Abstract
Viruses frequently infect the heart but clinical myocarditis is rare, suggesting that the cardiac antiviral response is uniquely effective. Indeed, the Type I interferon (IFN) response is cardiac cell type-specific and provides one integrated network of protection for the heart. Here, a proteomic approach was used to identify additional proteins that may be involved in the cardiac antiviral response. Reovirus-induced murine myocarditis reflects direct viral damage to cardiac cells, and offers an excellent system for study. Primary cultures of murine cardiac myocytes were infected with myocarditic or non-myocarditic reovirus strains, and whole cell lysates were compared by two-dimensional difference gel electrophoresis (2D-DIGE) and matrix-assisted laser desorption / ionization – time-of-flight (MALDI-TOF/TOF) tandem mass spectrometry. Results were quantitative and reproducible, and demonstrated that whole proteome changes clustered according to viral pathogenic phenotype. Moreover, the data suggest that the heat shock protein Hsp25 is modulated differentially by myocarditic and non-myocarditic reoviruses and may play a role in the cardiac antiviral response. Members of seven virus families modulate Hsp25 or Hsp27 expression in a variety of cell types, suggesting that Hsp25 participation in the antiviral response may be widespread. However, results here provide the first evidence for a virus-induced decrease in Hsp25/27, and suggest that viruses may have evolved a mechanism to subvert this protective response, as they have for IFN.
Keywords: Cardiac myocyte, Myocarditis, Reovirus, Hsp25, Hsp27, Proteome, DIGE
Introduction
Viruses are the most common pathogen causing myocarditis, with random autopsies suggesting that between 5 and 20% of the population has suffered a viral infection in the heart 1–3. While most cases are asymptomatic, myocarditis is frequently fatal in infants, and adult infections can progress to dilated cardiomyopathy requiring heart transplantation 2, 4. Moreover, greater than 50% of sudden deaths in young adults are due to cardiac causes with greater than 10% of those due to myocarditis 5, 6. While immune-mediated pathology can occur, immunosuppressive therapy is not beneficial 7, suggesting that most damage reflects direct viral cytopathic effect (CPE). Most virus families have been implicated in human viral myocarditis, with adenoviruses and coxsackieviruses indicated most often 8, 9. However, coxsackieviruses induce both direct and immune-mediated pathology in mice 10, and adenovirus-induced myocarditis remains poorly studied in an animal model 11. In contrast, reovirus-induced myocarditis in neonatal mice involves only direct viral CPE reflecting virus-induced apoptosis or non-apoptotic cell death, providing a well-characterized model system for study 12–17.
Virus-induced pathology is particularly problematic in the heart because only 1% of cardiac myocytes are renewed in the average human lifetime 18. While the blood-brain barrier protects the similarly vulnerable central nervous system from pathogens, there is no identifiable cell architecture protecting the heart. Instead, it is likely that cardiac cells have evolved uniquely effective innate responses to limit virus spread through the heart until immune cells can react. Indeed, the cytokine IFN-β is a determinant of protection against reovirus-induced murine myocarditis 19. That is, virus strain-specific differences in the capacity to induce myocarditis correlate with differences in viral induction of IFN-β and viral sensitivity to the antiviral effects of IFN-α/β in primary cultures of cardiac myocytes. Moreover, a non-myocarditic reovirus induces cardiac lesions in mice lacking IFN-α/β function 19. It seems unlikely, however, that the heart would have evolved only a single antiviral strategy given its constant exposure to viral pathogens. To identify additional proteins that may be involved in the cardiac antiviral response, a proteomic approach was used here to compare cardiac myocyte responses to infection with myocarditic and non-myocarditic reoviruses. Assessing primary cardiac myocyte cultures rather than hearts from infected mice ensured measurement of the innate cardiac myocyte response rather than that of a mix of cardiac and inflammatory cells.
While microarray technology offers an excellent tool to compare mRNA levels between samples, the prevalence of post-transcriptional regulation in eukaryotic cells precludes accurate predictions about final protein expression. Accordingly, proteomic approaches have become increasingly popular and have provided a useful catalog of cardiac protein expression 20–22. Two-dimensional difference gel electrophoresis (2D-DIGE) 23 is a powerful tool that builds on traditional 2D polyacrylamide gel electrophoresis, which provided one of the earliest methods to separate closely-related proteins 24, 25. Indeed, 2D-DIGE has been used to investigate differences between healthy and diseased hearts for a number of conditions, however none have examined viral infections and none have investigated purified cardiac myocytes 26–31. The two major strengths of 2D polyacrylamide gel electrophoresis are that it allows both separation of thousands of proteins simultaneously and identification of post-translational modifications. In particular, the use of intermediate ranges of pH gradients (i.e. pH 4 to 7 or 6 to 9) in the isoelectric focusing (IEF) dimension of the gel greatly enhances separation of proteins which would otherwise co-migrate in a traditional 2D gel (ph 3 to 10). In 2D-DIGE, proteins are labeled with fluorescent cyanine (Cy) dyes before electrophoresis, to both increase the sensitivity and broaden the dynamic range of protein quantitation 32, 33. Because proteins are labeled after cells are harvested, there is no risk of perturbing the cell environment and affecting cell responses. Moreover, labeling samples from different treatments with Cy3 and Cy5 and labeling a pool of multiple samples with Cy2 as an internal control allows quantitative comparison among an unlimited number of samples on different gels 34. Once gel spots are selected, they are proteolytically digested and analyzed by tandem mass spectrometry (MS/MS) for protein identification, from a preparative-scale gel 35.
The work reported here demonstrates that 2D-DIGE analysis of primary cardiac myocyte cultures provides quantitative and reproducible results, and that proteome changes cluster according to viral pathogenic phenotype. Moreover, results suggest that the heat shock protein Hsp25 is modulated differentially by myocarditic and non-myocarditic reoviruses and may play a role in the cardiac antiviral response.
Materials and Methods
Mice and primary cell cultures
Timed-pregnant Cr:NIH(S) mice were purchased from the National Cancer Institute. IFN-α/β-receptor-null 36 mice were maintained as breeding colonies to generate neonates and fetuses for generation of primary cell cultures. Mice were housed according to the recommendations of the Association for Assessment and Accreditation of Laboratory Animal Care, and all procedures were approved by the North Carolina State University institutional animal care and use committee. Primary cardiac myocyte cultures were generated from 1- or 2-day old neonatal or term fetal mice as described previously 13. Briefly, the apical two-thirds of the hearts from euthanized neonates or fetuses were removed and trypsinized. Cells were suspended in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Gaithersburg, MD) supplemented with 7% fetal calf serum and 10 µg of gentamicin (Sigma Co.) per ml (completed DMEM; cDMEM). Cells were plated in 6-well clusters, and incubated for 2 hours for separation by differential adhesion. Cardiac myocytes were harvested from the supernatant and the adherent cardiac fibroblasts were harvested by trypsinization. Following centrifugation, cardiac myocytes were suspended in cDMEM supplemented with 0.06% thymidine (Sigma Co., St. Louis, MO), cardiac fibroblasts were suspended in cDMEM, and both cultures were incubated in a 37°C, 5% CO2 incubator. All infections and incubations were in these media at 37°C unless otherwise indicated. Cells were never passaged before use. By immunofluorescent staining, the myocyte cultures contained <5% fibroblasts 37.
Viruses and cell lines
Reovirus type 3 Dearing (T3D) and type 1 Lang (T1L) were plaque purified and amplified in mouse L929 cells, which are maintained in a suspension system in sMEM (SAFC Biosciences, Denver, PA) supplemented with 5% fetal calf serum (Atlanta Biologicals, Atlanta, GA) and 2mM L-glutamine (Mediatech Inc., Herndon, VA). Reovirus 8B is a reassortant virus derived from a mouse infected with T3D and T1L (BJS-17). DB93A was derived from mouse L929 cells infected with 8B and a reassortant virus (EB121) derived from T3D and T1L 17. All viruses were purified by CsCl gradient centrifugation 38.
Infections
All primary cardiac cell cultures were incubated for two days before infection. For 2D-DIGE, cardiac myocytes were plated at 2 × 106 cells per well in 12-well clusters. Triplicate wells were infected with media or virus at 10 pfu per cell in 1400 µl for 1 hour, and then supplemented with an additional 2 ml media. Cultures were harvested 12 hours post-infection. For MTT assays, cardiac myocytes were plated at 1.5 × 105 cells per well in 96-well clusters. Triplicate wells were treated with 50 µM p38 MAPK inhibitor SB203580 (EMD CalBiochem, cat #559389) or vehicle for 1 hour, infected with media or virus at 25 pfu per cell in 100 µl for 1 hour, and then supplemented with an additional 100 µl media. Cultures were harvested 48 to 72 hours post-infection. For Western blots, cardiac myocytes were plated at 1 × 106 cells and cardiac fibroblasts at 5 × 105 cells per well in 24-well clusters. Duplicate wells were infected with media or virus at 10 pfu per cell in 700µl for 1 hour, and then supplemented with an additional 1000 µl media. Cultures were harvested 8 to 48 hours post-infection. For qRT-PCR, cardiac myocytes were plated and infected as for Western blots, and harvested 30 minutes to 24 hours post-infection.
Reagents for 2-dimensional difference gel electrophoresis (2D-DIGE)
Sodium chloride, Trizma base, bromophenol blue, chloroform, DMF, phosphatase inhibitor, sodium carbonate, sodium bicarbonate, ammonium bicarbonate, ammonium monobasic phosphate, L-lysine, and α-cyano-4-hydroxycinnamic acid were obtained from Sigma Aldrich. Urea, thiourea, SDS, DTT, Tris, and iodoacetamide were purchased from GE Healthcare. CHAPS was obtained from USB. Complete Mini Protease Inhibitor Cocktail Tablets were obtained from Roche Applied Science. Methanol, acetonitrile, and HPLC grade water were obtained from Burdick & Jackson. Acetic acid was obtained from J.T. Baker.
Sample Preparation and Protein Labeling
Total cell protein harvest procedures and protein concentration determinations were as for SDS-PAGE. Protein disulfide bonds were reduced in 2mM DTT. For each sample, 85 µg protein was precipitated using 4:1 methanol:chloroform, washed once with methanol, and resuspended in L-buffer (7M Urea, 2M Thiourea, 4% CHAPS, 30 mM Tris, pH 8.5). An internal standard was created by pooling 35.3 µg of each of the 15 samples. In order to create an even number of samples, 26 µl of the pool was used to create a 16th sample. Then, 26 µl of each of the 16 samples was labeled with 4.25 pmol of either Cy3 or Cy5 dye per µg protein. In order to avoid dye binding bias, two samples from each virus infection were labeled with one of the two dyes and the third sample was labeled with the other dye. The internal pooled standard was labeled using 3.32 pmol / µg Cy2. Reactions were stopped by adding 10 mM lysine in a volume equal to that of the dye. For each gel, a Cy3- and a Cy5-labeled sample was mixed with an equal volume (30 µl) of Cy2-labeled standard. Samples were then reduced and denatured in R-buffer (7M urea, 2M Thiourea, 4% CHAPS, 13mM DTT) at 10:1 v:v with ampholytes (IPG Buffer, pH 4–7, GE Healthcare). The samples were then applied to a 24 cm, pH 4–7 Immobiline DryStrip (GE Healthcare).
