Abstract
Matrix metalloproteinase-7 (MMP-7) deletion has been showed to improve survival after myocardial infarction (MI). MMP-7 has a large array of in vitro substrates, but in vivo substrates for MMP-7 following MI have not been fully identified. Accordingly, we evaluated the infarct regions of wild-type (WT; n=12) and MMP-7 null (null; n=10) mice using a proteomic strategy. Seven days post-MI, infarct regions of the left ventricles were excised, homogenized, and protein extracts were analyzed by two-dimensional gel electrophoresis and mass spectrometry. Of 13 spots that showed intensity differences between WT and null, the intensities of eight spots were higher and five spots were lower in the null group (p<0.05). Fibronectin and tenascin-C, known in vitro substrates of MMP-7, were identified in spots that showed lower intensity in the null. Immunoblotting and in vitro cleavage assays confirmed reduced fibronectin and tenascin-C fragment generation in the null, and this effect was restored by exogenous administration of MMP-7. Lower levels of full-length peroxiredoxin-1 and -2 and higher levels of the full-length peroxiredoxin-3 were detected in the null group, suggesting MMP-7 deletion may also indirectly regulate protein levels through non-enzymatic mechanisms. In conclusion, this is the first study to identify fibronectin and tenascin-C as in vivo MMP-7 substrates in the infarcted left ventricle using a proteomic approach.
Keywords: cardiac remodeling, mice, MMP-7, myocardial infarction, proteomics
1. Introduction
Matrix metalloproteinases (MMPs) are a family of proteolytic enzymes that regulate remodeling of the left ventricle (LV) after myocardial infarction (MI) by processing extracellular matrix proteins. MMP-7, also called matrilysin, is secreted as a 28 kDa pro-enzyme and is activated upon the removal of the pro-domain to generate a 19 kDa active enzyme.1 Unlike the majority of other MMPs, MMP-7 lacks a hinge region and a C-terminal hemopexin-like domain, making it the smallest MMP identified to date.2 Macrophages and cardiomyocytes are rich sources of MMP-7,3, 4 and increased MMP-7 levels are detected in both the remote and infarct regions after MI.5 In vitro, MMP-7 cleaves a wide array of extracellular matrix (ECM) proteins, including collagen IV, fibronectin (Fn), laminin and tenascin-C (TN-C);6-9 and non-ECM proteins, including tumor necrosis factor-α and other MMPs.9-11 However, the identification of in vivo substrates for MMP-7 following MI has not been reported.
Fn is a polymorphic glycoprotein that exists in a soluble form in plasma and an insoluble form in tissue.12 After MI, Fn levels increase in both plasma and the infarct region.13, 14 TN-C is normally undetectable in the adult heart , but TN-C expression increases rapidly after MI and is mainly localized to the border zone.15 Recently, Imanaka-Yoshida and colleagues demonstrated that TN-C null mice have reduced end-diastolic pressure and dimension and decreased myocardial stiffness compared to wild-type (WT) mice after MI, suggesting TN-C may mediate adverse LV remodeling.16 Although Fn and TN-C are in vitro substrates of MMP-7, whether they are cleaved by MMP-7 in vivo after MI has not been addressed.
In this study, we used a proteomic approach to identify differences in protein quantities in the infarct regions of MMP-7 null (null) LV compared to WT LV. We identified two known in vitro substrates of MMP-7, Fn and TN-C, which validated the use of our approach method to identify in vivo MMP-7 substrates. We also observed changes in levels of different peroxiredoxin isoforms in MMP-7 null mice, suggesting MMP-7 may indirectly regulate protein expression in the MI setting.
2. Methods
Male C57/BL6 WT (n=12) and MMP-7 null (n=10) mice, 4-6 months of age, were used for this study. The MMP-7 null mice were a gift from Dr. Lynn Matrisian (Vanderbilt University).17 All animal procedures were conducted in accordance with the “Guide for the Care and Use of Laboratory Animals” (NIH Publication No. 85-23, revised 1996) and were approved by the UTHSCSA Institutional Animal Care and Use Committee. MI was induced by coronary artery ligation, as described previously.5 At seven days after MI, the mice were anesthetized with 2-5% isoflurane, the coronary vasculature was flushed with saline, and the hearts were excised. The LV was separated from RV and weighed. The hearts were stained with 1% 2,3,5-triphenyltetrazolium chloride (Sigma) and photographed to measure infarct size. The infarct tissue was excised from the remote tissue and snap-frozen in liquid nitrogen and stored at −80°C in individual tubes.
2.2 Protein extraction
The infarct regions of the LV were homogenized in extraction buffer (Sigma; Protein Extraction Reagent Type 4; 7 M urea, 2 M thiourea, 40 mM Trizma® base and the detergent 1% C7BzO) and 1× Complete Protease Inhibitor Cocktail (Roche). The protein concentration of each sample was determined by the Bradford assay. Due to the high urea concentration in the extraction buffer, the protein extracts were diluted 1:40 with water prior to Bradford assay.
2.3 Two-dimensional gel electrophoresis (2-DE)
Protein samples (500 μg; n=12 for WT and n=10 for null mice) were reduced in 2.5% tributylphosphine containing 1× Complete Protease Inhibitor Cocktail (Roche) at room temperature for 1 h. Iodoacetamide was added to a final concentration of 3% and the samples were incubated at room temperature for 1h. The samples were then centrifuged at 425 × g for 5 min to pellet debris. Supernatants were harvested and acetone was added to a final concentration of 80%; proteins were precipitated at room temperature for 30 min. The samples were centrifuged at 20,817 × g for 10 min and supernatants were discarded. The pellets were air-dried and resuspended in 200 μl of extraction buffer and 1× Complete Protease Inhibitor Cocktail. After incubation at 30°C for 30 min, 500 μg of each sample was loaded to an 11-cm pH 3 – 10 IPG strip (Proteome Systems), rehydrated overnight at room temperature, and then focused for 75,000 Vh. The focused-IPG strips were then equilibrated in 5 ml of Equilibration Buffer (Proteome Systems) for 20 min. Equilibrated strips were loaded on pre-cast gels (Criterion™ XT 4-12% Bis-Tris, 11cm, 1mm; Bio-Rad) for the second-dimension of electrophoresis. The gels were run at 200 V (25 – 50 mA/gel) in XT MOPS running buffer (Bio-Rad), fixed in 25% methanol/10% acetic acid at room temperature for 30 min, and stained overnight with the ProteomIQ™ Blue Gel Stain Kit (Proteome Systems). After staining the gels were scanned using a Kodak Image Station 4000MM camera interfaced with Molecular Imaging Software, version 4.0 (Eastman Kodak Company). Images were saved as 16-bit tiff files. The images were analyzed using the Progenesis PG240 software package (Nonlinear Dynamics). Using the PG240 software, we performed image quality control to optimize image capture, image alignment to match spots, and pre-filter to remove artifacts (e.g., damage or noise areas). The volume of an individual spot, the integrated intensity within the spot area, was normalized to the total spot volume of the whole gel to give a normalized spot volume of the spot. Spots that exhibited significant differences between the two groups (p<0.05) were selected for protein identification by mass spectrometry.
