Abstract
The profile of protein sorting into multivesicular bodies (MVBs) has risen recently with the identification of three heteromeric complexes known as ESCRT-I,-II,-III (Endosomal Sorting Complex Required for Transport). Genetic analyses in yeast have identified up to 15 soluble class E VPS (vacuolar protein sorting) proteins that have been assigned to the ESCRT machinery and function in cargo recognition and sorting, complex assembly, vesicle formation and dissociation. Despite their functional importance in yeast and mammalian cells, little is known about their presence and function in other organisms including plants. We have made use of the fully sequenced genomes of Arabidopsis thaliana and Oryza sativa, Drosophila melanogaster and Caenorhabditis elegans to explore the identity, structural characteristics and phylogenetic relationships of proteins assigned to the ESCRT machinery.
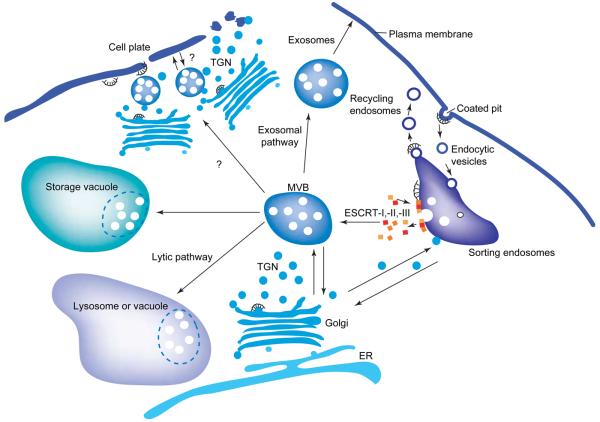
Multivesicular bodies (MVBs) are late endosomal organelles with up to several hundred interluminal vesicles that originate by invagination and budding of the limiting endosomal membrane (Figure 1) [1,2]. MVBs are also prevacuolar compartments involved in the retrograde endocytotic pathway where, upon fusion to vacuoles or lysosomes, the interluminal vesicles are released [3-6]. In yeast and mammals, MVBs are involved in sorting and degradation of transmembrane surface proteins as transporters (e.g. GAP1, STE6 and PDR5), receptors (e.g. EGFR, GHR, STE3 and STE2), in budding of viruses (e.g. HIV, MuLV and RSV) and in lipid partitioning [7,8]. However, MVBs are also central for sorting vacuolar proteins directly from the late Golgi to the anterograde and biosynthetic pathway (Figure 1) [9]. Furthermore, MVBs operate in the exocytotic pathway and the secreted interluminal vesicles are known as exosomes (Figure 1) [10-12]. Thus, MVBs act as an intermediate station for cargo on the way to degradation, intracellular recycling and secretion (Figure 1). Although MVBs have been described in plants, little is known about their biogenesis, cargos and function [2,4,13-16]. These studies show that plant MVBs are prevacuolar compartments and are labeled by vacuolar sorting receptor (VSR) antibodies such as BP80 [5,17]. Plant MVBs are not only involved in the transport of proteins to lytic but also to protein-storage vacuoles. As plasmalemmasomes in tobacco, MVBs have been implicated in the internalization of arabinogalactanrich plasma membrane proteins that are sequestered within the vacuole [18]. However, Eliot Herman and Chris Lamb [18] did not exclude the possibility that MVBs might be involved in secretion as well.
Figure 1.
Characteristics and roles of multivesicular bodies (MVBs). MVBs are endocytic intermediates that enclose, in a limiting membrane, luminal vesicles that originate by invagination at sorting endosomes. MVBs function at the cross road between endocytosis, exocytosis and transport to lysosomes or vacuoles and serve as prevacuolar compartments and precursors of exosomes. Sorting into MVBs can determine the delivery of transmembrane proteins into secretory lysosomes, and thus MVBs are involved in the activity control of endocytosed receptors. Furthermore, MVBs might also serve as storage compartments and carriers for morphogens. Invagination and protein sorting to MVBs requires the coordinated action of three multiprotein complexes, ESCRT-I, -II, -III (see Figure 2).
Protein sorting into MVBs is highly regulated, and genetic analyses in yeast have assigned 15 soluble class E VPS genes to this protein degradation, recycling and secretion pathway [19]. Biochemical and two-hybrid protein interaction studies in mammalian cells [20] and yeast [21-25] support their assembly into three heteromeric complexes named ESCRT-I, -II,–III (Endosomal Sorting Complex Required for Transport), which are involved in cargo recognition sorting and concentration, complex assembly, vesicle formation and dissociation [26,27].
