vector in appropriate orientation for subsequent manipulations. A digoxigenin-labeled CBP-specific antisense riboprobe was generated from this clone and used for hybridization to cellular RNA. RISH was carried out essentially as described earlier (Prasanth et al. 2000).
Whole organ immunostaining and confocal microscopy:
For antibody staining, eye discs from wandering third instar larvae of the desired genotypes were dissected, fixed in freshly prepared 4% paraformaldehyde in PBS for 20 min, and processed for immunofluorescence staining as described earlier (Prasanth et al. 2000). The following primary antibodies were used: (1) 1:30 dilution of a rabbit polyclonal antibody to hemagglutinin (HA) (sc-805, Santa Cruz) for HA-tagged 127Q and MJDtr-Q78(S) polyglutamine proteins, (2) 1:50 dilution of a polyclonal goat huntingtin antibody (sc-8767, Santa Cruz) for the human huntingtin protein, (3) 1:10 dilution of the P11 mouse monoclonal antibody (Saumweber et al. 1980) for Hrb87F hnRNP, (4) 1:10 dilution of the Q18 mouse monoclonal antibody (Saumweber et al. 1980) for Hrb57A hnRNP, and (5) 1:800 dilution of an affinity-purified guinea-pig polyclonal CBP antibody (Lilja et al. 2003).
Appropriate secondary antibodies conjugated either with Cy3 (1:200, Sigma-Aldrich, India) or Alexa Fluor 488 (1:200; Molecular Probes, Eugene, OR) were used to detect the given primary antibody. Chromatin was counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (1 μg/ml). Confocal imaging was carried out on a Zeiss LSM 510 Meta laser scanning confocal microscope at appropriate settings, using a plan-apo 63× oil immersion objective.
Reverse transcription–PCR:
Total RNA was purified from eye discs of third instar larvae or heads of adult flies of the desired genotype, using TRIzol reagent following the manufacturer's recommended protocol (Sigma-Aldrich, India). cDNA was synthesized for semiquantitative PCR as previously described (Mallik and Lakhotia 2009a). One-tenth volume of the reaction mixture was subjected to PCR using the following primers: hsrω-n forward 5′-GGCAGACATACGTACACGTGGCAGCAT-3′, hsrω-n reverse 5′-TTGCGCTCACAGGAGATCAA-3′; CBP-forward 5′-GGCCGATCACTTAGACGAAC-3′, CBP-reverse 5′-CTGGAGTAGGTGCTGCAGTT-3′; 127Q-forward 5′-CAGCGTCCTGATAAGTGAATTCG-3′, 127Q-reverse 5′-TACGTACGACTAGTCTGTTGCTG-3′; and GFP-forward 5′-CATGAAGCAGCACGACTT-3′, GFP-reverse 5′-TGTTCTGCTGGTAGTGGT-3′. Amplicons obtained following reverse transcription (RT)–PCR for GPDH [primer pair described earlier (Mallik and Lakhotia 2009a)] were used as loading controls. The PCR products were electrophoresed on a 2% agarose gel with appropriate molecular weight markers. Gel pictures were recorded using the G:BOX bioimaging system (Syngene).
Western blotting:
The Hrb87F protein levels in +/+ and w; P{w+ tsr+}/+; Df(3R)Hrb87F/+ were compared by homogenizing 30 pairs of eye imaginal discs from third instar larvae of each genotype in sample buffer as described earlier (Prasanth et al. 2000). The lysates were electrophoretically separated on a 12% SDS–polyacrylamide gel (with 5% stacking gel) and transferred to an immobilon-P membrane (Millipore, Bedford, MA). The primary antibodies used were 1:200 dilution of mouse β-tubulin antibody (E7, Developmental Studies Hybridoma Bank) and 1:100 dilution of Hrb87F antibody. Immunoblots were developed with 1:1000 dilution of goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Bangalore Genei, India) using the SuperSignal West Pico chemiluminescence reagent (Pierce, Rockford, IL). Blots were reprobed, as required, after stripping the earlier bound antibodies as described previously (Mallik and Lakhotia 2009a).
To compare CBP levels in different genetic backgrounds, extracts from 50 pairs of eye-antennal discs of the following genotypes were lysed in sample buffer and homogenized as above: (1) w; GMR-GAL4/GMR-GAL4; +/+; (2) w; GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3; (3) w; GMR-GAL4/GMR-GAL4; EP93D/EP93D; (4) w; GMR-GAL4/UAS-MJDtr-Q78(S); and (5) w; GMR-GAL4/UAS-MJDtr-Q78(S); hsrω-RNAi3/hsrω-RNAi3 (Prasanth et al. 2000). The lysates were slot blotted on PVDF membrane (Millipore), using a Millipore-S slot blot apparatus. They were probed with CBP (1:8000) and β-tubulin (1:200) antibodies. Blots were developed using HRP-conjugated secondary antibodies [1:5000 dilution of goat anti-guinea pig, (KPL) or 1:1000 dilution of goat anti-mouse (Bangalore Genei)] and SuperSignal West Pico chemiluminescence reagent (Pierce).
For relative quantitation of the levels of the desired protein in Western blots, the ratio of band densities of the target and the reference (β-tubulin) proteins for each sample on the blot was calculated. Means (±SD) of ratios from three independent blots for experimental and control samples were compared.
Immunoprecipitation:
CBP complexes were immunoprecipitated with the affinity-purified CBP antibody from extracts of 50 pairs of wild-type and GMR-GAL4-driven hsrω-RNAi3-expressing third instar eye imaginal discs. Approximately 1 μg of affinity-purified CBP antibody was used for each immunoprecipitation as described earlier (Prasanth et al. 2000). Recovered proteins were resolved by SDS–PAGE, transferred to PVDF membrane, and probed sequentially with the Hrb87F and Hrb57A antibodies at a dilution of 1:100 each.
All images were assembled using Adobe Photoshop 7.0.
RESULTS
As reported earlier (Mallik and Lakhotia 2009a,b), we established several independent hsrω-RNAi transgenic lines. The hsrω-RNAi3 line, in which the UAS-hsrω-RNAi transgene is inserted on chromosome 3, was used for most of the experiments reported in this study. Another line, hsrω-RNAi2, carrying the UAS-hsrω-RNAi transgene on chromosome 2 and being more effective in reducing hsrω transcript levels than the hsrω-RNAi3 line (Mallik and Lakhotia 2009b), was used in certain cases as noted later. The hsrω-RNAi3 line is referred to in the following as hsrω-RNAi. For ectopic overexpression of the hsrω gene, two EP lines, EP93D and EP3037, with the EP transposon (Rorth 1996) inserted at −130- and −144-bp positions, respectively, in the hsrω gene promoter (Mallik and Lakhotia 2009a), were used. Targeted expression of the desired UAS transgenes in developing eye discs/eyes was achieved by using the GMR-GAL4 driver (Hay et al. 1994). In certain cases, the Act5C-GAL4 driver (Ekengren et al. 2001) was used for ubiquitous expression of the desired UAS transgene(s).
Enhancement of poly(Q) toxicity by reduced levels of Hrb87F is ameliorated by hsrω-RNAi:
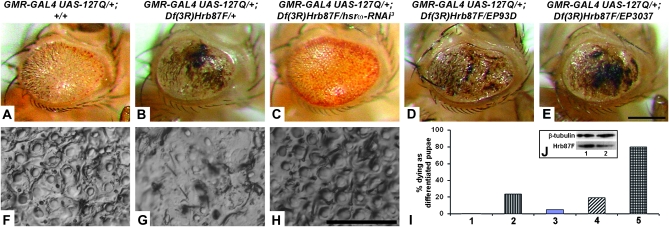
The Hrb87F protein, a Drosophila homolog of the mammalian hnRNP A2/B1 (Haynes et al. 1991), is associated with the omega speckles (Prasanth et al. 2000). As already reported (Sengupta and Lakhotia 2006), a null allele, Df(3R)Hrb87F (Zu et al. 1996), dominantly enhances the 127Q-induced toxicity leading to formation of extensive black lesions on the eye surface (compare Figure 1B and 1G, N = 392, with 1A and 1F, N = 608). In the present study, we found that it also results in dominant lethality since 24% of progeny carrying one copy of this deletion in a GMR-GAL4:127Q background died as differentiated pupae (Figure 1I, bar 2; N = 514). Downregulation of hsrω-n RNA level with one copy of hsrω-RNAi not only improved the external eye morphology relative to those expressing GMR-GAL4-driven 127Q in a Df(3R)Hrb87F/+ background (Figure 1, C and H, N = 326) but also substantially reduced the mortality such that only 4.9% differentiated pupae died (Figure 1I, bar 3; N = 343). That the Df(3R)Hrb87F/+ heterozygotes have reduced levels of the Hrb87F protein was confirmed by semiquantitative Western blotting. Compared to wild-type larval eye discs, the Hrb87F level in Df(3R)Hrb87F/+ heterozygotes was found to be ∼20% less [mean of ratio of Hrb87F level in Df(3R)Hrb87F/+ and wild-type eye discs = 0.797 ± 0.07; N = 3, Figure 1J].
Figure 1.—
Enhancement of 127Q-mediated eye degeneration by depletion of the Hrb87F hnRNP is modulated by levels of hsrω-n RNA. (A–E) Photomicrographs of eyes of adults whose genotypes are indicated at the top; (F–H) nail polish imprints of eyes of genotypes as in A–C, respectively. Bars for A–E in E and F–H in H: 20 μm. (I) Histograms of frequencies (%) of different genotypes that die as differentiated pupae: bar 1, GMR-GAL4 UAS-127Q/+; +/+ (N = 608); bar 2, GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/+ (N = 514); bar 3, GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/hsrω-RNAi3 (N = 343); bar 4, GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/EP93D (N = 466); bar 5, GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/EP3037 (N = 166); the frequencies of dying pupae in genotypes 2, 3, 4 and 5 are significantly different (P < 0.05, χ2-test) from that in 1. (J) A Western blot to show the reduced level of Hrb87F protein (bottom panel) in Df(3R)Hrb87F/+ (lane 2) larval eye discs compared to that in wild type (lane 1). The top panel shows β-tubulin levels as a loading control.
An increase in the level of hsrω transcripts by coexpression of EP93D or EP3037 in the Df(3R)Hrb87F/+ and 127Q background significantly enhanced the appearance of black lesions on the eye surface (Figure 1, D, N = 378, and E, N = 33, respectively) and resulted in death of a greater number (80.1%, N = 166) of w/w; GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/EP3037 (Figure 1I, bar 5) pupae. Intriguingly, however, expression of the EP93D allele with 127Q in the Hrb87F depleted background (N = 466) did not increase the frequency of dying pupae (Figure 1I, compare bar 4 with bar 2). The reason for this difference in the survival of w/w; GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/EP3037 and w/w; GMR-GAL4 UAS-127Q/+; Df(3R)Hrb87F/EP93D genotypes is not clear but it may be related to different locations of the two EP insertions in the hsrω gene promoter that may affect its developmental expression in some critical cells/tissues. This needs further examination.