2.4 Identification of spots by mass spectrometry
Gel spots that were exhibited statistically-significant differences in intensity between the two groups were manually excised using a One Touch™ 1.5 mm 2-DE spot picker (The Gel Company) and digested in situ with trypsin (Promega modified) in 40 mM NH4HCO3 at 37°C for 4 h. Digests were analyzed by capillary HPLC-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher LTQ linear ion trap mass spectrometer fitted with a New Objective PicoView 550 nanospray interface. On-line HPLC separation of the digests was achieved with an Eksigent NanoLC micro HPLC: column, PicoFrit™ (New Objective; 75 μm i.d.) packed to 10 cm with C18 adsorbent (Vydac; 218MSB5, 5 μm, 300 Å); mobile phase A, 0.5% acetic acid (HAc)/0.005% trifluoroacetic acid (TFA); mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA; gradient 2 to 42% B in 30 min; flow rate, 0.4 μl/min. The following MS conditions were used: ESI voltage, 2.9 kV; isolation window for MS/MS, 3; relative collision energy, 35%; scan strategy, survey scan followed by acquisition of data dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan above a set threshold. The uninterpreted CID spectra were searched using Mascot (Matrix Science) against the NCBInr_20070216 database. Peak lists were created using extract_msn.exe. Trypsin was specified as the proteolytic enzyme and up to two missed cleavages were allowed. The precursor and fragment ion mass tolerances were both set at ± 0.8 Da. Methionine oxidation and cysteine carbamidomethylation were considered as variable modifications for all searches. Cross correlation of the Mascot results with X! Tandem and determination of protein identity probabilities was accomplished with Scaffold™ (Proteome Software).
2.5 Immunoblotting
To confirm changes in specific proteins, immunoblotting was performed using antibodies against the following proteins: apolipoprotein A-I (Abcam ab20453), collagen VI (Abcam ab6588), Fn (Chemicon Ab1954), glyceraldehyde-3-phosphate dehydrogenase (Abcam ab9485), myosin light chain 2 (Abcam 48003), Prx-1 (Abcam ab15571), Prx-2 (Abcam ab16820), Prx-3 (Sigma P-1247), Prx-6 (Abcam ab59543), TN-C (Abcam ab6346), and voltage-dependent anion channel 2 (VDAC2; Abcam ab47104). For every sample, 10 μg of total protein was loaded in one lane of a 26-well 4–12% Criterion Bis-Tris gel (Bio-Rad) and the gels were run in XT MES buffer (Bio-Rad). After electrophoresis, protein samples in the gel were transferred to a nitrocellulose membrane (Bio-Rad). After blocking with 5% blotting grade blocker non-fat dry milk (Bio-Rad) in 1XPBS (Sigma) for 1 hr at room temperature, the membrane was incubated with primary antibody (1:1000 dilution) for overnight at 4°C. After washing three times with 1XPBS with 1% Tween-20 (PBS-T) for 15 min each, the membrane was incubated in secondary antibody (1:5000 dilution) for 1 hr at room temperature. Anti-rabbit IgG (Vector PI-1000) and anti-rat IgG (Santa Cruz sc-2006) antibodies were used. The membrane was washed three times with PBS-T for 15 min each and was detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientifics). Molecular Imaging Software (Kodak) was used for image analysis to quantify the densitometry. With this software, every blot was checked to confirm that no band was saturated. This ensures that the blot was not overexposed and that the signal was in the dynamic range. To confirm equal loading, immunoblotting of β-actin (Sigma A-2103) was performed to confirm loading and sample transfer was equal between the WT and MMP-7 null groups. The densitometry values for WT was 902±24 and for MMP-7 null was 865±22 (p=0.27). Densitometry values for individual samples on the immunoblots were normalized to the densitometry value of β-actin for the sample.
2.6 In vitro MMP-7 cleavage assay
Extracted total protein (10ug) from LV infarct of MMP-7 null mice was incubated with 10pg, 100pg or 10ng of recombinant human MMP-7 enzyme (Calibiochem Cat. No. 444270) at 37°C for 3 h in 1X zymogram developing buffer (Invitrogen) to determine if exogenous MMP-7 could restore the WT phenotype. After 3h, SDS loading buffer was added to stop the reaction. Each reaction was then loaded in one lane of a 26-well 4–12% Criterion Bis-Tris gel (Bio-Rad), and immunoblotting of Fn and TN-C were performed as described above to evaluate the action of exogenous MMP-7 enzyme on Fn and TN-C degradation in the MMP-7 null infarct extract. Untreated extract from MMP-7 null infarct was the negative control, and untreated extract from WT infarct was the positive control.
2.7 Statistical analyses
Data are reported as mean±SEM. All samples were analyzed individually and were not pooled at any step. Normalized spot volumes of 2-DE gels and normalized intensities for the immunoblots between WT and null groups were analyzed by an unpaired Student’s t-test. A p<0.05 was considered significant.
3. Results
3.1 Morphometric analysis
Necropsy values for WT and null mice are shown in Table 1. Both groups showed similar infarct sizes, 48±3% for WT and 54±3% for null (p=0.15), indicating that an equal injury stimulus was given. Both body mass and LV mass were lower in the null group, so that LV to body weight ratio was similar between the two groups.
Table 1.