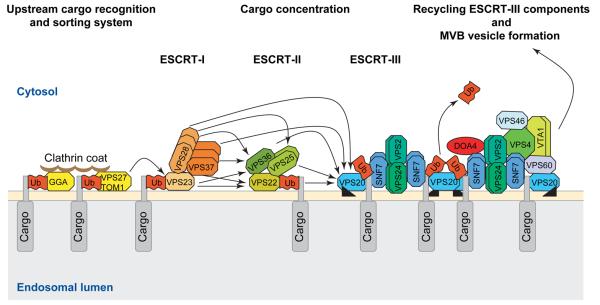
Current models for the function of the ESCRT machinery propose that monoubiquitinated membrane proteins are recognized by ubiquitin-binding proteins such as GGAs, VPS27 (yeast) or the mammalian ortholog HRS, HSE or STAM and the TOM1 families of proteins [28] (Figure 2). These proteins together with the endosomal clathrin-coat are likely to recognize cargo proteins and recruit ESCRT-I components from the cytoplasm. The association of the ESCRT-I complex with endosomal membranes activates the ESCRT-II complex, transfers the cargo to VPS23 (yeast) or to TSG101 (mammals) and allows subsequent takeover of the monoubiquitinated cargo by VPS36 (yeast) or by EAP45 (mammals), a component of the ESCRT-II complex. ESCRT-II recruits and initiates the oligomerization of ESCRT-III via the interaction of VPS22 (yeast) or EAP30 (mammals) and VPS20 (yeast) or CHMP6 (mammals). In two-hybrid analyses, the ESCRT-III component VPS20 is also able to interact with the ESCRT-I components VPS28 and VPS37, suggesting an association of all ESCRT complexes at the outer membrane of MVBs. ESCRT-III concentrates the MVB cargos and recruits additional ESCRT-III associated proteins as VPS46 or CHMP1A,B, VPS60 or CHMP5, a deubiquitination enzyme DOA4 and the AAA-type ATPase VPS4 or SKD1,2. VPS4 is necessary for the dissociation and recycling of the ESCRT-III components. Finally, invagination of interluminal MVB vesicles is facilitated by local differences in the lipid composition of membrane areas where cargo proteins were previously concentrated through the action of ESCRT-III.
Figure 2.
Model of the ESCRT-dependent protein sorting and concentration machinery in the MVB vesicle formation pathway. Monoubiquitinated transmembrane proteins are recognized by the upstream cargo sorting system of endosomes via a series of ubiquitin- and clathrin-binding proteins (GGAs, VPS27 and TOM1) that recruit ESCRT-I. ESCRT-I is able to bind ubiquitin via VPS23. The formation of ESCRT-I activates ESCRT-II, which initiates the formation of the ESCRT-III complex through the interaction of VPS22, VPS25 and VPS36 (ESCRT-II) with VPS20 (ESCRT-III). Concurrent with inward membrane budding the ubiquitin tags of the cargo proteins are removed by the deubiquitinating enzyme DOA4 and the ESCRT-III complex components dissociate from the endosomal membrane through the AAA-type ATPase activity of VPS4. Experimentally proven protein–protein interactions are indicated by arrows. The model also shows the known protein–protein interactions within the ESCRT complex. Dimerizations are symbolized as double boxes. The myristoylation of VPS20 is symbolized with a black triangle.
The aim of this comparative genome analysis is to identify and characterize homologs of the ESCRT-I, -II, -III components and the associated proteins that are necessary to feed and dissociate the ESCRT machinery in the plant and animal kingdoms. We have restricted our survey to the fully sequenced genomes such as Arabidopsis thaliana, Oryza sativa, Drosophila melanogaster and Caenorhabditis elegans and compared them to the functionally characterized yeast and human components.
Upstream cargo recognition and sorting system of the ESCRT machinery
The entry of membrane proteins into the MVB pathway starts with the binding of monoubiquitinated cargos to the outer endosomal membrane and the recruitment of the ESCRT-I complex. This membrane association is mediated by ubiquitin-binding proteins that contain one or two UIMs or a GAT domain in combination with protein domains that had been implicated in binding to membranes such as the phosphatidylinositol-3 phosphate (PI3P) binding FYVE finger domain (Figure 3) [29]. However, Akira Hayakawa and Naomi Kitamura [30] showed that the FYVE domain, which is only present in VPS27 or HRS homologs, is not essential and even PI3P binding is not necessary for the localization of HRS to endosomal membranes. However, Hayakawa and Kitamura have identified a 100 amino acid Pro- and Gln-rich region as important for binding HRS to membranes.
Figure 3.
