Abstract
Tissue engineering for in vitro drug-screening applications based on tissue function is an active area of translational research. Compared to targeted high-throughput drug-screening methods that rapidly analyze hundreds of thousands of compounds affecting a single biochemical reaction or gene expression, high-content screening (HCS) with engineered tissues is more complex and based on the cumulative positive and negative effects of a compound on the multiple pathways altering tissue function. It may therefore serve as better predictor of in vivo activity and serve as a bridge between high-throughput drug screening and in vivo animal studies. In the case of the musculoskeletal system, tissue function includes determining improvements in the mechanical properties of bone, tendon, cartilage, and, for skeletal muscle, contractile properties such as rate of contraction/relaxation, force generation, fatigability, and recovery from fatigue. HCS of compound banks with engineered tissues requires miniature musculoskeletal organs as well as automated functional testing. The resulting technologies should be rapid, cost effective, and reduce the number of small animals required for follow-on in vivo studies. Identification of compounds that improve the repair/regeneration of damaged tissues in vivo would have extensive clinical applications for treating musculoskeletal disorders.
Introduction
Tissue engineering of functional musculoskeletal organs as a tool for in vitro drug-screening applications has been envisioned since the field's inception1,2 and may contribute to the identification of new drugs for the treatment of numerous clinical disorders.3,4 High-throughput drug-screening (HTS) technologies use biochemical, gene expression, or single-cell assays, and with today's computerized robotic technologies, they are capable of screening millions of compounds in a rapid and cost-effective manner.5 Nevertheless, it has proven difficult to predict from these highly targeted HTS assays the ultimate functional activity of a compound in vivo since all compounds have effects on multiple intracellular second messenger pathways, known as a compound's side activities.6–8 This type of targeted pathway approach also has the limitation of possibly missing important unknown pathways that improve functional characteristics of the tissue. This assay method also misses potential negative effects a compound may have on tissue function.
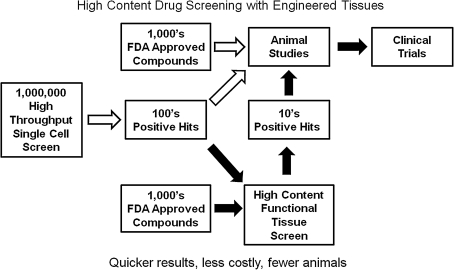
HTS assays often identify hundreds of positive hits, and it is difficult to know which should be carried forward into animal studies. High-content drug screening (HCS) with engineered tissues is based on physiological function and is the culmination of all the complex interactions of compounds on multiple intracellular pathways; it therefore may be a better predictor of a compound's ultimate in vivo effect at the tissue level (but not necessarily at the whole animal level). Using engineered tissue HCS as a secondary screen on the positive hits obtained from HTS assays may therefore save significant time, expense, and animals in moving a compound forward toward the clinic (Fig. 1). In addition, screening small compound banks of existing Food and Drug Administration–approved drugs (e.g., National Institutes of Health Clinical Collection, www.nihclinicalcollection.com) with engineered tissues for new indications, that is, drug repurposing,9,10 might identify compounds with established human safety and pharmacokinetic data that could cost effectively move rapidly into clinical trials for musculoskeletal disorders. For example, in Duchenne muscular dystrophy (DMD), a genetic disease affecting the skeletal muscle cells' cytoskeleton,11 an antibiotic (gentamicin)12 and an antihypertensive drug (Losartan)13 may be clinically useful in maintaining muscle strength in this devastating and fatal disease. Other examples include the depression drug buproprion now approved for smoking cessation, the depression drug duloxetine for urinary incontinence, and thalidomide, originally developed for morning sickness during pregnancy but now used for multiple myeloma treatment.14 Engineered tissues will be more difficult to establish for HCS applications than HTS assays because of the complexity of the engineering process, the physiological assay methods, and cost per compound screen, but it should help in identifying new therapeutics in the future. While more costly and difficult to establish, functional tissue screening has the additional advantage over single-cell assays in that the resulting data are the average signal of a large population of cells; many times single-cell assays require analysis of 100–1000 individual cells to obtain a significant result. This brief review will cover several recent advances in the use of engineered musculoskeletal tissues for drug-screening applications and suggested future directions for the field. Recent articles cover other HCS technologies for liver,15,16 cardiac tissue,17 tumors,18 and cell–biomaterial interactions.19–21
FIG. 1.
High-throughput drug screening versus high-content drug screening of bioactive compounds.
Engineering Bone, Cartilage, Ligaments, and Intervertebral Discs for Functional Drug Screening
A recent conference report on the commercial development of tissue engineering for the repair/regeneration in the musculoskeletal field enumerated many of the mechanical properties of musculoskeletal tissues necessary for successful tissue repair.22 For the tissues covered by the conference (anterior cruciate ligament, articular cartilage, long bones, intervertebral disc, rotator cuff, meniscus, and temporomandibular joint) mechanical testing for stress–strain, compression, viscoelastic, and shear stress properties were identified as paramount to successful in vivo applications. Numerous methods currently exist for the in vitro engineering of tendons, ligaments, and bones (reviewed in Ref.23). To develop useful secondary functional screening with these tissues for the analysis of hundreds of compounds, engineered tissue HCS technologies will first require a reliable source of primary cells, banked cells, stem cells, or immortalized cell lines. Immortalized cells are the preferred cell source for HCS since they are more cost effective than performing primary preparations from an animal source and theoretically would reduce variability between experiments. Immortalized cells have been described for chondrocytes,24 bone cells,25,26 and ligament cell lines.27 The disadvantage of immortalized cells (and stem cells) is the possibility of their dedifferentiation, or change of specific tissue function.