Necropsy Data
| Wild-type (n=12) |
MMP-7 null (n=10) |
|
|---|---|---|
| Age (months) | 5.0±0.2 | 4.7±0.2 |
| Body Weight (g) | 28.7±0.7 | 25.0±1.0a |
| LV mass (mg) | 127±4 | 103±3a |
| LV/BW | 4.4±0.1 | 4.1±0.1 |
| Infarct size (%) | 48±3 | 54±3 |
Data are mean ± SEM
p<0.05 vs WT
3.2 Differences in protein quantity assessed by 2-DE analysis and mass spectrometry
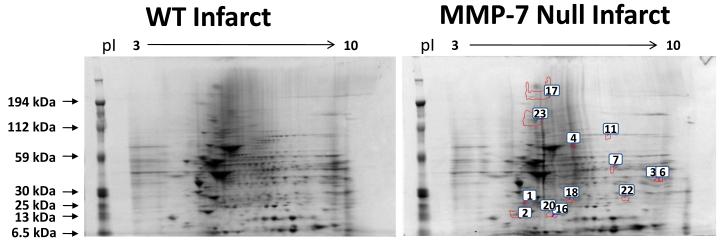
From image analysis of the 2-DE gels, we detected 13 spots that showed significant differences in normalized spot volume between WT and null infarct groups. Representative gels from each group are shown in Figure 1. Out of the 13 spots, 8 were higher and 5 were lower in intensity in the null group. A total of 142 proteins were identified in these spots, for an average of 11 proteins per spot. Several extracellular matrix proteins, including Fn, collagen type VI, and TN-C were identified in the spots that exhibited intensity differences between WT and null. The complete list of proteins identified by mass spectrometry can be found in Supplementary Table 1. Proteins that may be potential MMP-7 substrates are shown in Table 2. For inclusion in Table 2, the protein had to meet one of the following criteria: 1) the protein was an ECM protein; 2) the protein was identified in multiple spots or spots of decreasing molecular weight (suggesting the protein may have undergone proteolysis); 3) the protein was the only protein identified in the spot; or 4) the protein was previously associated with MMP activity or post-MI remodeling.
Figure 1.
Representative 2-DE gels of WT infarct extract (left) and MMP-7 null infarct extract (right). Spot numbers are indicated in the MMP-7 null gel.
Table 2.
Selected proteins identified in 2-DE gel spots differentially expressed in MMP-7 null infarcts (all p<0.05).
| Spot | PI | Observed MW (kDa) |
Wild-type Normalized Volume |
MMP-7 Null Normalized Volume |
Change compared to WT |
Protein Identified | Sequence Coverage (%) |
Total assigned peptidesa |
Theoretical MW (kDa) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.5 | 21.6 | 135±8 | 266±45 | ↑ | Myosin light polypeptide 3c,e | 54.4 | 11 | 22 |
| EF hand domain containing 2c | 31.7 | 6 | 27 | ||||||
| Apolipoprotein A-Ic,f | 18.9 | 5 | 31 | ||||||
| Peroxiredoxin-2e,f | 18.2 | 3 | 22 | ||||||
| Desminc | 6.4 | 3 | 53 | ||||||
| 2 | 4.3 | 16.1 | 580±42 | 946±160 | ↑ | Myosin light chain 2c,e,f | 50 | 11 | 19 |
| Myosin light polypeptide 3c,e | 14.7 | 3 | 22 | ||||||
| 3 | 9.4 | 37.9 | 287±94 | 466±67 | ↑ | Malate dehydrogenase 2c | 50.9 | 15 | 36 |
| Glyceraldehyde-3-phosphate dehydrogenasec,f | 21.9 | 7 | 36 | ||||||
| Voltage-dependent anion channel 2e,f | 14.6 | 4 | 32 | ||||||
| 4 | 6.2 | 78.4 | 300±15 | 439±52 | ↑ | Acyl-coenzyme A dehydrogenase, very long chaine | 37.3 | 22 | 71 |
| 6 | 9.5 | 37.7 | 468±57 | 673±64 | ↑ | Malate dehydrogenase 2c | 53 | 16 | 36 |
| Glyceraldehyde-3-phosphate dehydrogenasec,f | 10.2 | 4 | 36 | ||||||
| 7 | 7.8 | 49 | 672±60 | 949±58 | ↑ | Acyl-coenzyme A dehydrogenase, medium chaine | 27.3 | 11 | 46 |
| Creatine kinase , mitochondrial 2e | 25.8 | 10 | 47 | ||||||
| 11 | 7.5 | 96.6 | 476±51 | 645±52 | ↑ | Aconitase 2d | 26.2 | 19 | 85 |
| 16 | 5.5 | 15.5 | 228±16 | 171±12 | ↓ | Transgelin 2c,e | 35.7 | 7 | 22 |
| Myosin light polypeptide 3c,e | 21.6 | 3 | 22 | ||||||
| Myosin light chain 2c,e,f | 15.7 | 3 | 19 | ||||||
| Peroxiredoxin-3e,f | 11.3 | 3 | 28 | ||||||
| 17 | 5.4 | 217.7 | 10609±393 | 8370±477 | ↓ | Fibronectinb,c,e,f | 7.02 | 16 | 273 |
| Tenascin-Cb,e,f | 4.21 | 10 | 260 | ||||||
| Collagen, type VIb,c,e,f | 1.8 | 2 | 140 | ||||||
| Myosin heavy chain 6c | 1.4 | 5 | 424 | ||||||
| 18 | 6.1 | 23.4 | 654±28 | 524±54 | ↓ | Peroxiredoxin-6e,f | 22.3 | 6 | 25 |
| Peroxiredoxin-1e,f | 22.1 | 4 | 22 | ||||||
| Peroxiredoxin-4e | 16.8 | 5 | 31 | ||||||
| Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1c |
6.2 | 3 | 29 | ||||||
| 20 | 5.5 | 15.6 | 747±43 | 603±30 | ↓ | Myosin light chain 2c,e,f | 39.8 | 7 | 19 |
| Transgelin 2c,e | 27.1 | 7 | 22 | ||||||
| EF hand domain containing 2c | 8.8 | 2 | 27 | ||||||
| Peroxiredoxin-3c,e,f | 7.4 | 2 | 28 | ||||||
| 22 | 8.2 | 24.4 | 424±42 | 519±11 | ↑ | Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1c |
14.2 | 5 | 29 |
| 23 | 4.6 | 133 | 3973±119 | 3329±162 | ↓ | Myosin heavy chain 6c | 9.4 | 36 | 424 |
| Desminc | 8.3 | 4 | 53 | ||||||
| Collagen, type VIb,c,e,f | 6.4 | 6 | 140 | ||||||
| Vimentine | 5.8 | 3 | 52 | ||||||
| Fibronectinb,c,e,f | 2.8 | 6 | 273 |
total number of peptides assigned with a confidence level of ≥ 95%; all proteins listed were ≥ 99.0% confidence based on analysis by Scaffold 2; only proteins with at least two high-confidence assigned peptides are listed; the protein was listed because
it was an ECM protein
it was identified in multiple spots or spots of decreasing molecular weight
it was the only protein identified in the spot
it was previously associated with MMP activity or post-MI remodeling
examined further by immunoblotting.