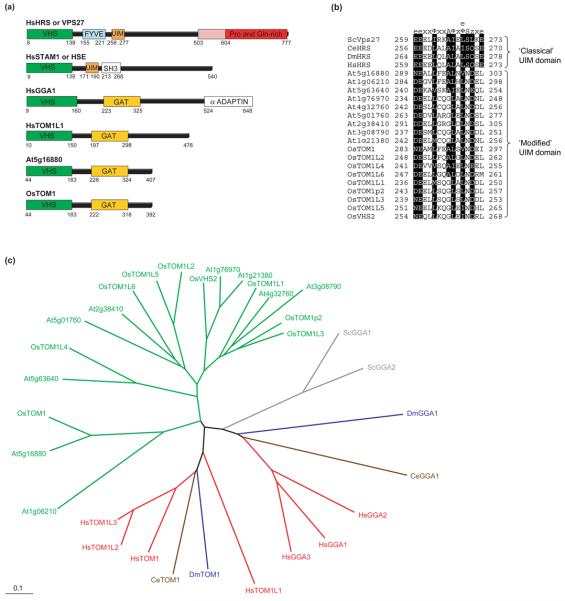
Structural similarities and phylogenetic analysis of components of the upstream cargo recognition and sorting system. (a) Domain organization of the human HRS or VPS27, STAM1, GGA1 and TOM1L1 proteins compared with a representative VHS-GAT domain protein of Arabidopsis (At5g16880) and rice (OsTOM1). The 100 amino acids of the Pro- and Gln-rich domain of HRS, which is essential for the localization to endosomes, are depicted in pink. (b) Comparison of the UIM domain of VPS27 and HRS proteins and the equivalent domain of VHS-GAT proteins of Arabidopsis and rice. Highly conserved residues are highlighted in black. Note the deviations from the ‘classical’ UIM domain with the consensus of e-e-x-x-φ-x-x-A-φ-x-e/φ-S-z-x-e, where e stands for negatively charged, φ for hydrophobic, x for helix-favoring and z for polar or hydrophobic residues. Ala and Ser are invariant. The VHS-GAT-containing putative plant homologs have an altered UIM motif at the same position: the invariant Ala and Ser are exchanged to Gly and Asp, respectively and the last negatively charged amino acid is replaced by a hydrophobic one. (c) Neighbor-joining phylogenetic tree generated with the VHS-GAT domains showing the predicted relationship between the yeast, Drosophila, Caenorhabditis elegans and human GGA and TOM1 proteins and their homologs in plants. The phylogram is based on an alignment prepared using Clustal X 1.81 and was drawn using Tree-View 1.6.6. The color-coded nomenclature is similar to that used in Table S1 in the Supplementary material and the first two letters of each protein name represent the organism: Sc, Saccharomyces cerevisiae (gray); Dm, Drosophila melanogaster (blue); Ce, Caenorhabditis elegans (brown); At, Arabidopsis thaliana (green); Os, Oryza sativa (green); and Hs, Homo sapiens (red).
GGAs, VPS27, HRS, HSE and TOM1 proteins all contain an N-terminal VHS domain of ~140 amino acids. The VHS domain consists of eight helices arranged in a superhelix and is considered to have a general membrane targeting and cargo recognition role in vesicular trafficking [31] (Figure 3, and see Supplementary material Figure S1). They suggested that the conserved positively charged surface of helix 3 in the VHS domain plays also a role in membrane binding. However, only the VHS domain of GGAs binds an acidic di-leucine (DxxLL) sorting signal found in proteins known to cycle between the trans-Golgi Network (TNG) and endosomes [32,33].
Given that monoubiquitination is the signal for cargo protein entry into the MVB pathway, all components of the upstream recognition and sorting system have domains known to bind to ubiquitin. The consensus of the UIM motif in VPS27 and its mammalian (HRS), Drosophila and C. elegans homologs is e-e-x-x-φ-x-x-A-φ-x-e/φ-S-z-x-e, where e is negatively charged, φ is hydrophobic, x is a helix-favoring residue and z a polar or hydrophobic residue (Figure 3). Ala and Ser are invariant and mutation of Ser to Asp or Glu greatly diminishes binding, whereas mutation of Ser to Ala has a modest effect on the interaction [29]. However, the VHS domain-containing putative plant homologs have an altered UIM motif at the same position, where invariant Ala and Ser are changed to Gly and Asp, respectively, and the last negatively charged amino acid is replaced by a hydrophobic one (Figure 3). Experimental evidence is needed to determine whether the modified UIM motif of the Arabidopsis and rice VHS domain proteins is functional (see Supplementary material Table S1).
Ubiquitin binding and delivery of cargo proteins to the putative plant ESCRT-machinery might also be mediated by proteins with a GAT (GGA and TOM1) domain, which has been shown to bind ubiquitin in a similar mode as the UIM and is necessary for the transport of monoubiquitinated cargo proteins to the MVB pathway [34-36]. The proximity of a VHS domain is necessary for efficient binding of ubiquitin to GAT domains. Two small protein families exhibit this domain arrangement – the GGAs and the TOM1s. Although GGAs are not found in plants, the TOM1 proteins are present in all the multicellular systems in our survey (i.e. Arabidopsis, rice, Drosophila, C. elegans and human). Our analysis reveals that the TOM1 protein family has significantly expanded in plants because we identified nine genes in Arabidopsis and rice that share putative clathrin-boxes as well as the VHS and GAT domains (see Supplementary material Figure S1 and Table S1). Also VPS27 and HRS contain clathrin-binding motifs and colocalize with flat ‘bilayered’ endosomal clathrin coats [26,37,38]. TOM1L1 interacts with HSR and the ubiquitin-binding protein TSG101 of the ESCRT-I complex [28]. Therefore, VSP27, HRS and TOM1 proteins might act as adaptor and concentrator molecules that interact with the flat endosomal clathrin coat and bind to both the monoubiquitinated transmembrane proteins and an ESCRT-I component. Thus, TOM1 proteins in plants might contribute to the loading of the ESCRT machinery, and the expansion of this gene family might be because of functional diversification. Experimental evidence is needed to support the hypothesis that the plant-specific VHS-GAT proteins exhibit the same functions as TOM1 proteins in recruiting the ESCRT-I complex to the monoubiquitinated cargo protein.
