Abstract
A detailed error analysis is presented for the computation of protein-ligand interaction energies. In particular, we show that it is probable that even highly accurate computed binding free energies have errors that represent a large percentage of the target free energies of binding. This is due to the observation that the error for computed energies quasi-linearly increases with the increasing number of interactions present in a protein-ligand complex. This principle is expected to hold true for any system that involves an ever increasing number of inter or intra-molecular interactions (e.g. ab initio protein folding). We introduce the concept of best-case scenario errors (BCSerrors) that can be routinely applied to docking and scoring exercises and used to provide errors bars for the computed binding free energies. These BCSerrors form a basis by which one can evaluate the outcome of a docking and scoring exercise. Moreover, the resultant error analysis enables the formation of an hypothesis that defines the best direction to proceed in order to improve scoring functions used in molecular docking studies.
Introduction
Since Paul Dirac noted in 1929, "The fundamental laws necessary for the mathematical treatment of a large part of physics and the whole of chemistry are thus completely known, and the difficulty lies only in the fact that application of these laws leads to equations that are too complex to be solved."1 theoretical chemistry has evolved to the point that in some instances tractable equations are utilized create a computational method that can routinely reach what is termed chemical accuracy or ±1kcal/mol from experiment.2,3 This accuracy is achieved for small interacting molecular systems like the water dimer or other related small molecule complexes.2,3 The extension of this result to macromolecular systems, however, is less clear. In principle one would like to believe that as chemically accurate models are used on ever-larger systems that the same level of accuracy would be possible. It is likely, though, that this is not the case and, indeed, we argue below that the expected errors in energies calculated on macromolecular systems are likely not to reach this level of accuracy. However, what level of accuracy would be expected is unclear. In this note we describe a "gedanken" experiment for protein-ligand scoring that addresses this very issue by delving deeper into the expected errors in energy computation in macromolecules.
Protein-ligand docking and scoring has been an active field of investigation for the last several decades.4–8 The concept is that given a small molecule compound we can computationally pose or dock it into a receptor site such that we obtain the correct orientation relative to experiment while simultaneously predicting a binding free energy in good agreement with experiment. This has proven to be a difficult task6,7, which is best captured by: "Accurate prediction of binding affinities for a diverse set of molecules turns out to be genuinely difficult.5". Indeed, extensive validation studies have shown how challenging this problem is9–12, but it is still largely uncertain why. Arguments including sampling13, structural water molecules, tautomeric states and conformational strain14 have all been put forward as (partial) explanations. Nonetheless, the way to significantly improve the current state-of-the-art still has to be delineated. Extensive work using free energy perturbation or alchemical methods have shown some promise relative to traditional docking approaches13,15, but still yield results with fairly large errors in terms of both binding orientation and the free energy of binding in prospective or blind studies.13
Types of errors
When deciding upon what type of error model to use there are two extremes that need to be considered. The first is determinate or systematic errors, which are errors that have a value that can be assigned and corrected for when obtaining insight into the reliability of a measurement. The second extreme is random or indeterminate errors whose sources are not certain, do not have a definite value, but do fluctuate in a random way. In each case it is possible to propagate the errors over a series of measurements or in our case interactions in, for example, a protein-ligand complex. In the present work we will assume that we will be accumulating errors via sums of interactions, hence, systematic errors are propagated as a simple sum of the individual errors in the interactions, while random errors are accumulated as the square root of the sum of the squares of the individual errors.16 The sum of errors used to propagate systematic errors can also be shown to represent the upper limit for random errors as well.16
The Variation Principle and Error
The Variation Principle, given as:
| (1) |
where Φ is the wavefunction and H is our Hamiltonian. This principle states that as the wavefunction Φ is improved at a defined Hamiltonian H (e.g. Hartree-Fock) that we should asymptotically approach the ground state energy Eo appropriate to that Hamiltonian.17 Hence, for the computation of Eo we should expect the error to be above the "true" Eo suggesting that we are dealing with a systematic error. Critically, for the present analysis, if we compute the interactions embodied within a protein-ligand complex using a variational method we would expect our computed interaction energy to be above or below the expected value since we do not know the magnitude of the difference (or error) between the individual Eo's computed for the interacting partners. This, of course, goes away when the equality in equation 1 is satisfied and the true Eo is reached. Hence, all interactions computed by a series of variational methods (e.g., HF/aug-cc-pVTZ, CASSCF, full CI, etc.) can be described as having a random error relative to our reference interaction energy value. Thus, we conclude that for variational methods error accumulation appears to behave as if we are working with a systematic or determinate error for the electronic energy, Eo, while for the computation of the interaction energy these methods behave as if one is dealing with a random error.