A second requirement is the adaption of computerized robotic liquid-handling HTS hardware to engineer musculoskeletal tissues. This automation has revolutionized HTS, allowing the rapid and reproducible screening of large compound banks. Automation will be important for several aspects of HCS: engineering the tissues themselves in a 96-well (7–8-mm-diameter wells) or smaller microwell plate format in a manner compatible with automated tissue culture medium changes, compound additions, and functional testing. Many macro tissue engineering protocols have been described over the past decade for cartilage, bone, and tendon, but many of these utilize a preformed rigid scaffold that would make mechanical testing of the engineered tissues difficult. Engineering in flexible extracellular gel matrices such as collagen or fibrin28 or in a scaffold-free manner29 would be more compatible with subsequent mechanical testing of the engineered tissues. Micromass cultures of mesenchymal limb bud cells have been developed using no extracellular matrix scaffolding for studying chondrocyte differentiation30 in a 24-well format, but this procedure requires a high number of primary cells (250,000/micromass) for hypertrophic chondrogenesis to occur. To be cost effective for HCS, the fewer the cells/tissue the better, but there will also be a lower limit to the number of cells/tissue required for functional mechanical testing, depending on the sensitivity of the screening assays. Another approach has been taken by Huang et al.31 using mesenchymal stem cells (MSCs) to study compound banks that modulate stem cell chondrogenesis in micro-MSC pellets. The authors have utilized 96- and 384-well plates to form pellets of MSC with as few as 10,000 cells/pellet. Targeted assays rather than functional assays were used on the pelleted constructs (e.g., glycosaminoglycan production, and collagen type II and aggrecan mRNA expression) but may be adaptable to functional HCS in the future.
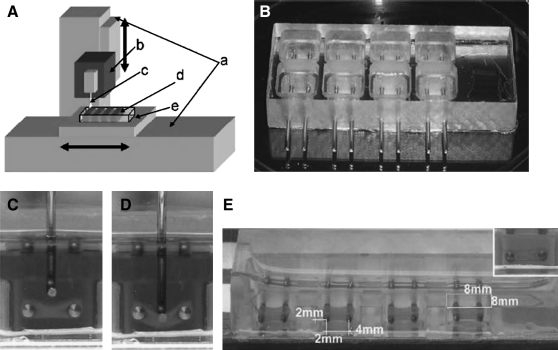
Once engineered, automated (most likely customized) hardware and analysis software will be required for mechanical testing of the miniature constructs. Numerous genetic and biochemical assays exist for assessing the differentiation of bone, cartilage, ligaments, and intervetebral discs that are amenable to compound screening (e.g., Refs.31–33), but this review will be limited to functional assays. Mechanical testing of large tissue constructs has existed for many years,34,35 and recently, a prototype miniaturized robotic mechanical testing device (MTD) was described that fits many of the criteria for HCS.36 Miniature hydrogel tissue constructs were engineered around two fixed posts in 8 × 8 mm wells in a similar manner to cardiac tissue constructs engineered by Eschenhagen's group.17 A customized automated force measurement device was built to deform the hydrogel tissue constructs in a reproducible manner and characterize their mechanical properties (Fig. 2). Construct pretension forces and elastic stress–strain characteristics could be rapidly measured sterilely, allowing repetitive long-term studies. Validation of the ability to measure changes in mechanical properties of the constructs was successfully performed after incubation with several compounds known to influence the mechanical properties of tissues. While the tissues used in this study were engineered from fibroblasts, it is likely that the technology can be readily adapted to engineered cartilage, bone, or ligament cells and used to screen for compounds that improve their mechanical properties. The MTD utilized electronic force transducers with background noise of <100 μN and accurately measured forces up to 10 μN. Sensitivity of the automated mechanical testing equipment will need to be adjusted to the force generated by the miniature organs and will depend on the number of cells/construct and the cell/extracellular matrix ratio. In the MTD study, the hydrogel fibroblast constructs contained approximately 200,000 cells/construct, which translates into nearly 20 million cells per 96-well plate. The cost of cell preparation and growth medium is therefore quite high compared with single-cell screening assays for even a modestly sized compound bank. As more sensitive electronic force transducers are developed that are capable of measuring smaller forces, developing tissue engineering procedures for constructs with fewer cells will be useful in this and other automated functional screening designs for identifying new treatments for tendon, ligament, and bone repair in a more cost-effective manner.
FIG. 2.
Hydrogel tissue construct (HTC) mechanics. (A) Schematic of the force measurement device. a, Linear actuator moved by a servo motor (±70 μm accuracy); b, isometric force transducer; c, force probe; d, tissue chamber; e, warming plate (37°C) connected to a water circulation bath. The device can automatically indent four samples in one row, and the operator manually translates the mold to the next row. A computer records the force response measured by the isometric transducer and regulates the speed of indentation. (B) Tissue chamber for producing and testing engineered tissues. (C) A force probe approaching an HTC formed between two stainless steel bars. (D) The probe indents the HTC vertically and stretches it longitudinally. (E) The typical size of HTCs was approximately 4 × 4 × 0.8 mm (length × width × thickness), and they are formed in 8 × 8 mm (opening) square wells in the tissue chamber. Reprinted with permission from Ref.36
Skeletal Muscle Tissue Engineering for Functional Drug Screening
Contractile skeletal muscle tissue engineered from precursor muscle cells (myoblasts) has a number of potential applications, including tissue repair,37 therapeutic protein delivery,38 mechanotransduction mechanistic studies,39 and in vitro drug screening.1 Many issues still need to be addressed for the in vivo repair applications, including efficient transfer of the contractile force from the engineered tissue through appropriate elastic scaffolds to bone to generate host movement. Improvements in in vivo cell survival after engineering in an artificial ex vivo environment will also be required.40 In vitro studies on mechanotransduction processes in skeletal muscle are well advanced41 since the first report that such studies could be performed in cell culture.42 These in vitro studies have contributed significantly to our understanding of how skeletal muscle senses mechanical forces and the multitude of second messenger pathways that translate those signals into biochemical changes leading to hyperplasia, hypertrophy, and metabolic alterations.43
Skeletal muscle, unlike cardiac muscle, has a readily available source of stem cells from both embryonic and adult tissue. In adults, these have traditionally been identified as satellite cells, located between the muscle fiber's plasma membrane and its basement membrane. While other sources of MSCs capable of regenerating skeletal muscle have been identified,44 the satellite cell alone is capable of regenerating new muscle tissue once activated to proliferate as myoblasts and self-renew as new quiescent satellite cells.45 Tissue culture conditions have been developed over the years to expand the proliferating myoblasts from rodent and human muscle to the large numbers required for HCS applications, and to differentiate them into contractile postmitotic muscle fibers. Myogenic cell lines suitable for HCS applications also exist from both rodents46 and humans.47
Tissue-engineered muscle organs offer a physiologic approach to HCS, providing a holistic approach for determining a compound's effect on the multiple pathways regulating important physiological parameters such as muscle strength, fatigability, and contraction-induced injury in both normal and diseased phenotypes. Existing two-dimensional monolayer drug-screening assays for skeletal muscle strength–inducing compounds are based on single targeted pathways such as elevated expression of a single gene like myosin heavy chain,46 or proteins such as muscle-specific insulin-like growth factor-1, α 7 integrin,48 or the dystrophin-like protein utrophin.49 Increased myofiber size (hypertrophy) is also a common two-dimensional drug-screening indicator of compounds that stimulate muscle growth.50–52 Other approaches focus on a compound's effects on muscle differentiation, for example, the fusion of myoblasts into new myofibers.53 Nevertheless, despite the development of these two-dimensional in vitro screening assays, the preferred methods are still in vivo models.54–56 This is not surprising as skeletal muscle function results from the complex interaction of multiple second-messenger pathways and structural proteins; even myofiber size is a poor indicator of muscle strength.57 Thus, the effect of a compound on overall muscle function may be missed by screening for effects on a single genetic pathway, protein target, or histological analyses. The search for drugs targeting muscle function in vitro will benefit from functional analyses and could serve as an important bridge between current targeted assays and follow-on animal studies.
