Abstract
Recent studies suggest that the human Kinesin-8 molecular motor Kif18A has a role in chromosome congression. Specifically, these studies find that Kif18A promotes chromosome congression by attenuating chromosome oscillation magnitudes. Together with recent modeling work, in vitro studies, and the analysis of in vivo yeast data, these reports reveal how Kinesin-8 molecular motors might control chromosome oscillation amplitudes by spatially regulating the dynamic instability of microtubule plus-ends within the mitotic spindle.
Introduction
The mitotic spindle is a microtubule-based structure that controls the proper segregation of chromosomes during mitosis [1]. Within the mitotic spindle, microtubules coordinate the congression of chromosomes to the metaphase plate before their ultimate segregation during anaphase. Although chromosomes generally align at the spindle equator, this alignment is dynamic, with chromosomes oscillating about this position throughout metaphase. Because the microtubules that coordinate chromosome congression are dynamic – growing and shortening through a behavior termed ‘dynamic instability’ [2] – it is likely that chromosome oscillations reflect the growth and shortening dynamics at microtubule ends. Therefore, a central issue in the study of mitosis is describing how the dynamics of microtubule ends are regulated spatially within the mitotic spindle such that chromosomes are congressed to the spindle equator.
Microtubules are composed of αβ-tubulin heterodimers that are stacked end-on, typically into 13 individual protofilaments. Growth and shortening at microtubule ends takes place primarily at the β-tubulin exposed ‘plus-end’ of microtubules, through the addition of GTP-tubulin heterodimers [3]. After their addition, lattice-bound GTP–tubulin subunits are hydrolyzed into GDP–tubulin subunits through nucleotide exchange on the β subunit. Importantly, GDP–tubulin subunits exposed at the tips of microtubules are unstable owing to a conformational change in the tubulin. This promotes the curling and peeling of individual microtubule protofilaments, thus stretching and destabilizing bonds between adjacent tubulin subunits. Stochastic switching between stable, straight, growing GTP–tubulin-capped microtubules and unstable, peeling, shortening GDP–tubulin microtubules forms the basis of microtubule dynamic instability. The frequency at which microtubules switch from the shortening to the growing state is termed the ‘rescue’ frequency; and the frequency at which the reverse switch – from growing to shortening – occurs is termed the ‘catastrophe’ frequency.
Microtubule-associated proteins (MAPs) associate with the microtubule lattice and can modulate the dynamics of microtubule ends. Members of the Kinesin-8 family of molecular motors are MAPs that have been classified as catastrophe promoters based on the observation that motor deletion mutants exhibit abnormally long and stable microtubules [4-7]. A recent in vitro study with GTP–tubulin-stabilized microtubules [or guanylyl-(alpha, beta)-methylene-diphosphonate (GMPCPP) microtubules] suggested that the yeast Kinesin-8 motor Kip3p promotes shortening of microtubule plus-ends in a length-dependent manner [8]. In this study, GMPCPP microtubules were used because GMPCPP mimics the GTP ‘cap’ found at the ends of growing microtubules. Here, the presence of Kip3p at moderate concentrations promoted rapid depolymerization of longer microtubules and promoted slower depolymerization of shorter microtubules (Table 1) [8]. Because Kip3p is a molecular motor, this result probably originates from the ability of Kip3p to specifically motor towards the plus ends of microtubules, concentrating in a length-dependent manner on the plus ends of longer microtubules. Upon arrival at the plus end, the motor then destabilizes interactions between neighboring tubulin dimers. Because longer microtubules have more potential Kip3p-binding sites, they will tend to attract more motors, so that there will be a higher concentration of microtubule-destabilizing Kip3p motors on the ends of longer microtubules, and a lower concentration at the ends of shorter microtubules. A recent study by Mayr and co-workers [9] described a similar property for the human Kinesin-8 motor Kif18A (Table 1).
Table 1.