2-dimensional difference gel electrophoresis (2D-DIGE)
Gels were loaded with different combinations of the 16 samples such that no two infections were ever directly compared more than once. First dimension isoelectric focusing was performed using an Ettan IPGphor II (GE Healthcare) for a total focusing time of 64.5kVHrs. at 20°C and 75µA / strip max (using Ettan IPGphor II Control Software). After focusing was complete, each strip was equilibrated for 15 minutes with 10 ml equilibration buffer (6M Urea, 75 mM Tris pH 8.8, 29.3% Glycerol, 2% SDS, 0.002% bromophenol blue) supplemented with 10 mg/ml DTT. The strips were then equilibrated for 15 minutes with 10 ml equilibration buffer supplemented with 25 mg/ml iodoacetamide. The strips were briefly washed with 1× Running Buffer (NextGen Sciences) to remove any excess equilibration buffer, and then loaded onto 12% Homogenous SDS-PAGE gels (NextGen Sciences). Electrophoresis was performed on an Ettan DALTtwelve System Separation Unit (Amersham Biosciences) at 0.5W/gel until the bromophenol blue dye front had just run off the bottom of the gel. Gels were scanned at 488 nm(Cy2), 532 nm(Cy3), and 633 nm(Cy5) using a Typhoon Trio Variable Mode Imager (GE Healthcare).
DIGE image analysis
DeCyder 2D Software, Version 6.5 from GE Healthcare was applied to visualize the digitalized images from the scanner 32, 33. Protein spots were identified in differential in-gel analysis (DIA) module, and gels were matched to the master gel in the biological variation analysis (BVA) module. DeCyder Extended Data Analysis (EDA) module Version 1.0 was applied to carry out the DIA analysis, principle component analysis (PCA) and pattern analysis. Protein spots were selected for identification based on their differential expression among different groups (P < 0.05 using one-way ANOVA) and their relative abundance in 3D visualization.
Preparative Gel Electrophoresis/ Protein Identification
A fraction of each of the 15 samples was used to generate a pool of 356 µg protein for preparative 2D-gel electrophoresis. The gel was stained for 1 hour with Deep Purple Total Protein Stain (1:200, GE Healthcare), scanned on the Typhoon Trio, and the image was imported for DIGE analysis using DeCyder software. After matching back to the master gel, gel plugs corresponding to differentially expressed proteins were isolated using an Ettan Spot Picker (GE Healthcare). Destained and dehydrated gel spots were digested overnight in Trypsin Gold, and then desalted and concentrated using C18 P10 ZipTips (Millipore). One third of each sample was loaded onto a ZipTip by pipetting up and down five times using a fresh 96-well plate. Samples (in ZipTips) were washed four times with 10 µL of 0.1% TFA and eluted with 2 µL of 50% acetonitrile / 0.1% trifluoroacetic acid. A total of 1 µL of eluate was pipetted onto a clean MALDI plate and covered with 1µL of α-cyano-4-hydroxycinnamic acid MALDI matrix.
Mass spectra for each spot were acquired using an Applied Biosystems 4700 Proteomics Analyzer MALDI-TOF/TOF, running version 3.0 software. Data-dependent MS/MS analysis was performed on the top 10 peaks from each MS spectrum. Peaklists for MS/MS database searching were generated using the “Peaks to Mascot” application included with the 4700 database search software. The data were searched using Mascot (v1.9.05) against the Swiss-Prot database (version 200403115, 146193 entries) using trypsin as the enzyme with three missed cleavage sites considered. Carbamidomethylation of cystienes and oxidation of methionine were considered as fixed modifications and precursor/product ion mass tolerances were set at ± 0.5 Da. Proteins identified with two or more unique individual peptide scores above the 95% confidence level cut-off were accepted as positive identifications. Protein-protein interaction and pathway analysis were assessed using Ingenuity Pathways Analysis (IPA 5.0; Ingenuity® Systems, www.ingenuity.com). Theoretical protein digestion was performed using ProteinProspector from University of California, San Francisco (http://169.230.19.26:8080/cgi-bin/msform.cgi?form=msdigest). Predictions of protein phosphorylation sites were performed using NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/).
SDS-PAGE and Western blot analysis
Infected cardiac myocyte cultures were washed four times with ice-cold 1× PBS. Total cell protein was harvested at the indicated times by incubation in TNE lysis buffer (50 mM Tris HCl pH 7.6, 150 mM NaCl, 2 mM EDTA pH 8.0, 1% [v/v] NP-40 containing a cocktail of protease and phosphatase inhibitors [Sigma Co.; P8340 and P2850]) on ice for 30 minutes with shaking. Lysates were centrifuged at 10,000 × g for 10 minutes at 4°C to remove cellular debris. For the data shown in Figure 7, cytoplasmic and nuclear protein was harvested at 17 hours post-infection by NE-PER kit (Pierce, Rockford, IL). Protein concentrations were determined using a bicinchoninic acid kit (Pierce, Rockford, IL), and 40 µg was boiled for 5 minutes in 1× Laemmli sample buffer and subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred to nitrocellulose membranes, and membranes were blocked for 1 hour at room temperature in 3% milk in Tris-buffered saline (20 mM Tris [pH 7.6], 137 mM NaCl) containing 0.05% Tween 20 (TBS-T) and then probed overnight at 4°C with the indicated primary antibody (1:1000 of Hsp25 polyclonal antibody, cat. # SPA-801; 1:1000 of Hsp27-phospho-Ser82 polyclonal antibody, cat. # SPA-524; 1:500 of Hsp27-phospho-Ser15 polyclonal antibody, cat. # SPA-525; Assay Designs, Ann Arbor, Michigan). Membranes were washed for 5 minutes twice in TBS-T, incubated for 1.5 hours at room temperature in goat anti-rabbit HRP-conjugated secondary antibody (Millipore, Catalog # AP132P), and then washed for 10 minutes three times in TBS-T. Antibody-labeled proteins were detected according to manufacturer’s recommendations (Amersham™ ECL™ or ECL™ Plus Western Blotting Detection reagent, GE Healthcare, Buckinghamshire, United Kingdom). Western blots were exposed to film and converted to digital format using an HP Scanjet 5470c.
Figure 7. Reovirus does not induce nuclear translocation of Hsp25.
Primary cardiac myocyte cultures were mock- or reovirus-infected (moi 10 pfu per cell), and cytoplasmic and nuclear lysates were harvested from duplicate wells at 17 hours post-infection for SDS-PAGE and Western blot to detect total Hsp25 (short film exposure to emphasize cytoplasmic Hsp25 differences; long film exposure to detect nuclear Hsp25). Results are representative of 3 experiments.
Cell viability (MTT assay)
At 48 or 72 hours post-infection, 20 µl of 0.6% MTT(catalog #M-5655; Sigma) in cDMEM was added to each well and cultures were incubated for 4 hours at 37°C. Culture plates were centrifuged at 750 × g for 8 minutes, supernatants were removed, and 100 µl of 0.04 N HCl in isopropanol was added to each well. Plates were incubated at room temperature for 15 minutes, 100 µl H2O was added to each well and the optical densities at 570 and 630 nm were determined on a Tecan Sunrise Microplate Reader (Tecan Systems Inc., San Jose, CA). Results are expressed as signal (OD570) minus background (OD630).
Indirect immunofluorescence
Procedure was as described previously 37. In brief, hearts from adult Cr:NIH(S) mice were washed in ice-cold 1× PBS and snap frozen in liquid nitrogen-cooled isopentane (Fisher Scientific Co.). Tissue samples were mounted frozen on a metal chuck using a small volume of Tissue-Tek OCT (Ted Pella, Inc., Redding, CA) in a cryostat (Leica CM1850) at −20°C. Transverse cryosections (1 µm) were collected on SuperFrost/Plus slides (Fisher Scientific) and stored at −80°C. Sections were thawed and blocked for 1 hour at room temperature with 10% donkey serum (Sigma) and 10% goat serum (Sigma Co.) in 1× phosphate-buffered saline (PBS) plus 0.1% Triton X-100 (PBST) for vimentin, or with 10% donkey serum in PBST for myomesin, and then probed overnight at 4°C with a mix of rabbit anti-Hsp25 (1:1000) and goat anti-myomesin (1:50, cat. # 30384, Santa Cruz Biotechnology, Inc.) or chicken anti-vimentin (1:5,000, Affinity Bioreagents Inc., Golden, CO) antibodies in 1 × PBS with 0.3% IgG-free, protease-free bovine serum albumin (BSA)(Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Slides were washed three times in 1 × PBS, and then incubated for 2 hours at room temperature with a mix of Alexa fluor 594 goat anti-chicken IgG (Invitrogen, SKU# A-11042, 1:1000) plus Alexa fluor 488 donkey anti-rabit IgG (Invitrogen, SKU# A-21206,1:1000) for detection of vimentin and Hsp25, or alexa fluor 594 donkey anti-goat IgG (Invitrogen, SKU# A-11058, 1:1000) plus Alexa fluor 488 donkey anti-rabbit IgG (1:1000) in 1×PBS with 0.3% IgG-free, protease-free BSA for detection of myomesin and Hsp25. Slides were washed in 1 × PBS, coverslips were mounted with Prolong Gold reagent (Invitrogen), and slides were analyzed using a Nikon TE-200 inverted epifluorescence microscope. Images were captured using a 100× objective under oil immersion, and were collected digitally (SPOT, Jr.; Diagnostics Instruments, Inc., Sterling Heights, MI) and processed using the manufacturer’s instructions and Adobe Photoshop (with no change in content).
Reverse transcription (RT) and Quantitative (q)RT-PCR
Cells were harvested at the indicated times post-infection using cell lysis buffer from an RNeasy Kit (Qiagen, Inc., Valencia, CA) supplemented with 1% β-mercaptoethanol, and homogenized using Qiashredders (Qiagen, Inc.), and total RNA was isolated using the RNeasy Kit according to the manufacturer’s instructions. Genomic DNA was removed using RNase-free DNase I (Qiagen, Inc.). The isolated RNA was stored at −80°C. To generate cDNA, one third of the RNA harvested from each well in a 24-well cluster or two thirds from a 48-well cluster was used as template in a 100 µl reaction containing 5 µM oligo(dT) (Invitrogen Corp., Carlsbad, CA), 1× Taq buffer (Promega corp., Madison, WI), 7.5 mM MgCl2 (Promega corp.), 1 mM dithiothreitol (Promega corp.), 1 mM each dNTP (Roche, Indianapolis, IN), 0.67 U/µl RNasin (Promega Corp., Madison, WI), and 0.20 U/µl of AMV reverse transcriptase (Promega Corp.). Gene expression was quantified using a Sybergreen system on an iCycler iQ fluorescence thermocycler (Bio-Rad Laboratories, Hercules, CA). Each 25 µl reaction contained 5% of the RT product, 1× Quantitech master mix (Qiagen, Inc.), 10 nM fluorescein (Invitrogen Corp., Carlsbad, CA), and 0.3µM each of the forward and reverse primers (GAPDH forward: 5’-GGGTGTGAACCACGAGAAAT-3’ and reverse: 5’-CCTTCCACAATGCCAAAGTT-3’; HSP25 Forward: 5’-GAAGAAAGGCAGGACGAACA-3’ and reverse: 5’-CTCAGGGGATAGGGAAGAGG-3’ ). iCyclerTM iQ Optional System Software, version 3.0 (Bio-Rad Laboratories) was used to analyze the data. Gene-of-interest expression was normalized to GAPDH gene expression.