Genetic interactions between CBP, hsrω transcripts, and poly(Q) proteins:
As noted in the Introduction, the sequestration of CBP into poly(Q) aggregates, and the consequent reduction in soluble CBP levels, results in transcriptional dysregulation as part of the poly(Q) pathogenesis (Nucifora et al. 2001; Jiang et al. 2003; Taylor et al. 2003). It is also reported that overexpression of CBP reduces the poly(Q) damage (Taylor et al. 2003; Rouaux et al. 2004). With a view to examine if hsrω-RNAi-mediated suppression of the poly(Q) damage involved CBP, we examined genetic interactions between CBP, hsrω transcripts, and poly(Q) proteins using the UAS-CBP FL-AD transgene, which carries a dominant-negative mutation in the acetyltransferase domain of CBP (Ludlam et al. 2002), or the UAS-CBP RNAi transgene (Kumar et al. 2004), using the eye-specific GMR-GAL4 driver.
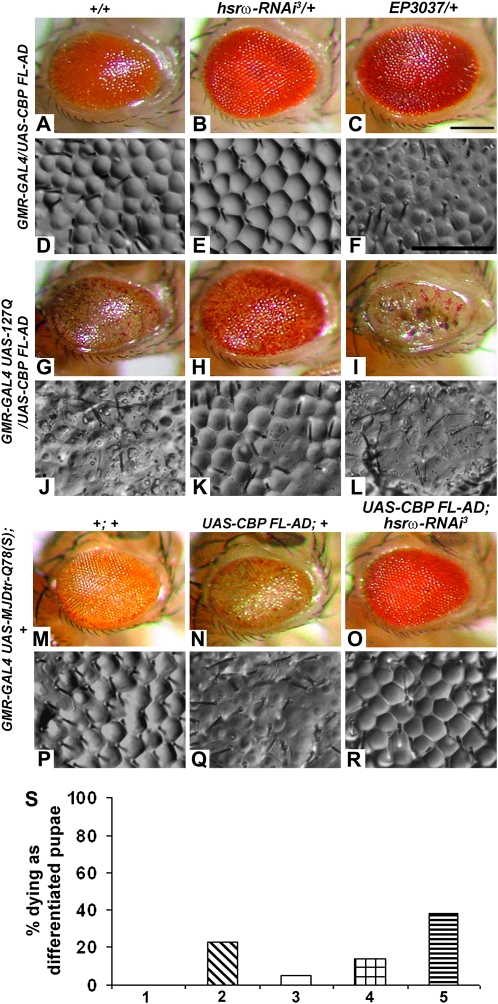
Eyes of GMR-GAL4/UAS-CBP FL-AD flies (N = 589) were moderately rough with significant reduction in the number of interommatidial bristles (Figure 2, A and D). Coexpression of one copy of the hsrω-RNAi transgene in this background (N = 523) resulted in substantial improvement in eye morphology and ommatidial arrays (Figure 2, B and E). On the other hand, enhancement of hsrω transcript levels through expression of EP3037 (Figure 2, C and F) or EP93D (not shown) exaggerated the eye damage (N = 499 and N = 466, respectively).
Figure 2.—
Depletion of hsrω-n transcripts overcomes the deleterious effects of mutant forms of CBP in absence as well as presence of pathogenic poly(Q) expression. Genotypes common to rows are indicated on the left while those for columns are indicated at the top. A–C, G–I, and M–O are photomicrographs and D–F, J–L, and P–R are nail polish imprints of adult eyes. Bars for A–C, G–I, and M–O in C and D–F, J–L, and P–R in F: 20 μm. (S) Histograms of frequency (%) of differentiated pupae of the indicated genotypes that die before emergence: bar 1, GMR-GAL4 UAS-127Q/+; +/+ (N = 608); bar 2, GMR-GAL4 UAS-127Q/UAS-CBP FL-AD; +/+ (N = 503); bar 3, GMR-GAL4 UAS-127Q/UAS-CBP FL-AD; hsrω-RNAi3/+ (N = 334); bar 4, GMR-GAL4 UAS-127Q/UAS-CBP FL-AD; EP93D/+ (N = 649); bar 5, GMR-GAL4 UAS-127Q/UAS-CBP FL-AD; EP3037/+ (N = 295); the frequencies of dying pupae in all the genotypes are significantly different (P < 0.01, χ2-test) from that in GMR-GAL4 UAS-127Q/+; +/+.
Expression of the CBP FL-AD transgene in 127Q or MJDtr-Q78(S) background resulted in varying degrees of lethality at the pupae stage (see below). The surviving flies in either case showed reduction in eye size [compare Figure 2G (N = 382) with Figure 1A (N = 608) and Figure 2N (N = 335) with 2M (N = 835)] and a greater disruption in ommatidial arrays (compare Figure 2J with Figure 1F and Figure 2Q with 2P). Coexpression of one copy of the hsrω-RNAi transgene improved the eye morphology and size (Figure 2, H and K, N = 316, O and R, N = 481). On the other hand, elevation of hsrω transcript levels in GMR-GAL4 UAS-127Q/UAS-CBP FL-AD; EP3037/+ (Figure 2, I and L, N = 182) or GMR-GAL4 UAS-127Q/UAS-CBP FL-AD; EP93D/+ (N = 558, not shown) flies resulted in greater reduction in the eye size along with near complete loss of ommatidial arrays and bristles and appearance of black lesions on the eye surface.
Expression of the dominant-negative CBP in concert with 127Q under GMR-GAL4 control caused 23% (N = 503) lethality at the differentiated pupae stage (Figure 2S, bar 2). Introduction of one copy of the hsrω-RNAi transgene in this background reduced the differentiated pupal death to only 5.4% (Figure 2S, bar 3; N = 334). On the other hand, coexpression of EP3037 enhanced differentiated pupal lethality to ∼38% (Figure 2S, bar 5; N = 295). However, GMR-GAL4-driven coexpression of dominant-negative CBP, 127Q, and EP93D resulted in only 14% pupal death (Figure 2S, bar 4; N = 649). As noted above (Figure 1I), this difference between the two EP alleles may be related to the different locations of the two EP transposons in the hsrω promoter.
Expression of the CBP FL-AD transgene in the MJDtr-Q78(S) background also resulted in 15% pupal death (N = 394), which was significantly (P < 0.01, χ2-test) reduced to 3% (N = 495) following coexpression of a single copy of the hsrω-RNAi transgene.
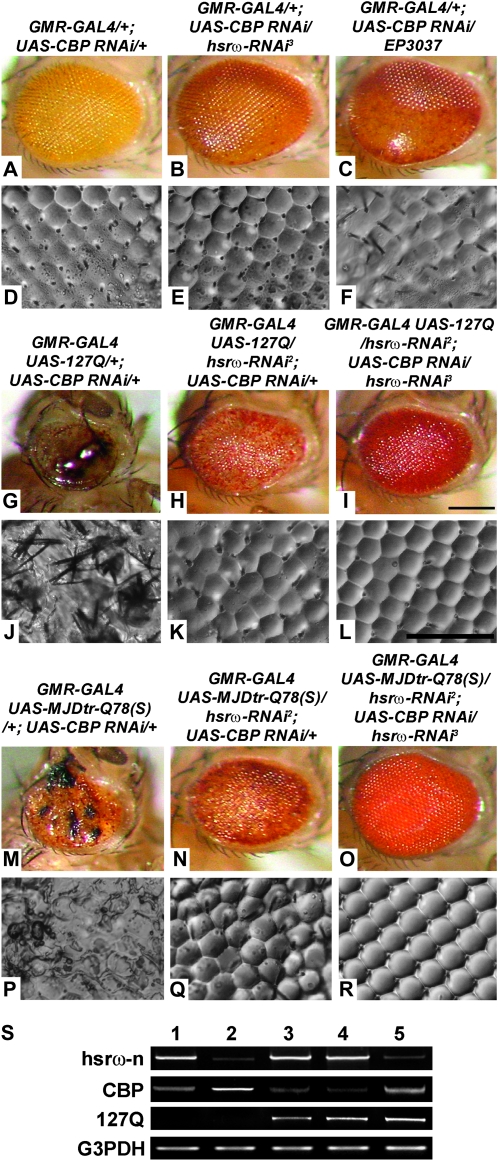
To further examine interactions between CBP and the noncoding hsrω transcripts, we downregulated CBP through RNA interference, using a UAS-CBP RNAi transgene (Kumar et al. 2004). GMR-GAL4-driven expression of UAS-CBP RNAi resulted in depigmented and somewhat smooth eyes with individual facets being less distinct, especially in the posterior region (Figure 3, A and D; N = 487). Depletion of hsrω-n transcripts in the CBP RNAi background (N = 513) brought about significant improvement in pigmentation and eye morphology, including in the posterior part (Figure 3, B and E). Augmentation of hsrω transcript levels using EP3037 (Figure 3, C and F; N = 433) or EP93D (not shown; N = 495) in a CBP depleted background enhanced eye degeneration with the loss of ommatidial integrity extending to most of the eye.
Figure 3.—
hsrω-RNAi suppresses deleterious effects of CBP RNAi in absence as well as presence of pathogenic poly(Q) expression. A–C, G–I, and M–O are photomicrographs and D–F, J–L, and P–R are nail polish imprints of adult eyes of the genotypes indicated above the columns. Bars for A–C, G–I, and M–O in I and D–F, J–L, and P–R in L: 20 μm. (S) Semiquantitative RT–PCR amplicons for hsrω-n, CBP, and 127Q transcripts in total RNA from heads of GMR-GAL4/+; UAS-CBP RNAi/+ (lane 1), GMR-GAL4/+; UAS-CBP RNAi/hsrω-RNAi3 (lane 2), GMR-GAL4 UAS-127Q/+; +/+ (lane 3), GMR-GAL4 UAS-127Q/+; UAS-CBP RNAi/+ (lane 4), and GMR-GAL4 UAS-127Q/hsrω-RNAi2; UAS-CBP RNAi/+ (lane 5) adult flies show that multiplicity of UAS target sequences in different genotypes does not limit the GMR-GAL4 activity. G3PDH was coamplified as a loading control.
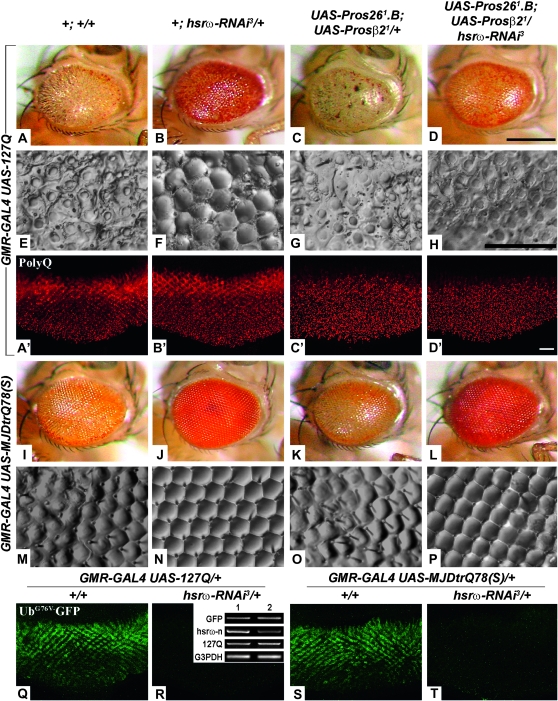
Downregulating CBP levels in the 127Q or MJDtr-Q78(S) background caused high incidence of late pupal lethality (see below) and dramatically enhanced poly(Q)-mediated eye degeneration in the few surviving flies (Figure 3, G and J, N = 22, and M and P, N = 2, respectively). Expression of the hsrω-RNAi transgene not only robustly suppressed the enhanced eye damage following depletion of CBP but also substantially reversed the 127Q- or MJDtr-Q78(S)-induced eye degeneration in a dose-dependent manner since the rescue with one copy each of hsrω-RNAi2 and hsrω-RNAi3 (Figure 3, I and L, N = 137, and O and R, N = 576) was better than that seen with one copy of hsrω-RNAi2 (Figure 3, H and K, N = 166, and N and Q, N = 174). Eyes of adult flies coexpressing one copy each of the hsrω-RNAi2 and hsrω-RNAi3 transgenes with 127Q or MJDtr-Q78(S) appeared near normal.