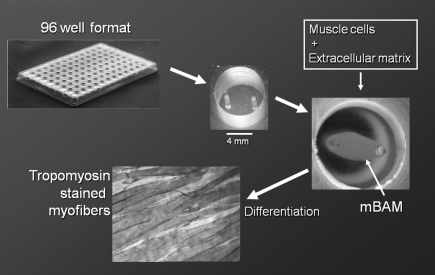
Tissue engineering of skeletal muscle from rodent and human muscle precursor cells is well advanced. Numerous methods have been developed to engineer three-dimensional skeletal muscle in vitro capable of generating contractile forces in a directed manner (reviewed in Refs.58,59). One of the simplest used in our laboratory60 is based on the original observation of Eugene Bell that most cells will align parallel to the passive tensions developed in a collagen gel when it coalesces.61 Mixing proliferating myoblasts with extracellular matrix solutions such as collagen or fibrin and casting in a well with attachment posts results in a tubular structure attached to the two posts as the collagen coalesces away from the well sides (Fig. 3). The significant passive forces within the matrix cause the myoblasts to align in the long axis of the construct. When the tissue culture medium is switched to differentiation medium, the myoblasts fuse into muscle fibers aligned with the long axis of the construct, and these miniature bioartificial muscles (mBAMs) generate directed forces when electrically stimulated. These contractile muscle fibers can be maintained under tension for weeks in vitro,58,62 and the forces generated by engineered contractile tissues can be measured using electronic force transducers.62–65 Current transducers, however, are difficult to scale down to reproducibly measure the small forces generated by miniature contractile tissues necessary for microwell plate formats used in HCS assays. Newer non-transducer-based methods to measure cellular forces on a smaller scale have been developed.66–69 These methods, based on optical imaging of cell contraction, were adapted to HCS applications for skeletal70 and cardiac muscles71 as described next.
FIG. 3.
Tissue engineering miniature bioartificial muscles (mBAMs) on μposts in a 96-microwell plate. Posts are molded from flexible polydimethylsiloxane 4 mm apart and 7 mm high in 7-mm-diameter wells. mBAM shown is day 4–5 postcasting in the 7-mm-diameter well as described in the text. A 7–8-day postcasting mBAM was stained for sarcomeric tropomyosin and shows well-aligned myofibers.
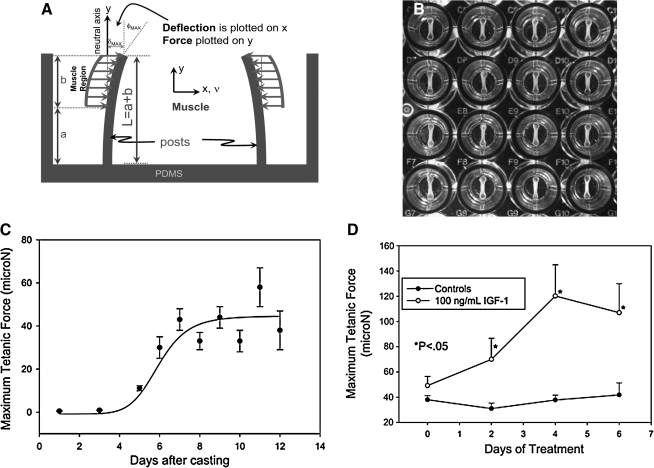
mBAMs were tissue engineered around flexible μposts (Fig. 3) cast from elastic polydimethylsiloxane (PDMS) in a 96-microwell format.70 In this system, measurement of force is based on the degree of flexion of the attachment posts captured by motion detection imaging. Medical-grade PDMS is nontoxic to skeletal muscle cells38 and commonly used for in vitro tissue engineering and in vivo reconstructive surgical applications; it shows minor hysteresis (<5%) and creep when used in force-sensing applications.72 For a μpost of radius R and length L (Fig. 4A), cast from PDMS material with a known elastic modulus (E), the moment of inertia (I) is given by I = ¼πR4 and force based on μpost deflection δ by the formula73:
FIG. 4.