ESCRT-I
ESCRT-I is a heterotrimeric 350 kDa complex comprising a single copy VPS23 (yeast) or TSG101 (human) and multiple copies of VPS28 and VPS37 [39]. ESCRT-I transiently associates with endosomal membranes and acts in the recognition of monoubiquitinated cargo proteins. VPS23 and the human homolog TSG101 contain a ubiquitin-conjugating (UBC)-like domain also known as UEV domain. The UEV domain is similar to the ubiquitin conjugating enzyme E2 UBC but lacks the cysteine in the active site. Therefore, VPS23 does not function as an ubiquitin conjugating enzyme and yet it is still able to bind ubiquitinated proteins. Via its C-terminal coiled coil structure, the human VPS23 homolog TSG101 interacts with VPS27, VPS28 [40] and VPS37 [41]. In rice and Arabidopsis, one and two putative VPS23 homologs are present, respectively (see Supplementary material Table S2 and Figure S2A).
VPS28s are small proteins of about 200 aa and their region of high similarity is called the VPS28 domain. Arabidopsis has two putative VPS28 proteins whereas the rice genome codes for only one (see Supplementary material Table S2 and Figure S2B). VPS28 interacts with components of all three ESCRT complexes (ESCRT-I, VPS23 or TSG101, and VPS37; ESCRT-II, VPS22, VPS25 and VPS36; ESCRT-III, VPS20) (Figure 2).
The most divergent component of the ESCRT-I complex is VPS37, which is characterized by a coiled coil and Mod_r domain important for protein–protein interactions. The N-terminal half of this 150-amino acid Mod_r domain is acidic, whereas the C-terminal half is basic [42]. Proteins with a Mod_r domain are only present in eukaryotes. This shared protein motif and the low level of sequence identity were not significant enough to identify the VPS37 homologs in human, Drosophila, C. elegans and plants. However, recent experimental interaction studies have identified the four human orthologs of VPS37 (see Supplementary material Table S2 and Figure S2C) [40,43]. Using the human VPS37 sequences, we identified putative homologs in Drosophila and C. elegans and two putative Arabidopsis and rice homologs (see Supplementary material Table S2 and Figure S2C).
VPS37C can form a ternary complex with TSG101 and VPS28 by binding to a domain located at the C-terminus of TSG101. Furthermore, the yeast VPS37 protein has been shown to interact with VPS20 [25], thus linking ESCRT-I with the ESCRT-III complex (Figure 2). Whether the putative plant homologs share similar functions has still to be determined experimentally.
ESCRT-II
ESCRT-II is a heterotrimeric 138 kDa complex that transiently associates with endosomal membranes downstream of ESCRT-I and recruits ESCRT-III. Analysis of the crystal structure has revealed that the core ESCRT-II complex forms a ‘Y’ structure composed of one VPS22 molecule, the C-terminal 171 amino acids of VPS36 and two molecules of VPS25 (see Supplementary material Table S3 and Figure S3) [44,45].
The human homologs of VPS22, VPS25 and VPS36, namely EAP25, EAP45 and EAP30, were originally identified as ELL-associated proteins (EAP) that form a multiprotein complex with the RNA polymerase II elongation factor ELL [46,47]. Consistent with its central role in the MVB sorting machinery, ESCRT-II components interact with ESCRT-I and ESCRT-III [25,48,49].
Our analyses show that three VPS22 homologs are present in the Arabidopsis genome and two in the rice genome. However, the current annotation of Arabidopsis lists At3g30867 as a pseudogene and At3g31960 is annotated as an expressed protein missing half of the conserved N-terminal coiled coil and the C-terminal EAP30 domain (see Supplementary material Figure S3A). Based on TBLASTN analyses and reconstruction of intron–exon junctions we propose that At3g31960 is also a pseudogene with several frame shifts and additional introns (see Supplementary material Figure S3A). For both pseudogenes, ESTs and full-length cDNA sequences are lacking (see Supplementary material Table S3). Thus, only At4g27040 can serve as a functional VPS22 homolog in Arabidopsis.
VPS25 and its human homolog EAP25 are small proteins of about 20 kDa that are able to dimerize. Each monomer consists of two winged tandemly arranged helixes referred to as the DUF852 domain, which is important for the interaction with the C-terminal domain of EAP30 and VPS36 (see Supplementary material Figure S2B). At the N-terminus of VPS25, two PPXY motifs, PPVY (amino acid 5–8) and PPXY (amino acid 11–14), are important for the interaction with VPS22 and VPS36. Whereas the first motif exists only in yeast, the second PPXY motif is highly conserved through the kingdoms (see Supplementary material Figure S3B). VPS25 is the main subunit responsible for the recruitment of ESCRT-III via its interaction with VPS20 [44,45].
EAP45 is part of the ESCRT-II component. It binds to a subset of phosphoinositides [PtdIns(3,4,5)P3; PtdIns(3,4)P2; PtdIns(3,5)P2] in the endosomal membrane and accepts the monoubiquitinated MVB-cargos from the ESCRT-I complex. This phosphoinositide and ubiquitin binding is mediated by the N-terminal GLUE domain [50]. Whereas the yeast VPS36 contains two additional N-terminal zinc fingers (NZF), neither of the animal nor the single Arabidopsis and rice homologs possess this NZF domain (see Supplementary material Figure S2C) [51,52]. The most conserved domain, the VPS36 domain, is at the C-terminus.