GMR-GAL4-driven expression of 127Q or MJDtr-Q78(S) in the CBP RNAi background caused near complete lethality (>97%) at the pharate stage (Table 1). Introduction of a single copy of the hsrω-RNAi3 transgene did not improve adult emergence since the pharate stage lethality remained at 97.2 and 86% in 127Q and MJDtr-Q78(S) backgrounds, respectively (Table 1). However, expression of a single copy of the hsrω-RNAi2 transgene improved their emergence to adult stage [28% survivors in 127Q and 86% in MJDtr-Q78(S), Table 1]. When both the hsrω-RNAi2 and the hsrω-RNAi3 transgenes were coexpressed with 127Q or MJDtr-Q78(S) in the CBP RNAi background, 32 and 95% pupae, respectively, emerged as adult flies (Table 1). Following coexpression of either EP93D or EP3037 with 127Q and CBP RNAi, lethality at the differentiated pupae stage remained 100% (Table 1).
TABLE 1.
Downregulation of hsrω-n RNA suppresses CBP-RNAi-mediated pupal lethality of 127Q- or MJDtr-Q78(S)-expressing individuals
| Genotype | No. of pupae | % dead differentiated pupae |
|---|---|---|
| 127Q model | ||
| GMR-GAL4 UAS-127Q/+; UAS-CBP RNAi/+ | 933 | 97.0 |
| GMR-GAL4 UAS-127Q/+; UAS-CBP RNAi/hsrω-RNAi3 | 457 | 97.2 |
| GMR-GAL4 UAS-127Q/hsrω-RNAi2; UAS-CBP RNAi/+ | 584 | 71.6 |
| GMR-GAL4 UAS-127Q/hsrω-RNAi2; UAS-CBP RNAi/hsrω-RNAi3 | 432 | 68.3 |
| GMR-GAL4 UAS-127Q/+; UAS-CBP RNAi/EP93D | 461 | 99.6 |
| GMR-GAL4 UAS-127Q/+; UAS-CBP RNAi/EP3037 | 157 | 100.0 |
| MJDtr-Q78(S) or SCA3 model | ||
| GMR-GAL4 UAS-MDtr-Q78(S)/+; UAS-CBP RNAi/+ | 627 | 99.7 |
| GMR-GAL4 UAS-MDtr-Q78(S)/+; UAS-CBP RNAi/hsrω-RNAi3 | 608 | 86.3 |
| GMR-GAL4 UAS-MDtr-Q78(S)/hsrω-RNAi2; UAS-CBP RNAi3/+ | 201 | 13.4 |
| GMR-GAL4 UAS-MDtr-Q78(S)/hsrω-RNAi2; UAS-CBP RNAi/hsrω-RNAi3 | 606 | 5.0 |
Since in several of the above genotypes, only one copy of the GMR-GAL4 driver is available for activating the multiple UAS-regulated transgenes, it remains possible that the different UAS promoters may not be adequately stimulated to transcribe the downstream gene. We confirmed that the multiple UAS transgenes in different genotypes were indeed activated as expected in response to the GMR-GAL4 driver through RT–PCR (see Figure 3S) with total RNA from heads of GMR-GAL4/+; UAS-CBP RNAi/+ (lane 1), GMR-GAL4/+; UAS-CBP RNAi/hsrω-RNAi3 (lane 2), GMR-GAL4 UAS-127Q/+; +/+ (lane 3), GMR-GAL4 UAS-127Q/+; UAS-CBP RNAi/+ (lane 4), and GMR-GAL4 UAS-127Q/hsrω-RNAi2; UAS-CBP RNAi/+ (lane 5) adult flies using appropriate primers (see materials and methods) for hsrω-n, CBP, and 127Q transcripts. Relative abundance of RT–PCR amplicons in the different genotypes in Figure 3S reflects the expected expression levels of the corresponding transcripts: (i) the hsrω-n transcript levels were reduced in genotypes carrying the hsrω-RNAi transgene (lanes 2 and 5); (ii) CBP transcript levels following expression of the CBP-RNAi transgene varied in relation to the presence or absence of the hsrω-RNAi and/or 127Q transgenes (lanes 1–5); and (iii) the 127Q transcripts were equally abundant in all three genotypes in which the 127Q transgene was present irrespective of the presence of other transgenes (lanes 3–5 in Figure 3S). In agreement with results presented later, expression of the hsrω-RNAi transgene is associated with an increase in CBP transcript levels (lanes 2 and 5 in the CBP row in Figure 3S). These results, together with our earlier report (Mallik and Lakhotia 2009a), thus clearly establish that under our experimental conditions, the GAL4 produced by one copy of the GMR-GAL4 transgene was adequate to activate the multiple UAS targets present in a given genotype.
With a view to further examine genetic interaction between CBP and hsrω transcripts, we used the Act5C-GAL4 driver to ubiquitously express UAS-CBP RNAi in absence (Act5C-GAL4/+; UAS-CBP RNAi/+) or presence of the hsrω-RNAi (Act5C-GAL4/hsrω-RNAi2; UAS-CBP RNAi/hsrω-RNAi3) transgenes. Results presented in Table 2 reveal that global expression of CBP RNAi caused some degree of embryonic and larval lethality (Table 2, column 2), which was enhanced in presence of the hsrω-RNAi transgenes (Table 2, column 4). It may be noted that ubiquitous expression of the hsrω-RNAi transgene by itself also caused significant embryonic and larval lethality (Table 2, column 3) and, therefore, ubiquitous coexpression of CBP RNAi and hsrω-RNAi is expected to result in greater early lethality. The high frequency of unfertilized eggs in all these crosses reflects some perturbations in gametogenesis because of Act5C-GAL4-driven CBP RNAi or hsrω-RNAi (our unpublished observations). The high frequency of embryonic and larval lethality in the cross in column 3 of Table 2 is also contributed by death of CyO or Act5C-GAL4 homozygous progeny.
TABLE 2.
hsrω-RNAi rescues Act5C-GAL4-driven UAS-CBP RNAi-mediated pupal lethality
| Crosses | Act5C-GAL4/CyO; +/+ × +/+; UAS-CBP RNAi/UAS-CBP RNAi | Act5C-GAL4/CyO; hsrω-RNAi3/hsrω-RNAi3 × Act5C-GAL4/CyO; hsrω-RNAi3/hsrω-RNAi3 | Act5C-GAL4/CyO; hsrω-RNAi3/hsrω-RNAi3 × hsrω-RNAi2/hsrω-RNAi2; UAS-CBP RNAi/UAS-CBP RNAi |
|---|---|---|---|
| 1. Total no. of eggs examined | 1009 | 1088 | 1953 |
| 2. No. (%) of unfertilized eggs | 147 (14.6%) | 177 (16.3%) | 680 (34.8%) |
| 3. No. (%) of fertilized eggs | 862 (85.4%) | 911 (83.7%) | 1273 (65.2%) |
| 4. No. (%) of dead embryosa | 42 (4.9%) | 246 (27.0%) | 78 (6.1%) |
| 5. No. (%) of dead first and second instar larvaea | 116 (13.4%) | 344 (37.8%) | 294 (23.1%) |
| 6. No. (%) of pupaea | 704 (81.7%) | 321 (35.2%) | 901 (70.8%) |
| 7. No. (%) of dead undifferentiated pupaeb | 354 (50.3%) | 5 (1.6%) | 72 (8.1%) |
| 8. No. (%) of dead differentiated pupaeb | 13 (1.8%) | 20 (6.2%) | 8 (0.9%) |
| 9. No. (%) of Act5C-GAL4-driven CBP RNAi-expressing flies that eclosedb | 0 | NA | NA |
| 10. No. (%) of Curly winged flies (expressing Act5C-GAL4-driven hsrω-RNAi)b | NA | 296 (92.2%) | NA |
| 11. No. (%) of Act5C-GAL4-driven CBP RNAi and hsrω-RNAi-expressing flies that eclosedb | NA | NA | 260 (28.8%) |
| 12. No. (%) of Curly winged flies (without Act5C-GAL4 driver)b | 337 (47.9%) | NA | 561 (62.3%) |
The percentage values in rows 4–6 were calculated with respect to the total number of developing eggs (row 3).
The percentage values in rows 7–11 were calculated with respect to the total number of pupae (row 6); NA, not applicable since these genotypes are not produced in the given crosses.
All of the surviving progeny larvae in the Act5C-GAL4/CyO; +/+ X +/+; UAS-CBP RNAi/UAS-CBP RNAi cross pupated but nearly 50% of them died as early (majority) or differentiated (a few) pupae (rows 7 and 8 in column 2, Table 2). The Act5C-GAL4/+; UAS-CBP RNAi/+ genotype was completely absent in eclosing flies while the CyO/+; UAS-CBP RNAi/+ flies emerged in the expected (∼50%) proportion. Significantly, Act5C-GAL4-driven coexpression of one copy of CBP-RNAi with one copy each of the hsrω-RNAi2 and hsrω-RNAi3 transgenes dramatically reduced pupal lethality to just ∼9% so that 29% of the pupae eclosed as phenotypically normal, fertile Act5C-GAL4/hsrω-RNAi2; UAS-CBP RNAi/hsrω-RNAi3 (row 11 in column 4, Table 2) flies. It may be noted that Act5C-GAL4-driven expression of two copies of the hsrω-RNAi3 transgene by itself resulted in death of ∼8% pupae (rows 7 and 8 in column 3, Table 2). These results, therefore, indicate that hsrω-RNAi can substantially overcome the lethal consequences of global depletion of CBP.
Cellular levels of hsrω transcripts reciprocally affect CBP mRNA and protein levels in eye discs:
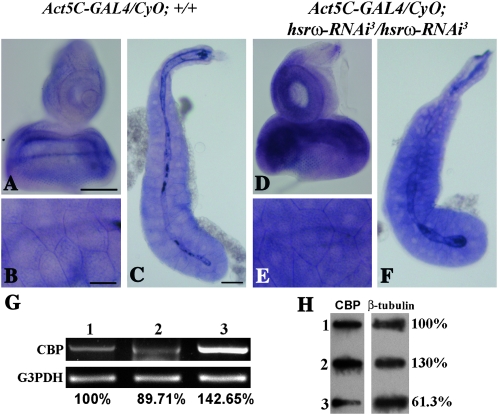
To understand the basis of the above noted genetic interaction between CBP and hsrω, we examined if altered levels of the hsrω transcripts affected CBP gene expression. We estimated CBP transcript levels in Act5C-GAL4/CyO; +/+ and Act5C-GAL4-driven hsrω-RNAi-expressing larval tissues (Figure 4, A–F). RNA:RNA in situ hybridization performed with a CBP-specific riboprobe demonstrated that Act5C-GAL4-mediated depletion of hsrω-n RNA dramatically increased the level of CBP transcripts in third instar larval eye imaginal discs (Figure 4D) as well as salivary glands (Figure 4, E and F) when compared with only GAL4-expressing eye discs (Figure 4A) or salivary glands (Figure 4, B and C), respectively.