Effect of Kinesin-8 on microtubule-shortening rate and catastrophe frequency
| Parameter | Experiment type and organism | With Kinesin-8a | Without Kinesin-8b |
|---|---|---|---|
| Microtubule-shortening rate (μm/min) | In vivo: budding yeast astral microtubules | 1.91 ± 0.69 [16] | 2.82 ± 0.95 [16] |
| In vivo, human (HeLa cells) | 1.92 ± 0.06 [15] (metaphase) | 2.80 ± 0.08 [15] (metaphase) | |
| 1.68 ± 0.08 [15] (anaphase) | 2.12 ± 0.14 [15] (anaphase) | ||
| In vitro, GMPCPP microtubules | ~0.10 [16] (50 nM Kip3p) | ~0.00 [16] | |
| ~0.50 [8] (3.3 nM Kip3p) | ~0.00 [8] | ||
| In vitro, GMPCPP microtubules | ~1.25 [9] (100 nM Kif18A) | ~0.00 [9] | |
| Microtubule catastrophe frequency (min−1) | In vivo, budding yeast astral microtubules | 0.48 [16] | 0.27 [16] |
Kinesin-8 molecular motors are present in the assay, either endogenously as in the in vivo studies, or added to the assay in the noted concentrations as in the in vitro studies.
Kinesin-8 molecular motors are either deleted or knocked down as in the in vivo studies, or absent as in the in vitro studies.
Kinesin-8 motors are localized to microtubules in the mitotic spindle [10], and deletion of Kinesin-8 motors prevents proper chromosome congression in some organisms [9,11-14]. Thus, in this review we discuss how the principles revealed through these in vitro studies might provide a mechanism by which chromosome oscillations and the congression of chromosomes to the spindle equator take place during mitosis.
Kinesin-8 motors and the mitotic spindle
Recently, Stumpff et al. [15] addressed this question by carefully quantifying kinetochore oscillations in mitotic HeLa cells that had varying expression levels of the Kinesin-8 molecular motor Kif18A. Surprisingly, the in vivo effect of Kinesin-8 molecular motors on microtubule depolymerization rates appears to be the opposite of the reported in vitro effect on GMPCPP-stabilized microtubules. Although the presence of Kinesin-8 motors results in more rapid in vitro depolymerization of longer GMPCPP-stabilized microtubules, the presence of Kif18A in vivo appears to slow the rate of microtubule-shortening during both metaphase and anaphase (Table 1) [15]. This result is consistent with recent in vivo measurements of yeast cytoplasmic microtubule dynamics, in which shortening rates were slowed by ~48% in the presence of Kip3p (Table 1) [16]. Mayr et al. [9] reported slower in vivo chromosome oscillation velocities with Kif18A depletion. However, these velocities were probably underestimated, because the observed frame-rate (0.91 frames/min9) was similar to the kinetochore-switching rate (in cells depleted of Kif18A, the switching rate was ~1.23 switches/min15). Thus, at least in part, the loss of chromosome congression in human HeLa cells depleted of Kif18A is the result of larger chromosome oscillation amplitudes that originate from rapid microtubule depolymerization in the absence of Kif18A.
Depolymerization at the microtubule plus-end
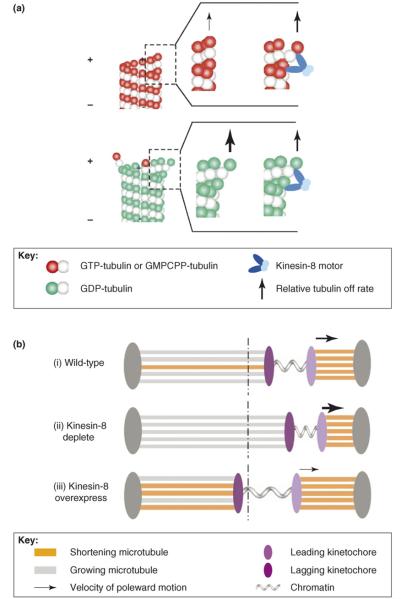
It is interesting to consider how these opposing effects of Kinesin-8 on in vitro GMPCPP microtubule depolymerization rates and on in vivo GDP-tubulin microtubule depolymerization rates might arise. Given that stable GMPCPP microtubules are composed primarily of GTP–tubulin subunits, in vitro GMPCPP microtubule protofilaments prefer to be relatively straight. This means that the strain between the motor domains of the plus-end-directed Kinesin-8 molecular motor might force outward protofilament curling (Figure 1a, upper panel). This curling would destabilize the lateral bonds with neighboring tubulin subunits, which would then increase the dissociation rate of the most distal tubulin subunit, provided that the lost lateral-bond energy and the gained curling-energy together exceed the motor-cross-linking energy. Based on our previous modeling of microtubule disassembly [17,18], we estimate that the lost lateral-bond energy in the presence of the motor is ~+5 kBT (this is for a single bond, where 1 kBT/single bond=0.6 kcal/mol of bonds; note that a high-affinity bond, KD~1 nM, has a standard Gibbs