ESCRT-III
ESCRT-III is composed of at least four highly charged, small coiled coil proteins with an extremely basic (pI 11) N-terminal half (~125 amino acids) and an exceptionally acidic (pI 4) C-terminus. These ‘classical’ ESCRT-III components share the ~160-amino acid SNF7 domain, which is a succession of distinctive coiled coil protein–protein interaction domains. The four ESCRT-III components in yeast are VPS20, VPS32 or SNF7, VPS2 and VPS24, and their mammalian homologs are CHMP6, CHMP4, CHMP2 and CHMP3, respectively. However, two other SNF7 domain proteins, VPS46 or CHMP1 and VPS60 or CHMP5, have been identified in the yeast or mammalian genomes, respectively, and also in our analyses of plants, Drosophila and C. elegans. A CHMP1 homolog has been identified in maize by using a genetic approach to identify regulators that determine the number of aleurone layers [53]. Whereas the homozygous sal1 mutants of maize do not germinate, the depletion of the Nicotiana benthamiana CHMP1 homolog NbCHMP1 resulted in only subtle changes in leaf morphology and color [54].
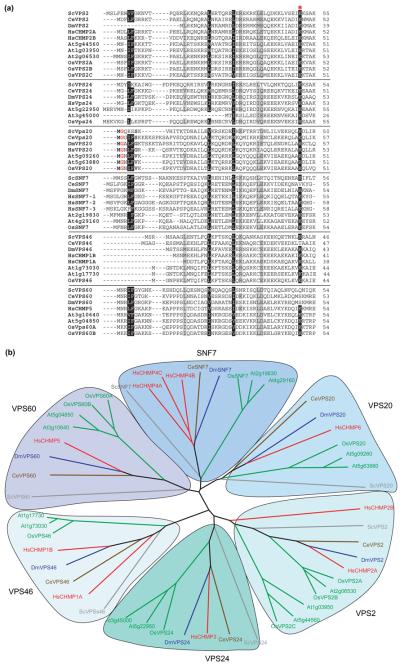
Phylogenetic analyses have grouped the SNF7 proteins into six classes, termed after their yeast protagonist VPS2, SNF7, VPS20, VPS24, VPS46 and VPS60 (Figure 4, and see Supplementary material Table S4). Within a class, plant and human SNF7 proteins share between 30% and 57% amino acid identity, and Arabidopsis and rice share between 63% and 85% amino acid identity.
Figure 4.
Protein alignments and phylogenetic trees of the ESCRT-III complex components. (a) Alignment of the first 50 amino acids of all the ‘classical (VPS2, SNF7, VPS24, VPS20)’ and nonclassical (VPS46, VPS60) proteins of the ESCRT-III complex. Highly conserved residues are highlighted in black. The Gly highlighted in red font and the surrounding residues of the VPS20 class highlighted in bold mark the N-terminal myristoylation site. The red asterisk marks the conserved Lys (K49), which is important for lipid and membrane interactions. (b) Neighbor-joining phylogenetic tree with 1000 bootstrap replicates generated with all the SNF7 domain-containing ESCRT-III complex components. The tree is based on a protein alignment prepared with Clustal × 1.81 and was drawn using Tree-View 1.6.6. The color-coded nomenclature is similar to that used in Table S4 in the Supplementary material. The six SNF7-domain protein subclasses are highlighted and named after their yeast protagonists. Note the expansion of members in the plant lineages and the plant-specific VPS2 subclass.
Although the genomes of yeast, Drosophila and C. elegans code for only one member of each SNF7 protein class, up to three members per class have been identified in the Arabidopsis, rice and human genomes (Figure 4, and see Supplementary material Table S4). Interestingly, the VPS2 class can be further subdivided into a plant-specific subclass with two members in Arabidopsis and one member in rice (Figure 4).
The ESCRT-III complex consists of at least two subcomplexes, VPS20 or CHMP6 and SNF7 or CHMP4, which are also known as the ‘core’; VPS2 or CHMP3 and VPS24 or CHMP2 are designated as the ‘coat’ complex [48]. The endosomal membrane association of the ‘core’ subcomplex depends on the myristoylation of VPS20 or CHMP6 [48] whereas the ‘coat’ complex association is mediated through the PtdIns(3,5)P2 effector protein function of VPS24 or CHMP6. Paul Whitley et al. [55] showed the binding of the N-terminus of a mammalian VPS24 to phosphoinositides and in particular to the endosomal membrane-specific PtdIns(3,5)P2. However, the normally cytosolic and nuclear VPS24 only becomes stably associated with late endosomal membranes in vivo if its C-terminus is missing or if the AAA-ATPase VPS4 or SKD1 is defective and unable to disassemble the ESCRT-III complex.
ESCRT associated proteins: VPS46, VPS60 and VTA1
The stoichiometry and size of the ESCRT-III complex has not been determined and additional components might be present, for example, the highly charged, small coiled coil SNF7 domain proteins VPS60 or CHMP5 and VPS46 or CHMP1 and the larger coiled coil protein VTA1 or SBP1 [25,27,56,57] (see Supplementary material Table S5). Direct and indirect associations with ESCRT-III proteins have been suggested for these ‘nonclassical’ ESCRT-III components [25] (N. Schlager and M.T. Hauser, unpublished). Further protein interaction studies with the yeast and mammalian ESCRT-III components indicate that as well as heterodimerization they can also homodimerize [21-25] (N. Schlager and M-T. Hauser, unpublished).