Force measurement with flexible μposts. (A) Mechanical model correlating μpost displacement (δ) with force (F). (B) Section of 96-microwell plate with mBAMs attached to flexible posts. (C) Time course for active force development. mBAMs were electrically stimulated every 2–3 days, μpost deflection was measured optically, and active force generation was calculated. Results are means + SE (standard error) from four separate experiments with n = 8–18 per time point. Error bars are smaller than symbols where not seen. (D) Anabolic effect of insulin-like growth factor-1 (IGF-1) on active force. mBAMs were allowed to differentiate for 6–7 days postcasting and then insulin-like growth factor-1 (100 ng/mL) added to the wells. Each point is the mean + SE; n = 6 mBAMs per group. Adapted from Refs.70,75
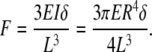 |
Since all of the parameters in the equation are known, it can be used to fit the force–displacement curves using E as the only fitting parameter. A software algorithm written in MatLab was developed to automatically determine the maximal μpost deflections in images captured with a CMOS camera and converts the μm of movement to μN of force. Forces can be calculated with an accuracy of 5–6 μN, and both passive and active electrically stimulated forces accurately measured.70 Recently, a similar approach was reported that significantly reduces the size of the wells, engineered tissue constructs, and sensitivity of the force measurements down to the nN range.74
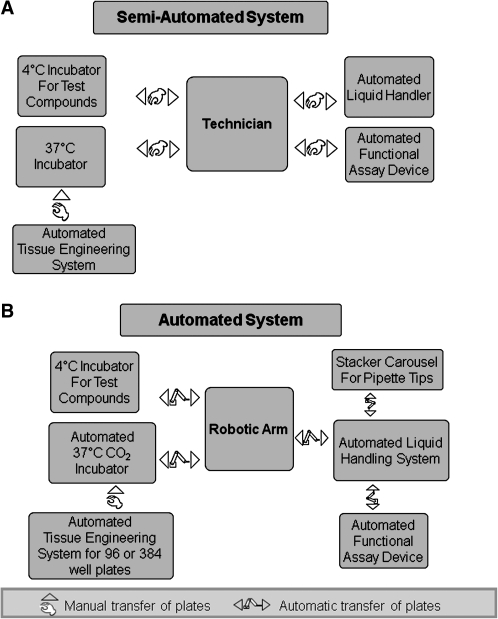
A 96-microwell plate format was designed (Fig. 4B) for tissue engineering mBAMs that enables the use of standard robotic liquid-handling systems.75 The liquid handler mixes a cell suspension with soluble extracellular matrix in each well, a process that takes approximately 7–8 min per 96-well plate. Each construct is engineered with 50,000–100,000 cells/mBAM and, after differentiation, contains several hundred aligned striated muscle fibers. When electrically stimulated, they bend the μposts (Supplemental Video S1, available online at www.liebertonline.com), and the forces generated are then calculated from the captured images. A customized Myoforce Analysis Device (MAD™) was designed and built, which automatically places electrodes into each well, electrically stimulates the mBAMs, and captures images of the flexing μposts while maintaining sterility.75 MAD is encased in an insulated box so that temperature, CO2 level, and humidity can be maintained in a similar manner to a standard tissue culture incubator. Once loaded onto the MAD on a heated stage, the device automatically transports the microwell plate to the stimulating electrodes and electrically stimulates each well for 1–2 s with preselected parameters for tetanic force generation, and 40–60 images of the μpost deflections are captured. A 96-well plate requires just under 20 min to stimulate and collect the images. After electrical stimulation, the plates are transferred to the liquid handler, the tissue culture medium changed, and the plates returned to a humidified 5% CO2 incubator for 24 h before restimulating in MAD. This mBAM HCS technology, aimed at identifying compounds that alter muscle strength, is termed MyoForce Analysis System (MFAS™), and is the first HCS technology available for assaying skeletal muscle function in vitro. It is currently a semiautomated rather than fully automated procedure, requiring technical assistance to move the assay plates between the various pieces of hardware (Fig. 5). Because of the complexity of functional HCS with contractile tissues, fully automated functional screening may not be worth the extra costs associated with complete automation, especially if hundreds rather than tens of thousands of compounds are tested.
FIG. 5.
(A) Semiautomated versus. (B) automated high-content screening with engineered tissues.
mBAMs generate increasing levels of active force when electrically stimulated as the myoblasts differentiate into organized postmitotic muscle fibers and begin expressing sarcomeric proteins. mBAM contractions generally begin 5–6 days postcasting, that is, 3–4 days after switching to differentiation medium (Fig. 4C). Maximal active tetanic force reaches a plateau after 7–9 days postcasting and can be maintained at this level for at least another 5–7 days. Screening of compounds that affect muscle fiber strength is initiated once this plateau had been reached. To validate the assay, the effect of a well-established skeletal muscle anabolic factor (insulin-like growth factor-1)76 was tested. Active mBAM force generation was significantly increased by 2 days after addition of insulin-like growth factor-1 to the tissue culture medium and continued to increase for the next 2–4 days to levels two- to threefold greater than control mBAMs (Fig. 4D). Identifying components that have detrimental effects on the musculoskeletal system is also an important role for MFAS, and the effect of a skeletal muscle catabolic factor was therefore tested. Dose-dependent skeletal muscle weakness is a main side effect of cholesterol lowering statins, affecting 7% of patients on statin monotherapy, and causing serious myositis and rhabdomyolysis in 0.2–0.3% of the patient population.77,78 When the statin atorvastatin was added to the mBAMs there was a concentration- and time-dependent decrease in active force generation,70 even at levels as low as 0.01 μM, a level close to the reported circulating plasma levels for the drug. MFAS thus appears to be a reliable HCS technology for assaying compounds that affect skeletal muscle strength and identifying new potential treatments for muscle weakness resulting from various disorders such as cancer cachexia, aging, and various neuromuscular diseases.
A significant advantage of drug screening with engineered tissues is the ability to utilize cells from either humans with a neuromuscular disorder affecting muscle function or equivalent rodent models of the diseases. DMD is a progressive, lethal, muscle-wasting disease resulting from a defect in the dystrophin gene.79 A genetic murine homolog of DMD exists (mdx), and the myoblasts from the mdx mouse have been conditionally immortalized,80 providing an ideal cell source to screen for compounds that might attenuate the progressive muscle weakness associated with the disease. While not a cure, identification of factors that improve muscle function in boys with DMD could lead to an improved quality of life. mdx mBAMs were tissue engineered from mdx myoblasts and generated active forces comparable to control murine mBAMs. Thirty-one compounds of interest as potential treatments for patients with DMD were tested with MFAS at three to six concentrations.75 Eleven of the compounds significantly increased mdx mBAM tetanic force relative to placebo-treated controls. The assay also identified beneficial compound interactions as well as deleterious interactions of combinatorial therapies taken by some DMD patients. In vitro mdx muscle and DMD patients thus responded in a similar manner to many of these compounds, and the in vitro assay will be a useful tool for the rapid identification of new potential treatments for muscle weakness in DMD and other muscle disorders. More recently, an immortalized human myoblast from a DMD patient has been described47 and may eventually serve as a useful screen directly on diseased human muscle tissue.