Variational methods are very particular in the sense that most modern computational methods are non-variational. This includes force fields, semiempirical QM (e.g., AM1, PM3, SCCDFTB, etc.), density functional theory (DFT), MPX (X=2–4) theory and coupled cluster theory (e.g. CCSD(T)).17 Hence, in contrast to variational methods, non-variational methods, in principal, are expected to display random errors (in that we do not know if the computed energy is above or below Eo) for both the computation of Eo and for the interaction energy that are propagated as the square root of the sum of squares (see equation 9 below) rather than as a simple sum. Interestingly, the accumulation of systematic errors will yield a larger overall error for Eo than the random analysis given the same absolute magnitudes of the individual errors. The difference between systematic and random uncertainties has to do with cancellation of errors that can happen when one deals with random errors. Nonetheless, as noted above it can be proven that the sum of error model represents the upper limit for random errors.16 Hence, the use of more theoretically grounded variational methods over non-variational methods, does not appear to offer a benefit when it comes to error propagation or the computation of interaction energies.
Protein-ligand Binding Free Energies
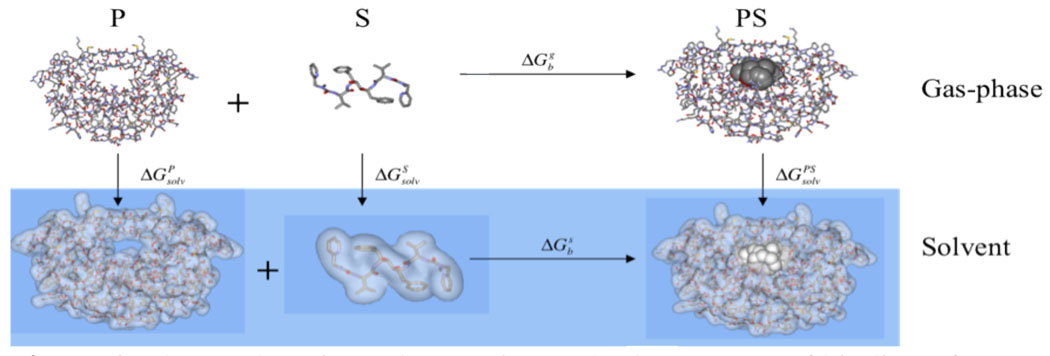
Consider the standard thermodynamic cycle shown in Figure 1.15,18 Using this we can write the following standard expressions:
| (2) |
where ΔG indicates free energy changes in the gas-phase (ΔGg) or solution (ΔGs) and the subscript b denotes binding. Terms associated with changes in the solvation free energy are also indicated by solv. What we want to do next is to break this down into individual components of the total energy, enthalpy and entropy in a compact form, which we briefly outline below. Expanding out the free energy of binding in the gas-phase term and consolidating the solvation free energies we can write:
| (3) |
where the Etotal terms are the individual electronic energies, Hcorr are the individual enthalpy correction terms to the electronic energies and the S terms are the respective the entropies. The latter two terms can be computed using the rigid-rotor harmonic approximation.19,20 ΔΔGsol is given as:
| (4) |
and the individual terms can be obtained from explicit or implicit solvation models or experiment.21,22 Rearranging and collecting terms together that represent the change in the total energy, enthalpy and entropy, respectively, we obtain:
| (5) |
which, when consolidated, leads us to a compact representation23 of the four key terms we have to evaluate in order to obtain the free energy of binding of a ligand to a protein in aqueous solution:
| (6) |
where the first three of the individual terms are defined as:
| (7) |
Figure 1.
Thermodyamic cycle to estimate the free energy of binding of a drug molecule to a protein receptor. P=Protein, S=small molecule/substrate and PS=protein-small molecule/substrate complex.