Figure 4.—
Depletion of hsrω-n RNA upregulates CBP expression. Light microscope images (A–F) show localization of CBP RNA in Act5C-GAL4/CyO; +/+ (A–C) and Act5C-GAL4-driven hsrω-RNAi-expressing (D–F) third instar larval eye imaginal discs (A and D) and salivary glands (B, C and E, F); higher magnification images of the basal regions of salivary glands shown in C and F are shown in B and E, respectively. Bars for A and D in A and B and E in B, 50 μm; and for C and F in C, 100 μm. (G) Levels of CBP transcripts following a semiquantitative RT–PCR with total RNA from larval eye discs of different genotypes using CBP-specific primers (lane 1, GMR-GAL4/GMR-GAL4; +/+; lane 2, GMR-GAL4/GMR-GAL4; EP93D/EP93D; lane 3, GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3). The bottom panel shows coamplified G3PDH as a loading control. (H) A Western slot blot showing the amount of CBP (left) in extracts from GMR-GAL4/GMR-GAL4; +/+ (slot 1), GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3 (slot 2), and GMR-GAL4/GMR-GAL4; EP93D/EP93D (slot 3) eye imaginal discs. The right panel, showing amounts of β-tubulin in each of the extracts, was used as a loading control; values on the right show CBP levels in each sample relative to that in wild type (slot 1), which was taken as 100%.
We also estimated CBP transcript levels by semiquantitative RT–PCR with RNA isolated from eye discs of third instar larvae expressing GMR-GAL4-driven EP93D or hsrω-RNAi. As shown in Figure 4G, compared to the CBP levels in GMR-GAL4/GMR-GAL4; +/+ eye imaginal discs (lane 1), a distinct increase (∼42%) in transcript level was seen in hsrω-RNAi-expressing discs (Figure 4G, lane 3). On the other hand, the level of CBP transcripts was detectably reduced (to ∼90%) in GMR-GAL4/GMR-GAL4; EP93D/EP93D eye discs (Figure 4G, lane 2). The earlier noted (Figure 3S) results of RT–PCR of total RNA from adult heads of different genotypes also showed that even in the presence of CBP-RNAi, hsrω-RNAi significantly increased the CBP transcript levels (Figure 3S, lane 2). These results further showed that, as reported earlier (Taylor et al. 2003), expression of 127Q resulted in decreased levels of CBP transcripts (Figure 3S, lane 3), which were further reduced by coexpressing the CBP-RNAi transgene; significantly, hsrω-RNAi in this case also enhanced the CBP transcript levels (Figure 3S, lane 5).
To check if the enhanced level of CBP transcripts in hsrω-RNAi-expressing cells was accompanied by increase in the protein also, we compared levels of CBP in Western slot blots of total proteins from GMR-GAL4/GMR-GAL4; +/+, GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3, and GMR-GAL4/GMR-GAL4; EP93D/EP93D eye discs (slots 1, 2, and 3, respectively, in Figure 4H). Analysis of three independent blots showed that compared to wild type, the CBP level in hsrω-RNAi-expressing discs was, on average, enhanced by 17.6% (mean ratio of CBP level in GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3 and GMR-GAL4/GMR-GAL4: +/+ eye discs = 1.176 + 0.04); on the other hand in GMR-GAL4/GMR-GAL4; EP93D/EP93D discs it was reduced, on an average, by 30% (mean = 0.698 ± 0.04; Figure 4H).
Cellular levels of hsrω transcripts reciprocally affect CBP levels in poly(Q)-expressing eye discs also:
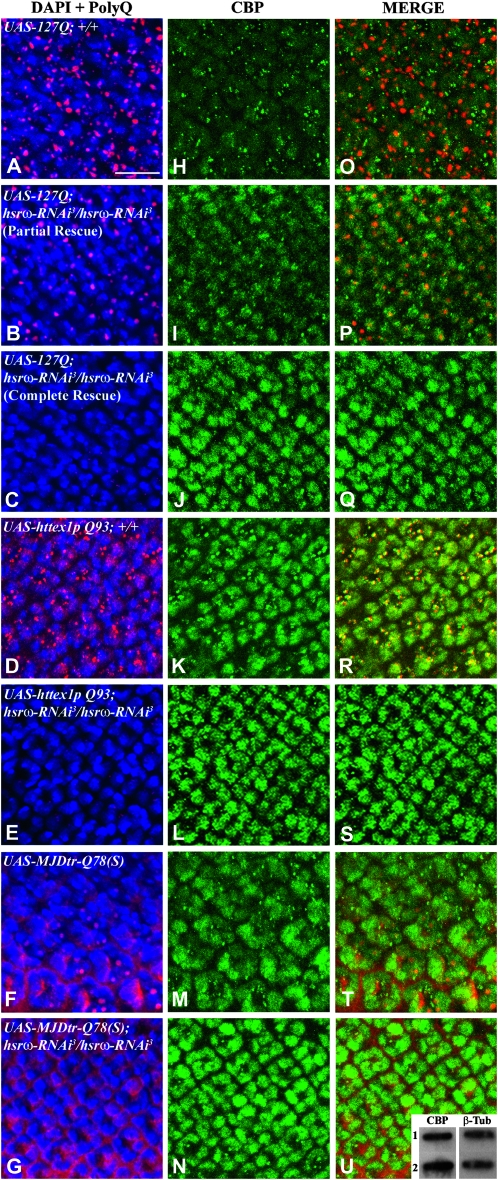
In view of the above results, we examined if downregulation of hsrω-n transcripts affected the cellular distribution and levels of CBP in poly(Q)-expressing eye discs also. Poly(Q) aggregation and levels of CBP were assayed by co-immunostaining eye discs from late third instar larvae expressing 127Q, httex1p Q93, or MJDtr-Q78(S) alone or in combination with two copies of hsrω-RNAi (Figure 5). As described earlier (Mallik and Lakhotia 2009a), the poly(Q) protein and the IBs were distributed in characteristic patterns in the different disease models (Figure 5, A–G). Likewise, the patterns of CBP distribution also varied in the three models (Figure 5, H–N). In 127Q- (Figure 5H, N = 10) or MJDtr-Q78(S)- (Figure 5M, N = 8), but not in httex1p Q93 (Figure 5K, N = 7) -expressing eye disc cells, the diffuse CBP staining on chromatin appeared to be less than that in discs not expressing the mutant poly(Q) proteins (Figure 6B). On the other hand, the 127Q- or httex1p Q93-expressing discs showed many cytoplasmic and some nuclear granules of CBP (Figure 5, H and K, respectively), but in those expressing MJDtr-Q78(S), such granules were very few (Figure 5M). Most of the nuclear and cytoplasmic granules of CBP in the 127Q discs did not colocalize with the poly(Q) IBs (Figure 5, H and O) but in the httex1p Q93-expressing discs, the CBP aggregates (Figure 5K) almost always colocalized (Figure 5R) with the huntingtin IBs (Figure 5D). Coexpression of two copies of hsrω-RNAi with the expanded poly(Q) transgenes not only reduced poly(Q) IBs and CBP aggregates (Figure 5, B, C, E, and G and I, J, L, and N) but also substantially increased the levels of diffuse CBP staining on chromatin (Figure 5, I and J, N = 11; L, N = 11; and N, N = 6). As reported earlier (Mallik and Lakhotia 2009a), hsrω-RNAi in 127Q-expressing eye discs results in either partial or complete rescue of neurodegeneration. Accordingly, some eye discs coexpressing 127Q and hsrω-RNAi (N = 11) showed reduced levels of poly(Q) aggregates (Figure 5B, 54.5% eye discs) while others showed little or no poly(Q) staining (Figure 5C, 45.5% discs). Very significantly, the levels of CBP in these discs were, respectively, marginally (Figure 5, I and P; N = 6, 54.5% eye discs) or significantly (Figure 5, J and Q; N = 5, 45.5% discs) elevated. Consistent with the above immunofluorescence results, analysis of three independent Western slot blots demonstrated that third instar larval eye imaginal discs coexpressing MJDtr-Q78(S) and hsrω-RNAi (inset in Figure 5U, slot 2) showed, on an average, ∼24% increase (mean ratio = 1.244 ± 0.06) in the CBP level compared with eye discs expressing MJDtr-Q78(S) alone (inset in Figure 5U, slot 1).
Figure 5.—
Depletion of hsrω transcripts elevates CBP levels and inhibits aggregation of poly(Q) proteins. Confocal projections of some optical sections of eye imaginal discs, excluding the peripodial membrane, co-immunostained for poly(Q) protein (red, A–G) and CBP (green, H–N); genotype for each row is indicated in the first column; all of them also carry one copy of GMR-GAL4. In each case, chromatin is counterstained with DAPI (blue, A–G). Merged images of poly(Q) and DAPI-stained nuclei are shown in A–G and of poly(Q) and CBP in O–U. The morphogenetic furrow in all cases is toward the top. Bar, 10 μm. The inset in U is a slot blot showing CBP levels (left pane) from GMR-GAL4/UAS-MJDtr-Q78(S); +/+- (slot 1) and GMR-GAL4/UAS-MJDtr-Q78(S); hsrω-RNAi3/hsrω-RNAi3 (slot 2) -expressing larval eye imaginal discs. The right pane in the inset shows similar levels of β-tubulin in both the slots.
CBP and hnRNPs show physical association in eye disc cells:
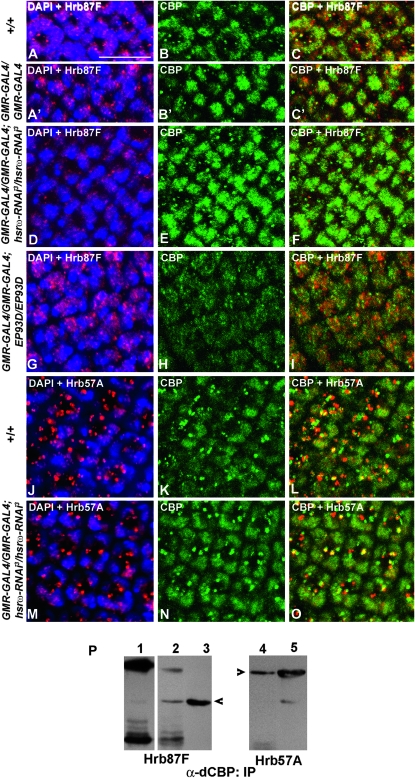
With a view to understand the basis of the above noted interactions between hsrω transcripts and CBP, we immunostained eye discs of various genotypes to examine the distribution of CBP and the hnRNPs like Hrb57A or Hrb87F that are normally associated with the hsrω transcripts in omega speckles (Prasanth et al. 2000). Nuclei of the photoreceptor cells of late third instar wild-type (Figure 6, A–C, N = 15; J–L, N = 8) as well as GMR-GAL4/GMR-GAL4; +/+ (Figure 6, A′–C′, N = 7) larval eye discs showed CBP and both the hnRNPs to be distributed in diffuse as well as speckled/granular patterns; the cytoplasm in these cells also showed distinct granules of CBP and the hnRNPs. Interestingly, in the peripodial cells of eye discs, CBP and Hrb87F, but not Hrb57A, were much more abundant than in the disc proper cells but were restricted mostly to the nucleus in diffuse and granular domains (not shown). The Hrb57A-containing cytoplasmic granules were distinctly larger than those stained by the Hrb87F antibody (compare Figure 6A and 6J). The diffuse nuclear CBP staining overlapped with that for Hrb87F (Figure 6C) and Hrb57A (Figure 6L). On the other hand, the CBP granules, especially those in the photoreceptor cell cytoplasm, were often adjacent to or partially or fully overlapped with those of Hrb87F (Figure 6C) and Hrb57A (Figure 6L).