free-energy change of −21 kBT, and that a moderate affinity bond, KD~1 μM, has an energy change of −14 kB T). Furthermore, we estimate that the curling energy penaly is ~+3 kBT, for a total of +8 kBT of unfavorable energy. Given that this is less than the Gibbs free-energy of GTP hydrolysis (i.e. ~−20 kBT), it is feasible for the motor to generate such strains. By comparison, the longitudinal cross-linking energy of XMAP215 – in this case, it is considered to be a MAP that can cross-link tubulin dimers in a manner similar to Kif18A – was estimated to be ~−4kBT17. Thus, there is a possible net increase of ~+4 kBT of unfavorable energy due to outward protofilament-curling generated from strain between the motor-walking domains. This would increase the tubulin dissociation rate by more than 50-fold. A similar Kinesin-8-mediated mechanism would then presumably apply to growing microtubules in vivo with a stabilizing GTP–tubulin cap. Curling of GTP–tubulin subunits at growing microtubule tips might promote removal of the stable GTP–tubulin cap and this in turn would initiate microtubule catastrophe by exposing unstable GDP–tubulin subunits at the microtubule tip. The difference between GMPCPP-stabilized microtubules in vitro and GDP-tubulin microtubules in vivo is that, once uncapped, depolymerization of GDP–tubulin microtubules then proceeds spontaneously through the curling and peeling of unstable GDP–tubulin protofilaments. In this case, Kinesin-8 motors might act passively to cross-link and stabilize the bonds between neighboring tubulin subunits (Figure 1a, lower panel), thereby slowing the in vivo depolymerization rate. Thus, it might be that the microtubuledestabilizing effect of Kinesin-8 is manifested in vivo by an increase in microtubule catastrophe frequency – as measured recently in experiments with cytoplasmic microtubules in yeast (Table 1) [16] – even though Kinesin-8 motors actually slow the microtubule-shortening rate in vivo. This behavior is consistent with the observation that the Drosophila Kinesin-8 motor Klp67A tends to destabilize dynamic microtubules during metaphase, while stabilizing the formation of the central spindle during anaphase. These effects might be caused by increased microtubule catastrophe during metaphase with a reduced interpolar microtubule depolymerization rate during anaphase [4]. It would be interesting to test the effect of Kinesin-8 on dynamic GTP–tubulin microtubule depolymerization rates in vitro, in an effort to determine whether these contrasting behaviors can be observed.
Figure 1.
Kinesin-8 motors and microtubules. (a) In vitro microtubule-shortening rates are higher in the presence of Kinesin-8 molecular motors, whereas the opposite is observed in vivo. Stable GMPCPP microtubules used in the in vitro experiments (upper panel) might be more sensitive to mechanical tip destabilization through Kinesin-8-mediated protofilament kinking, whereas GDP–tubulin microtubules in vivo (lower panel) show rapid, spontaneous depolymerization; thus Kinesin-8 molecular motors might act to passively slow the natural depolymerization process (see text for more quantitative details). (b) A model for Kif18A-mediated kinetochore oscillation amplitude. (i) In wild type spindles, longer microtubules attached to lagging kinetochores are selectively targeted by Kif18A for catastrophe and subsequent depolymerization, decreasing coordination in the polymerization state and slowing the velocity of pole-ward motion (dotted line represents the spindle equator). (ii) In Kif18A depletion, longer, polymerizing microtubules are not targeted for catastrophe and depolymerization. In addition, rapid microtubule-shortening results in faster pole-ward movement of the leading kinetochore. (iii) Conversely, overexpression of Kif18A might result in increased catastrophe of microtubules attached to the lagging kinetochore, decreasing the coordination in polymerization state. Combined with a slower microtubule depolymerization rate, pole-ward kinetochore motion is slowed, decreasing oscillation amplitude, and resulting in increased tension between sister kinetochores.