For our purposes a key result of this simple analysis is that the master equation (equation 6) yields a ΔE term, which indicates the change in the electronic energy of a molecule in the gas-phase. This term, along with the second term in equation 6 (the ΔH term), are quantities for which modern electronic structure theory can obtain highly reliable values for using appropriate methodologies.2,19 Given that our analysis yields terms involving the change in electronic energy and the vibrational enthalpy correction term upon binding in the gas-phase we can ask ourselves how can we make use of this in an error analysis? Herein we make the assumption that the change in electronic energy and the vibrational enthalpy correction term can be estimated as a linear combination of the individual interactions. We define "individual interaction" not in a pairwise or atom-by-atom sense, but in a chemical sense. Hence, two carbon atoms interacting does not satisfy our definition, but an hydroxyl hydrogen bonding to a carbonyl or a valine side chain forming a van der Waals complex with an phenyl ring from an inhibitor does fit our definition. Furthermore, we can envision these interactions as not being isolated in the gas-phase, but as being in the context of the protein-ligand environment in a combined quantum mechanical/molecular mechanical (QM/MM) sense.17 This is an approximation, but as noted by Zhou and Gilson24 it has some justification, while doing the same for the entropy term (ΔS) is not due to standard state concentration and translational entropy considerations. Experimentally, this is borne out by recent work of Klebe and co-workers25, that show for a series of thrombin inhibitors the overall free energy and entropy terms show significant cooperative effects, but the enthalpy term alone, within the experimental error bars, show little or no cooperativity. Thus we can approximate interaction energy and enthalpy terms as:
| (8) |
The score function given in equation 6 has a total of four terms in the expression and we have discussed two of the four (ΔE and ΔH). For the TΔS terms we cannot decompose them24, which presently leaves us at a loss regarding how to best estimate the expected overall error for this term. Intuitively, we expect the error to be larger given the need for extensive sampling and the accurate computation of translational, rotational and vibrational components of the total entropy, but ultimately quantitatively expressing this error contribution will be essential.20,24 Hence, at this juncture estimating the expected uncertainty in the entropy component would involve significant guesswork and will be left for future analysis.
The approximation of equation 8 assumes that each interaction pair, while "seeing" all other interactions (in a QM/MM sense), behaves independently from the other pairs in terms of their overall interaction energy, vibrational enthalpy correction and their associated errors. For two "interaction pairs" that are far away from each other this is not an unreasonable approximation, but for two interaction pairs that are closer to one another one might imagine this to be more of a significant approximation especially if QM effects like polarization and charge transfer play a role. For each of these intermolecular interactions we can compare various computational approaches for obtaining interaction energies or enthalpies with experiment or with "chemically" accurate quantum mechanical methods in order to obtain an error estimate for each interaction. Hence, the hypothesis here is that through the careful analysis of these interactions we can identify errors in computationally less expensive methods and improve them to derive better simple models that can be used in more extensive drug design applications. Assuming that we are dealing with random errors we can propagate the errors using the following expression:
| (9) |
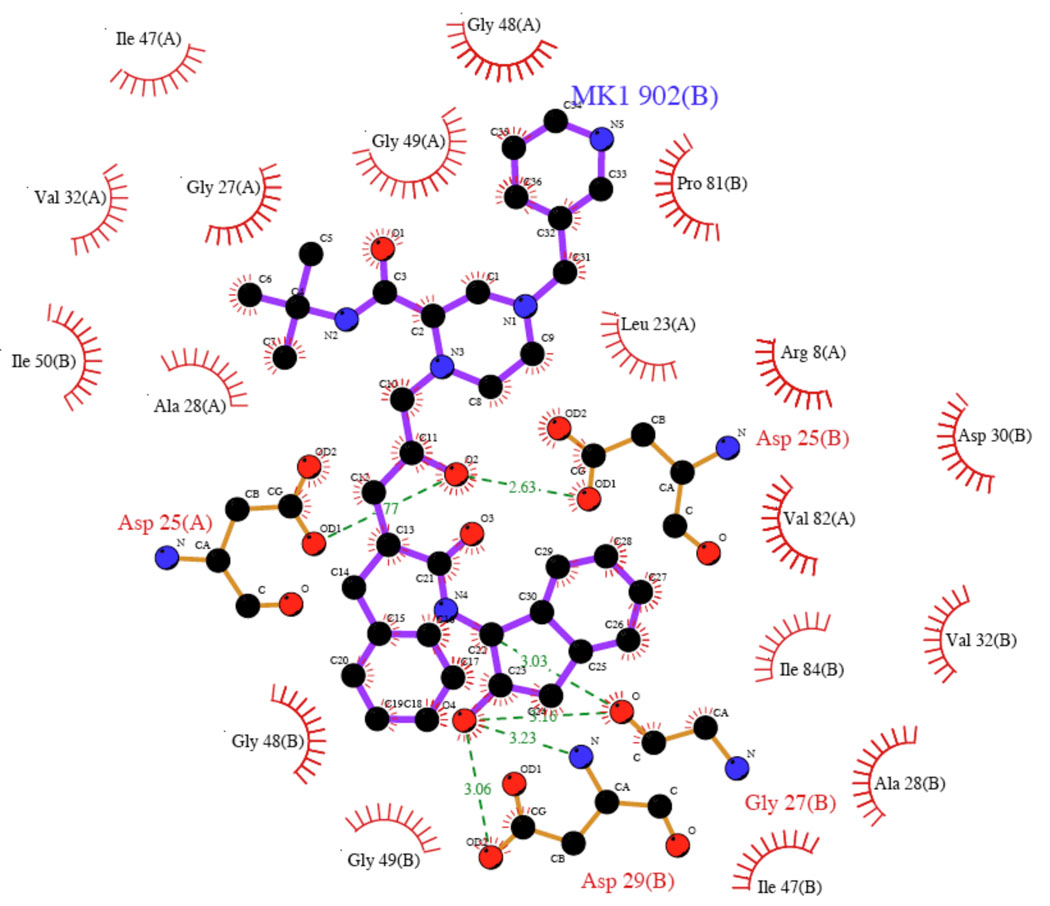