Results
2D-DIGE analysis
The four reoviruses selected for comparison are comprised of two prototype strains (T1L and T3D 39), a potently myocarditic reassortant virus generated in vivo during a mixed infection with T1L and T3D (8B 17), and a non-myocarditic reassortant virus derived from 8B (DB93A 17) (Table 1). Together, they allow comparisons relating to both viral myocarditic potential and viral induction of IFN-β. A preliminary experiment comparing mock-and T3D-infected cultures at 8, 12 and 18 hours post-infection indicated that the greatest differences between mock- and virus-infected cultures were captured at 12 hours post-infection (data not shown), and therefore this single time-point was selected for further 2D-DIGE studies. Triplicate lysates from mock- or virus-infected primary cardiac myocyte cultures were labeled and electrophoresed with an internal control on a total of 8 gels (Fig. 1). The scanned 2D-DIGE gel images were analyzed using DeCyder 2D Software with exclusion filters set manually. The total number of protein spots detected on the 8 gels ranged from 5,617 to 7,408 per gel, with the master gel selected as the one with the greatest number of protein spots. A total of 3,000 protein spots were selected for comparisons based on their abundance exceeding a filter-set threshold, and 197 differentially expressed protein spots were detected in the EDA module with One-Way ANOVA and P < 0.01 as the threshold.
Table 1.
Reovirus strains selected for infection of primary cardiac myocyte cultures.
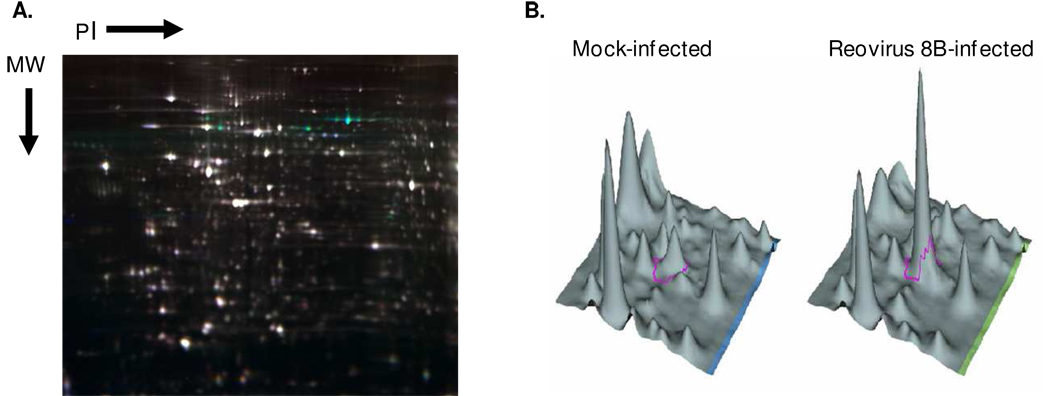
Figure 1. Representative 2D-DIGE gel image and 3D-view.
Triplicate wells of primary cardiac myocyte cultures were mock- or reovirus-infected (moi 10 pfu per cell, Table 1), and whole cell lysates were harvested 12 hours post-infection for 2D-DIGE. A) Representative gel containing mock- (Cy3-labeled) and 8B- (Cy5-labeled) infected cardiac myocyte samples as well as an internal standard (Cy2-labeled) composed of a pool of all samples. Green indicates greater expression in the 8B- than in mock-infected sample, red indicates the converse, white indicates equivalent expression, and blue indicates presence in the pooled internal control but neither of the two samples. B) Example of 3D-view for Cy3 or Cy5 intensity for a selected protein spot (ringed in pink).
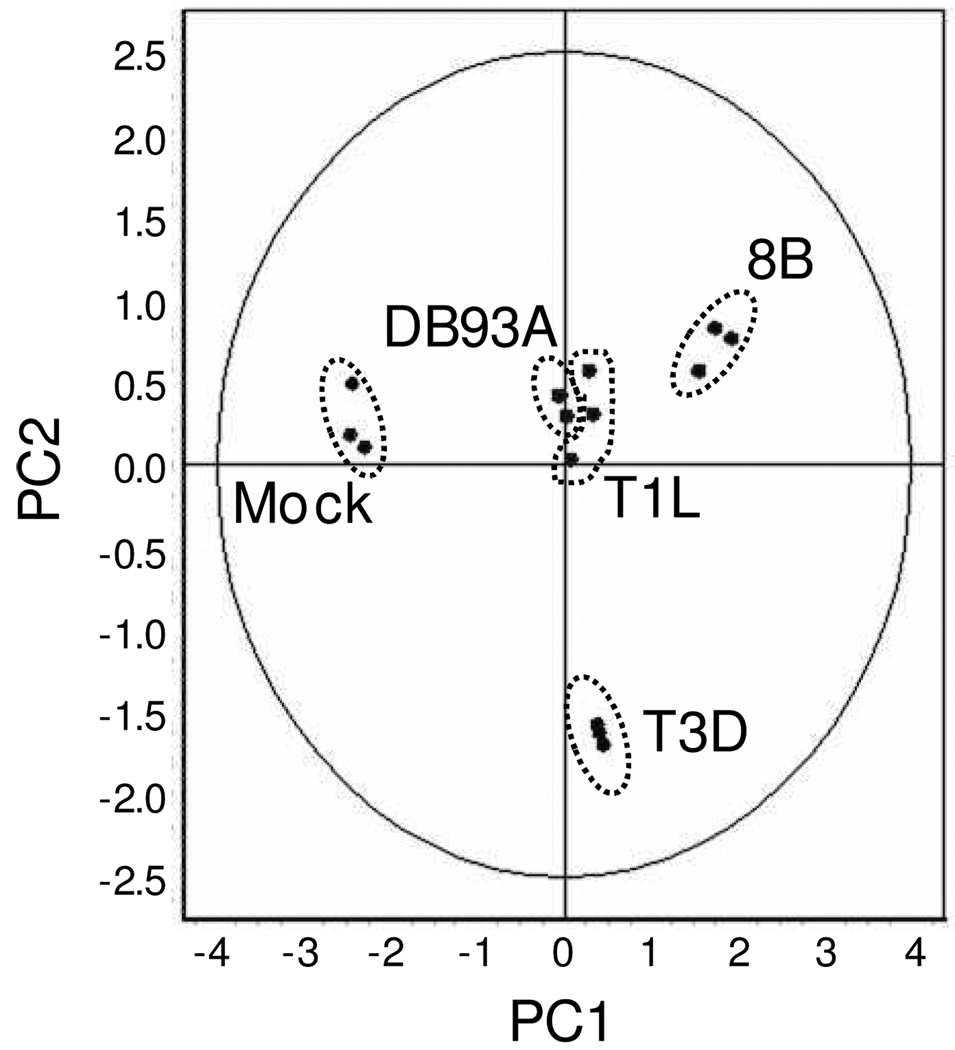
Principle component analysis (PCA), which is a statistical method to eliminate redundant variables and reduce data complexity, was performed on the 197 differentially expressed proteins (Fig. 2). Triplicate samples for each infection were tightly clustered, indicating high reproducibility between primary cardiac myocyte culture wells and between gels. Reovirus-infected samples were most distantly segregated from mock-infected samples. The two viruses that induce IFN-β poorly (T1L and DB93A) were clustered closely. The two other viruses, 8B and T3D, were segregated by myocarditic potential. Thus, global proteome changes correlated well with viral phenotypes.
Figure 2. Principle Component Analysis (PCA).
PCA was performed on the triplicate samples for the 197 differentially expressed proteins identified by 2D-DIGE. Each data point represents all 197 proteins for one sample. PC1 accounted for 55.8% of the variance, while PC2 accounted for 23.3% of the variance.
Protein identification by MALDI-TOF/TOF and pathway analysis
The 3000 proteins were re-analyzed by One-way Anova at a lower threshold of significance (P < 0.05) to increase the pool of proteins for subsequent analyses. This identified 227 differentially expressed protein spots, which were then picked for identification by MALDI-TOF/TOF and database searches. Of the 227 protein spots, 71 met the identification threshold criteria and corresponded to unique proteins (rather than hypothetical proteins or post-translational modifications of the same protein). These proteins are listed along with the number of unique peptides identified and sequence coverage in Table 2. Proteins fell into many groupings, based on whether they were up- or down-regulated in viral infections, and whether accumulation was virus strain-specific or segregated by previously characterized viral phenotypes (e.g. induction of IFN, or induction of myocarditis). Supplemental Table 1 provides an example of proteins that are uniquely altered in 8B-infected cultures, grouped by biological function.
Table 2.
List of Proteins Identified from Preparative Gel Scoring Above Thresholds
| Spot | Protein Name | # of Peptides Identified |
% Seq Coverage |
|---|---|---|---|
| 2301 | (P24142) Prohibitin (B-cell receptor associated protein | 9 | 49.1 |
| 1583 | (P29758) Ornithine aminotransferase; mitochondrial | 9 | 20.9 |
| 1392 | (P56480) ATP synthase beta chain; mitochondrial | 9 | 22.3 |
| 874 | (P08003) Protein disulfide isomerase A4 precursor | 9 | 14.5 |
| 852 | (P20029) 78 kDa glucose-regulated protein precursor | 9 | 17.0 |
| 689 | (P11499) Heat shock protein HSP 90-beta (HSP 84) | 9 | 15.0 |
| 199 | (P11087) Collagen alpha 1(I) chain precursor | 9 | 12.3 |
| 2024 | (Q61792) LIM and SH3 domain protein 1 (LASP-1) | 8 | 31.6 |
| 2017 | (P16125) L-lactate dehydrogenase B chain | 8 | 21.7 |
| 2002 | (O35639) Annexin A3 (Annexin III) (Lipocortin III) | 8 | 27.2 |
| 1705 | (P07940) RNA-directed RNA polymerase (EC 2.7.7.48) | 8 | 21.6 |
| 1153 | (P27773) Protein disulfide isomerase A3 precursor (EC | 8 | 14.0 |
| 558 | (Q9WU78) Programmed cell death 6 interacting protein | 8 | 12.8 |
| 1581 | (Q9CZ13) Ubiquinol-cytochrome C reductase complex | 7 | 23.6 |
| 1440 | (P09456) cAMP-dependent protein kinase type I-alpha | 7 | 24.3 |
| 1182 | (P19226) 60 kDa heat shock protein; mitochondrial | 7 | 22.4 |
| 677 | (Q9R0E1) Procollagen-lysine;2-oxoglutarate | 7 | 11.3 |
| 470 | (Q61316) Heat shock 70-related protein APG-2 | 7 | 11.0 |
| 462 | (Q01853) Transitional endoplasmic reticulum ATPase | 7 | 9.6 |
| 2525 | (O08807) Peroxiredoxin 4 (EC 1.11.1.-) (Prx-IV) | 6 | 28.0 |
| 2246 | (O35459) Delta3;5-delta2;4-dienoyl-CoA isomerase | 6 | 23.5 |
| 1812 | (Q99LC3) NADH-ubiquinone oxidoreductase 42 kDa | 6 | 28.5 |
| 1584 | (P35486) Pyruvate dehydrogenase E1 component | 6 | 13.2 |
| 1554 | (P21550) Beta enolase (EC 4.2.1.11) | 6 | 16.0 |
| 1464 | (Q99KJ8) Dynactin complex 50 kDa subunit | 6 | 22.0 |
| 1283 | (O08749) Dihydrolipoyl dehydrogenase; mitochondrial | 6 | 17.9 |
| 1219 | (P09103) Protein disulfide isomerase precursor | 6 | 16.2 |
| 1082 | (Q60715) Prolyl 4-hydroxylase alpha-1 subunit | 6 | 14.3 |
| 869 | (P38647) Stress-70 protein; mitochondrial precursor | 6 | 9.6 |
| 801 | (P15690) NADH-ubiquinone oxidoreductase 75 kDa | 6 | 10.0 |
| 666 | (P13020) Gelsolin precursor; plasma | 6 | 11.3 |
| 535 | (P56399) Ubiquitin carboxyl-terminal hydrolase 5 | 6 | 9.3 |
| 2585 | (Q60932) Voltage-dependent anion-selective channel | 5 | 24.8 |
| 2550 | (P14602) Heat shock 27 kDa protein (HSP 27) | 5 | 28.7 |
| 2448 | (P57759) Endoplasmic reticulum protein ERp29 | 5 | 16.8 |
| 2114 | (P14152) Malate dehydrogenase; cytoplasmic | 5 | 18.2 |
| 2074 | (P14869) 60S acidic ribosomal protein P0 (L10E) | 5 | 20.9 |
| 1784 | (P50752) Troponin T; cardiac muscle isoforms (TnTC) | 5 | 18.5 |
| 1563 | (Q64345) Interferon-induced protein | 5 | 15.6 |
| 954 | (P17879) Heat shock 70 kDa protein 1 (HSP70.1) | 5 | 10.3 |
| 860 | (P11078) Major virion structural protein Mu-1/Mu-1C | 5 | 11.5 |
| 776 | (P21981) Protein-glutamine | 5 | 10.7 |