3.3 Immunoblotting analysis to determine protein levels in post-MI LV
As an additional method to evaluate the levels of candidate MMP-7 substrates identified in the LV post-MI, extracts of the infarct region of WT and null mice were analyzed by immunoblotting. Eleven proteins were selected for this analysis, based on the criteria that an antibody was commercially available and the protein either had an extracellular localization or was a candidate substrate. The results of immunoblotting are summarized in Table 3. Of the 11 proteins analyzed, differences in the levels of three (Fn, TN-C and Prx-1) agreed with the 2-DE gel results. For four proteins (Prx-2, Prx-3, GAPDH and VDAC2), the immunoblot results were in the opposite direction compared to the 2-DE gel analysis. Apolipoprotein A1, myosin light chain 2, Prx-6 and collagen VI showed no difference in intensity between the two groups by immunoblotting.
Table 3.
Agreement between the results observed by 2-DE gel analysis and immunoblotting
| 2-DE Gels |
Immunoblotting |
|||||
|---|---|---|---|---|---|---|
| Protein | Spot # | MW (kDa) |
Changea | MW (kDa) |
Changea | Concordance |
| Fibronectin | 17 | 218 | ↓ | 273 | ↓ | Yes |
| Fibronectin | 23 | 133 | ↓ | 166 | ↓ | Yes |
| Peroxiredoxin-1 | 18 | 23 | ↓ | 23 | ↓ | Yes |
| Tenascin-C | 17 | 218 | ↓ | 260 | ↓ | Yes |
| 200 | – | No | ||||
| ↓ | 170 | ↓ | Yes | |||
| ↓ | 133 | ↓ | Yes | |||
| GAPDH | 3 | 38 | ↑ | 36 | ↓ | No |
| Peroxiredoxin-2 | 1 | 22 | ↑ | 23 | ↓ | No |
| Peroxiredoxin-3 | 16 | 16 | ↓ | 23 | ↑ | No |
| 20 | ↓ | No | ||||
| VDAC2 | 3 | 38 | ↑ | 38 | ↓ | No |
| Apolipoprotein A1 | 1 | 22 | ↑ | 30 | – | No |
| Collagen VI | 23 | 133 | ↓ | 100 | – | No |
| myosin light chain 2 | 2 | 16 | ↓ | 20 | – | No |
| Peroxiredoxin-6 | 18 | 23 | ↓ | 25 | – | No |
Direction of difference in spot and band intensity compared to WT; GAPDH – glyceraldehyde-3-phosphate dehydrogenase; VDAC2 – voltage-dependent anion channel 2
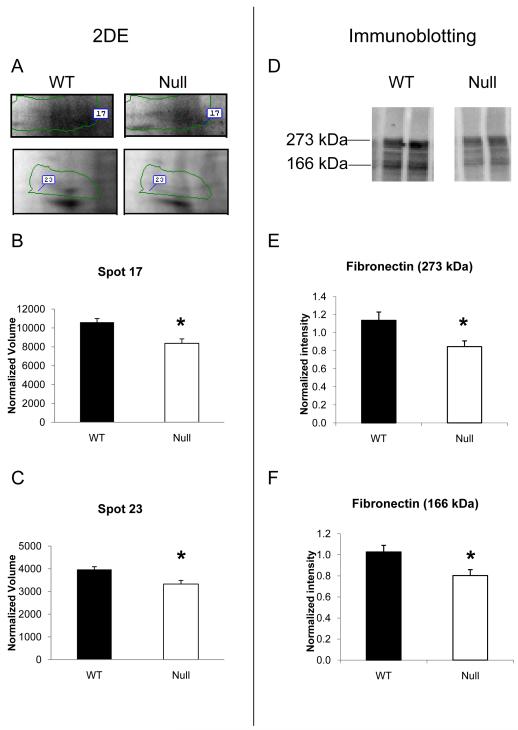
Fn, a known in vitro substrate for MMP-7, was identified in two spots (spot 17 at 218 kDa and spot 23 at 133 kDa) and exhibited significantly lower levels in the null group compared to WT by 2-DE gel analysis. In general agreement with these findings, by immunoblotting, Fn was detected at 273 kDa and 166 kDa and both bands were significantly less intense in the MMP-7 null infarct compared to WT (Figure 2).
Figure 2.
MMP-7 null infarcted LV showed lower levels of fibronectin by 2-DE and immunoblotting. (A) Fibronectin was identified in spot 17 and spot 23 of the 2-DE gels. Both spot 17 (B) and spot 23 (C) showed significant lower intensity in infarct LV of MMP-7 null mice compared to WT. (D) Immunoblotting of fibronectin in WT and MMP-7 null LV extracts were performed. The densitometry of the 273 kDa full-length (E) and the 166 kDa fragments (F) of fibronectin indicated both bands showed significantly lower intensity in MMP-7 null LV infarct when compared with the wt (p<0.05).
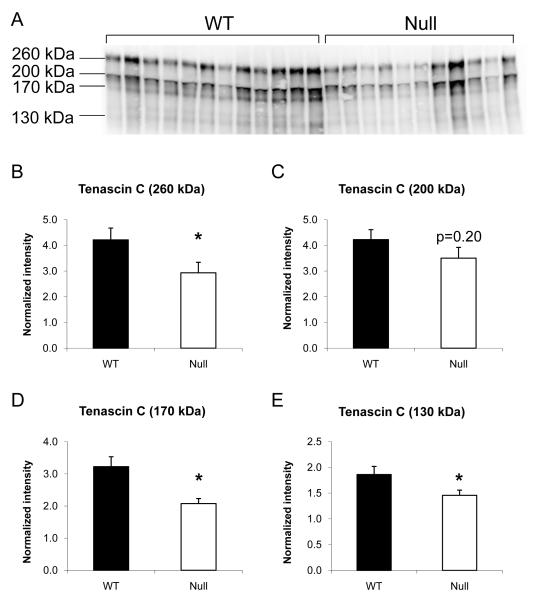
TN-C was identified in spot 17 (218 kDa) and this spot exhibited significantly lower intensity in the null group compared to WT. By immunoblotting, 4 major bands were detected using the anti-TN-C antibody; the 133 kDa, 170 kDa and 260 kDa forms were significantly lower in the MMP-7 null group, while the 200 kDa form did not show any significant difference between groups (Figure 3).
Figure 3.