While it is attractive to think about doing this computationally26 or experimentally, the fact is this is difficult to do, so initially it is better to examine the boundary conditions of our approximations and the resultant ramifications using a gedanken experiment. Let's pick a concrete example of Indinavir (crixivan, Ki = 0.358nM or −12.8kcal/mol binding free energy)27,28 bound to the HIV-1 protease (PDBID: 1HSG).29 The LigPlot30 diagram is given in Figure 2 highlighting both hydrophobic and hydrophilic contacts. From the LigPlot analysis and a graphical one, 18 hydrophobic contacts were identified (Gly 48 and Gly 49 on chains A and B are involved in carbonyl---H-C interactions31) and 6 hydrogen bonds along with 5 hydrogen bonds between the ligand and crystallographic waters for a total of 29 individual protein-ligand contacts in our "pharamcophore" that we imagine, for the sake of our gedanken argument, that we have obtained experimental values for each individual interaction energy (29 total) in the context of the ligand. It may not be possible to do this experimentally, but again we are constructing a gendaken experiment to help us understand the boundary conditions of our error hypothesis. Further let's imagine that we will test the performance of two computational methods (these could be ab initio, semiempirical or force field based calculations) for their ability to compute interaction energies and that they are found to have error bars relative to our experimental values for each enthalpy of interaction of ±1kcal/mol and ±0.5kcal/mol, respectively. By each enthalpy of interaction we mean chemically distinct pairs of molecules that in part represent each of the 29 interactions identified in this protein-ligand complex. For example, the side chain of Val33 places a methyl group into the face of an aromatic ring of the inhibitor (in Figure 2 the ring adjacent to the MK1 label). This interaction along with the remaining 28 we assume to each have individual errors of ±1kcal/mol or ±0.5kcal/mol with respect to experiment to give us a range of individual errors to evaluate when we do our error propagation. The use of this error range per interaction was chosen because this can be achieved, with great effort, using modern converged QM calculations and, hence, represents error bars that could be realized today.2 Error varies along the reaction coordinate for the formation of each individual interaction26, so by error we specifically mean the deviation from an established value at the minimum.
Figure 2.
The LigPlot diagram of Indinavir bound to HIV-1 protease.
In the two cases outlined above the total error between experiment and the two model Hamiltonians is ±5.4kcal/mol for the case where we have 29 interaction each with an error of ±1 kcal/mol and ±2.7kcal/mol for the case where we have 29 interaction each with an error of ±1 kcal/mol both propagating the error as given by equation 9. An individual error of ±1 kcal/mol to ±0.5kcal/mol is quite good, but imagine if one of the twenty-nine interactions is off by ±5kcal/mol with respect to experiment, while the remaining 28 stay at ±1 and ±0.5kcal/mol, respectively. This leads to predicted errors of ±7.3kcal/mol and ±5.6kcal/mol for the two cases. In these cases the one badly modeled interaction makes the single largest contribution to the error suggesting that by simply improving this one bad interaction we can significantly improve the approximate model. This points out one of the advantages of this approach in that we can focus on interaction classes that are poorly modeled and expend our parameterization efforts on these problem areas first in order to realize the largest improvement in simpler models.
The next term that we need to consider is the change in the solvation free energy, ΔΔGsol. From equation 4 we see that the solvation term consists of terms involving the solvation free energy of the ligand, the protein and the protein-ligand complex. First we will consider the expected error for the solvation free energy of the ligand and then explore how to estimate this for the protein calculations. For small drug like fragments the SAMPL prospective32–34 validation has demonstrated that the deviation from experiment is of the order of ±1.8–±2.5kcal/mol. For the sake of our analysis we propose that an expected error of ±2.0kcal/mol for the computed value for the solvation free energy of Indinavir is appropriate.