Figure 6.—
CBP shows association with hnRNPs and its cellular levels are reciprocally modulated by levels of the hsrω transcripts. Shown are projections of confocal images of late third instar larval eye discs (genotypes indicated on the left) co-immunostained with Hrb87F (red, A, A′, D, and G) and CBP (green, B, B′, E, and H) or with Hrb57A (red, J and M) and CBP (green, K and N) antibodies. Merged images of hnRNPs and DAPI-stained nuclei (blue) are shown in A, A′, D, G, J, and M; of CBP and Hrb87F in C, C′, F, and I; and of CBP and Hrb57A in L and O. Bar, 10 μm. (P) CBP antibody co-immunoprecipitated endogenous Hrb87F (arrowhead in lanes 1–3) from wild-type (lane 1) and from GMR-GAL4-driven hsrω-RNAi (lane 2) third instar larval eye-imaginal discs. Likewise, immunoprecipitation with CBP also pulled down Hrb57A from GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3 eye discs (lane 4). Lanes 3 and 5 show Hrb87F and Hrb57A proteins, respectively, in the crude extracts used for immunoprecipitation; the filter in lane 3 was reprobed with the Hrb57A antibody to produce the image in lane 5 and, therefore, a residual signal corresponding to Hrb87F is also seen in this lane. Lane 1 was overdeveloped compared to lanes 2–5 to enable visualization of the Hrb87F band in this lane.
As seen in Figure 6, A′–C′, the distribution of Hrb87F and CBP in photoreceptor cells in GMR-GAL4 homozygous eye discs was more or less comparable to that in wild type (Figure 6, A–C). However, GMR-GAL4-driven expression of the hsrω-RNAi transgene resulted in significant redistribution of both hnRNPs and CBP (Figure 6, D–F, N = 14; M–O, N = 12). Nuclear granules of the hnRNPs (Figure 6, D and M) and CBP (Figure 6, E, F, N, and O) were nearly absent while cytoplasmic granules of the hnRNPs, especially those of Hrb57A (Figure 6M) and of CBP, were more abundant. It is interesting to note that the cytoplasmic granules of Hrb57A and CBP also showed a greater overlap following expression of hsrω-RNAi (Figure 6O). As noted above (Figure 5), the overall staining for CBP was enhanced in eye discs with reduced hsrω transcripts (Figure 6, E and N). In contrast, GMR-GAL4-driven expression of EP93D in larval eye discs resulted in enhanced nuclear accumulation of the Hrb87F protein and an overall reduced staining for CBP with the nuclear as well as cytoplasmic granules also being much less prominent (Figure 6, G–I; N = 9). In these discs, the ommatidial arrays were less organized as revealed by DAPI staining of photoreceptor nuclei (Figure 6G).
The apparent association between CBP and the hnRNPs was confirmed by immunoprecipitation followed by Western blotting. As shown in Figure 6P, immunoprecipitation of crude extracts of eye imaginal discs from wild-type (lane 1) and GMR-GAL4/GMR-GAL4; hsrω-RNAi3/hsrω-RNAi3 (lanes 2 and 4) third instar larvae with the CBP antibody pulled down Hrb87F (lanes 1 and 2) as well as Hrb57A (lane 4).
hsrω-RNAi-mediated rescue of eye degeneration due to dominant-negative CBP mutants requires the CBP transactivation domain:
To identify the domain(s) through which the hsrω transcripts interact with CBP, we overexpressed a series of CBP loss-of-function mutants (Kumar et al. 2004; Anderson et al. 2005) in conjunction with single copies of hsrω-RNAi or EP93D or EP3037 in the developing eye and looked for modulation of the retinal phenotypes associated with the expression of the CBP variants alone.
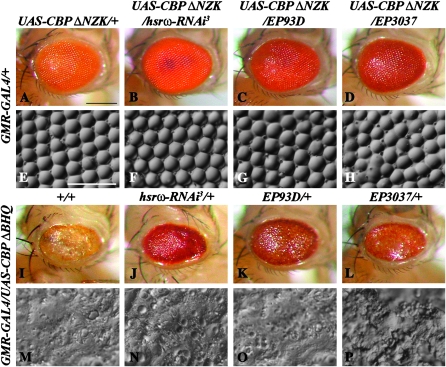
Flies expressing the CBP ΔNZK variant, lacking the N-terminal half of the protein that includes the domains for binding to CREB and to nuclear hormone receptors (Kumar et al. 2004; Anderson et al. 2005), exhibited a slight roughening of the external retinal surface and disruption of the ommatidial bristle arrangement because of missing or extra bristles at some places (Figure 7, A and E). Downregulation of hsrω-n RNA in this background resulted in near wild-type eye morphology (Figure 7, B and F). On the other hand, enhancement of hsrω transcripts through expression of EP93D (Figure 7, C and G) or EP3037 (Figure 7, D and H) enhanced the eye damage (Figure 7, C, D, G, and H). The EP3037 allele had a slightly greater enhancing effect.
Figure 7.—
Effect of altered levels of hsrω transcripts on eye degeneration following expression of individual CBP variants. In all cases, the CBP variant was expressed from a UAS construct driven by GMR-GAL4. Genotypes common to rows are indicated on the left while those for columns are indicated at the top. A–D and I–L are photomicrographs and E–H and M–P are nail polish imprints of adult eyes. Bars for A–D and I–L in A and for E–H and M–P in E: 20 μm.
As reported earlier (Kumar et al. 2004), GMR-GAL4-driven expression of the CBP ΔQ construct that deletes the alanine- and glutamine-rich transactivation domain alone resulted in 100% lethality (N = 594) at the early pupae stage before eye differentiation. The eye phenotype following expression of CBP ΔBHQ, which removes the BROMO, HAT, and poly(Q) domains (Kumar et al. 2004; Anderson et al. 2005), was very severe (N = 1086). The external surface of the adult eye was flattened and reduced in size (Figure 7, I and M). Interestingly, the early pupal lethality resulting from the expression of CBP ΔQ or the eye phenotype due to CBP ΔBHQ was not affected by either hsrω-RNAi (Figure 7, J and N; N = 1024) -mediated down regulation or EP-mediated overexpression (EP93D, Figure 7, K and O, N = 877; or EP3037, Figure 7, L and P, N = 826) of hsrω transcripts. Thus, any variations in the cellular hsrω transcript levels failed to modulate phenotypes caused by ectopic expression of CBP loss-of-function mutants that lack the poly(Q) domain.
UPP activity is modulated by hsrω transcripts:
In our earlier study (Mallik and Lakhotia 2009a) we raised the possibility that the pathogenic poly(Q) polypeptides that fail to aggregate following hsrω-RNAi are degraded by the proteasomal machinery of the cell so that the source of toxicity is substantially eliminated. Therefore, we first examined if the proteasome activity is affected by the hsrω transcripts, using two different approaches.
In the first approach, we used the UAS-Pros261.B and UAS-Prosβ21 transgenes that carry temperature-sensitive missense mutations in the 20S proteasome subunits β6 and β2, respectively (Belote and Fortier 2002). Although these temperature-sensitive mutant alleles have maximum effect at 29° (Belote and Fortier 2002), we reared the larvae and flies carrying these transgenes at 25° since expression of the mutant proteasome subunits even at 25° affects eye structure but the additional poly(Q) toxicity due to elevated temperature is avoided.
Externally, eyes of flies coexpressing the two proteasome mutant transgenes under control of GMR-GAL4 at 25° appeared near normal (Figure 8, A and B; N = 225); coexpression of hsrω-RNAi along with the proteasome mutant transgenes also did not alter the external eye morphology (Figure 8, D and E; N = 281). However, the pseudopupil images of eyes (Figure 8C, N = 9) of flies expressing the proteasome mutants exhibited severe retinal damage, which was substantially, although not completely, rescued by coexpression of one copy of hsrω-RNAi (Figure 8F, N = 12).
Figure 8.—
Retinal damage due to impaired proteasomal function is suppressed by hsrω-RNAi but overexpression of hsrω transcripts per se compromises the proteasome activity. Genotypes are indicated at the top of each row in A–F. For G–L the genotype for chromosome 2 is indicated on the left while that for chromosome 3 is at the top of each column. A and D are photomicrographs, B and E are nail polish imprints, and C and F are pseudopupil images of adult eyes. G–I are confocal projections of larval eye discs of the indicated genotypes showing the extent of proteasomal dysfunction through GFP expression (green, G–I). J–L show DAPI-stained nuclei in the same sets of confocal projections shown in G–I, respectively; compared to the wild-type-like distribution of nuclei of developing ommatidial units in a typical ring-like arrangement in ordered diagonal arrays in third instar eye imaginal discs from GMR-GAL4/UAS-UbG76V-GFP; hsrω-RNAi3/+ (K), the nuclei of ommatidial units in eye discs from GMR-GAL4/UAS-UbG76V-GFP; UAS-DTS5-11/+ (J) as well as GMR-GAL4/UAS-UbG76V-GFP; EP3037/+ (L) appear disorganized. Insets in J–L show higher magnification images of a single ommatidial cluster in the corresponding disc. Bars for A and D in A, for B and E in B, and for G–L in G, 20 μm; and for C and F in C, 10 μm.
In another approach, we used the UAS-UbG76V-GFP transgenic line (Dantuma et al. 2000) to examine if misexpression of hsrω transcripts compromises the endogenous UPP activity. The UAS-UbG76V-GFP transgene is a fluorescent reporter of UPP function carrying an uncleavable ubiquitin-conjugated GFP variant under control of the UAS promoter. This modified protein has an extremely short half-life and therefore any dysfunction of the endogenous UPP results in GFP fluorescence in the transgene-expressing cells while in cells with normal proteasome activity, the GFP-tagged ubiquitin is efficiently degraded so that very low GFP fluorescence is seen (Dantuma et al. 2000). When UAS-UbG76V-GFP was coexpressed with a single copy of the dominant-negative proteasome subunit transgene, UAS-DTS5-11, at 29° using the GMR-GAL4 driver, a robust GFP fluorescence was seen in eye imaginal discs as expected (Figure 8G, N = 5); the ommatidial units in these discs were also considerably disorganized as revealed by the DAPI fluorescence of nuclei (Figure 8J). This validated the effectiveness of the GFP-reporter assay in this transgenic line. Eye imaginal discs from flies expressing the UAS-UbG76V-GFP reporter along with one copy of hsrω-RNAi at 25° showed a very low GFP fluorescence signal (Figure 8H, N = 7) and the DAPI staining (Figure 8K) revealed well organized ommatidial units. On the other hand, GMR-GAL4-driven expression of a single copy of EP3037 at 25° resulted in a strong UbG76V-GFP (Figure 8I, N = 12) fluorescence together with disruption in the organization of ommatidial units (Figure 8L).