VPS46 was originally identified as a class A VPS gene and is identical to DID2 and FTI1 [25]. VPS60 was isolated in a genetic screen for mutants that block transportation of the ABC-transporter STE6 to the vacuole [58]. vps60 and vps46 mutants exhibit less severe phenotypes suggesting that these genes have redundant functions, possess a different selectivity to transport cargo proteins or play a regulatory role in the ESCRT-III activity [48].
VTA1 associates to a homomultimeric complex and its late endosomal localization and membrane association depends on the ESCRT-machinery [56]. If VTA1 is deleted, the resulting phenotypes are not typical of class E VPS phenotypes and, thus, VTA1 might function in a subset of sorting events. VTA1 and SBP1 homologs are single-copy genes in all analyzed genomes (see Supplementary material Table S5) and their gene products interact with VPS4 (SNF7) and VPS60, respectively [25,56,57,59]. The overall shared amino acid identity between yeast and the putative human and plant homologs is low (15% and 14%, respectively). The protein can be subdivided into three parts: at the N-terminal, 150 amino acids form an as yet uncharacterized DUF605 and a coiled coil domain; at the C-terminus, a second coiled coil domain shows a high level of sequence identity between yeast and the human and plant homologs (52.8% and 40.0%, respectively). Together with the N-terminal coiled coil domain, the short C-terminal domain is responsible for the interaction with VPS4 [57]. The two terminal domains are separated by a highly diverse stretch of between 100 (human) and 300 (rice) amino acids.
The human gene was identified in several different studies and thus has many synonymous names, such as LIP5 [60], SBP1, DRG-1, MY012 and HSPC228 [61]. The Arabidopsis homolog At4g26750 is currently wrongly annotated as member of a hydroxyproline-rich glycoprotein.
VPS4
VPS4, an AAA-ATPase, is one of the last factors to be recruited by ESCRT-III proteins. [62]. VPS4 proteins act by binding, catalyzing and energizing the dissociation of the ESCRT-III complex and the release of the membrane-associated SNF7 domain proteins into the cytoplasm for further rounds of sorting [57,63]. In diverse protein–protein interaction studies, it has been shown that VPS4 and its two mammalian homologs VPS4B or SKD1 and VPS4A or SKD2 interact with ‘classical’ ESCRT-III complex components (SNF7 or CHMP4, VPS2 or CHMP2, VPS20 or CHMP6) and the associated proteins VPS46 or CHMP1, and VTA1 or SBP1 [20-22,24,25,63-66].
The two highly related human VPS4 proteins are 80% identical and share approximately 60% amino acid identity with the yeast VPS4 protein (see Supplementary material Table S5 and Figure S4). Apart from the catalytic AAA-ATPase domain of VPS4 proteins, their substrate specificity is defined by a 70-amino acid N-terminal MIT domain (see Supplementary material Figure S4). Our analysis revealed that the Arabidopsis and rice genomes code for >170 proteins with an AAAT-ATPase domain, but only one of these also had an N-terminal MIT domain. The Arabidopsis and rice VPS4 homologs are approximately 50% and 55% identical to the human and yeast VPS4 proteins, respectively. Recently, Yingtzy Jou et al. [67] identified mcSKD1, a homolog of VPS4, in a screen for salt-induced genes of the halophyte Mesembryanthemum crystallinum. They showed that mcSKD1 functionally complements the yeast potassium-uptake mutant skd1 and propose that mcSKD1 plays a dual role in facilitating potassium-uptake under unstressed conditions and in promoting sodium sequestration upon elevated salinity.
The MIT domain forms an antiparallel three helical bundle. Five Ala and/or Gly residues (9, 16, 28, 35) are highly conserved and are involved in the formation of an ‘Alanine zipper’ between the first two helixes (see Supplementary material Figure S4) [64]. The ‘Alanine zipper’ also participates in the coiled coil formation as a result of its interaction with aromatic and/or large hydrophobic side-chain residues as Leu, Tyr and Ile. A highly conserved neighboring Leu [64] and Glu [68] patch binds directly to the C-terminal acidic region of CHMP1B, one of the human VPS46 homologs [64].
Conclusions
In yeast and mammalian cells, the ESCRT machinery is crucial for the formation of MVBs and for sorting cargo proteins to them. MVBs might also be involved in recycling membranes. Although the presence of MVBs and their roles as prevacuolar compartments for lytic and storage vacuoles, in vacuolar receptor recycling, and in internalization and secretion of arabinogalactan-rich glycoproteins have been described [2,4,5,13-16,18], only a few reports have identified components of the ESCRT-machinery in plants [53,54,67]. Here we show by cross-species comparisons that most protein entry components and all the ESCRT-components are present in Arabidopsis and rice. However, there is an urgent need for in planta research. Preliminary results from our laboratory and Martin Hülskamp's group suggest that putative components of the ESCRT machinery are necessary for the proper execution of cytokinesis in Arabidopsis (V. Winter et al., unpublished; C. Spitzer et al., unpublished). This putative ESCRT function is corroborated by recent findings that MVBs are enriched at the newly formed cell plate during cytokinesis [15,16]. The identification of mutants that affect components of the ESCRT-machinery, of suitable markers for the characterization of MVB maturation, and of their endogenous cargos should shed light on the cellular processes that MVB are involved with. It is expected that novel connections between ESCRT-dependent protein sorting and MVB formation and other fields of plant biology will emerge in the future.