Immunoblotting showed lower levels of full-length and fragments of tenascin-C in MMP-7 null LV infarct. (A) 4 major bands of 133 kDa, 170 kDa, 200 kDa and 260 kDa were observed by immunoblotting of TN-C in WT and MMP-7 null LV. (B) Densitometry showed significantly lower levels of full length (260 kDa) TN-C in MMP-7 null LV infarct when compared to WT. (C) Although the 200 kDa form did not show a significant difference, both 170 kDa (D) and 130kDa (E) fragments of TN-C showed significantly lower intensity in the MMP-7 null group.
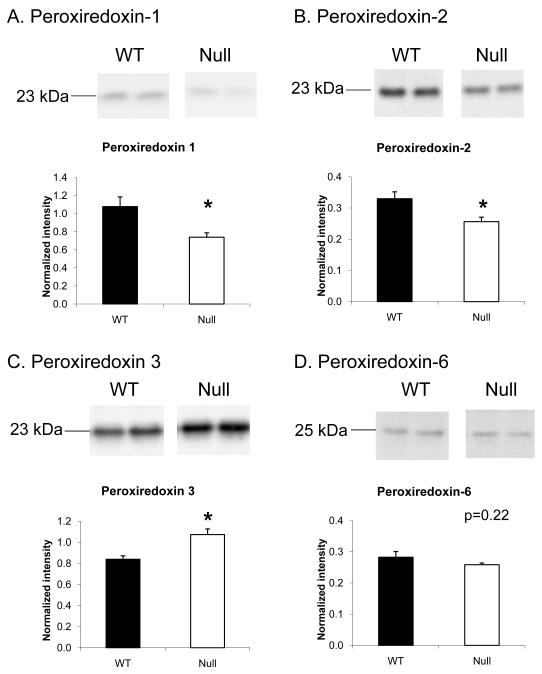
Several Prx isoforms (1, 2, 3, 4 and 6) were identified in the 2-DE gel analysis. For example, Prx-1, -4 and -6 were identified in spot 18, and this spot was significantly less intense in the null group. Since the predicted molecular weights of Prx-1 and Prx-6 matched the observed molecular weight of spot 18, immunoblotting for Prx-1 and Prx-6 were performed. We did not evaluate Prx-4, because the observed molecular weight of the spot did not match the expected molecular weight for this peroxiredoxin family member. In the immunoblot analysis of Prx-1, there was a 23 kDa band that agreed with the differences detected by 2-DE gel analysis (Figure 4A), showing lower intensity in the null group compared to WT. However, no significant differences in Prx-6 were detected between the two groups by immunoblotting (Figure 4D), which suggests Prx-1 but not Prx-6 contributed to the lower intensity of spot 18 in the 2-DE gel.
Figure 4.
Levels of different Prx isoforms were differentially regulated by MMP-7 deletion. Immunoblotting showed significantly lower intensity of Prx-1 (A) and Prx-2 (B) in the MMP-7 null LV infarct when compared to WT. (C) However, higher levels of Prx-3, a mitochondrial Prx, were detected in the MMP-7 null group. (D) No significant difference of Prx-6 between WT and MMP-7 null groups were observed.
Prx-2 was identified in spot 1. This spot was shown to be significantly higher in intensity in the null group compared to WT, but this protein exhibited significantly lower levels in the null group by immunoblotting (Figure 4B). This indicates that other proteins identified in spot 1, not Prx-2, likely contribute to the observed intensity difference in the 2-DE gels.
Similarly, Prx-3 was identified in spots 16 and 20, and both spots exhibited significantly lower intensities in the null group. By immunoblotting, however, Prx-3 levels were found to be significantly higher in the null group (Figure 4C), suggesting that other proteins in these spots are likely to have contributed to the observed differences in spot intensity.
3.4 In vitro MMP-7 cleavage generates Fn and TN-C fragments from MMP-7 null extracts
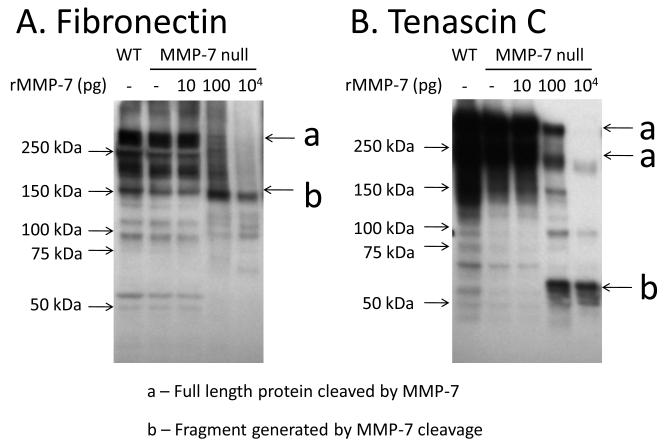
To determine if exogenous MMP-7 could rescue the null phenotype, we performed an in vitro MMP-7 cleavage assay. As shown in Figure 5, incubation with recombinant MMP-7 enzyme generated fragments of Fn and TN-C that were similar to the WT patterns. For Fn, MMP-7 null extract treated with 100 pg or 10 ng of recombinant MMP-7 showed processing of full-length Fn (273 kDa) compared to the untreated control. At the same time, MMP-7 treatment resulted in increased levels of the 166 kDa Fn fragment compared to the untreated control. Exogenous MMP-7 increased the levels of this Fn fragment to levels similar to the WT extract (Figure 5A), suggesting this fragment was an in vivo cleavage product of MMP-7. Similarly, MMP-7 treatment reduced the levels of full-length TN-C (260 kDa and 200 kDa) in the null extract, but increased the levels of 65 kDa fragment in the MMP-7 null extract to levels similar to the WT extract (Figure 5B).
Figure 5.
The addition of exogenous MMP-7 generated Fn and TN-C fragments in MMP-7 null infarct extract. (A) MMP-7 null extract treated with 100 pg or 10 ng of recombinant MMP-7 showed reduced levels of full-length Fn (273 kDa) and increased levels of the 166 kDa Fn fragment compared to the untreated control. (B) MMP-7 null extract treated with 100 pg or 10 ng of MMP-7 showed reduced levels of full-length TN-C (260 kDa and 200 kDa) in the null extract, but increased the levels of 65 kDa fragment in the MMP-7 null extract.