Together, these results show that overexpression of hsrω transcripts compromises UPP while their depletion through hsrω-RNAi may actually help restore the compromised proteasome functions.
Suppression of poly(Q) protein toxicity by hsrω-RNAi requires a functional proteasome:
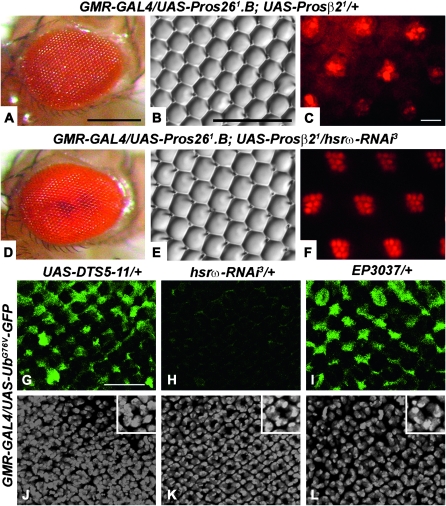
We next expressed the UAS-Pros261.B and UAS-Prosβ21 transgenes either in 127Q or in 127Q and hsrω-RNAi backgrounds using the GMR-GAL4 driver. Expression of 127Q in the compromised proteasome background enhanced the poly(Q) eye phenotype (compare Figure 9A and 9E, N = 286, with 9C and 9G, N = 293). Interestingly, unlike the substantial suppression of poly(Q)-induced eye damage by coexpression of one copy of hsrω-RNAi in a genetically functional proteasome background (Figure 9, B and F, N = 244; also see Mallik and Lakhotia 2009a), suppression of the 127Q damage by hsrω-RNAi was significantly less when the UPP activity was compromised by expression of the dominant-negative transgenes (Figure 9, D and H; N = 231).
Figure 9.—
Functional proteasomal activity is required for suppression of poly(Q) damage to eyes and for suppression of formation of IBs in eye discs. A–D and I–L are photomicrographs and E–H and M–P are nail polish imprints of the eye surface. Genotypes common to rows are indicated on the left while those for columns are indicated at the top. (A′–D′) Confocal projections of third instar larval eye imaginal discs immunoassayed for the poly(Q) protein (red, A′–D′). (Q–T) Confocal projections showing UbG76V-GFP reporter activity in eye imaginal discs from GMR-GAL4 UAS-127Q/+; +/+ (Q), GMR-GAL4 UAS-127Q/+; hsrω-RNAi/+ (R), GMR-GAL4 UAS-MJDtrQ78(S)/+; +/+ (S), and GMR-GAL4 UAS-MJDtrQ78(S)/+; hsrω-RNAi3/+ (T) third instar larvae. Bars for A–D and I–L in D, for E–H and M–P in H, and for A′–D′ and Q–T in D′: 20 μm. Inset in R shows semiquantitative RT–PCR levels of EGFP, hsrω-n, and 127Q transcripts in total RNA from heads of w/w; GMR-GAL4 UAS-127Q/+; +/+ (lane 1) and w/w; GMR-GAL4 UAS-127Q/+; hsrω-RNAi3/+ (lane 2) adult flies, respectively. G3PDH was coamplified and used as a loading control.
We have shown earlier (Mallik and Lakhotia 2009a) that expression of two copies of hsrω-RNAi with the 127Q transgene resulted in ∼50% discs with reduced IBs and 50% with no IBs. In the present study, we examined the effect of one copy of the hsrω-RNAi transgene in 127Q-expressing eye discs with normal and ectopically compromised UPP activity. As expected, one copy of the hsrω-RNAi transgene in the absence of any mutant UPP activity also reduced the IBs but fewer (∼30%) discs exhibited nearly complete disappearance of the IBs (Table 3). Coexpression of the proteasome mutants in the poly(Q) background increased aggregate formation at the expense of the diffuse poly(Q) staining in rows nearer to the morphogenetic furrow (Table 3; compare Figure 9A′ with 9C′). Significantly, discs coexpressing 127Q and hsrω-RNAi in concert with the proteasome mutants also exhibited enhanced incidence of poly(Q) aggregates than those with normal UPP activity (Table 3; compare Figure 9B′ with 9D′).
TABLE 3.
Suppression of poly(Q) aggregation by hsrω-RNAi is inhibited by coexpression of dominant-negative proteasome subunits
| No. of discs | Relative level of poly(Q) IBsa (% discs) |
||||
|---|---|---|---|---|---|
| Genotype | +++ | ++ | + | − | |
| GMR-GAL4/UAS-127Q; +/+ | 17 | 100 | |||
| GMR-GAL4/UAS-127Q; hsrω-RNAi3/+ | 13 | 69.2 | 30.8 | ||
| GMR-GAL4 UAS-127Q/UAS-Pros261.B; UAS-Prosβ21/+ | 9 | 100 | |||
| GMR-GAL4 UAS-127Q/UAS-Pros261; UAS-Prosβ21/hsrω-RNAi3 | 8 | 100 | |||
The levels are expressed in arbitrary categories with +++ indicating maximum and – indicating few or no inclusion bodies (IBs).
Coexpression of the two transgenes encoding mutant proteasomal subunits with MJDtr-Q78(S) under control of the GMR-GAL4 driver did not further enhance the MJDtr-Q78(S) eye degenerative phenotype (compare Figure 9I and 9M with 9K and 9O). However, coexpression of the mutant proteasomal transgenes in the SCA3 fly model also abrogated the ability of hsrω-RNAi to rescue MJDtr-Q78(S)-induced eye damage (compare Figure 9J and 9N with 9L and 9P).
Expression of the UbG76V-GFP reporter in flies expressing 127Q and MJDtrQ78 alone or in conjunction with one copy of the hsrω-RNAi transgene revealed that as reported earlier (Chan and Bonini 2000; Venkatraman et al. 2004; Bennett et al. 2005), poly(Q) expression by itself disrupted UPP activity, resulting in substantial UbG76V-GFP reporter fluorescence (Figure 9, Q and S; N = 8 in both cases). Interestingly, GFP fluorescence in eye imaginal discs from third instar larvae coexpressing one copy of the hsrω-RNAi transgene and 127Q (Figure 9R, N = 8) or MJDtrQ78 (Figure 9T, N = 12) was very low, suggesting substantial restoration of normal proteasome function despite expression of the expanded poly(Q) protein. Results of RT–PCR presented in the inset in Figure 9R clearly show that the absence of GFP fluorescence following hsrω-RNAi in any of these genotypes is not due to inhibition of the expression of the UbG76V-GFP transgene by reduced levels of hsrω transcripts since the UbG76V-GFP as well as the 127Q transcripts were equally abundant whether the hsrω transcript levels were normal (lane 1) or lowered by hsrω-RNAi (lane 2).
Taken together, these observations show that poly(Q) expression disrupts normal UPP function that is restored by hsrω-RNAi. However, if the cell's proteasomal activity is impaired by expression of dominant-negative subunits, reduced levels of hsrω transcripts fail to ameliorate the poly(Q) toxicity.
DISCUSSION
Earlier reports from our laboratory showed that while overexpression of the noncoding hsrω transcripts enhanced (Sengupta and Lakhotia 2006), reducing the cellular levels of these transcripts through RNAi suppressed the neurodegeneration caused by mutant proteins with expanded poly(Q) stretches (Mallik and Lakhotia 2009a). As shown earlier (Mallik and Lakhotia 2009a), expression of the hsrω-RNAi transgene had no effect on poly(Q) transcription but it diminished/eliminated the source of toxicity by inhibiting formation of the IBs and by enhancing clearance of the mutant poly(Q) proteins. The present study provides useful insights into the possible mechanisms through which these noncoding transcripts modulate cellular toxicity of the mutant poly(Q) proteins.
Studies in a variety of poly(Q) model systems have reported that many essential cellular proteins, e.g., transcription factors like CBP, TBP, etc. (Li and Li 2004; Bae et al. 2005), chaperone proteins (Cummings et al. 1998; Chan et al. 2000; Waelter et al. 2001), and proteasome subunits (DiFiglia et al. 1997; Cummings et al. 1998; Waelter et al. 2001), etc., are sequestered by the expanded poly(Q) proteins. In agreement with earlier reports (Chai et al. 1999; Chan et al. 2002; Jiang et al. 2003; Taylor et al. 2003; Sengupta and Lakhotia 2006), the present study shows that the poly(Q) damage is enhanced by functional depletion of hnRNPs, CBP, or proteasome components because of expression of dominant-negative mutants or RNAi or null mutations. We show that hsrω-RNAi substantially rescued the poly(Q) toxicity even when additional damage was caused by the presence of mutant alleles of Hrb87F or CBP. On the other hand, compromised proteasome activity affected the rescue of poly(Q) damage by hsrω-RNAi.
It is significant that while complete absence of Hrb87F does not affect normal development of Drosophila melanogaster (Zu et al. 1996), ∼20% reduction in cellular levels of Hrb87F, as seen in the Df(3R)Hrb87F/+ eye discs (Figure 1J), resulted in a significant enhancement in the poly(Q) eye phenotype (Figure 1, also see Sengupta and Lakhotia 2006). As noted earlier, the mutant poly(Q) proteins deplete the functional availability of many essential proteins and thus disrupt cellular homeostasis. Therefore, even a 20% depletion of the otherwise dispensable Hrb87F exaggerates the poly(Q) toxicity. We have shown (Mallik and Lakhotia 2009a) earlier that hsrω-RNAi results in disappearance of the omega speckles so that the various proteins, including Hrb87F, sequestered in them (Lakhotia et al. 1999; Prasanth et al. 2000) become available in the soluble cellular pool. The increased availability of such essential proteins in the functional pool following hsrω-RNAi compensates not only for the genetic deficiency of Hrb87F but also for the functional depletion of this and other proteins by the poly(Q) IBs. This finds support in the fact that targeted overexpression of hnRNP A2/B1 and its Drosophila homologs, Hrb87F and Hrb98DE, suppresses CGG repeat-induced neurodegeneration in the FXTAS fly model (Sofola et al. 2007). It has recently been shown (Ji and Tulin 2009) that poly(ADP) ribosylation and deglycosylation of hnRNPs modulate their activity and their binding with the hsrω transcripts; the authors suggested that only nonribosylated hnRNPs can be sequestered by these transcripts. It is, therefore, likely that the release of hnRNPs from the omega speckles following hsrω-RNAi provides for a greater pool of the hnRNPs being available for ribosylation and thus activity.
CBP is one of the important regulators of chromatin structure and transcription and its sequestration by the mutant poly(Q) proteins is believed to be a major cause for neurodegeneration (Li and Li 2004; Bae et al. 2005). It is also reported that overexpressing CBP or enhancing its activity suppresses poly(Q) IB formation and neurodegeneration (Taylor et al. 2003). Our findings that developmental defects in eyes caused by expression of dominant-negative forms of CBP or by its depletion through RNAi are rescued by hsrω-RNAi clearly show that the hsrω transcripts can modulate CBP metabolism in eye disc cells. This possibility is confirmed by our finding that levels of CBP transcripts and that of the CBP protein are elevated following hsrω-RNAi and are lowered by hsrω overexpression.