The protein and protein-ligand cases are trickier to evaluate, but in the present case we only need to know how many side chains are exposed in the protein and the protein-ligand complex. This has to do with the way in which these methods work. They are surface or reaction field based algorithms21,22 and the expectation is that the surface residues will have the greatest impact on the solvation free energy while the buried residues contribute significantly less. In the free protein we estimate that there are 100 exposed side chains, which is reduced to 88 upon complexation of Indinavir. However, the only part of the protein that matters is the part that becomes buried (or in other words is experiencing a change) upon ligand complexation in an error analysis because the remaining exposed residues contribute a constant error that is cancelled out in the final expression for ΔΔGsol. Thus, in order to better estimate the error in the solvation free energy calculations for the protein and protein-ligand complex we adopt the approach where only the associated buried (exposed) part of the protein is considered in the complexation process. In the case of 1HSG, we estimate that 12 active site residues are buried (or exposed) as the result of the complexation (disassociation) process.
How can we estimate the expected error in our solvation free energy computations? We propose that we can adopt an approach similar to what we did for the interaction energy component of the score function in that we view the solvation process and being broken down into individual solvation free energy calculations for each amino acid side chain.
| (10) |
Hence, the problem becomes a case of estimating the expected error for the solvation free energy of each individual side chain that becomes buried or exposed. A broad range of solvation models have been proposed21,22 each with its own average error from experiment, but the very best models reach errors on the order of ±1.0 kcal/mol or less for small neutral species and above this for charged molecules. Given that we have a mixture of charged and neutral species in the present case we will adopt ±1kcal/mol as a good estimate for the individual errors given in equation 10. We note that we are using a different error for this situation when compared to the error in the solvation free energy of the ligand molecule (estimated as ±2.0kcal/mol above). The reason for this is that we view the side chains to be simple organic molecules like benzene (Phe), phenol (Tyr), propane (Val), etc. for which typical solvation models21,22 give smaller errors than for larger drug like molecules analyzed, for example, in the SAMPL prospective study.33 Larger or smaller errors can be adopted and the reader can do the simple mathematics to see how this affects the outcome of the error estimation. For the protein-ligand case we assume the error is ~±0.0kcal/mol because as noted above only the residues that are buried or exposed contribute to the overall error estimation. This is not to say that the surface residues that are solvent exposed in both the free and complexed protein state have no error it simply acknowledges that because these exposed residues appear in both of these states that their errors largely cancel. For the free protein we estimate the error to be ~±3.5kcal/mol, which is the square root of 12 (12 residues being buried/exposed each with a solvation free energy error of ±1 kcal/mol). Finally, the ligand is proposed to have a solvation free energy error of ±2.0kcal/mol as outlined above. To estimate the total error for the ΔΔGsol term in equation 6 we take the three components (protein-ligand = ~±0.0kcal/mol, protein ~±3.5kcal/mol and ligand ±2.0kcal/mol) square them and take the square root to yield a total error estimate of ±4.0kcal/mol.
Putting all of this together we can now estimate a lower bound for the expected error range using equation 6 to compute binding free energies. It is a lower limit because we have not attempted to estimate the expected errors in the entropy component of equation 6. The estimated error for the electronic energy and enthalpic term was ±5.4kcal/mol (assuming ±1kcal/mol error for each of the 29 individual the computed interaction energies) and ±2.7kcal/mol (assuming a ±0.5kcal/mol uncertainty), while for the solvation term we arrived at a value of ±4.0 kcal/mol. Propagating these two terms again as the square root of the sum of the squares yields values of ±6.7kcal/mol and ±4.8kcal/mol as the expected uncertainty computed given the parameters we have chosen. For Indinavir the experimental binding free energy is −12.8 kcal/mol27,28 so our estimated errors represent up to one half of the binding free energy of this molecule. Put another way, if we use a computational scoring function that has the error parameters described above and it predicts that the binding free energy of Indinavir is 1nM or −12.3kcal/mol we would have an uncertainty that would suggest that our predicted binding affinity actually ranges from better than picomolar to sub-micromolar, which is a wide binding affinity range. In the sub-micromolar case you would decide to not to further study the molecule in question, while in the better than picomolar instance the compound would almost certainly be further examined.