Kinesin-8 mediated regulation of chromosome oscillations
As shown by Stumpff et al. [15], the increased microtubule-shortening velocity is a major factor in the increased kinetochore oscillation amplitude in Kif18A-depleted cells. In these cells, it is likely that shortening microtubules drive pole-ward motion of leading kinetochores [19]. As described above, Kif18A might also promote the length-dependent catastrophe of longer microtubules [8,9]. The promotion of microtubule plus-end catastrophe based on the length of the microtubule might provide an efficient mechanism for establishing a spatial gradient in spindle microtubule dynamics [20] that would regulate microtubule lengths and thus promote chromosome congression during mitosis [21]. Recent modeling studies provide a framework for integrating collective microtubule dynamics to better understand their role in chromosome oscillations and congression [22-24]. Specifically, modeling work by Civelekoglu-Scholey et al. [22] predicts that, for chromosome oscillations to occur, the polymerization state of microtubule ends must be at least partially synchronized, and that pole-ward chromosome motion is dependent on depolymerization of microtubules attached to the leading kinetochore (Figure 1b(i)). Thus, the attenuation of the microtubule depolymerization rate in wild type spindles would be predicted to reduce oscillation amplitude (Figure 1b(i)). In addition, microtubule catastrophe promotion, as mediated by Kif18A, would be predicted to exert its effect primarily on the longer microtubules attached to lagging kinetochores (Figure 1b(i)), increasing tension between sister kinetochores and, through reduced synchronization of the polymerization state, limiting oscillation amplitude. As shown in Figure 1b(ii), depletion of Kif18A would thus increase shortening velocity for microtubules attached to the leading kinetochore, and, through a lowered catastrophe frequency for the longer microtubules attached to the lagging kinetochores, increase the synchronization of the polymerization state. Overexpression of Kif18A would have the opposite effect, as shown in Figure 1b(iii). This model predicts that Kif18A accumulates preferentially on longer microtubules attached to lagging kinetochores, which is consistent with the asymmetric distribution of Kif18A on sister kinetochores, as was observed by Stumpff et al. [15]. Although a more complete description of kinetochore directional instability probably involves both the coordination of microtubule plus-end polymerization state and the dynamic turnover of microtubule-kinetochore attachments [22-25], the results reported by Stumpff et al. [15] suggest that Kinesin-8 regulates microtubule dynamics to limit kinetochore oscillation amplitude and increase tension between sister kinetochores.
Conclusions
Although the recent in vitro and in vivo Kinesin-8 studies shed significant light on the contribution that this molecular motor makes to mitotic chromosome congression, open questions remain. For example, in budding yeast, in which each kinetochore is associated with a single microtubule, depletion of Kinesin-8 has no apparent effect on proper kinetochore congression [10]. In addition, as reported by Stumpff et al. [15], the increase in kinetochore oscillation amplitude in HeLa cells is limited to two-fold, even with >90% depletion of Kif18A. This suggests that additional mechanisms, such as polar ejection forces, contribute significantly to the oscillation and centering of chromosomes during mitosis.
References
- 1.Mitchison TJ, Salmon ED. Mitosis: a history of division. Nat. Cell Biol. 2001;3:E17–E21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 3.Walker RA, et al. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatt MK, et al. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J. Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- 5.West RR, et al. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luboshits G, Benayahu D. MS-KIF18A, new kinesin; structure and cellular expression. Gene. 2005;351:19–28. doi: 10.1016/j.gene.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varga V, et al. Yeast Kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 9.Mayr MI, et al. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia MA, et al. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- 12.Savoian MS, et al. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J. Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- 13.Gandhi R, et al. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West RR, et al. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- 15.Stumpff J, et al. The Kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta ML, Jr, et al. Plus end-specific depolymerase activity of Kip3, a Kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 17.VanBuren V, et al. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys. J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanBuren V, et al. Estimates of lateral and longitudinal bond energies within the microtubule lattice. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6035–6040. doi: 10.1073/pnas.092504999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Gardner MK, et al. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civelekoglu-Scholey G, et al. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys. J. 2006;90:3966–3982. doi: 10.1529/biophysj.105.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joglekar AP, Hunt AJ. A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys. J. 2002;83:42–58. doi: 10.1016/S0006-3495(02)75148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner MK, Odde DJ. Modeling of chromosome motility during mitosis. Curr. Opin. Cell Biol. 2006;18:639–647. doi: 10.1016/j.ceb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Cimini D, et al. Anaphase spindle mechanics prevent missegregation of merotelically oriented chromosomes. Curr. Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]