| 759 | (P12419) Major nonstructural protein mu-NS | 5 | 8.3 |
| 746 | (P11077) Major virion structural protein Mu-1/Mu-1C | 5 | 11.5 |
| 2325 | (P09495) Tropomyosin alpha 4 chain (Tropomyosin 4) | 4 | 14.7 |
| 2097 | (P57776) Elongation factor 1-delta (EF-1-delta) | 4 | 20.8 |
| 1858 | (Q9CX34) Suppressor of G2 allele of SKP1 homolog | 4 | 16.4 |
| 1680 | (Q9QYR9) Acyl coenzyme A thioester hydrolase | 4 | 12.6 |
| 1592 | (P55264) Adenosine kinase (EC 2.7.1.20) (AK) | 4 | 13.0 |
| 1396 | (P47738) Aldehyde dehydrogenase; mitochondrial | 4 | 9.5 |
| 1280 | (Q61553) Fascin (Singed-like protein) | 4 | 10.1 |
| 1005 | (Q9EQP2) EH-domain containing protein 4 (mPAST2) | 4 | 5.7 |
| 918 | (P14733) Lamin B1 | 4 | 7.6 |
| 605 | (Q9R0B9) Procollagen-lysine;2-oxoglutarate | 4 | 7.7 |
| 441 | (Q02053) Ubiquitin-activating enzyme E1 1 | 4 | 4.1 |
| 2356 | (Q9WV35) Probable C->U editing enzyme APOBEC-2 | 3 | 17.6 |
| 1968 | (Q61937) Nucleophosmin (NPM) | 3 | 10.8 |
| 1964 | (P37140) Serine/threonine protein phosphatase | 3 | 11 |
| 1757 | (Q99KV1) DnaJ homolog subfamily B member 11 | 3 | 9.8 |
| 1614 | (Q07076) Annexin A7 (Annexin VII) (Synexin) | 3 | 7.3 |
| 1355 | (P30837) Aldehyde dehydrogenase X; mitochondrial | 3 | 8.5 |
| 1290 | (P54728) UV excision repair protein RAD23 homolog B | 3 | 6.4 |
| 1188 | (Q9CYT6) Adenylyl cyclase-associated protein 2 | 3 | 6.9 |
| 952 | (Q61233) L-plastin (Lymphocyte cytosolic protein 1) | 3 | 6.6 |
| 864 | (Q99MN1) Lysyl-tRNA synthetase (EC 6.1.1.6) | 3 | 5.5 |
| 797 | (O88487) Dynein intermediate chain 2 | 3 | 7.4 |
| 609 | (Q8K0D5) Elongation factor G 1; mitochondrial | 3 | 4.1 |
| 502 | (P08113) Endoplasmin precursor | 3 | 4 |
| 1894 | (P03527) Sigma 3 protein (Major outer capsid protein) | 2 | 6.4 |
| 1818 | (Q9JHJ0) Ubiquitous tropomodulin (U-Tmod) | 2 | 8.9 |
| 1025 | (Q9D0F9) Phosphoglucomutase (EC 5.4.2.2) | 2 | 5.7 |
These proteins also participate in diverse pathways, including those for calcium signaling, ERK/ MAPK signaling, protein ubiquitination, mitochondrial dysfunction, oxidative stress, and amino acid metabolism (data not shown). Of interest, the potently myocarditic reovirus 8B uniquely modulated proteins involved in mitochondrial dysfunction, endoplasmic reticulum stress, and phospholipid degradation, consistent with this virus’ extreme cytopathogenicity.
Analysis of post-translational modifications identifies Hsp25 phosphorylation
We found 18 cases where a single protein was identified in multiple spots with the same molecular weight but different pIs, suggestive of post-translational modifications. One of these proteins was Hsp25, the mouse homolog of human Hsp27 (Supplemental Fig. 1)
Three gel spots were identified as this protein from searches of the Swiss-Prot database . Each gel spot included high coverage for Hsp25 by peptide mass fingerprinting, and included sufficient MS-MS data for sequence confirmation. The individual Mascot results are shown as Supplemental Tables 2 – 4. By comparing the pool of un-matched peptide masses from the initial MS results to Hsp25 theoretical peptide masses using Protein Prospector, two un-matched peptides were identified as potentially phosphorylated (Supplemental Tables 3, 4). The predicted mono-phosphorylated peptide was absent from the most basic spot but present in the two more acidic spots, as would be predicted for successive phosphorylation. As would be expected, the potentially di-phosphorylated peptide was present in only the most acidic spot. Moreover, phosphorylation prediction software (NetPhos 2.0 Server) suggested phosphorylation of Hsp25 on those two peptides; on serine-15 (ser15), corresponding to the novel peptide in both mono- and di-phosphorylated Hsp25, and on ser86, corresponding to the novel peptide found only in di-phosphorylated Hsp25. Importantly, these two Hsp25 residues have previously been reported to be phosphorylated by various stimuli 40, 41. Western blot analyses confirmed these gel spot identifications (see below).
Quantification and confirmation of Hsp25
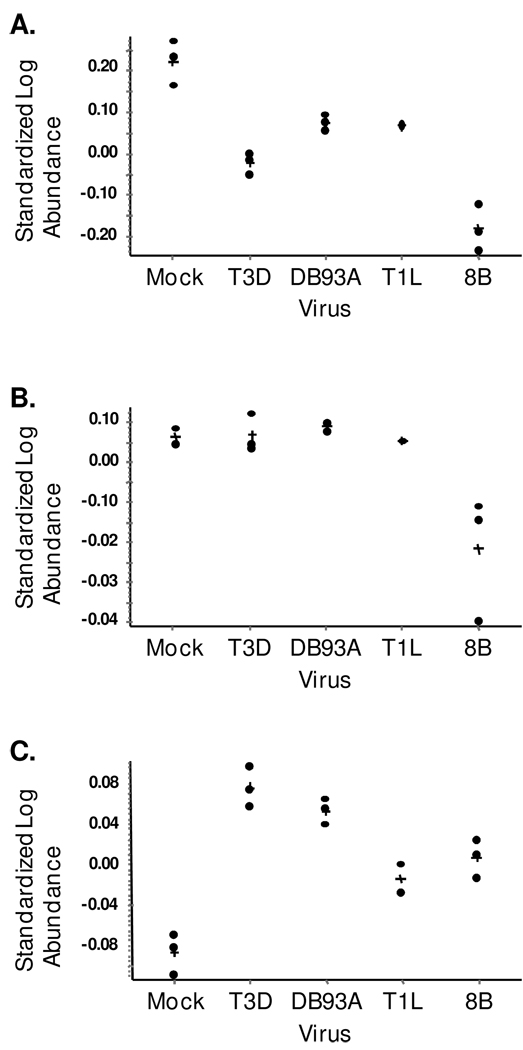
Protein abundance was determined for the predicted unphosphorylated, mono-phosphorylated and di-phosphorylated Hsp25 gel spots (Fig. 3). Unphosphorylated Hsp25 was most abundant in mock-infected samples (P < 0.03 to 0.001) and least abundant in 8B-infected samples (P < 0.01 to 0.002, Fig. 3A). Unphosphorylated Hsp25 was also less abundant in T3D- relative to DB93A- and T1L-infected samples (P < 0.02). One simple interpretation is that viral infection stimulates phosphorylation of Hsp25 resulting in decreased unphosphorylated Hsp25, and that the extent of phosphorylation is virus-specific. Indeed, di-phosphorylated Hsp25 was more abundant in reovirus- than mock-infected samples (P < 0.028 to 0.001 for the four viruses), and was more abundant in cultures infected with than non-myocarditic (T3D, DB93A) than myocarditic (8B, T1L) viruses (Fig. 3C, P < 0.02 for each of the four comparisons). Finally, mono-phosphorylated Hsp25 was similar between mock- and virus-infected cultures consistent with efficient further phosphorylation to the di-phosphorylated state, except for 8B (P < 0.04, Fig. 3B). The lower unphosphorylated and mono-phosphorylated Hsp25 in 8B-infected cultures without a corresponding increase in di-phosphorylated Hsp25 suggested that this potently myocarditic virus might induce degradation of Hsp25, or reduce its synthesis or stability.
Figure 3. Quantification of Hsp25 by 2D-DIGE.
Protein abundance was determined for each of the indicated gel spots using DeCyder software. A) Predicted unphosphorylated Hsp25 spot, B) Predicted mono-phosphorylated Hsp25 spot, C) Predicted di-phosphorylated Hsp25 spot. Results from triplicate samples are shown in each case except T1L, where only two (A and C) or one (B) protein spot was detectable on the gels. In some other cases, data-points lie directly on top of one another. “+” indicates mean.
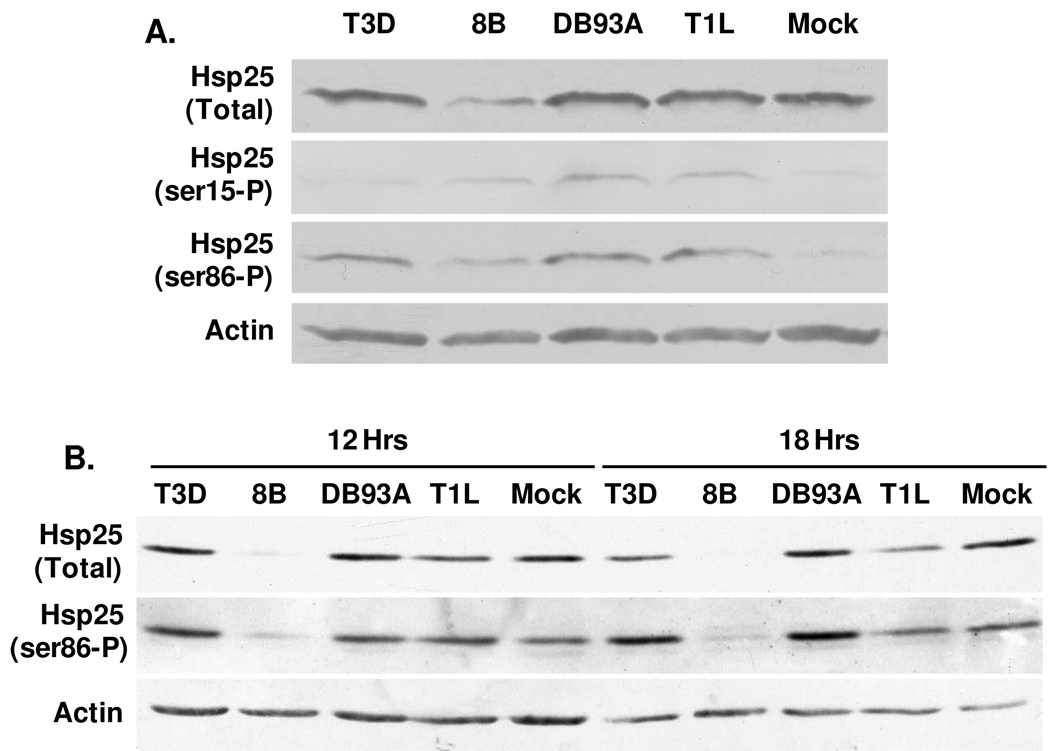
In order to confirm protein identifications, primary cardiac myocyte cultures were infected as for 2D-DIGE and lysates were electrophoresed and probed by Western blot (Fig. 4A). Antisera recognizing total Hsp25 confirmed Hsp25 expression in cardiac myocytes, and confirmed that total Hsp25 was least abundant in 8B-infected cultures. Since this antibody recognizes both unphosphorylated and phosphorylated Hsp25, it is not surprising that relative Hsp25 levels here were not the same as those for unphosphorylated Hsp25 by 2D-DIGE (Fig. 3). Antisera specific for Hsp25 phosphorylated on ser15 and ser86 confirmed virus-induced phosphorylation for each of those residues. Moreover, Hsp25-ser86-P levels were greater for all four virus- relative to mock-infections, consistent with 2D-DIGE results for di-phosphorylated Hsp25 (Fig. 3). The experiment was repeated in cardiac myocytes generated from mice lacking the IFN-α/β-receptor to increase viral replication. Results emphasized the reduction of Hsp25 in 8B-infected cultures, and the increased Hsp25 phosphorylation specifically in cultures infected with non-myocarditic reoviruses T3D and DB93A (Fig. 4B). Quantitative RT-PCR confirmed that reovirus modulation of Hsp25 levels is post-transcriptional (Supplemental Fig. 2). Together the data confirm both 2D-DIGE identification and quantification of Hsp25 and phosphorylated forms, and indicate that stimulation of Hsp25 phosphorylation or reduction of Hsp25 accumulation is virus strain-specific.