Discussion
From this analysis we estimate that the error in our model scoring function with its associated uncertainties would yield not particularly satisfactory results for an absolute binding free energy determination. The choice of Indinavir is a real challenge since this molecule is quite large and has many protein-ligand contacts one has to consider, so for simpler molecules or cases where fewer contacts are present one would expect the error to decrease. Thus, one important conclusion from this analysis is that as the molecular size increases or the number of contacts increases we would expect a quasi-linear increase in the expected error. In retrospect, this is not that unexpected of a conclusion.
Does our conclusion bear up against the current results available in the literature? In a detailed recent analysis of docking success Kolb and Irwin concluded that: "When they work do docking screens really discover ligands for the right reasons? For simple models systems with very small ligands, docking appears to work amazingly well."7 They go on to discuss that for large drug like molecules success has been tough to come by. From our analysis we would conclude that the uncertainty in studies of small molecules interacting with a receptor would be significantly less than that for a molecule like Indinavir. Thus, an important conclusion of Kolb and Irwin's work and the present analysis is that working with small ligand scaffolds or fragments as done in fragment drug design35 is a more prudent approach than doing in silico screens through large numbers of higher molecular weight drug-like molecules. Success with smaller ligands is also seen in the extensive and careful work of Gilson and co-workers on host guest systems, where they have been able to achieve errors in the 1–2kcal/mol range for the estimation of binding free energies.15,36,37 Shoichet, Dill and co-workers have also had good success with free energy perturbation methodologies when prospectively applied to small aromatic organic molecules binding to an engineered binding site in T4 Lysozyme38,39 where they realized errors of ~2kcal/mol. The most complex system reported on to date are the retrospective FKBP studies of Roux and co-workers40 and Pande and Shirts and co-workers.41 42 In these studies errors for the prediction of the binding free energy for a series of 8 FKBP inhibitors was between 1.4–2.5kcal/mol depending on the choice of protocols utilized. This level of agreement is outstanding, especially for the larger molecules studied in these efforts. In a final prospective study carried out by Roux and co-workers13 on 50 compounds to JNK kinase the agreement was far less satisfactory, but Roux and his team noted that longer simulations might be needed to obtain converged results.
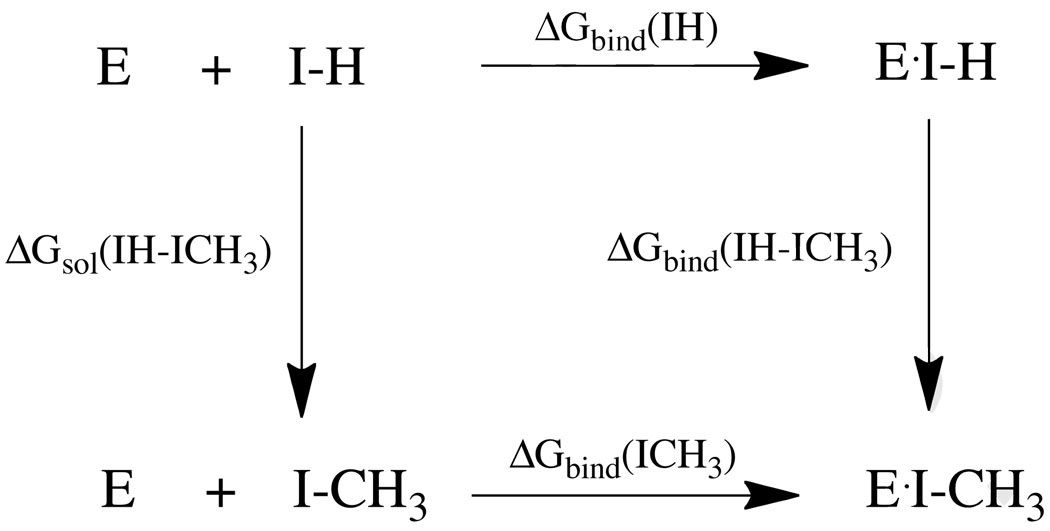
Overall, our hypothesis is borne out by the available literature, with the possible exception of the FKBP results. However, clearly more work of this sort is needed to sort out the issues raised herein. An interesting question arises with regards to thermodynamic cycle-free energy perturbation (TC-FEP) methods.43–46 These have been very successful in examining relative free energies for many series of compounds, which appears to fly in the face of our error hypothesis. However, upon careful inspection it is clear that relative methods like TC-FEP have distinct advantages that reduce the expected error. Taking the TC shown in Scheme 1 we arrive at the following well-known relationship:
| (11) |
This relates that the relative binding free energy can be computed via two alchemical transformations rather than by computing two absolute free energies (ΔGbind(IH) and ΔGbind(ICH3)) and taking their difference. For the first term on the right hand of equation 11 (ΔGsol(IH-ICH3)) we simulate the conversion of the free ligand in aqueous solution by converting a hydrogen (H) into a methyl (CH3) group. This involves the alteration of only one interaction site exposed to solvent and as a result the error in this computation is likely to be quite small, but for the sake of argument we will assume it is ±1kcal/mol. For the conversion of a hydrogen atom into a methyl group within the protein (ΔGbind(IH-ICH3)) this involves the conversion of only one interaction type, while all remaining ones remain the same. Hence, we can hypothesize that we can realize an error for this conversion of ±1 to ±0.5kcal/mol from experiment based on previous experience using this method.43–47 Thus, the fully propagated errors (assuming random errors) is ±1.12kcal/mol to ±1.4kcal/mol. The expected error for the absolute free energy of binding computations are expected to be much larger (as outlined above), but through the clever use of a TC we cancel out many of the