Limitations of In Vitro Functional Screening with Engineered Musculoskeletal Tissues
The pathophysiology of most musculoskeletal disorders is complicated, involving a multitude of organs and tissues in the body. For example, the loss of the dystrophin protein in boys with DMD can impact not only the muscle fibers directly, but also the supporting connective tissue, its vasculature, and innervation.11 Compound screening with tissue-engineered muscle constructs therefore has significant limitations as to its overall relevance in predicting a drug's effect in vivo. In mBAMs the muscle fibers are noninnervated and not fully differentiated, expressing a mixture of embryonic, neonatal, and adult myosin isoforms.70 The striated myofibers in the mBAMs are thus of a more neonatal than adult fiber type and may prove to be a better predictor of compounds for use in younger DMD patients who may have different therapeutic requirements than older patients.81 The surrounding extracellular environment in mBAMs is also quite different from in vivo muscle, with a lack of blood vessels and complex extracellular matrix components. Beneficial compounds acting as antifibrotic agents, through the vasculature, or on the inflammatory response will thus be missed in the in vitro mBAM assay. Amino acid supplements may be effective in vivo but ineffective in vitro since the tissue culture medium has been optimized previously for cell growth. Other compounds such as those aimed at minimizing muscle contraction–induced membrane damage will also be missed with the active tetanic force assay. The mBAM-screening technology is currently being expanded to measure other physiological parameters such as muscle contraction rate, relaxation rate, muscle fatigue/damage, and recovery from fatigue/damage.
Future Directions
Drug screening with engineered tissues is still in its early stages of development. Microfluidic bioreactors,82,21 microfluidic scaffolds,83 and microfluidic patterning4 may all contribute to a more in vivo–like environment, especially for pharmacokinetic studies. Applied repetitive mechanical loading on three-dimensional tissue effects tissue function62,84 and will better reflect the complex interactions of drugs on tissue mechanics. The screening of new potential drug therapies directly against a specific disease along with pharmacogenetics, that is, individual patient tissue screens,85 is also an exciting future direction for the field. Skeletal muscle stem cells are easily obtained by a simple 10-min outpatient biopsy procedure38 and expandable to the numbers required for limited screening for optimal individual response to drug treatments. Physiologic-based HCS technologies assaying skeletal muscle function with invertebrate and vertebrate models are under development [Caenorhabditis elegans86,87 and zebrafish88–90], and these assays may detect compound effects on organ systems not present in the tissue engineered constructs. Finally, induced pluripotent stem (iPS) cells,91 and embryonic stem cells92,93 are potential future sources of cells for engineered tissue drug-screening applications.
Footnotes
Based on a presentation given at the TERMIS Workshop on “Translational Models for Musculoskeletal Tissue Engineering and Regenerative Repair“ on December 7, 2008, in San Diego, California.
Acknowledgments
The author thanks David Mooney and Janet Shansky for critically reading the manuscript. Supported by the National Institutes of Health (AG029705, NS059098, and HL HL093939) and the National Science Foundation (Award ID 0724445).
Disclaimer
Any opinions, findings, and conclusions or recommendations expressed in this article are those of the author and not necessarily those of the granting agencies.
Disclosure Statement
The author declares competing financial interests: Dr. Vandenburgh owns stock in Myomics, Inc.
References
- 1.Vandenburgh H.H. Swasdison S. Karlisch P. Computer aided mechanogenesis of skeletal muscle organs from single cells in vitro. FASEB J. 1991;5:2860. doi: 10.1096/fasebj.5.13.1916108. [DOI] [PubMed] [Google Scholar]
- 2.Langer R. Vacanti J.P. Tissue engineering. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 3.Griffith L.G. Naughton G. Tissue engineering—current challenges and expanding opportunities. Science. 2002;295:1009. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 4.Khademhosseini A. Langer R. Borenstein J. Vacanti J.P. Tissue engineering special feature: microscale technologies for tissue engineering and biology. Proc Natl Acad Sci USA. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terstappen G.C. Schlupen C. Raggiaschi R. Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 6.Dove A. Screening for content—the evolution of high throughput. Nat Biotech. 2003;21:859. doi: 10.1038/nbt0803-859. [DOI] [PubMed] [Google Scholar]
- 7.Wermuth C.G. Selective optimization of side activities: another way for drug discovery. J Med Chem. 2004;47:1303. doi: 10.1021/jm030480f. [DOI] [PubMed] [Google Scholar]
- 8.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov Today. 2005;10:139. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 9.O'Connor K.A. Roth B.L. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat Rev Drug Discov. 2005;4:1005. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- 10.Ashburn T.T. Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain J.S. Rando T.A. Advances in Therapeutics. Informa Healthcare; New York, NY: 2006. Duchenne muscular dystrophy. [Google Scholar]
- 12.Barton-Davis E.R. Cordier L. Shoturma D.I. Leland S.E. Sweeney H.L. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohn R.D. van Erp C. Habashi J.P. Soleimani A.A. Klein E.C. Lisi M.T. Gamradt M. Ap Rhys C.M. Holm T.M. Loeys B.L. Ramirez F. Judge D.P. Ward C.W. Dietz H.C. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat Med. 2007;13:204. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boguski M.S. Mandl K.D. Sukhatme V.P. Repurposing with a difference. Science. 2009;324:1394. doi: 10.1126/science.1169920. [DOI] [PubMed] [Google Scholar]