Figure 4. Confirmation of Hsp25 expression and phosphorylation by Western blot.
Primary cardiac myocyte cultures generated from wild type (A) or IFN-α/β-receptor-null (B) mice were mock- or reovirus-infected as for Figure 1, and harvested 12 hours (A) or 12 and 18 hours (B) post-infection for SDS-PAGE and Western blot analysis using the indicated antibody probes. Results are representative of at least 3 independent experiments.
Reovirus-induced Hsp25 phosphorylation and reduction of Hsp25 accumulation is virus strain-specific, p38-MAPK-dependent, and IFN-independent
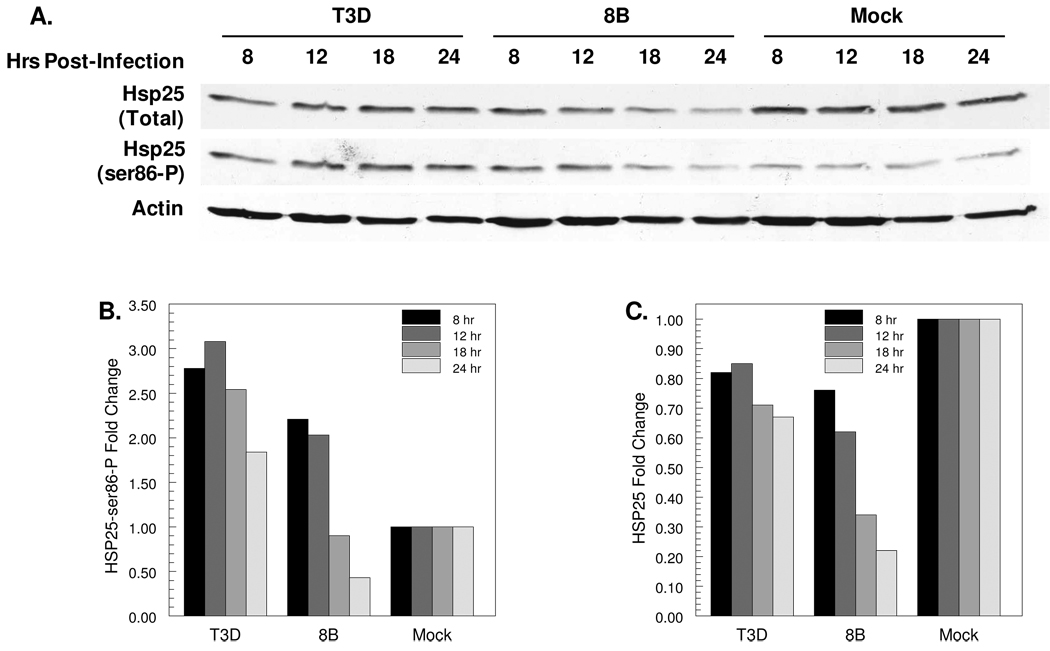
To confirm that differences in virus-induced Hsp25 phosphorylation and reduction of Hsp25 accumulation are virus strain-specific, lysates from cardiac myocyte cultures were probed at a range of times post-infection (Fig. 5). Both T3D and 8B induced phosphorylation of Hsp25 at 8 and 12 hours post-infection, but while phosphorylated Hsp25 remained elevated in T3D-infected cultures at 18 and 24 hours post-infection, it was reduced in 8B-infected cultures to levels below those in mock-infected cultures (Fig. 5B). While levels of total Hsp25 remained relatively constant in T3D-infected cultures over time, Hsp25 was successively reduced in 8B-infected cultures (Fig. 5C). Together, these results suggest that both reovirus strains induce initial Hsp25 phosphorylation, but then 8B reduces accumulation of Hsp25.
Figure 5. Reovirus-induced phosphorylation and degradation of Hsp25 is virus strain-specific.
Primary cardiac myocyte cultures were mock-, T3D- or 8B-infected at an moi of 10 pfu per cell, and cultures were harvested for SDS-PAGE and Western blot at the indicated times post-infection (A). For each sample, the band intensity for Hsp25-Ser86-P (B) or Hsp25 (C) was first normalized to that for actin, and then normalized to the mock-infected sample at that same time-point. Each bar represents the single samples shown, and is representative of similar results from at least 3 independent experiments.
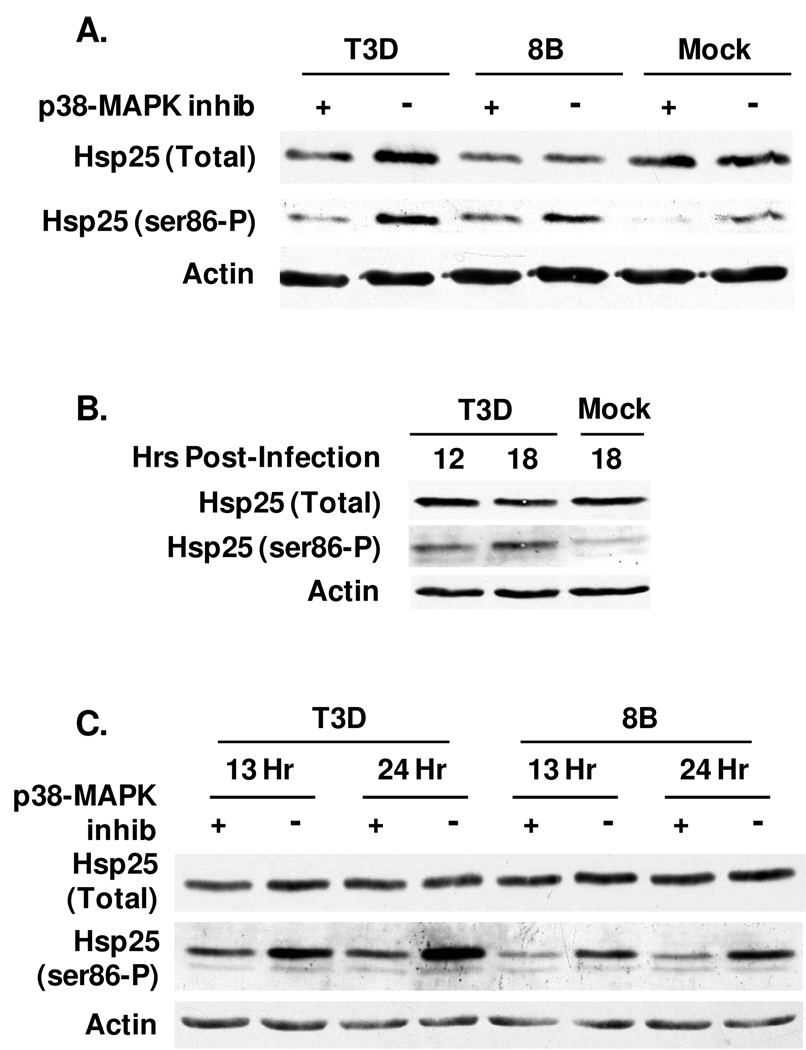
Hsp25 phosphorylation can be mediated through the p38-MAPK pathway. To determine whether reovirus-induced phosphorylation of Hsp25 is p38-MAPK-dependent, primary cardiac myocyte cultures were treated with the p38-MAPK inhibitor SB203580 for 1 hour and then mock- or reovirus-infected for 13 hours (Fig. 6). As expected in the absence of inhibitor, T3D and 8B induced Hsp25 phosphorylation relative to mock-infected cultures (Fig. 6A). The p38-MAPK inhibitor reduced Hsp25 phosphorylation in both T3D and 8B-infected cultures, indicating reovirus use of this pathway. Residual detectable phosphorylation suggested reovirus may use pathways in addition to p38-MAPK. To determine the role of Type I IFN, cardiac myocytes were generated from mice lacking the IFN-α/β-receptor. Efficient reovirus-induced phosphorylation of Hsp25 in these cells demonstrates that it is IFN-independent (Fig. 4B and Fig. 6B). IFN can stimulate p38-MAPK 42 , however the p38-MAPK inhibitor reduced Hsp25 phosphorylation, confirming that reovirus-induced Hsp25 phosphorylation is p38-MAPK-dependent and IFN-independent (Fig. 6C).
Figure 6. Reovirus-induced phosphorylation of Hsp25 is p38-MAPK-dependent and IFN-independent.
Primary cardiac myocyte cultures were generated from wild type (A) or IFN-α/β-receptor-null (B, C) mice. Cultures were treated with 50 µM p38-MAPK inhibitor SB203580 for 1 hour as indicated (A, C) or left untreated (B), and then mock- or reovirus-infected (moi 10 pfu per cell) for 13 hours (A) or indicated hours. Total cell lysates were subjected to SDS-PAGE and then transferred to nitrocellulose for Western Blots. Results are representative of 2 experiments.
Reovirus does not induce Hsp25 nuclear translocation
Stress can induce Hsp25 nuclear translocation. Whole cell lysates can under-represent the nuclear fraction, and therefore separate cytoplasmic and nuclear fractions were generated. Neither T3D nor 8B induced nuclear accumulation of Hsp25 (Fig. 7), confirming that the reduced accumulation of Hsp25 in 8B-infected cells was not due to nuclear sequestration.