errors giving results that can be in excellent agreement with experiment. In a related approach where a ligand "core" is fixed and R groups are systematically added is another way in which error can be reduced because in this model changes in the R group are incurring errors while the fixed "core" represents a constant error that can be ignored when looking at the relative efficacy of ligands in this so-called congeneric series. Clearly, developing technologies or approaches based off of relative energy computations can afford significant error reductions assuming that major conformational changes are not realized as a result of the perturbation. If the latter occurs, more interactions come into play increasing the expected uncertainty.
Scheme 1.
Thermodynamic Cycle
Sampling is one of the two major issues facing computational biology along with accurate energy computation. For complex systems that undergo many conformational changes thorough sampling is an absolute necessity along with accurate energy computations. In many cases by carrying out a molecular dynamics (MD) simulation or via exhaustive sampling of a protein-ligand complex better estimates of the binding free energy can be obtained even though only a local sampling in the binding pocket is realized, while no major conformational changes are observed.23,46,48 Furthermore, carrying out consensus scoring studies across a range of scoring functions can achieve a similar result.49,50 Using our error analysis it is clear these effects have their roots in the fact that we are dealing with random errors in typical score functions as also noted by Wang and Wang51 in a purely statistical docking/scoring experiment. If we take one docking pose the positioning may optimize or minimize the expected uncertainty, but both estimates are misleading and by sampling over many local conformations we can obtain a Gaussian distribution in the error function and thereby minimize the uncertainty.16,51 Hence, by MD or exhaustive sampling we are achieving the same outcome that replicate analytical measurements do to calibrate an instrument subject to random errors. Based on this idea it would be always prudent to consensus score over a range of local "poses" (generated by MD simulations, for example) after docking a given protein-ligand complex in order to obtain the best estimate for the computed binding energy for a given score function.50 If large-scale conformational changes are important then this would not be particularly beneficial, but sampling a ±1Å root-mean square deviation around the initial pose could afford some benefit. This is what consensus scoring12,48–51 and MM-PBSA like methods18,23 have achieved with positive benefits
Conclusions
Through simple error propagation analysis we show that as the size of a molecule increases the expected error in free energy computation for protein-ligand complexation would increase quasi-linearly to a point that the error bar is a large fraction of the expected binding free energy. This conclusion is largely supported by the available literature, which shows that, for small molecules, docking and scoring or absolute free energy computation does yield excellent results, but that as the system size increases the evidence for success in the use of these methods is sparse. Usually, sampling effects are put forth as the major reason for this behavior and the present analysis absolutely does not eliminate this is a serious issue faced when using these techniques. It is clearly important to sample extensively, but the modeling of complex phenomenon also requires that the energy be computed with extreme accuracy as shown via our simple error analysis. Moreover, we suggest that sampling local structure around an initial pose would have benefits like those seen in consensus scoring.49
Through the use of the principles outlined herein, one can formulate an error estimate for the free energy of binding of any given protein-ligand docking pose. We term this the best-case scenario error (BCSerror). By simply counting up the number of interactions being made as a result of complexation and through an estimate of the burial of residues within an active site, error estimates for three of the four terms given in equation 6 can be produced, which ultimately, via further error propagation yields an estimate of the error in the predicted free energy of binding. In the preceding we gave error estimates that we think represents the "best case scenario", but one can use any set of values one prefers based on personal biases. The utility of this comes when it comes time for data reduction from a large scale docking effort. Many hits are reported and via estimates of the expected error for each case in a hit list better "educated" decisions can be made.