- 15.Sivaraman A. Leach J.K. Townsend S. Iida T. Hogan B.J. Stolz D.B. Fry R. Samson L.D. Tannenbaum S.R. Griffith L.G. A microscale in vitro physiological model of the liver: predictive screens for drug metabolism and enzyme induction. Curr Drug Metab. 2005;6:569. doi: 10.2174/138920005774832632. [DOI] [PubMed] [Google Scholar]
- 16.Khetani S.R. Bhatia S.N. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 17.Eschenhagen T. Zimmermann W.H. Engineering myocardial tissue. Circ Res. 2005;97:1220. doi: 10.1161/01.RES.0000196562.73231.7d. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach C. Chen R. Matsumoto T. Schmelzle T. Brugge J.S. Polverini P.J. Mooney D.J. Engineering tumors with 3D scaffolds. Nat Meth. 2007;4:855. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 19.Yliperttula M. Chung B.G. Navaladi A. Manbachi A. Urtti A. High-throughput screening of cell responses to biomaterials. Eur J Pharm Sci. 2008;35:151. doi: 10.1016/j.ejps.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Jongpaiboonkit L. King W.J. Murphy W.L. Screening for 3D environments that support human mesenchymal stem cell viability using hydrogel arrays. Tissue Eng Part A. 2009;15:343. doi: 10.1089/ten.tea.2008.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters A. Brey D.M. Burdick J.A. High-throughput and combinatorial technologies for tissue engineering applications. Tissue Eng Part B. 2009;15:225. doi: 10.1089/ten.TEB.2009.0049. [DOI] [PubMed] [Google Scholar]
- 22.Evaluation criteria for musculoskeletal and craniofacial tissue engineering constructs: a conference report. Tissue Eng Part A. 2008;14:2089. doi: 10.1089/ten.tea.2007.0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikos A.G. Herring S.W. Ochareon P. Elisseeff J. Lu H.H. Kandel R. Schoen F.J. Toner M. Mooney D. Atala A. Van Dyke M.E. Kaplan D. Vunjak-Novakovic G. Engineering complex tissues. Tissue Eng. 2006;12:3307. doi: 10.1089/ten.2006.12.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu S.H. Tsai C.L. Tang C.M. Evaluation of cellular affinity and compatibility to biodegradable polyesters and Type-II collagen-modified scaffolds using immortalized rat chondrocytes. Artif Organs. 2002;26:647. doi: 10.1046/j.1525-1594.2002.06889.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang J. Cao C. Wang W. Tong X. Shi D. Wu F. Zheng Q. Guo C. Pan Z. Gao C. Wang J. Proliferation and osteogenesis of immortalized bone marrow-derived mesenchymal stem cells in porous polylactic glycolic acid scaffolds under perfusion culture. J Biomed Mater Res A. [Epub ahead of print]. 2009 doi: 10.1002/jbm.a.32378. [DOI] [PubMed] [Google Scholar]
- 26.Stringer B. Waddington R. Sloan A. Phillips I. Telford G. Hughes D. Craig G. Gangemi L. Brook I. Freeman C. Cao X. Gosal M. Smith S. Russell G. Foster G. Bespoke human hypertrophic chondrocytic cell lines provide the osteoinductive signals required for vascularized bone formation. Tissue Eng. 2007;13:133. doi: 10.1089/ten.2006.0111. [DOI] [PubMed] [Google Scholar]
- 27.Kubota M. Chiba M. Obinata M. Ueda S. Mitani H. Establishment of periodontal ligament cell lines from temperature-sensitive Simian virus 40 large T-antigen transgenic rats. Cytotechnology. 2004;44:55. doi: 10.1023/B:CYTO.0000043412.08814.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler D.L. Juncosa-Melvin N. Boivin G.P. Galloway M.T. Shearn J.T. Gooch C. Awad H. Functional tissue engineering for tendon repair: a multidisciplinary strategy using mesenchymal stem cells, bioscaffolds, and mechanical stimulation. J Ortho Res. 2008;26:1. doi: 10.1002/jor.20456. [DOI] [PubMed] [Google Scholar]
- 29.Napolitano A.P. Dean D.M. Man A.J. Youssef J. Ho D.N. Rago A.P. Lech M.P. Morgan J.R. Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. BioTechniques. 2007;43:494. doi: 10.2144/000112591. [DOI] [PubMed] [Google Scholar]
- 30.Stanton L. Sabari S. Sampaio A.V. Underhill T.M. Beier F. p38 MAP kinase signalling is required for hypertrophic chondrocyte differentiation. Biochem J. 2004;378:53. doi: 10.1042/BJ20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang A. Motlekar N. Stein A. Diamond S. Shore E. Mauck R. High-throughput screening for modulators of mesenchymal stem cell chondrogenesis. Ann Biomed Eng. 2008;36:1909. doi: 10.1007/s10439-008-9562-4. [DOI] [PubMed] [Google Scholar]
- 32.Vunjak-Novakovic G. Altman G. Horan R. Kaplan D.L. Tissue engineering of ligaments. Annu Rev Biomed Eng. 2004;6:131. doi: 10.1146/annurev.bioeng.6.040803.140037. [DOI] [PubMed] [Google Scholar]
- 33.Frohlich M. Grayson W.L. Wan L.Q. Marolt D. Drobnic M. Vunjak-Novakovic G. Tissue engineered bone grafts: biological requirements, tissue culture and clinical relevance. Curr Stem Cell Res Ther. 2009;3:254. doi: 10.2174/157488808786733962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buma P. Schreurs W. Verdonschot N. Skeletal tissue engineering—from in vitro studies to large animal models. Biomaterials. 2004;25:1487. doi: 10.1016/s0142-9612(03)00492-7. [DOI] [PubMed] [Google Scholar]
- 35.Petrigliano F.A. McAllister D.R. Wu B.M. Tissue engineering for anterior cruciate ligament reconstruction: a review of current strategies. Arthroscopy. 2006;22:441. doi: 10.1016/j.arthro.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 36.Marquez J.P. Legant W. Lam V. Cayemberg A. Elson E. Wakatsuki T. High-throughput measurements of hydrogel tissue construct mechanics. Tissue Eng Part C Methods. 2009;15:181. doi: 10.1089/ten.tec.2008.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill E. Boontheekul T. Mooney D.J. Regulating activation of transplanted cells controls tissue regeneration. Proc Natl Acad Sci USA. 2006;103:2494. doi: 10.1073/pnas.0506004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shansky J. Ferland P. McGuire S. Powell C. Del Tatto M. Nachman M. Hennessey J. Vandenburgh H. Tissue engineering human skeletal muscle for clinical applications. In: Vunjak G., editor; Freshney I., editor. Culture of Cells for Tissue Engineering. Hoboken NJ: Wiley; 2006. pp. 239–257. [Google Scholar]