Supplementary Material
Acknowledgements
We thank Martin Hülskamp, Christoph Spitzer and Swen Schellmann for sharing unpublished results, Juan Antonio Torres Acosta for helping with bioinformatic analyses and valuable suggestions. We thank the anonymous referees for their valuable suggestions and comments. We are grateful to Christian Schlötterer for comments on the manuscript. This project is supported by the Austrian Science Fund (FWF grant P16420-B12).
Abbreviations and glossary
- AAA-type
ATPases associated with a variety of cellular activities.
- ABC
ATP-binding cassette.
- ARF
ADP-ribosylation factor.
- CHMP
chromatin modifying protein, later renamed charged multivesicular body proteins.
- DRG-1
dopamine responsive protein.
- EAP
ELL-associated protein.
- EGFR
epidermal growth factor receptor.
- ELL
Eleven-nineteen lysine-rich leukemia complex.
- ESCRT
endosomal sorting complex required for transport.
- FYVE
this phosphatidylinositol-3 phosphate binding motif is named after four proteins – Fab1b, YOTB, Vac1p, EEA1 domain.
- GAP1
general amino acid permease1.
- GAT
GGA and TOM1.
- GGA
Golgi-localizing, Gamma-adaptin ear domain homology, ADP-ribosylation factor (ARF)-binding proteins.
- GLUE
GRAM-like ubiquitin-binding domain in EAP45 domain.
- HIV
human immunodeficiency virus.
- HSE
HRS-binding protein.
- HSPC228
hematopoietic stem/progenitor cell open reading frame 228.
- HRS
hepatocyte growth factor-regulated tyrosine kinase substrate.
- LIP
LYST interacting protein.
- LYST
lysosomal trafficking regulator.
- MIT
microtubule interacting and trafficking molecule domain.
- Mod(r)
modifier of rudimentary domain.
- MuLV
murine leukemia virus.
- MVB
multivesicular bodies.
- My012
chromosome 6 open reading frame 55.
- Nmyr
N-terminal myristoylation signal: ^MG|^G[^EDRKHPFYW]..[STAGCN][^P].
- NZF
N-terminal zinc fingers.
- PI3P
phosphatidylinositol-3 phosphate.
- RSV
respiratory syncytial virus.
- SAL1
supernumerary aleurone 1.
- SBP1
SKD-binding protein1.
- SDK
Suppressor of K+ transport growth defect.
- SH3
Src homology 3 domain.
- SNF
sucrose nonfermenting.
- STAM
signal transducing adaptor molecule.
- STE2
α-factor receptor.
- STE3
a factor receptor.
- STE6
ABC-transporter STE6.
- TNG
trans-Golgi network.
- TOM1
target of Myb1.
- TSG101
tumor-susceptibility gene 101.
- UBC
ubiquitin-conjugating enzyme.
- UEV
ubiquitin E2 variant.
- UIM
ubiquitin-interacting motif.
- VHS
VPS27, HRS, STAM domain.
- VPS
vacuolar protein sorting.
- VTA1
VPS twenty associated.
Footnotes
Supplementary data
Supplementary data associated with this article can be found at doi:10.1016/j.tplants.2006.01.008
References
- 1.Palade GEA. A small particulate component of the cytoplasm. J. Biophys. Biochem. Cytol. 1955;1:59–67. doi: 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanchak MA, Fowke LC. The morphology of multivesicular bodies in soybean protoplasts and their role in endocytosis. Protoplasma. 1987;138:173–182. [Google Scholar]
- 3.Marcote MJ, et al. Membrane transport in the endocytic pathway: animal versus plant cells. Protoplasma. 2000;210:123–132. [Google Scholar]
- 4.Jiang L, et al. Multivesicular bodies: a mechanism to package lytic and storage functions in one organelle? Trends Cell Biol. 2002;12:362–367. doi: 10.1016/s0962-8924(02)02322-x. [DOI] [PubMed] [Google Scholar]
- 5.Tse YC, et al. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell. 2004;16:672–693. doi: 10.1105/tpc.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 7.Piper RC, Luzio JP. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic. 2001;2:612–621. doi: 10.1034/j.1600-0854.2001.20904.x. [DOI] [PubMed] [Google Scholar]
- 8.Katzmann DJ, et al. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 9.Raiborg C, et al. Hrs recruits clathrin to early endosomes. EMBO J. 2001;20:5008–5021. doi: 10.1093/emboj/20.17.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denzer K, et al. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000;113:3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 11.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Fevrier B, et al. Traffic exosomes: a bubble ride for prions? Traffic. 2005;6:10–17. doi: 10.1111/j.1600-0854.2004.00247.x. [DOI] [PubMed] [Google Scholar]
- 13.Samuels AL, et al. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J. Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otegui MS, et al. Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell. 2001;13:2033–2051. doi: 10.1105/TPC.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segui-Simarro JM, et al. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segui-Simarro JM, Staehelin LA. Cell cycle-dependent changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: a quantitative and spatial analysis. Planta. 2005;223:223–236. doi: 10.1007/s00425-005-0082-2. [DOI] [PubMed] [Google Scholar]
- 17.Robinson DG, et al. Vesicle transfer of storage proteins to the vacuole: the role of the Golgi apparatus and multivesicular bodies. J. Plant Physiol. 1998;152:659–667. [Google Scholar]
- 18.Herman EM, Lamb CJ. Arabinogalactan-rich glycoproteins are localized on the cell surface and in intravacuolar multivesicular bodies. Plant Physiol. 1992;98:264–272. doi: 10.1104/pp.98.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond CK, et al. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 21.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 22.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 23.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowers K, et al. Protein–protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 26.Raiborg C, et al. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- 27.Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 28.Puertollano R. Interactions of TOM1L1 with the multivesicular body sorting machinery. J. Biol. Chem. 2005;280:9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- 29.Swanson KA, et al. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayakawa A, Kitamura N. Early endosomal localization of Hrs requires a sequence within the Proline-and Glutamine-rich region but not the FYVE finger. J. Biol. Chem. 2000;275:29636–29642. doi: 10.1074/jbc.M002696200. [DOI] [PubMed] [Google Scholar]