Expression of Hsp25 is cell type-specific in the heart
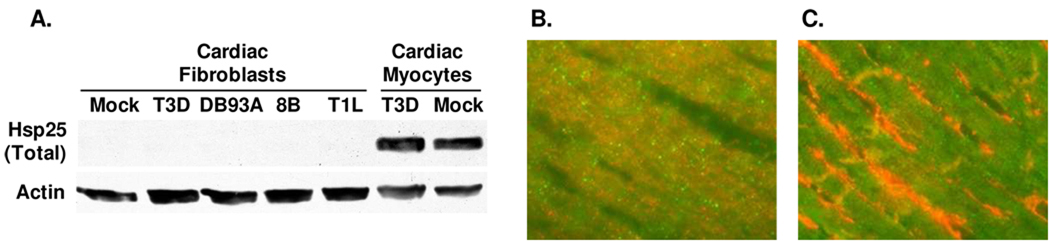
Cardiac myocytes are surrounded by cardiac fibroblasts in the heart, and their IFN responses are cell type-specific for an integrated antiviral response 37. To determine possible cardiac cell type-specificity for reovirus modulation of Hsp25, primary cultures of cardiac myocytes and cardiac fibroblasts were infected and lysates were subjected to SDS-PAGE and Western blot analysis (Fig. 8A). Surprisingly, Hsp25 was undetectable in either mock- or reovirus-infected cardiac fibroblast cultures, regardless of the reovirus strain tested. To confirm that this did not reflect alterations in Hsp25 during cell culture, mouse cardiac sections were probed for Hsp25 by immunofluorescent microscopy (Fig. 8B). Indeed, Hsp25 was readily detected in cardiac myocytes, identified by co-expression of myomesin, but was not detected in cardiac fibroblasts, identified by strong co-expression of vimentin. Together, results indicate that Hsp25 expression is cell type-specific in the heart.
Figure 8. Expression of Hsp25 is cell type-specific in the heart.
A) Primary cultures of cardiac myocytes and cardiac fibroblasts were mock- or reovirus-infected (moi 10 pfu per cell), and cultures were harvested at 12 hours post-infection for SDS-PAGE and Western blot analysis to detect total Hsp25. B) Cardiac sections from adult mice were stained for Hsp25 (green) and myomesin (red) to detect cardiac myocytes. C) As for (B), but stained for Hsp25 (green) and vimentin (red) to detect cardiac fibroblasts.
Inhibition of p38-MAPK increases T3D- and 8B-induced CPE in cardiac myocytes
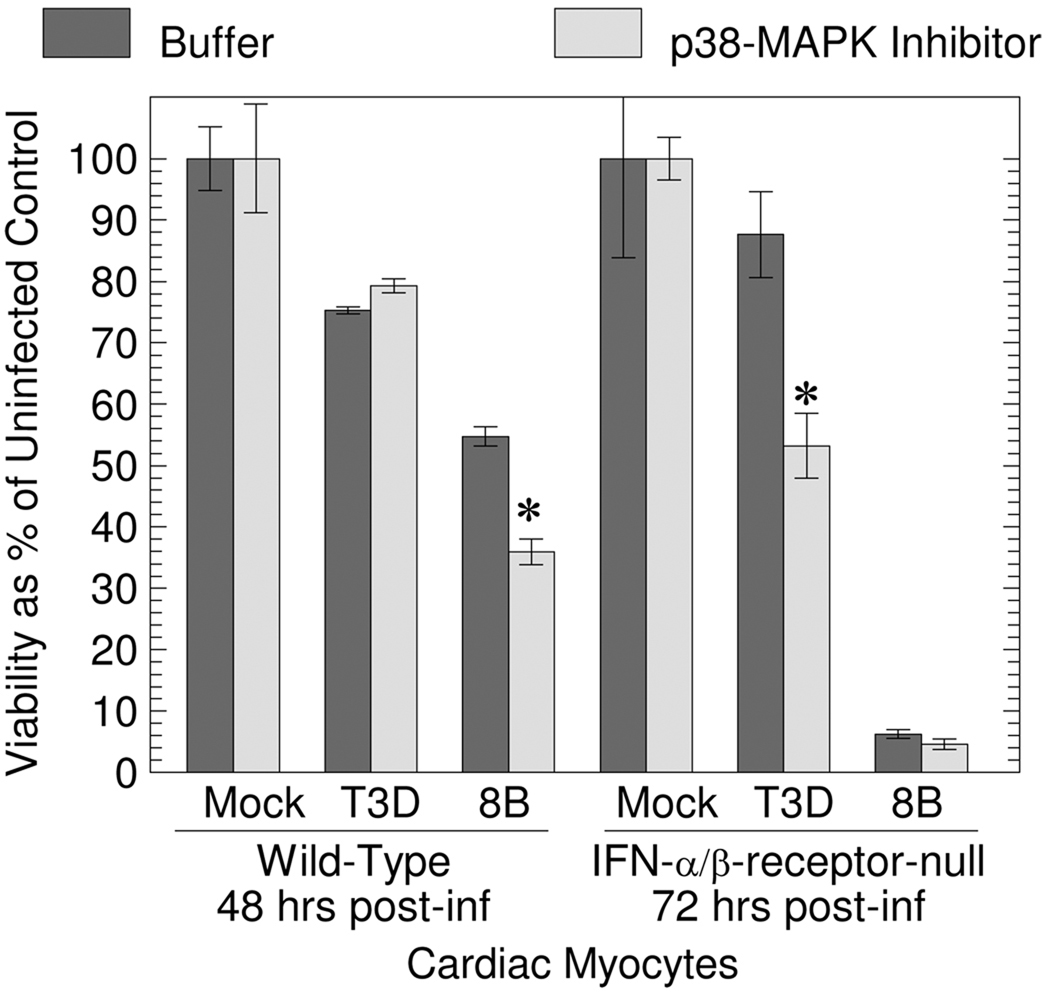
Phosphorylation of Hsp25 can increase or decrease Hsp25 anti-apoptotic function. To probe the role of Hsp25 phosphorylation in reovirus-induced CPE, cardiac myocyte cultures generated from wild type or IFN-α/β-receptor-null mice were treated with the p38-MAPK inhibitor SB203580 for 1 hour, and then mock- or reovirus-infected. T3D induces high levels of IFN-β and is highly sensitive to the antiviral effects of IFN-α/β in cardiac myocytes 19, therefore potential antiviral effects of Hsp25 against this virus might be more apparent in IFN-α/β-receptor-null cells. Inhibition of p38-MAPK increased 8B-induced CPE in wild type cells, and increased T3D-induced CPE in IFN-α/β-receptor-null cells (Fig. 9). Results are consistent with an antiviral role for phosphorylated Hsp25.
Figure 9. Inhibition of p38-MAPK increases T3D- and 8B-induced CPE in cardiac myocytes.
Primary cardiac myocyte cultures were generated from wild type or IFN-α/β-receptor null mice, treated with buffer or the p38-MAPK inhibitor S203580 (50 µM) for 1 hour, and then mock- or reovirus-infected (moi 25 pfu per cell). At the indicated time post-infection, cell viability was determined by MTT assay. Results represent the average of triplicate wells ± standard deviation, and are representative of 3 experiments.
Discussion
To identify novel proteins involved in the cardiac antiviral response, the proteomes of primary cultures of cardiac myocytes infected with different reovirus strains were compared by 2D-DIGE. Results were reproducible between replicate cultures and provided a good correlation between known viral phenotypes and whole proteome changes. Hsp25 was identified as one candidate antiviral protein. While non-myocarditic reovirus strains induced Hsp25 phosphorylation, which is a well-characterized protective response in stressed cardiac myocytes (see below), a potently myocarditic reovirus strain decreased Hsp25 abundance. Hsp25 abundance is not decreased in stressed cardiac myocytes or failing hearts, indicating that this effect is specific to viral infection, possibly reflecting degradation. Viral activation and then degradation of a cell response is reminiscent of the interplay between viruses and cells during the protective IFN response, and suggests that Hsp25 may serve not just as a marker of virus-induced cytopathology, but as an active participant in the antiviral response.
Members of seven different virus families induce Hsp25/Hsp27 expression or phosphorylation, including Herpesviridae 43–46, Papillomaviridae 47, 48, Hepadnaviridae 49, 50, Paramyxoviridae 51, 52, Flaviviridae 53–55, Togaviridae 56, and Retroviridae 57, 58. However, only one study has addressed possible Hsp25/27 antiviral activity, finding that while HIV1 induces Hsp27 expression, Hsp27 can inhibit HIV1-induced cell cycle arrest and apoptosis 58. HIV1 induction of Hsp27 and Hsp27 inhibition of HIV1 damage to the cell are similar to virus induction of IFN and IFN inhibition of viral damage. While viral infection of most cells can induce expression and secretion of Type I IFN 59–62, viruses have evolved many mechanisms to sabotage this potent protective response 63–65. Indeed, reoviruses have evolved a novel mechanism to repress IFN signaling 66. However to date, there has been no evidence that, by analogy, viruses might subvert the Hsp25/Hsp27 response. Results here demonstrate that while all four reovirus strains tested, regardless of potential to induce disease, induce phosphorylation of Hsp25, only the potently myocarditic reovirus 8B decreases Hsp25 abundance. Together, the data suggest that Hsp25 participates in the antiviral response and that some reovirus strains have evolved to subvert this Hsp25 function.
Murine Hsp25 and its human homolog Hsp27 are members of the small HSP family, and can inhibit apoptosis both up- and downstream of cytochrome c release from mitochondria 67. Hsp25 is expressed at greater basal levels in muscle (heart, skeletal muscle) and the gastrointestinal and respiratory tracts than in other tissues 68–70. However, a variety of stimuli induce Hsp25/27 expression or phosphorylation as a critical protective response in many cell types. The frequency with which microarray and proteomic approaches have identified variations in Hsp25/27 expression raises questions about the validity of these identifications, and emphasizes the importance of confirmation using complementary techniques such as qRT-PCR and Western blots 71. Results here demonstrate that reovirus modulation of Hsp25 expression in cardiac myocytes is post-transcriptional (qRT-PCR, Supplemental Fig. 2), unlike reovirus modulation of other HSPs in other cell types 72, 73. Indeed, reovirus modulation of Hsp25 would have remained undetected without proteomic approaches.
Hsp27 phosphorylation is increased in hearts from patients with ischemia (oxygen deprivation) / reperfusion (oxygen restoration) damage or dilated cardiomyopathy 74, and in hearts from dogs with congestive heart failure 75. Ischemia / reperfusion is modeled in vitro in hearts and in primary cardiac myocyte cultures, and induces apoptosis similar to that in patients. Oxidative stress activates p38-MAPK to phosphorylate and activate MAPKAPK2 (MK2), which then phosphorylates Hsp25/27 in cardiac myocytes 76. The p38-MAPK / MK2 pathway also mediates Hsp25/27 phosphorylation following other stresses such as TNF-α addition to cardiac cells 77, and stresses in other cell types 78–81. Hsp25/27 can also be phosphorylated through other pathways 80, and Type I IFN can activate p38-MAPK 42, providing IFN-mediated mechanisms for Hsp25/27 phosphorylation. Thus the mechanism for Hsp25/27 phosphorylation is both stimulus- and cell type-specific. Results here demonstrate that reovirus-induced Hsp25 phosphorylation in cardiac myocytes is p38-MAPK-dependent and IFN-independent, but do not exclude involvement of other pathways (Fig. 6). Inhibition of p38-MAPK increased reovirus-induced CPE in cardiac myocytes (Fig. 9), consistent with impairment of a protective response normally mediated by phosphorylated Hsp25.
Over-expressed Hsp25/27 is protective against ischemic damage in cardiac myocytes 82–84. Stress induces phosphorylation of two Hsp25 residues (ser15 and -86) 40, 41, and while some studies suggest that phosphorylation is protective 85, 86 and implicate the p38-MAPK pathway 76, 87, others suggest phosphorylation is irrelevant to protection 88–90. The relationship between phosphorylation and protection is likely to be cell type-specific, and to depend on the Hsp25/27 function assayed. Results here demonstrate that both non-myocarditic and myocarditic reoviruses induce phosphorylation of Hsp25 in cardiac myocytes, likely reflecting a general stress response in those cells. However both unphosphorylated and phosphorylated Hsp25 were decreased in cardiac myocyte cultures infected with the potently myocarditic reovirus 8B, suggesting that all forms can be viral targets.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 AI062657 and by graduate student support from the North Carolina State University Genomics Graduate Program (L.L.). We thank Susan Irvin and Jennifer Zurney for insightful discussions, and Wrennie Edwards for technical assistance.