- 39.Vandenburgh H. Chromiak J. Shansky J. DelTatto M. LeMaire J. Space travel directly induces skeletal muscle atrophy. FASEB J. 1999;13:1031. doi: 10.1096/fasebj.13.9.1031. [DOI] [PubMed] [Google Scholar]
- 40.Mooney D.J. Vandenburgh H. Cell delivery mechanisms for tissue repair. Cell Stem Cell. 2008;2:205. doi: 10.1016/j.stem.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Bader D.L. Knight M.M. Biomechanical analysis of structural deformation in living cells. Med Biol Eng Comput. 2008;46:951. doi: 10.1007/s11517-008-0381-4. [DOI] [PubMed] [Google Scholar]
- 42.Vandenburgh H.H. Kaufman S. In vitro model for stretch-induced hypertrophy of skeletal muscle. Science. 1979;203:265. doi: 10.1126/science.569901. [DOI] [PubMed] [Google Scholar]
- 43.Burkholder T.J. Mechanotransduction in skeletal muscle. Front Biosci. 2007;12:174. doi: 10.2741/2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peault B. Rudnicki M. Torrente Y. Cossu G. Tremblay J.P. Partridge T. Gussoni E. Kunkel L.M. Huard J. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol Ther. 2007;15:867. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 45.Sacco A. Doyonnas R. Kraft P. Vitorovic S. Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cross-Doersen D. Isfort R.J. A novel cell-based system for evaluating skeletal muscle cell hypertrophy-inducing agents. In Vitro Cell Dev Biol Anim. 2003;39:407. doi: 10.1290/1543-706X(2003)039<0407:ANCSFE>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 47.Zhu C.H. Mouly V. Cooper R.N. Mamchaoui K. Bigot A. Shay J.W. Di Santo J.P. Butler-Browne G.S. Wright W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 48.Friesen W. Acharjee S. Zhuo J. Baiazitov R. Lee S. Moon Y.C. Sweeney H.L. Welch E. Identification and characterization of small molecules for the treatment of Duchenne muscular dystrophy. Neuromuscul Disord. 2008;18:812. [Google Scholar]
- 49.Courdier-Fruh I. Barman L. Briguet A. Meier T. Glucocorticoid-mediated regulation of utrophin levels in human muscle fibers. Neuromuscul Disord. 2002;12(Suppl 1):S95. doi: 10.1016/s0960-8966(02)00089-5. [DOI] [PubMed] [Google Scholar]
- 50.Vandenburgh H.H. Karlisch P. Shansky J. Feldstein R. Insulin and insulin-like growth factor-1 induce pronounced hypertrophy of skeletal myofibers in tissue culture. Am J Physiol Cell Physiol. 1991;260:C475. doi: 10.1152/ajpcell.1991.260.3.C475. [DOI] [PubMed] [Google Scholar]
- 51.Semsarian C. Wu M.J. Ju Y.K. Marciniec T. Yeoh T. Allen D.G. Harvey R.P. Graham R.M. Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature. 1999;400:576. doi: 10.1038/23054. [DOI] [PubMed] [Google Scholar]
- 52.Bodine S.C. Latres E. Baumhueter S. Lai V.K. Nunez L. Clarke B.A. Poueymirou W.T. Panaro F.J. Na E. Dharmarajan K. Pan Z.Q. Valenzuela D.M. DeChiara T.M. Stitt T.N. Yancopoulos G.D. Glass D.J. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 53.Jiang Z. Clemens P.R. Cellular caspase-8-like inhibitory protein (cFLIP) prevents inhibition of muscle cell differentiation induced by cancer cells. FASEB J. 2006;20:2570. doi: 10.1096/fj.06-6347fje. [DOI] [PubMed] [Google Scholar]
- 54.Granchelli J.A. Pollina C. Hudecki M.S. Pre-clinical screening of drugs using the mdx mouse. Neuromuscul Disord. 2000;10:235. doi: 10.1016/s0960-8966(99)00126-1. [DOI] [PubMed] [Google Scholar]
- 55.Glass D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 56.Payne E.T. Yasuda N. Bourgeois J.M. Devries M.C. Rodriguez M.C. Yousuf J. Tarnopolsky M.A. Nutritional therapy improves function and complements corticosteroid intervention in mdx mice. Muscle Nerve. 2006;33:66. doi: 10.1002/mus.20436. [DOI] [PubMed] [Google Scholar]
- 57.Hoffman E.P. Escolar D. Translating mighty mice into neuromuscular therapeutics: is bigger muscle better? Am J Pathol. 2006;168:1775. doi: 10.2353/ajpath.2006.060270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kosnik P.E. Dennis R.G. Vandenburgh H.H. Tissue engineering skeletal muscle. In: Guilak F., editor; Butler D.L., editor; Goldstein S.A., editor; Mooney D.J., editor. Functional Tissue Engineering. New York: Springer; 2003. pp. 377–392. [Google Scholar]
- 59.Liao H. Zhou G.-Q. Development and progress of engineering of skeletal muscle tissue. Tissue Eng Part B. 2009;15 doi: 10.1089/ten.teb.2009.0092. [DOI] [PubMed] [Google Scholar]
- 60.Shansky J. Del Tatto M. Chromiak J. Vandenburgh H. A simplified method for tissue engineering skeletal muscle organoids in vitro. In Vitro Cell Dev Biol Anim. 1997;33:659. doi: 10.1007/s11626-997-0118-y. [DOI] [PubMed] [Google Scholar]
- 61.Bell E. Ivarsson B. Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci USA. 1979;76:1274. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powell C. Smiley B. Mills J. Vandenburgh H. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am J Physiol Cell Physiol. 2002;283:C1557. doi: 10.1152/ajpcell.00595.2001. [DOI] [PubMed] [Google Scholar]
- 63.Dennis R.G. Kosnik P.E. Gilbert M.E. Faulkner J.A. Excitability and contractility of skeletal muscle engineered from primary cultures and cell lines. Am J Physiol Cell Physiol. 2001;280:C288. doi: 10.1152/ajpcell.2001.280.2.C288. [DOI] [PubMed] [Google Scholar]
- 64.Wakatsuki T. Fee J.A. Elson E.L. Phenotypic screening for pharmaceuticals using tissue constructs. Curr Pharm Biotechnol. 2004;5:181. doi: 10.2174/1389201043376940. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann W.H. Schneiderbanger K. Schubert P. Didie M. Munzel F. Heubach J.F. Kostin S. Neuhuber W.L. Eschenhagen T. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90:223. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- 66.Feinberg A.W. Feigel A. Shevkoplyas S.S. Sheehy S. Whitesides G.M. Parker K.K. Muscular thin films for building actuators and powering devices. Science. 2007;317:1366. doi: 10.1126/science.1146885. [DOI] [PubMed] [Google Scholar]