4. Discussion
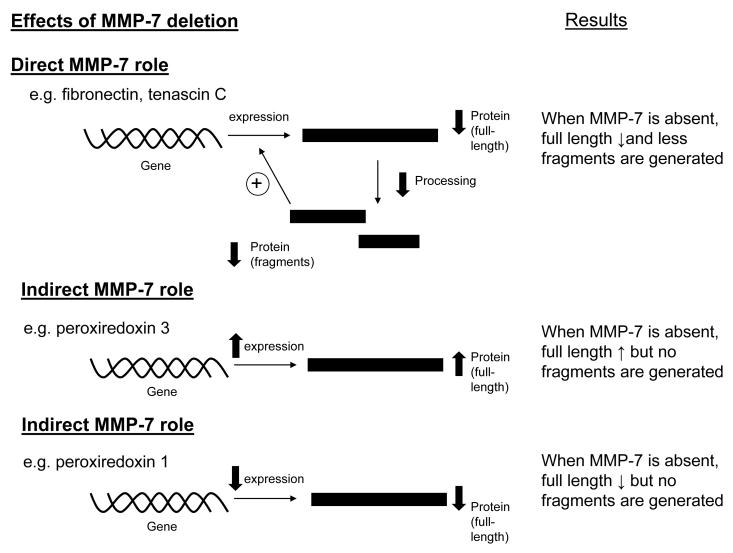
The objective of this study was to identify candidate MMP-7 substrates in the infarct region of mice LV. The most significant findings of this study were: 1) Fn, a known in vitro and in vivo substrate for MMP-7, was found at lower levels (both full length and proteolytic fragments) in the MMP-7 null mice; 2) TN-C, which has been shown to be cleaved by MMP-7 in vitro, was processed by MMP-7 in vivo in WT post-MI LV; and 3) the protein levels of multiple Prx isoforms were differentially regulated by MMP-7 after MI. Together, these results demonstrate that this proteomic strategy can be used to identify and catalogue potential in vivo MMP-7 substrates and better understand MMP-7 roles in the post-MI setting. As illustrated in Figure 6, the observed changes in the absence of MMP-7 suggest both direct and indirect roles of MMP-7.
Figure 6.
Schematic diagram of the direct and indirect effects of MMP-7 deletion in infarcted left ventricles. When MMP-7 plays a proteolytic role and cleaves the full-length protein and to fragments, the deletion of MMP-7 is expected to result in lower levels of fragments. If the fragments can feedback induce the expression of the protein, lower levels of fragments due to MMP-7 deletion will reduce expression and therefore, result in lower levels of full length proteins. MMP-7 also indirectly regulates the expression of proteins, including Prx-1 and -3. If MMP-7 indirectly suppresses the expression of one protein, MMP-7 deletion will result in increased expression of that protein and result in higher levels of full-length protein. On the other hand, if MMP-7 indirectly induces the expression of one protein, MMP-7 deletion will result in reduced expression of that protein and result in lower levels of full-length protein.
Fn is induced after MI and is a known in vivo substrate for MMP-7.18, 19 In diabetic nephropathy, advanced glycation end products (AGEs) suppress MMP-7 expression and contribute to the accumulation of full-length Fn in the diabetic kidney.19 However, MMP-7 regulation of Fn processing post-MI has not been previously demonstrated. By 2-DE, we identified Fn in two spots that were both lower in normalized volume in the MMP-7 null infarct. Immunoblotting confirmed lower levels of full-length (273 kDa) and the 166 kDa fragment of Fn in MMP-7 null mice compared to WT. Because MMP-7 is known to process Fn, we expected lower levels of Fn fragments in the MMP-7 null mice, and this served as a validation of our proteomic approach to identify MMP-7 substrates. We confirmed that adding exogenous MMP-7 to MMP-7 null infarct extract proteolyzed full-length Fn to generate Fn fragments. The fact that exogenous MMP-7 rescued the MMP-7 null phenotype confirms Fn can be cleaved by MMP-7 in the infarct myocardium. We were surprised to observe reduced levels of full-length Fn in the MMP-7 null group. One possible explanation is that Fn fragments can induce Fn expression in a feedback manner. Other ECM proteins have been found to be positively or negatively regulated by their own ECM fragments.20 The reduced generation of Fn fragments in MMP-7 null mice, therefore, likely resulted in diminished Fn expression (Figure 6A).
There are two major TN-C isoforms, a 260 kDa large TN-C isoform and a 200 kDa small TN-C isoform, and both of them can be cleaved by MMP-7 in vitro.7 Keeling and Herrera recently showed reduced levels of extracellular MMP-7 in light chain deposition disease and suggested that impaired MMP-7 secretion from mesangial cells may contribute to accumulation of TN-C in an in vitro model.21 Expression of TN-C is high during embryogenesis, but is undetectable in the adult heart.22 After MI, TN-C is re-expressed to promote myofibroblast recruitment and to regulate post-MI wound healing.15 In a time course evaluation of acute MI patients, significantly higher peak serum TN-C levels were observed at day 5 post-MI in the group that showed ≥20% increase in end-diastolic volume 6 months post-MI.23 Based on the results of that study, Sato and colleagues proposed that the peak serum TN-C level predicts the risk of major adverse cardiac events, including cardiac death and hospitalization for congestive heart failure, in acute MI patients.23 In our study, the levels of the 170 kDa TN-C fragment, a major cleavage product of large TN-C isoform by MMP-7 previously reported by Zardi’s laboratory,7 were lower in the infarct of MMP-7 null mice compared to WT mice. This is possibly due to less processing of TN-C in absence of MMP-7 and indicates that TN-C in the infarct myocardium is likely an in vivo substrate of MMP-7. The MMP-7 cleavage assay confirmed that exogenous MMP-7 can process full-length TN-C (260 kDa and 200 kDa forms) in the null extract to generate the 65 kDa TN-C fragment, which is a major MMP-7 cleavage product of large TN-C isoform.7 In the cleavage assay, no increased in levels of 170 kDa TN-C fragment was detected, which may be due to further processing of the 170 kDa fragment into smaller size degradation products. Similar to Fn, lower levels of full-length TN-C were observed in MMP-7 null mice, suggesting a possible feedback regulation of TN-C expression by its fragments (Figure 6A).
MMP-7 cleavage of Fn and Tn-C have previously been demonstrated in vitro.6-9 Together with the findings in this study, this suggests that MMP-7 directly cleaves Fn and Tn-C in the LV infarct. However, as MMP-7 can activate other MMPs by cleavage of their pro-forms,9-11 it is highly possible that MMP-7 also exerts indirect effects on cleavage. For example, MMP-7 cleaves and activates pro-MMP-9,9, 24 which also cleaves Fn.25 At the same time, MMP-7 can activate pro-MMP-2,10, 24 which also cleaves Tn-C.7 Therefore, the net effect of MMP-7 on Fn and Tn-C in the LV infarct is likely due to both direct cleavage and indirect action on MMP-2 and MMP-9 activation.