References
- 1.Cooper LT., Jr Myocarditis. N Engl J Med. 2009;360(15):1526–1538. doi: 10.1056/NEJMra0800028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 3.Woodruff JF. Viral myocarditis. A review. Am J Pathol. 1980;101(2):425–484. [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy RE, 3rd, Boehmer JP, Hruban RH, Hutchins GM, Kasper EK, Hare JM, Baughman KL. Long-term outcome of fulminant myocarditis as compared with acute (nonfulminant) myocarditis. N Engl J Med. 2000;342(10):690–695. doi: 10.1056/NEJM200003093421003. [DOI] [PubMed] [Google Scholar]
- 5.Puranik R, Chow CK, Duflou JA, Kilborn MJ, McGuire MA. Sudden death in the young. Heart Rhythm. 2005;2(12):1277–1282. doi: 10.1016/j.hrthm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Doolan A, Langlois N, Semsarian C. Causes of sudden cardiac death in young Australians. Med J Aust. 2004;180(3):110–112. doi: 10.5694/j.1326-5377.2004.tb05830.x. [DOI] [PubMed] [Google Scholar]
- 7.Mason JW, O'Connell JB, Herskowitz A, Rose NR, McManus BM, Billingham ME, Moon TE. A clinical trial of immunosuppressive therapy for myocarditis. The Myocarditis Treatment Trial Investigators. N Engl J Med. 1995;333(5):269–275. doi: 10.1056/NEJM199508033330501. [DOI] [PubMed] [Google Scholar]
- 8.Bowles NE, Ni J, Kearney DL, Pauschinger M, Schultheiss HP, McCarthy R, Hare J, Bricker JT, Bowles KR, Towbin JA. Detection of viruses in myocardial tissues by polymerase chain reaction. evidence of adenovirus as a common cause of myocarditis in children and adults. J Am Coll Cardiol. 2003;42(3):466–472. doi: 10.1016/s0735-1097(03)00648-x. [DOI] [PubMed] [Google Scholar]
- 9.Martin AB, Webber S, Fricker FJ, Jaffe R, Demmler G, Kearney D, Zhang YH, Bodurtha J, Gelb B, Ni J, et al. Acute myocarditis. Rapid diagnosis by PCR in children. Circulation. 1994;90(1):330–339. doi: 10.1161/01.cir.90.1.330. [DOI] [PubMed] [Google Scholar]
- 10.Huber S. Host immune responses to coxsackievirus B3. Curr Top Microbiol Immunol. 2008;323:199–221. doi: 10.1007/978-3-540-75546-3_9. [DOI] [PubMed] [Google Scholar]
- 11.Blailock ZR, Rabin ER, Melnick JL. Adenovirus myocarditis in mice. An electron microscopic study. Exp Mol Pathol. 1968;9(1):84–96. doi: 10.1016/0014-4800(68)90053-1. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto SD, Brown RD, Robinson BA, Tyler KL, Long CS, Debiasi RL. Cardiac cell specific apoptotic and cytokine response to reovirus infection: determinants of myocarditic phenotype. Journal of cardiac failure. 2009 doi: 10.1016/j.cardfail.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baty CJ, Sherry B. Cytopathogenic effect in cardiac myocytes but not in cardiac fibroblasts is correlated with reovirus-induced acute myocarditis. J Virol. 1993;67(10):6295–6298. doi: 10.1128/jvi.67.10.6295-6298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBiasi RL, Edelstein CL, Sherry B, Tyler KL. Calpain inhibition protects against virus-induced apoptotic myocardial injury. J Virol. 2001;75(1):351–361. doi: 10.1128/JVI.75.1.351-361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBiasi RL, Robinson BA, Sherry B, Bouchard R, Brown RD, Rizeq M, Long C, Tyler KL. Caspase inhibition protects against reovirus-induced myocardial injury in vitro and in vivo. J Virol. 2004;78(20):11040–11050. doi: 10.1128/JVI.78.20.11040-11050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherry B, Li XY, Tyler KL, Cullen JM, Virgin HWt. Lymphocytes protect against and are not required for reovirus-induced myocarditis. J Virol. 1993;67(10):6119–6124. doi: 10.1128/jvi.67.10.6119-6124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry B, Fields BN. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J Virol. 1989;63(11):4850–4856. doi: 10.1128/jvi.63.11.4850-4856.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry B, Torres J, Blum MA. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J Virol. 1998;72(2):1314–1323. doi: 10.1128/jvi.72.2.1314-1323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber MJ, Agnetti G, Bezstarosti K, Lankhuizen IM, Dalinghaus M, Guarnieri C, Caldarera CM, Helbing WA, Lamers JM. Recent developments in proteomics: implications for the study of cardiac hypertrophy and failure. Cell Biochem Biophys. 2006;44(1):11–29. doi: 10.1385/CBB:44:1:011. [DOI] [PubMed] [Google Scholar]
- 21.Jager D, Jungblut PR, Muller-Werdan U. Separation and identification of human heart proteins. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771(1–2):131–153. doi: 10.1016/s1570-0232(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 22.McGregor E, Dunn MJ. Proteomics of the heart: unraveling disease. Circ Res. 2006;98(3):309–321. doi: 10.1161/01.RES.0000201280.20709.26. [DOI] [PubMed] [Google Scholar]
- 23.Timms JF, Cramer R. Difference gel electrophoresis. Proteomics. 2008;8(23–24):4886–4897. doi: 10.1002/pmic.200800298. [DOI] [PubMed] [Google Scholar]
- 24.O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 25.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4(12):3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 26.Mayr M, Liem D, Zhang J, Li X, Avliyakulov NK, Yang JI, Young G, Vondriska TM, Ladroue C, Madhu B, Griffiths JR, Gomes A, Xu Q, Ping P. Proteomic and metabolomic analysis of cardioprotection: Interplay between protein kinase C epsilon and delta in regulating glucose metabolism of murine hearts. J Mol Cell Cardiol. 2009;46(2):268–277. doi: 10.1016/j.yjmcc.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekkers DH, Bezstarosti K, Gurusamy N, Luijk K, Verhoeven AJ, Rijkers EJ, Demmers JA, Lamers JM, Maulik N, Das DK. Identification by a differential proteomic approach of the induced stress and redox proteins by resveratrol in the normal and diabetic rat heart. J Cell Mol Med. 2008;12(5A):1677–1689. doi: 10.1111/j.1582-4934.2008.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Page B, Young R, Iyer V, Suzuki G, Lis M, Korotchkina L, Patel MS, Blumenthal KM, Fallavollita JA, Canty JM., Jr Persistent regional downregulation in mitochondrial enzymes and upregulation of stress proteins in swine with chronic hibernating myocardium. Circ Res. 2008;102(1):103–112. doi: 10.1161/CIRCRESAHA.107.155895. [DOI] [PubMed] [Google Scholar]
- 29.Sakai J, Ishikawa H, Satoh H, Yamamoto S, Kojima S, Kanaoka M. Two-dimensional differential gel electrophoresis of rat heart proteins in ischemia and ischemia-reperfusion. Methods Mol Biol. 2007;357:33–43. doi: 10.1385/1-59745-214-9:33. [DOI] [PubMed] [Google Scholar]
- 30.Hamblin M, Friedman DB, Hill S, Caprioli RM, Smith HM, Hill MF. Alterations in the diabetic myocardial proteome coupled with increased myocardial oxidative stress underlies diabetic cardiomyopathy. J Mol Cell Cardiol. 2007;42(4):884–895. doi: 10.1016/j.yjmcc.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng C, Jin X, Xia L, Shen SM, Wang XL, Cai J, Chen GQ, Wang LS, Fang NY. Alterations of mitochondrial enzymes contribute to cardiac hypertrophy before hypertension development in spontaneously hypertensive rats. J Proteome Res. 2009;8(5):2463–2475. doi: 10.1021/pr801059u. [DOI] [PubMed] [Google Scholar]
- 32.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1(3):377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Yan JX, Devenish AT, Wait R, Stone T, Lewis S, Fowler S. Fluorescence two-dimensional difference gel electrophoresis and mass spectrometry based proteomic analysis of Escherichia coli. Proteomics. 2002;2(12):1682–1698. doi: 10.1002/1615-9861(200212)2:12<1682::AID-PROT1682>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3(1):36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 35.Vestal ML, Campbell JM. Tandem time-of-flight mass spectrometry. Methods Enzymol. 2005;402:79–108. doi: 10.1016/S0076-6879(05)02003-3. [DOI] [PubMed] [Google Scholar]
- 36.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 37.Zurney J, Howard KE, Sherry B. Basal expression levels of IFNAR and Jak-STAT components are determinants of cell-type-specific differences in cardiac antiviral responses. J Virol. 2007;81(24):13668–13680. doi: 10.1128/JVI.01172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith RE, Zweerink HJ, Joklik WK. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39(4):791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 39.Schiff LA, Nibert ML, Tyler KL. Orthoreoviruses and their repliation. 5 ed. Vol. 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 1853–1915. [Google Scholar]
- 40.Gaestel M, Schroder W, Benndorf R, Lippmann C, Buchner K, Hucho F, Erdmann VA, Bielka H. Identification of the phosphorylation sites of the murine small heat shock protein hsp25. J Biol Chem. 1991;266(22):14721–14724. [PubMed] [Google Scholar]
- 41.Stokoe D, Engel K, Campbell DG, Cohen P, Gaestel M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 1992;313(3):307–313. doi: 10.1016/0014-5793(92)81216-9. [DOI] [PubMed] [Google Scholar]
- 42.Barca O, Costoya JA, Senaris RM, Arce VM. Interferon-beta protects astrocytes against tumour necrosis factor-induced apoptosis via activation of p38 mitogen-activated protein kinase. Exp Cell Res. 2008;314(11–12):2231–2237. doi: 10.1016/j.yexcr.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Forsman A, Ruetschi U, Ekholm J, Rymo L. Identification of intracellular proteins associated with the EBV-encoded nuclear antigen 5 using an efficient TAP procedure and FT-ICR mass spectrometry. J Proteome Res. 2008;7(6):2309–2319. doi: 10.1021/pr700769e. [DOI] [PubMed] [Google Scholar]
- 44.Fukagawa Y, Nishikawa J, Kuramitsu Y, Iwakiri D, Takada K, Imai S, Satake M, Okamoto T, Fujimoto M, Okita K, Nakamura K, Sakaida I. Epstein-Barr virus upregulates phosphorylated heat shock protein 27 kDa in carcinoma cells using the phosphoinositide 3-kinase/Akt pathway. Electrophoresis. 2008;29(15):3192–3200. doi: 10.1002/elps.200800086. [DOI] [PubMed] [Google Scholar]
- 45.Gober MD, Laing JM, Burnett JW, Aurelian L. The Herpes simplex virus gene Pol expressed in herpes-associated erythema multiforme lesions upregulates/activates SP1 and inflammatory cytokines. Dermatology. 2007;215(2):97–106. doi: 10.1159/000104259. [DOI] [PubMed] [Google Scholar]
- 46.Gober MD, Wales SQ, Aurelian L. Herpes simplex virus type 2 encodes a heat shock protein homologue with apoptosis regulatory functions. Front Biosci. 2005;10:2788–2803. doi: 10.2741/1736. [DOI] [PubMed] [Google Scholar]