- 67.Das M. Gregory C.A. Molnar P. Riedel L.M. Wilson K. Hickman J.J. A defined system to allow skeletal muscle differentiation and subsequent integration with silicon microstructures. Biomaterials. 2006;27:4374. doi: 10.1016/j.biomaterials.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 68.Discher D.E. Janmey P. Wang Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 69.Chaudhuri O. Parekh S.H. Lam W.A. Fletcher D.A. Combined atomic force microscopy and side-view optical imaging for mechanical studies of cells. Nat Meth. 2009;6:383. doi: 10.1038/nmeth.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vandenburgh H. Shansky J. Benesch-Lee F. Barbata V. Reid J. Thorrez L. Valentini R. Crawford G. A drug screening platform based on the contractility of tissue engineered muscle. Muscle Nerve. 2008;37:438. doi: 10.1002/mus.20931. [DOI] [PubMed] [Google Scholar]
- 71.Shansky J. Benesch-Lee F. Zhang P. Choi B.-R. Nath N. Mende U. Vandenburgh H. Screening drug activity with tissue engineered cardiac muscle. Abstract presented at the Keystone Symposia on Pathological and Physiological Cardiac Hypertrophy. Copper Mountain CO. 2008 [Google Scholar]
- 72.O'Sullivan S. Nagle R. McEwen J.A. Casey V. Elastomer rubbers as deflection elements in pressure sensors: investigation of properties using a custom designed programmable elastomer test rig. J Phys D Appl Phys. 2003;36:1910. [Google Scholar]
- 73.Crandall S.H. Dahl N.C. Lardner T.J. An Introduction to the Mechanics of Solids. New York, NY; McGraw Hill; 1999. [Google Scholar]
- 74.Legant W.R. Pathak A. Yang M.T. Deshpande V.S. McMeeking R.M. Chen C.S. Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proc Natl Acad Sci USA. 2009;106:10097. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vandenburgh H. Shansky J. Benesch-Lee F. Skelly K. Spinazzola J. Saponjian Y. Tseng B.S. Automated drug screening with contractile muscle tissue engineered from dystrophic myoblasts. FASEB J. 2009 doi: 10.1096/fj.09-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Musaro A. McCullagh K. Paul A. Houghton L. Dobrowolny G. Molinaro M. Barton E.R. Sweeney H.L. Rosenthal N. Localized IGF-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet. 2001;27:195. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- 77.Tobert J.A. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat Rev Drug Discov. 2003;2:517. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- 78.Thompson P.D. Clarkson P.M. Rosenson R.S. An assessment of statin safety by muscle experts. Am J Cardiol. 2006;97:69C. doi: 10.1016/j.amjcard.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 79.Matsumura K. Campbell K.P. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994;17:2. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- 80.Morgan J.E. Beauchamp J.R. Pagel C.N. Peckham M. Ataliotis P. Jat P.S. Noble M.D. Farmer K. Partridge T.A. Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: a model system for the derivation of tissue-specific and mutation-specific cell lines. Dev Biol. 1994;162:486. doi: 10.1006/dbio.1994.1103. [DOI] [PubMed] [Google Scholar]
- 81.Grounds M.D. Two-tiered hypotheses for Duchenne muscular dystrophy. Cell Mol Life Sci. 2008;65:1621. doi: 10.1007/s00018-008-7574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang S.T. Zhang X. Wen Y. Microbioreactors for high-throughput cytotoxicity assays. Curr Opin Drug Discov Dev. 2008;11:111. [PubMed] [Google Scholar]
- 83.Choi N.W. Cabodi M. Held B. Gleghorn J.P. Bonassar L.J. Stroock A.D. Microfluidic scaffolds for tissue engineering. Nat Mater. 2007;6:908. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 84.Raimondi M.T. Engineered tissue as a model to study cell and tissue function from a biophysical perspective. Curr Drug Discov Technol. 2006;3:245. doi: 10.2174/157016306780368126. [DOI] [PubMed] [Google Scholar]
- 85.Roses A.D. Pharmacogenetics in drug discovery and development: a translational perspective. Nat Rev Drug Discov. 2008;7:807. doi: 10.1038/nrd2593. [DOI] [PubMed] [Google Scholar]
- 86.Gaud A. Simon J.M. Witzel T. Carre-Pierrat M. Wermuth C.G. Segalat L. Prednisone reduces muscle degeneration in dystrophin-deficient Caenorhabditis elegans. Neuromuscul Disord. 2004;14:365. doi: 10.1016/j.nmd.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 87.Catoire H. Pasco M.Y. bu-Baker A. Holbert S. Tourette C. Brais B. Rouleau G.A. Parker J.A. Neri C. Sirtuin inhibition protects from the polyalanine muscular dystrophy protein PABPN1. Hum Mol Genet. 2008;17:2108. doi: 10.1093/hmg/ddn109. [DOI] [PubMed] [Google Scholar]
- 88.Steffen L.S. Guyon J.R. Vogel E.D. Beltre R. Pusack T.J. Zhou Y. Zon L.I. Kunkel L.M. Zebrafish orthologs of human muscular dystrophy genes. BMC Genomics. 2007;8:79. doi: 10.1186/1471-2164-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirata H. Zebrafish muscular disease models towards drug discovery. Expert Opin Drug Discov. 2009;4:507. doi: 10.1517/17460440902835483. [DOI] [PubMed] [Google Scholar]
- 90.Parng C. Seng W.L. Semino C. McGrath P. Zebrafish: a preclinical model for drug screening. Assay Drug Dev Technol. 2002;1:41. doi: 10.1089/154065802761001293. [DOI] [PubMed] [Google Scholar]
- 91.Webb S. iPS cell technology gains momentum in drug discovery. Nat Rev Drug Discov. 2009;8:263. doi: 10.1038/nrd2867. [DOI] [PubMed] [Google Scholar]
- 92.Pouton C.W. Haynes J.M. Embryonic stem cells as a source of models for drug discovery. Nat Rev Drug Discov. 2007;6:605. doi: 10.1038/nrd2194. [DOI] [PubMed] [Google Scholar]
- 93.Thomson H. Bioprocessing of embryonic stem cells for drug discovery. Trends Biotechnol. 2007;25:224. doi: 10.1016/j.tibtech.2007.03.003. [DOI] [PubMed] [Google Scholar]