Peroxiredoxins (Prx) are a family of antioxidant proteins ubiquitously expressed in multiple tissues types, including heart, liver, and kidney.26 Six Prx isoforms (1-6) have been identified in mammals.26 Overexpression of Prx-3 was shown to attenuate LV remodeling after MI,27 and targeted disruption of Prx-6 results in increased infarct size and reduced LV function after ischemia-reperfusion.28 These studies indicate that particular Prx isoforms play a protective role in ischemic myocardial injury, but the regulation of Prx after MI is unclear. Differences in the levels of several Prx isoforms were found by 2-DE and mass spectrometry analysis. Immunoblotting showed that Prx-1 and -2 levels were lower in the infarct region of MMP-7 null mice while Prx-3 levels were higher in the MMP-7 null group compared to WT. No lower molecular weight Prx fragments were observed in the WT samples, indicating that direct proteolysis of Prx by MMP-7 does not likely account for the differences in protein levels. In addition, there are no reports that Prx is an in vitro substrate of MMP-7. MMP-7, therefore, may indirectly regulate the protein levels of different Prx isoforms by mechanisms that have not been fully elucidated (Figure 6B and 6C). Prx-1 and -2 are localized in the cytoplasm and nucleus, and no association of these Prx and LV remodeling has been demonstrated. Prx-3 is the only isoform localized in mitochondria, and overexpression of Prx-3 can attenuate post-MI LV remodeling by reducing mitochondrial oxidative stress.27 Our data suggest that the higher levels of Prx-3 detected in the post-MI LV of MMP-7 null mice may be associated with attenuated mitochondrial oxidative stress. Our group has previously shown that in the LV of MMP-7 null mice, there is increased MMP-8 and reduced MMP-13 levels.29 However, even with the potential compensation of these other MMPs, MMP-7 deletion still has a net effect on post-MI response, indicating that compensation from other proteases is not complete.
In conclusion, we identified candidate MMP-7 substrates that are present at lower levels in the infarct of MMP-7 null mice compared to that of WT mice. Identification of known in vitro ECM substrates Fn and TN-C, and confirmation that exogenous MMP-7 can rescue the MMP-7 null phenotype validates this proteomic approach as a useful tool to explore MMP functions. Altered Prx levels in the MMP-7 null group in absence of proteolytic fragmentation indicate that MMP-7 deletion or inhibition may indirectly affect oxidative stress levels in post-MI myocardium, providing an additional protective effect of MMP-7 inhibition.
Supplementary Material
Acknowledgements
The authors acknowledge grant support from NIH T32 HL07446 and the American Heart Association POST2150178 (RZ) and from NIH HL75360, the American Heart Association AHA 0855119F, the Max and Minnie Tomerlin Voelcker Fund, and the Morrison Trust (MLL). The mass spectrometry analyses were conducted in the UTHSCSA Institutional Mass Spectrometry Laboratory.
References
- 1.Crabbe T, Willenbrock F, Eaton D, Hynds P, Carne AF, Murphy G, Docherty AJ. Biochemical characterization of matrilysin. Activation conforms to the stepwise mechanisms proposed for other matrix metalloproteinases. Biochemistry. 1992;31(36):8500–7. doi: 10.1021/bi00151a017. [DOI] [PubMed] [Google Scholar]
- 2.Gaire M, Magbanua Z, McDonnell S, McNeil L, Lovett DH, Matrisian LM. Structure and expression of the human gene for the matrix metalloproteinase matrilysin. J Biol Chem. 1994;269(3):2032–40. [PubMed] [Google Scholar]
- 3.Boixel C, Fontaine V, Rucker-Martin C, Milliez P, Louedec L, Michel JB, Jacob MP, Hatem SN. Fibrosis of the left atria during progression of heart failure is associated with increased matrix metalloproteinases in the rat. J Am Coll Cardiol. 2003;42(2):336–44. doi: 10.1016/s0735-1097(03)00578-3. [DOI] [PubMed] [Google Scholar]
- 4.Furman C, Copin C, Kandoussi M, Davidson R, Moreau M, McTaggiart F, Chapman MJ, Fruchart J-C, Rouis M. Rosuvastatin reduces MMP-7 secretion by human monocyte-derived macrophages: potential relevance to atherosclerotic plaque stability. Atherosclerosis. 2004;174(1):93–98. doi: 10.1016/j.atherosclerosis.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Lindsey ML, Escobar GP, Mukherjee R, Goshorn DK, Sheats NJ, Bruce JA, Mains IM, Hendrick JK, Hewett KW, Gourdie RG, Matrisian LM, Spinale FG. Matrix Metalloproteinase-7 Affects Connexin-43 Levels, Electrical Conduction, and Survival After Myocardial Infarction. Circulation. 2006;113(25):2919–2928. doi: 10.1161/CIRCULATIONAHA.106.612960. [DOI] [PubMed] [Google Scholar]
- 6.Agnihotri R, Crawford HC, Haro H, Matrisian LM, Havrda MC, Liaw L. Osteopontin, a novel substrate for matrix metalloproteinase-3 (stromelysin-1) and matrix metalloproteinase-7 (matrilysin) J Biol Chem. 2001;276(30):28261–7. doi: 10.1074/jbc.M103608200. [DOI] [PubMed] [Google Scholar]
- 7.Siri A, Knauper V, Veirana N, Caocci F, Murphy G, Zardi L. Different susceptibility of small and large human tenascin-C isoforms to degradation by matrix metalloproteinases. J Biol Chem. 1995;270(15):8650–4. doi: 10.1074/jbc.270.15.8650. [DOI] [PubMed] [Google Scholar]
- 8.von Bredow DC, Nagle RB, Bowden GT, Cress AE. Degradation of fibronectin fibrils by matrilysin and characterization of the degradation products. Exp Cell Res. 1995;221(1):83–91. doi: 10.1006/excr.1995.1355. [DOI] [PubMed] [Google Scholar]