- 47.Ciocca DR, Lo Castro G, Alonio LV, Cobo MF, Lotfi H, Teyssie A. Effect of human papillomavirus infection on estrogen receptor and heat shock protein hsp27 phenotype in human cervix and vagina. Int J Gynecol Pathol. 1992;11(2):113–121. doi: 10.1097/00004347-199204000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Lo WY, Lai CC, Hua CH, Tsai MH, Huang SY, Tsai CH, Tsai FJ. S100A8 is identified as a biomarker of HPV18-infected oral squamous cell carcinomas by suppression subtraction hybridization, clinical proteomics analysis, and immunohistochemistry staining. J Proteome Res. 2007;6(6):2143–2151. doi: 10.1021/pr060551+. [DOI] [PubMed] [Google Scholar]
- 49.Lim SO, Park SG, Yoo JH, Park YM, Kim HJ, Jang KT, Cho JW, Yoo BC, Jung GH, Park CK. Expression of heat shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in hepatitis B virus-related hepatocellular carcinomas and dysplastic nodules. World J Gastroenterol. 2005;11(14):2072–2079. doi: 10.3748/wjg.v11.i14.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han J, Yoo HY, Choi BH, Rho HM. Selective transcriptional regulations in the human liver cell by hepatitis B viral X protein. Biochem Biophys Res Commun. 2000;272(2):525–530. doi: 10.1006/bbrc.2000.2801. [DOI] [PubMed] [Google Scholar]
- 51.Singh D, McCann KL, Imani F. MAPK and heat shock protein 27 activation are associated with respiratory syncytial virus induction of human bronchial epithelial monolayer disruption. Am J Physiol Lung Cell Mol Physiol. 2007;293(2):L436–L445. doi: 10.1152/ajplung.00097.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokota S, Yokosawa N, Kubota T, Okabayashi T, Arata S, Fujii N. Suppression of thermotolerance in mumps virus-infected cells is caused by lack of HSP27 induction contributed by STAT-1. J Biol Chem. 2003;278(43):41654–41660. doi: 10.1074/jbc.M305701200. [DOI] [PubMed] [Google Scholar]
- 53.Choi YW, Tan YJ, Lim SG, Hong W, Goh PY. Proteomic approach identifies HSP27 as an interacting partner of the hepatitis C virus NS5A protein. Biochem Biophys Res Commun. 2004;318(2):514–519. doi: 10.1016/j.bbrc.2004.04.052. [DOI] [PubMed] [Google Scholar]
- 54.Fang C, Yi Z, Liu F, Lan S, Wang J, Lu H, Yang P, Yuan Z. Proteome analysis of human liver carcinoma Huh7 cells harboring hepatitis C virus subgenomic replicon. Proteomics. 2006;6(2):519–527. doi: 10.1002/pmic.200500233. [DOI] [PubMed] [Google Scholar]
- 55.Liew KJ, Chow VT. Microarray and real-time RT-PCR analyses of a novel set of differentially expressed human genes in ECV304 endothelial-like cells infected with dengue virus type 2. J Virol Methods. 2006;131(1):47–57. doi: 10.1016/j.jviromet.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Nakatsue T, Katoh I, Nakamura S, Takahashi Y, Ikawa Y, Yoshinaka Y. Acute infection of Sindbis virus induces phosphorylation and intracellular translocation of small heat shock protein HSP27 and activation of p38 MAP kinase signaling pathway. Biochem Biophys Res Commun. 1998;253(1):59–64. doi: 10.1006/bbrc.1998.9724. [DOI] [PubMed] [Google Scholar]
- 57.Wainberg Z, Oliveira M, Lerner S, Tao Y, Brenner BG. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology. 1997;233(2):364–373. doi: 10.1006/viro.1997.8618. [DOI] [PubMed] [Google Scholar]
- 58.Liang D, Benko Z, Agbottah E, Bukrinsky M, Zhao RY. Anti-vpr activities of heat shock protein 27. Mol Med. 2007;13(5–6):229–239. doi: 10.2119/2007-00004.Liang. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samuel CE. Innate immunity minireview series: making biochemical sense of nucleic acid sensors that trigger antiviral innate immunity. J Biol Chem. 2007;282(21):15313–15314. doi: 10.1074/jbc.R700013200. [DOI] [PubMed] [Google Scholar]
- 62.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282(28):20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 63.Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344(1):119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43(3):350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 66.Zurney J, Kobayashi T, Holm GH, Dermody TS, Sherry B. Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J Virol. 2009;83(5):2178–2187. doi: 10.1128/JVI.01787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arya R, Mallik M, Lakhotia SC. Heat shock genes - integrating cell survival and death. J Biosci. 2007;32(3):595–610. doi: 10.1007/s12038-007-0059-3. [DOI] [PubMed] [Google Scholar]
- 68.Wakayama T, Iseki S. Expression and cellular localization of the mRNA for the 25-kDa heat-shock protein in the mouse. Cell Biol Int. 1998;22(4):295–304. doi: 10.1006/cbir.1998.0252. [DOI] [PubMed] [Google Scholar]
- 69.Tanguay RM, Wu Y, Khandjian EW. Tissue-specific expression of heat shock proteins of the mouse in the absence of stress. Dev Genet. 1993;14(2):112–118. doi: 10.1002/dvg.1020140205. [DOI] [PubMed] [Google Scholar]
- 70.Kato K, Ito H, Iwamoto I, Lida K, Inaguma Y. Protein kinase inhibitors can suppress stress-induced dissociation of Hsp27. Cell Stress Chaperones. 2001;6(1):16–20. doi: 10.1379/1466-1268(2001)006<0016:pkicss>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, Zivny J, Vulpe CD. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8(9):1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 72.Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80(4):2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeBiasi RL, Clarke P, Meintzer S, Jotte R, Kleinschmidt-Demasters BK, Johnson GL, Tyler KL. Reovirus-induced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J Virol. 2003;77(16):8934–8947. doi: 10.1128/JVI.77.16.8934-8947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lutsch G, Vetter R, Offhauss U, Wieske M, Grone HJ, Klemenz R, Schimke I, Stahl J, Benndorf R. Abundance and location of the small heat shock proteins HSP25 and alphaB-crystallin in rat and human heart. Circulation. 1997;96(10):3466–3476. doi: 10.1161/01.cir.96.10.3466. [DOI] [PubMed] [Google Scholar]
- 75.Dohke T, Wada A, Isono T, Fujii M, Yamamoto T, Tsutamoto T, Horie M. Proteomic analysis reveals significant alternations of cardiac small heat shock protein expression in congestive heart failure. J Card Fail. 2006;12(1):77–84. doi: 10.1016/j.cardfail.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 76.Blunt BC, Creek AT, Henderson DC, Hofmann PA. H2O2 activation of HSP25/27 protects desmin from calpain proteolysis in rat ventricular myocytes. Am J Physiol Heart Circ Physiol. 2007;293(3):H1518–H1525. doi: 10.1152/ajpheart.00269.2006. [DOI] [PubMed] [Google Scholar]
- 77.Bellahcene M, Jacquet S, Cao XB, Tanno M, Haworth RS, Layland J, Kabir AM, Gaestel M, Davis RJ, Flavell RA, Shah AM, Avkiran M, Marber MS. Activation of p38 mitogen-activated protein kinase contributes to the early cardiodepressant action of tumor necrosis factor. J Am Coll Cardiol. 2006;48(3):545–555. doi: 10.1016/j.jacc.2006.02.072. [DOI] [PubMed] [Google Scholar]
- 78.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol. 1999;1(2):94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 79.Park JK, Ronkina N, Hoft A, Prohl C, Menne J, Gaestel M, Haller H, Meier M. Deletion of MK2 signalling in vivo inhibits small Hsp phosphorylation but not diabetic nephropathy. Nephrol Dial Transplant. 2008;23(6):1844–1853. doi: 10.1093/ndt/gfm917. [DOI] [PubMed] [Google Scholar]
- 80.Evans IM, Britton G, Zachary IC. Vascular endothelial growth factor induces heat shock protein (HSP) 27 serine 82 phosphorylation and endothelial tubulogenesis via protein kinase D and independent of p38 kinase. Cell Signal. 2008;20(7):1375–1384. doi: 10.1016/j.cellsig.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Su X, Ao L, Zou N, Song Y, Yang X, Cai GY, Fullerton DA, Meng X. Post-transcriptional regulation of TNF-induced expression of ICAM-1 and IL-8 in human lung microvascular endothelial cells: an obligatory role for the p38 MAPK-MK2 pathway dissociated with HSP27. Biochim Biophys Acta. 2008;1783(9):1623–1631. doi: 10.1016/j.bbamcr.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Efthymiou CA, Mocanu MM, de Belleroche J, Wells DJ, Latchmann DS, Yellon DM. Heat shock protein 27 protects the heart against myocardial infarction. Basic Res Cardiol. 2004;99(6):392–394. doi: 10.1007/s00395-004-0483-6. [DOI] [PubMed] [Google Scholar]
- 83.Lu XY, Chen L, Cai XL, Yang HT. Overexpression of heat shock protein 27 protects against ischaemia/reperfusion-induced cardiac dysfunction via stabilization of troponin I and T. Cardiovasc Res. 2008;79(3):500–508. doi: 10.1093/cvr/cvn091. [DOI] [PubMed] [Google Scholar]
- 84.Vander Heide RS. Increased expression of HSP27 protects canine myocytes from simulated ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2002;282(3):H935–H941. doi: 10.1152/ajpheart.00660.2001. [DOI] [PubMed] [Google Scholar]
- 85.Kwon JH, Kim JB, Lee KH, Kang SM, Chung N, Jang Y, Chung JH. Protective effect of heat shock protein 27 using protein transduction domain-mediated delivery on ischemia/reperfusion heart injury. Biochem Biophys Res Commun. 2007;363(2):399–404. doi: 10.1016/j.bbrc.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 86.Li G, Ali IS, Currie RW. Insulin-induced myocardial protection in isolated ischemic rat hearts requires p38 MAPK phosphorylation of Hsp27. Am J Physiol Heart Circ Physiol. 2008;294(1):H74–H87. doi: 10.1152/ajpheart.00675.2007. [DOI] [PubMed] [Google Scholar]
- 87.Clerk A, Michael A, Sugden PH. Stimulation of multiple mitogen-activated protein kinase sub-families by oxidative stress and phosphorylation of the small heat shock protein, HSP25/27, in neonatal ventricular myocytes. Biochem J. 1998;333(Pt 3):581–589. doi: 10.1042/bj3330581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hollander JM, Martin JL, Belke DD, Scott BT, Swanson E, Krishnamoorthy V, Dillmann WH. Overexpression of wild-type heat shock protein 27 and a nonphosphorylatable heat shock protein 27 mutant protects against ischemia/reperfusion injury in a transgenic mouse model. Circulation. 2004;110(23):3544–3552. doi: 10.1161/01.CIR.0000148825.99184.50. [DOI] [PubMed] [Google Scholar]
- 89.Martin JL, Hickey E, Weber LA, Dillmann WH, Mestril R. Influence of phosphorylation and oligomerization on the protective role of the small heat shock protein 27 in rat adult cardiomyocytes. Gene Expr. 1999;7(4–6):349–355. [PMC free article] [PubMed] [Google Scholar]
- 90.Rogalla T, Ehrnsperger M, Preville X, Kotlyarov A, Lutsch G, Ducasse C, Paul C, Wieske M, Arrigo AP, Buchner J, Gaestel M. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J Biol Chem. 1999;274(27):18947–18956. doi: 10.1074/jbc.274.27.18947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.