Our observations that cellular distributions of hnRNPs, like Hrb87F and Hrb57A, partially overlap with that of CBP and that these proteins are co-immunoprecipitated suggest that CBP interacts with Hrb57A and Hrb87F. Downregulation of hsrω-n RNA results in disappearance of the omega speckles and redistribution of the hnRNPs (Mallik and Lakhotia 2009a). In view of the physical association of these proteins, we speculate that the enhanced availability of hnRNPs in the diffuse cellular pool may pull more CBP into the diffuse fraction so that a greater amount of CBP becomes available for activity rather than remaining stored/sequestered. Additionally, caspase-6-mediated cleavage and degradation of CBP followed by a subsequent decrease in histone acetylation, another critical step common to several neuropathologies (Rouaux et al. 2004), may also be inhibited by hsrω-RNAi since our other studies showed that hsrω-RNAi inhibits caspase activity through stabilization of DIAP1 via its interaction with Hrb57A (Mallik and Lakhotia 2009b). The levels of hsrω-n transcripts may also affect CBP mRNA levels through the variety of RNA-processing and transcription factors that directly or indirectly associate with the hsrω transcripts (Jolly and Lakhotia 2006). As reported earlier (Taylor et al. 2003; Saha and Pahan 2006), the net increase in CBP levels, following hsrω-RNAi, would inhibit formation of poly(Q) IBs and restore the histone acetylation homeostasis.
It is remarkable that while hsrω-RNAi suppressed the eye phenotypes resulting from expression of CBP-FL AD or CBP RNAi or CBP ΔNZK, it failed to rescue the lethality or the eye damage following expression of CBP ΔQ or CBP ΔBHQ, respectively. This indicates that the transactivation domain of CBP is required for the suppressive action of hsrω-n RNAi. It is likely that the hnRNPs like Hrb87F, Hrb57A, etc., interact with CBP through its Q domain so that the hnRNPs released by disappearance of the omega speckles following hsrω-RNAi fail to compensate the damage caused by expression of dominant-negative CBP ΔQ or CBP ΔBHQ. Further studies are required to understand the mechanism(s) of these interactions.
Our studies also reveal interaction of UPP with hsrω transcripts and an important role of this interaction in the modulation of poly(Q) toxicity. Restoration of the eye phenotype following targeted disruption of the normal proteasomal activity by hsrω-RNAi indicates that the proteasome activity improves when levels of these noncoding transcripts are reduced. This is also confirmed by the direct demonstration, through the GFP-reporter expression, that the intrinsic UPP is compromised in cells overexpressing hsrω. Our finding that proteasome activity is impaired in 127Q-expressing flies is consistent with earlier reports that poly(Q) toxicity in vivo is enhanced by proteasome mutations or by inhibitors of proteasome activity (Chan et al. 2002; Venkatraman et al. 2004; Bennett et al. 2005; Diaz-Hernandez et al. 2006; Wang et al. 2008). It is significant that hsrω-RNAi ameliorated the proteasomal dysfunction due to poly(Q) expression, since the proteasome-GFP reporter expression was very low in cells coexpressing poly(Q) and hsrω-RNAi (Figure 9). Restoration of proteasome function in mutant poly(Q)-expressing cells is thus an additional pathway through which hsrω-RNAi suppresses the neurodegeneration. This finds further support in the observation that when the endogenous UPP function is intrinsically compromised by expression of dominant-negative mutants, hsrω-RNAi is no longer as effective in suppressing the poly(Q) damage as in cells with normal proteasome function. The mechanism(s) through which the hsrω transcripts regulate UPP pathways remain to be understood.
Many of the poly(Q) proteins involved in CAG repeat expansion disorders contain caspase consensus cleavage sites and caspase-mediated cleavage of the mutant protein appears necessary for pathogenesis (Evert et al. 2000). Inhibition of activity of caspases like caspase-1, caspase-3, or caspase-8 or alteration of the caspase cleavage sites in the mutated protein delays and reduces the expanded poly(Q) protein pathogenicity (Goldberg et al. 1996; Wellington et al. 1998, 2002; Ellerby et al. 1999; Ona et al. 1999; Sanchez et al. 1999; Berke et al. 2004). Our other studies show that in cells in which apoptosis is ectopically induced, hsrω-RNAi stabilizes DIAP1 through enhanced association with Hrb57A (Mallik and Lakhotia 2009b). Elevated levels of DIAP1 inhibit caspase activity and thus apoptosis (Hay 2000; Arya et al. 2007). Further, expression of expanded poly(Q) proteins brings about hyperactivation of JNK (Merienne et al. 2003; Morfini et al. 2006; Scappini et al. 2007), which contributes to neuronal dysfunction and cell death in neurodegenerative disorders. Significantly, hsrω-RNAi suppresses activation of the JNK pathway also (Mallik and Lakhotia 2009b). Inhibition of caspase and JNK activities thus appear to be other paths through which hsrω-RNAi suppresses the poly(Q) toxicity in the fly models.
In summary, we suggest that hsrω-RNAi suppresses poly(Q) toxicity by modulating several components involved in the pathogenesis of these debilitating diseases. First, hsrω-RNAi enhances the availability of hnRNPs and CBP in functional pools. This in turn would suppress IB formation and restore histone acetylation and transcriptional regulation in cells expressing the mutant poly(Q) proteins (Steffan et al. 2001; Taylor et al. 2003; Li and Li 2004; Iijima-Ando et al. 2005). Second, the proteasomal activity is improved when hsrω RNA levels are reduced and this helps the cells to get rid of toxic proteins. Third, the release of hnRNPs from omega speckles following hsrω-RNAi stabilizes DIAP1 (Mallik and Lakhotia 2009b), resulting in inhibition of apoptosis so that neuronal cells, that otherwise would have died, survive. Additionally, in view of the above noted role of JNK in poly(Q) damage, the suppression of JNK activation in eye disc cells following hsrω-RNAi (Mallik and Lakhotia 2009b) may also contribute to amelioration of the poly(Q) damage. Further, the hsrω transcripts are known to interact with several other proteins, including Hsp90 (Jolly and Lakhotia 2006), and therefore, it remains possible that other network effects also contribute to the observed suppression of the poly(Q) damage. The observed pleiotropic effects reflect involvement and, therefore, critical importance of the hsrω noncoding transcripts in cellular homeostasis. Since most of the wild-type poly(Q) proteins, whose mutations result in neurodegeneration, are themselves involved in diverse regulatory processes (Carlson et al. 2009), alterations in the noncoding hsrω transcript pool can be expected to bring about unpredictable and divergent consequences in cells with genetically compromised regulation. These transcripts apparently function as hubs for coordination of several cellular networks and thus ensure homeostasis. Such multiple networking interactions provide a basis for the context-dependent actions of the same molecule in different cells or in the same cell under different conditions (Arya et al. 2007). The multipronged action of these noncoding transcripts also provides a new paradigm for a therapeutic target for the human poly(Q) disorders.
Acknowledgments
We thank Parsa Kazemi-Esfarjani (UAS-127Q), Nancy M. Bonini [UAS-MJDtr-Q78(S) and UAS-DTS5-11], Leslie Thompson (UAS-httex1p Q93), Susan Haynes (Df(3R)Hrb87F), Justin P. Kumar (UAS-CBP FL-AD, UAS-CBP RNAi, UAS-CBP ΔNZK, UAS-CBP ΔQ, and UAS-CBP ΔBHQ), J. Paul Taylor (UAS-UbG76V-GFP), and the Bloomington Stock Center for providing different fly stocks (EP3037, UAS-Pros261.B; UAS-Prosβ21, GMR-GAL4, and Act5C-GAL4) used in the present study. We gratefully acknowledge Harold Saumweber and Mattias Mannervik for generously providing the P11 and Q18 and the CBP antibodies, respectively. We also thank the Developmental Studies Hybridoma Bank for the E7 antibody. This work was supported, in part, by a Shyama Prasad Mukherjee fellowship from the Council of Scientific and Industrial Research (New Delhi), India to M.M. and a research grant from the Department of Science and Technology, Government of India, to S.C.L. We acknowledge support from the Department of Science and Technology, Government of India, for the Confocal Microscope Facility in our laboratory.
References
- Anderson, J., R. Bhandari and J. P. Kumar, 2005. A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye. Genetics 171 1655–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate, M., S. Mitra, E. S. Schweitzer, M. R. Segal and S. Finkbeiner, 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431 805–810. [DOI] [PubMed] [Google Scholar]
- Arya, R., and S. C. Lakhotia, 2006. A simple nail polish imprint technique for examination of external morphology of Drosophila eyes. Curr. Sci. 90 1179–1180. [Google Scholar]
- Arya, R., M. Mallik and S. C. Lakhotia, 2007. Heat shock genes: integrating cell survival and death. J. Biosci. 32 595–610. [DOI] [PubMed] [Google Scholar]
- Bae, B. I., H. Xu, S. Igarashi, M. Fujimuro, N. Agrawal et al., 2005. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron 47 29–41. [DOI] [PubMed] [Google Scholar]
- Belote, J. M., and E. Fortier, 2002. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis 34 80–82. [DOI] [PubMed] [Google Scholar]
- Bennett, E. J., N. F. Bence, R. Jayakumar and R. R. Kopito, 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell 17 351–365. [DOI] [PubMed] [Google Scholar]
- Berke, S. J., F. A. Schmied, E. R. Brunt, L. M. Ellerby and H. L. Paulson, 2004. Caspase-mediated proteolysis of the polyglutamine disease protein ataxin-3. J. Neurochem. 89 908–918. [DOI] [PubMed] [Google Scholar]
- Bonini, N. M., and M. E. Fortini, 2003. Human neurodegenerative disease modeling using Drosophila. Annu. Rev. Neurosci. 26 627–656. [DOI] [PubMed] [Google Scholar]
- Branco, J., I. Al-Ramahi, L. Ukani, A. M. Perez, P. Fernandez-Funez et al., 2008. Comparative analysis of genetic modifiers in Drosophila points to common and distinct mechanisms of pathogenesis among polyglutamine diseases. Hum. Mol. Genet. 17 376–390. [DOI] [PubMed] [Google Scholar]
- Carlson, K. M., J. M. Andresen and H. T. Orr, 2009. Emerging pathogenic pathways in the spinocerebellar ataxias. Curr. Opin. Genet. Dev. 19 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y., S. L. Koppenhafer, S. J. Shoesmith, M. K. Perez and H. L. Paulson, 1999. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 8 673–682. [DOI] [PubMed] [Google Scholar]
- Chan, H. Y., and N. M. Bonini, 2000. Drosophila models of human neurodegenerative disease. Cell Death Differ. 7 1075–1080. [DOI] [PubMed] [Google Scholar]
- Chan, H. Y., J. M. Warrick, G. L. Gray-Board, H. L. Paulson and N. M. Bonini, 2000. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 9 2811–2820. [DOI] [PubMed] [Google Scholar]
- Chan, H. Y., J. M. Warrick, I. Andriola, D. Merry and N. M. Bonini, 2002. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum. Mol. Genet. 11 2895–2904. [DOI] [PubMed] [Google Scholar]
- Chen, S., V. Berthelier, W. Yang and R. Wetzel, 2001. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 311 173–182. [DOI] [PubMed] [Google Scholar]
- Chen, S., F. A. Ferrone and R. Wetzel, 2002. Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. USA 99 11884–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, C. J., M. A. Mancini, B. Antalffy, D. B. DeFranco, H. T. Orr et al., 1998. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19 148–154. [DOI] [PubMed] [Google Scholar]