- 31.Misra S, et al. Structure of the VHS domain of human Tom1 (target of myb 1): insights into interactions with proteins and membranes. Biochemistry. 2000;39:11282–11290. doi: 10.1021/bi0013546. [DOI] [PubMed] [Google Scholar]
- 32.Misra S, et al. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 2002;415:933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, et al. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science. 2001;292:1716–1718. doi: 10.1126/science.1060896. [DOI] [PubMed] [Google Scholar]
- 34.Scott PM, et al. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 2004;6:252–259. doi: 10.1038/ncb1107. [DOI] [PubMed] [Google Scholar]
- 35.Shiba Y, et al. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 2004;279:7105–7111. doi: 10.1074/jbc.M311702200. [DOI] [PubMed] [Google Scholar]
- 36.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 37.Raiborg C, et al. FYVE and coiled-coil domains determine the specific localization of Hrs to early endosomes. J. Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- 38.Sachse M, et al. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol. Biol. Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katzmann DJ, et al. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 40.Eastman SW, et al. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J. Biol. Chem. 2005;280:628–636. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- 41.Stuchell MD, et al. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 42.Begley D, et al. Modifier of rudimentary p1, mod(r)p1, a trans-acting regulatory mutation of rudimentary. Mol. Gen. Genet. 1995;248:69–78. doi: 10.1007/BF02456615. [DOI] [PubMed] [Google Scholar]
- 43.Bache KG, et al. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol. Biol. Cell. 2004;15:4337–4346. doi: 10.1091/mbc.E04-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo H, et al. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Wernimont AK, Weissenhorn W. Crystal structure of subunit VPS25 of the endosomal trafficking complex ESCRT-II. BMC Struct. Biol. 2004;4:10. doi: 10.1186/1472-6807-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt AE, et al. Cloning and characterization of the EAP30 subunit of the ELL complex that confers derepression of transcription by RNA polymerase II. J. Biol. Chem. 1999;274:21981–21985. doi: 10.1074/jbc.274.31.21981. [DOI] [PubMed] [Google Scholar]
- 47.Kamura T, et al. Cloning and characterization of ELL-associated proteins EAP45 and EAP20. A role for yeast EAP-like proteins in regulation of gene expression by glucose. J. Biol. Chem. 2001;276:16528–16533. doi: 10.1074/jbc.M010142200. [DOI] [PubMed] [Google Scholar]
- 48.Babst M, et al. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 49.Babst M, et al. Endosome associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 50.Slagsvold T, et al. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 51.Hierro A, et al. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004;431:221–225. doi: 10.1038/nature02914. [DOI] [PubMed] [Google Scholar]
- 52.Alam SL, et al. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen B, et al. sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6552–6557. doi: 10.1073/pnas.0732023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang KS, et al. Molecular characterization of NbCHMP1 encoding a homolog of human CHMP1 in Nicotiana benthamiana. Mol. Cells. 2004;17:255–261. [PubMed] [Google Scholar]
- 55.Whitley P, et al. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J. Biol. Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 56.Shiflett SL, et al. Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- 57.Yeo SC, et al. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- 58.Kranz A, et al. A family of small coiled coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell. 2001;12:711–723. doi: 10.1091/mbc.12.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward DM, et al. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- 60.Tchernev VT, et al. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol. Med. 2002;8:56–64. [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang QH, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Babst M, et al. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott A, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott A, et al. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujita H, et al. Mammalian class E Vps proteins, SBP1 and mVps2/CHMP2A, interact with and regulate the function of an AAA-ATPase SKD1/Vps4B. J. Cell Sci. 2004;117:2997–3009. doi: 10.1242/jcs.01170. [DOI] [PubMed] [Google Scholar]
- 66.Lin Y, et al. Interaction of the mammalian ESCRT-III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 67.Jou Y, et al. Tissue-specific expression and functional complementation of a yeast potassium-uptake mutant by a salt-induced ice plant gene mcSKD1. Plant Mol. Biol. 2004;54:881–893. doi: 10.1007/s11103-004-0335-7. [DOI] [PubMed] [Google Scholar]
- 68.daSilva LL, et al. Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell. 2005;17:132–148. doi: 10.1105/tpc.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.