- 9.Imai K, Yokohama Y, Nakanishi I, Ohuchi E, Fujii Y, Nakai N, Okada Y. Matrix metalloproteinase 7 (matrilysin) from human rectal carcinoma cells. Activation of the precursor, interaction with other matrix metalloproteinases and enzymic properties. J Biol Chem. 1995;270(12):6691–6697. doi: 10.1074/jbc.270.12.6691. [DOI] [PubMed] [Google Scholar]
- 10.Crabbe T, Smith B, O’Connell J, Docherty A. Human progelatinase A can be activated by matrilysin. FEBS Lett. 1994;345(1):14–6. doi: 10.1016/0014-5793(94)00412-9. [DOI] [PubMed] [Google Scholar]
- 11.Haro H, Crawford HC, Fingleton B, Shinomiya K, Spengler DM, Matrisian LM. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-alpha in a model of herniated disc resorption. J Clin Invest. 2000;105(2):143–50. doi: 10.1172/JCI7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stenman S, von Smitten K, Vaheri A. Fibronectin and atherosclerosis. Acta Med Scand Suppl. 1980;642:165–70. doi: 10.1111/j.0954-6820.1980.tb10949.x. [DOI] [PubMed] [Google Scholar]
- 13.Willems IE, Arends JW, Daemen MJ. Tenascin and fibronectin expression in healing human myocardial scars. J Pathol. 1996;179(3):321–5. doi: 10.1002/(SICI)1096-9896(199607)179:3<321::AID-PATH555>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Orem C, Celik S, Orem A, Calapoglu M, Erdol C. Increased plasma fibronectin levels in patients with acute myocardial infarction complicated with left ventricular thrombus. Thromb Res. 2002;105(1):37–41. doi: 10.1016/s0049-3848(01)00414-5. [DOI] [PubMed] [Google Scholar]
- 15.Imanaka-Yoshida K, Hiroe M, Nishikawa T, Ishiyama S, Shimojo T, Ohta Y, Sakakura T, Yoshida T. Tenascin-C modulates adhesion of cardiomyocytes to extracellular matrix during tissue remodeling after myocardial infarction. Lab Invest. 2001;81(7):1015–24. doi: 10.1038/labinvest.3780313. [DOI] [PubMed] [Google Scholar]
- 16.Nishioka T, Onishi K, Shimojo N, Nagano Y, Matsusaka H, Ikeuchi M, Ide T, Tsutsui H, Hiroe M, Yoshida T, Imanaka-Yoshida K. Tenascin-C may aggravate left ventricular remodeling and function after myocardial infarction in mice. Am J Physiol Heart Circ Physiol. doi: 10.1152/ajpheart.00255.2009. In Press. [DOI] [PubMed] [Google Scholar]
- 17.Rudolph-Owen LA, Hulboy DL, Wilson CL, Mudgett J, Matrisian LM. Coordinate Expression of Matrix Metalloproteinase Family Members in the Uterus of Normal, Matrilysin-Deficient, and Stromelysin-1-Deficient Mice. Endocrinology. 1997;138(11):4902–4911. doi: 10.1210/endo.138.11.5478. [DOI] [PubMed] [Google Scholar]
- 18.Stanton LW, Garrard LJ, Damm D, Garrick BL, Lam A, Kapoun AM, Zheng Q, Protter AA, Schreiner GF, White RT. Altered Patterns of Gene Expression in Response to Myocardial Infarction. Circ Res. 2000;86(9):939–945. doi: 10.1161/01.res.86.9.939. [DOI] [PubMed] [Google Scholar]
- 19.McLennan SV, Kelly DJ, Schache M, Waltham M, Dy V, Langham RG, Yue DK, Gilbert RE. Advanced glycation end products decrease mesangial cell MMP-7: a role in matrix accumulation in diabetic nephropathy? Kidney Int. 2007;72(4):481–8. doi: 10.1038/sj.ki.5002357. [DOI] [PubMed] [Google Scholar]
- 20.Davis GE. Matricryptic sites control tissue injury responses in the cardiovascular system: Relationships to pattern recognition receptor regulated events. J Mol Cell Cardiol. doi: 10.1016/j.yjmcc.2009.09.002. In Press (March 2010 publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeling J, Herrera GA. An in vitro model of light chain deposition disease. Kidney Int. 2009;75(6):634–45. doi: 10.1038/ki.2008.504. [DOI] [PubMed] [Google Scholar]
- 22.Imanaka-Yoshida K, Matsumoto K, Hara M, Sakakura T, Yoshida T. The dynamic expression of tenascin-C and tenascin-X during early heart development in the mouse. Differentiation. 2003;71(4-5):291–8. doi: 10.1046/j.1432-0436.2003.7104506.x. [DOI] [PubMed] [Google Scholar]
- 23.Sato A, Aonuma K, Imanaka-Yoshida K, Yoshida T, Isobe M, Kawase D, Kinoshita N, Yazaki Y, Hiroe M. Serum tenascin-C might be a novel predictor of left ventricular remodeling and prognosis after acute myocardial infarction. J Am Coll Cardiol. 2006;47(11):2319–25. doi: 10.1016/j.jacc.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 24.von Bredow DC, Cress AE, Howard EW, Bowden GT, Nagle RB. Activation of gelatinase-tissue-inhibitors-of-metalloproteinase complexes by matrilysin. Biochem J. 1998;331:965–972. doi: 10.1042/bj3310965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opdenakker G, Van den Steen PE, Van Damme J. Gelatinase B: a tuner and amplifier of immune functions. Trends Immunol. 2001;22(10):571–9. doi: 10.1016/s1471-4906(01)02023-3. [DOI] [PubMed] [Google Scholar]
- 26.Immenschuh S, Baumgart-Vogt E. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal. 2005;7(5-6):768–77. doi: 10.1089/ars.2005.7.768. [DOI] [PubMed] [Google Scholar]
- 27.Matsushima S, Ide T, Yamato M, Matsusaka H, Hattori F, Ikeuchi M, Kubota T, Sunagawa K, Hasegawa Y, Kurihara T, Oikawa S, Kinugawa S, Tsutsui H. Overexpression of Mitochondrial Peroxiredoxin-3 Prevents Left Ventricular Remodeling and Failure After Myocardial Infarction in Mice. Circulation. 2006;113(14):1779–1786. doi: 10.1161/CIRCULATIONAHA.105.582239. [DOI] [PubMed] [Google Scholar]
- 28.Nagy N, Malik G, Fisher AB, Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2006;291(6):H2636–40. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- 29.Dozier S, Escobar GP, Lindsey ML. Matrix metalloproteinase (MMP)-7 activates MMP-8 but not MMP-13. Med Chem. 2006;2(5):523–6. doi: 10.2174/157340606778250261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.