- Cummings, C. J., Y. Sun, P. Opal, B. Antalffy, R. Mestril et al., 2001. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10 1511–1518. [DOI] [PubMed] [Google Scholar]
- Dantuma, N. P., K. Lindsten, R. Glas, M. Jellne and M. G. Masucci, 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18 538–543. [DOI] [PubMed] [Google Scholar]
- Davies, S. W., M. Turmaine, B. A. Cozens, M. DiFiglia, A. H. Sharp et al., 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90 537–548. [DOI] [PubMed] [Google Scholar]
- Diaz-Hernandez, M., A. G. Valera, M. A. Moran, P. Gomez-Ramos, B. Alvarez-Castelao et al., 2006. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J. Neurochem. 98 1585–1596. [DOI] [PubMed] [Google Scholar]
- DiFiglia, M., E. Sapp, K. O. Chase, S. W. Davies, G. P. Bates et al., 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277 1990–1993. [DOI] [PubMed] [Google Scholar]
- Dunah, A. W., H. Jeong, A. Griffin, Y. M. Kim, D. G. Standaert et al., 2002. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296 2238–2243. [DOI] [PubMed] [Google Scholar]
- Ekengren, S., Y. Tryselius, M. S. Dushay, G. Liu, H. Steiner et al., 2001. A humoral stress response in Drosophila. Curr. Biol. 11 714–718. [DOI] [PubMed] [Google Scholar]
- Ellerby, L. M., R. L. Andrusiak, C. L. Wellington, A. S. Hackam, S. S. Propp et al., 1999. Cleavage of atrophin-1 at caspase site aspartic acid 109 modulates cytotoxicity. J. Biol. Chem. 274 8730–8736. [DOI] [PubMed] [Google Scholar]
- Everett, C. M., and N. W. Wood, 2004. Trinucleotide repeats and neurodegenerative disease. Brain 127 2385–2405. [DOI] [PubMed] [Google Scholar]
- Evert, B. O., U. Wullner and T. Klockgether, 2000. Cell death in polyglutamine diseases. Cell Tissue Res. 301 189–204. [DOI] [PubMed] [Google Scholar]
- Gatchel, J. R., and H. Y. Zoghbi, 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6 743–755. [DOI] [PubMed] [Google Scholar]
- Goldberg, Y. P., D. W. Nicholson, D. M. Rasper, M. A. Kalchman, H. B. Koide et al., 1996. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat. Genet. 13 442–449. [DOI] [PubMed] [Google Scholar]
- Gusella, J. F., and M. E. MacDonald, 2000. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat. Rev. Neurosci. 1 109–115. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., 2000. Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ. 7 1045–1056. [DOI] [PubMed] [Google Scholar]
- Hay, B. A., T. Wolff and G. M. Rubin, 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120 2121–2129. [DOI] [PubMed] [Google Scholar]
- Haynes, S. R., D. Johnson, G. Raychaudhuri and A. L. Beyer, 1991. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 19 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger, D., L. Tora and D. Devys, 2006. Transcriptional alterations and chromatin remodeling in polyglutamine diseases. Trends Genet. 22 562–570. [DOI] [PubMed] [Google Scholar]
- Iijima-Ando, K., P. Wu, E. A. Drier, K. Iijima and J. C. Yin, 2005. cAMP-response element-binding protein and heat-shock protein 70 additively suppress polyglutamine-mediated toxicity in Drosophila. Proc. Natl. Acad. Sci. USA 102 10261–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., and A. V. Tulin, 2009. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 37 3501–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., F. C. Nucifora, Jr., C. A. Ross and D. B. DeFranco, 2003. Cell death triggered by polyglutamine-expanded huntingtin in a neuronal cell line is associated with degradation of CREB-binding protein. Hum. Mol. Genet. 12 1–12. [DOI] [PubMed] [Google Scholar]
- Jolly, C., and S. C. Lakhotia, 2006. Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 34 5508–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement, I. A., P. J. Skinner, M. D. Kaytor, H. Yi, S. M. Hersch et al., 1998. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95 41–53. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., T. Jamal, A. Doetsch, F. R. Turner and J. B. Duffy, 2004. CREB binding protein functions during successive stages of eye development in Drosophila. Genetics 168 877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhotia, S. C., 2003. The noncoding developmentally active and stress inducible hsrω gene of Drosophila melanogaster integrates post-transcriptional processing of other nuclear transcripts, pp. 203–221 in Noncoding RNAs: Molecular Biology and Molecular Medicine, edited by J. Barciszewski and V. A. Erdmann. Kluwer Academic/Plenum Publishers, New York.
- Lakhotia, S. C., P. Ray, T. K. Rajendra and K. V. Prasanth, 1999. The non-coding transcripts of hsr-omega gene in Drosophila: Do they regulate trafficking and availability of nuclear RNA-processing factors? Curr. Sci. 77 553–563. [Google Scholar]
- Li, S. H., and X. J. Li, 2004. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 20 146–154. [DOI] [PubMed] [Google Scholar]
- Li, S. H., A. L. Cheng, H. Zhou, S. Lam, M. Rao et al., 2002. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell. Biol. 22 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja, T., D. Qi, M. Stabell and M. Mannervik, 2003. The CBP coactivator functions both upstream and downstream of Dpp/Screw signaling in the early Drosophila embryo. Dev. Biol. 262 294–302. [DOI] [PubMed] [Google Scholar]
- Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman et al., 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22 3832–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik, M., and S. C. Lakhotia, 2009. a RNAi for the large non-coding hsromega transcripts suppresses polyglutamine pathogenesis in Drosophila models. RNA Biol. 6 464–478. [DOI] [PubMed] [Google Scholar]
- Mallik, M., and S. C. Lakhotia, 2009. b The developmentally active and stress-inducible noncoding hsr{omega} gene is a novel regulator of apoptosis in Drosophila. Genetics 183 831–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne, K., D. Helmlinger, G. R. Perkin, D. Devys and Y. Trottier, 2003. Polyglutamine expansion induces a protein-damaging stress connecting heat shock protein 70 to the JNK pathway. J. Biol. Chem. 278 16957–16967. [DOI] [PubMed] [Google Scholar]
- Morfini, G., G. Pigino, G. Szebenyi, Y. You, S. Pollema et al., 2006. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat. Neurosci. 9 907–916. [DOI] [PubMed] [Google Scholar]
- Nelson, B., S. Nishimura, H. Kanuka, E. Kuranaga, M. Inoue et al., 2005. Isolation of gene sets affected specifically by polyglutamine expression: implication of the TOR signaling pathway in neurodegeneration. Cell Death Differ. 12 1115–1123. [DOI] [PubMed] [Google Scholar]
- Nucifora, Jr., F. C., M. Sasaki, M. F. Peters, H. Huang, J. K. Cooper et al., 2001. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291 2423–2428. [DOI] [PubMed] [Google Scholar]
- Ona, V. O., M. Li, J. P. Vonsattel, L. J. Andrews, S. Q. Khan et al., 1999. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington's disease. Nature 399 263–267. [DOI] [PubMed] [Google Scholar]
- Prasanth, K. V., T. K. Rajendra, A. K. Lal and S. C. Lakhotia, 2000. Omega speckles: a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 113 3485–3497. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux, C., J. P. Loeffler and A. L. Boutillier, 2004. Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem. Pharmacol. 68 1157–1164. [DOI] [PubMed] [Google Scholar]
- Saha, R. N., and K. Pahan, 2006. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 13 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, I., C. J. Xu, P. Juo, A. Kakizaka, J. Blenis et al., 1999. Caspase-8 is required for cell death induced by expanded polyglutamine repeats. Neuron 22 623–633. [DOI] [PubMed] [Google Scholar]
- Saudou, F., S. Finkbeiner, D. Devys and M. E. Greenberg, 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95 55–66. [DOI] [PubMed] [Google Scholar]
- Saumweber, H., P. Symmons, R. Kabisch, H. Will and F. Bonhoeffer, 1980. Monoclonal antibodies against chromosomal proteins of Drosophila melanogaster: establishment of antibody producing cell lines and partial characterization of corresponding antigens. Chromosoma 80 253–275. [DOI] [PubMed] [Google Scholar]
- Scappini, E., T. W. Koh, N. P. Martin and J. P. O'Bryan, 2007. Intersectin enhances huntingtin aggregation and neurodegeneration through activation of c-Jun-NH2-terminal kinase. Hum. Mol. Genet. 16 1862–1871. [DOI] [PubMed] [Google Scholar]
- Schaffar, G., P. Breuer, R. Boteva, C. Behrends, N. Tzvetkov et al., 2004. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15 95–105. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F., 1999. Dominant-negative mutation in the beta2 and beta6 proteasome subunit genes affect alternative cell fate decisions in the Drosophila sense organ lineage. Proc. Natl. Acad. Sci. USA 96 11382–11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, S., and S. C. Lakhotia, 2006. Altered expressions of the noncoding hsromega gene enhances poly-Q-induced neurotoxicity in Drosophila. RNA Biol. 3 28–35. [DOI] [PubMed] [Google Scholar]
- Shao, J., and M. I. Diamond, 2007. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 16(Spec. No. 2): R115–R123. [DOI] [PubMed] [Google Scholar]
- Sofola, O. A., P. Jin, Y. Qin, R. Duan, H. Liu et al., 2007. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan, J. S., L. Bodai, J. Pallos, M. Poelman, A. McCampbell et al., 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413 739–743. [DOI] [PubMed] [Google Scholar]
- Taylor, J. P., A. A. Taye, C. Campbell, P. Kazemi-Esfarjani, K. H. Fischbeck et al., 2003. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev. 17 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman, P., R. Wetzel, M. Tanaka, N. Nukina and A. L. Goldberg, 2004. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell 14 95–104. [DOI] [PubMed] [Google Scholar]
- Waelter, S., A. Boeddrich, R. Lurz, E. Scherzinger, G. Lueder et al., 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., C. E. Wang, A. Orr, S. Tydlacka, S. H. Li et al., 2008. Impaired ubiquitin-proteasome system activity in the synapses of Huntington's disease mice. J. Cell Biol. 180 1177–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick, J. M., H. L. Paulson, G. L. Gray-Board, Q. T. Bui, K. H. Fischbeck et al., 1998. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93 939–949. [DOI] [PubMed] [Google Scholar]
- Warrick, J. M., H. Y. Chan, G. L. Gray-Board, Y. Chai, H. L. Paulson et al., 1999. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23 425–428. [DOI] [PubMed] [Google Scholar]
- Wellington, C. L., L. M. Ellerby, A. S. Hackam, R. L. Margolis, M. A. Trifiro et al., 1998. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J. Biol. Chem. 273 9158–9167. [DOI] [PubMed] [Google Scholar]
- Wellington, C. L., L. M. Ellerby, C. A. Gutekunst, D. Rogers, S. Warby et al., 2002. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J. Neurosci. 22 7862–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., J. R. Dunlap, R. B. Andrews and R. Wetzel, 2002. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum. Mol. Genet. 11 2905–2917. [DOI] [PubMed] [Google Scholar]
- Yoo, S. Y., M. E. Pennesi, E. J. Weeber, B. Xu, R. Atkinson et al., 2003. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron 37 383–401. [DOI] [PubMed] [Google Scholar]
- Zu, K., M. L. Sikes, S. R. Haynes and A. L. Beyer, 1996. Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol. Biol. Cell 7 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]