The result of the present analysis provides an interesting hypothesis that, in principle, should allow us to better understand how to make the computation of absolute binding free energy more robust from an energetic perspective. The error propagation of the enthalpy of binding and the solvation free energy provide a framework from which we can address the errors expected using advanced quantum chemical techniques to force fields. This approach can be applied to related problems that involve the formation of multiple interactions like protein-folding and the protein-ligand problem highlighted herein. However, due to the complexity (dependence on standard state concentration) of the entropic part the use of the present approach does not afford a clear way in which we can get a better grasp of the errors expected in the computation of entropy. This is a subject for on-going analysis.
Acknowledgments
KMM thanks the NIH (GM044974 and GM066689) for funding the present research. The present work is the result of numerous discussions with a wide range of students and colleagues. Helpful conversations with Dr. Xiao He, Mr. John Faver, Professor Raphael Haftka, Professor Adrian Roitberg, Professor Erik Deumens, Professor Barry Honig, Dr. Marti Head and Dr. Greg Warren are gratefully acknowledged.
Select Literature
- 1.Dirac PAM. Proc. R. Soc. London Series a. 1929;123:714–733. [Google Scholar]
- 2.Helgaker T, Klopper W, Tew DP. Mol. Phys. 2008;106:2107–2143. [Google Scholar]
- 3.Bartlett RJ, Musial M. Reviews of Modern Physics. 2007;79:291–352. [Google Scholar]
- 4.Brooijmans N, Kuntz ID. Ann. Rev. Biophys. Biomolec. Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 5.Leach AR, Shoichet BK, Peishoff CE. J. Med. Chem. 2006;49:5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 6.Morra G, Genoni A, Neves MAC, Merz KM, Colombo G. Curr. Med. Chem. 2010;17:25–41. doi: 10.2174/092986710789957797. [DOI] [PubMed] [Google Scholar]
- 7.Kolb P, Irwin JJ. Curr. Top. in Med. Chem. 2009;9:755–770. doi: 10.2174/156802609789207091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guvench O, MacKerell AD. Curr. Opin. Struct. Biol. 2009;19:56–61. doi: 10.1016/j.sbi.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. J. Med. Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 10.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. J. Comput.-Aided Mol. Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 11.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 12.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 13.Deng YQ, Roux B. J. Phys. Chem. B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirado-Rives J, Jorgensen WL. J. Med. Chem. 2006;49:5880–5884. doi: 10.1021/jm060763i. [DOI] [PubMed] [Google Scholar]
- 15.Gilson MK, Zhou HX. Annu. Rev. Biophys. Biomol. Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 16.Taylor JR. An Introduction to Error Analysis. The Study of Uncertainties in Physical Measurements. Sausalito, CA: University Science Books; 1997. [Google Scholar]
- 17.Cramer CJ. Essentials of Computational Chemistry. New York, New York: John Wiley & Sons; 2002. [Google Scholar]
- 18.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 19.Hehre WJ, Radom L, Schleyer PvR, Pople JA. Ab Initio Molecular Orbital Theory. New York: John Wiley & Sons; 1986. [Google Scholar]
- 20.McQuarrie DA. Statistical Thermodynamics. New York, New York: Haroer & Row; 1973. [Google Scholar]
- 21.Cramer CJ, Truhlar DG. Chem. Rev. 1999;99:2161–2200. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 22.Tomasi J, Mennucci B, Cammi R. Chem. Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn B, Kollman PA. J. Med. Chem. 2000;43:3786–3791. doi: 10.1021/jm000241h. [DOI] [PubMed] [Google Scholar]
- 24.Zhou HX, Gilson MK. Chem. Rev. 2009;109:4092–4107. doi: 10.1021/cr800551w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baum B, Muley L, Smolinski M, Heine A, Hangauer D, Klebe G. J. Mol. Biol. 2010;397:1042–1054. doi: 10.1016/j.jmb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Molnar LF, He X, Wang B, Merz KM. J. Chem. Phys. 2009;131 doi: 10.1063/1.3173809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorsey BD, Levin RB, McDaniel SL, Vacca JP, Guare JP, Darke PL, Zugay JA, Emini EA, Schleif WA, Quintero JC, et al. J. Med. Chem. 1994;37:3443–3451. doi: 10.1021/jm00047a001. [DOI] [PubMed] [Google Scholar]
- 28.Vacca JP, Dorsey BD, Schleif WA, Levin RB, McDaniel SL, Darke PL, Zugay J, Quintero JC, Blahy OM, Roth E, et al. Proc. Natl. Acad. Sci. U S A. 1994;91:4096–4100. doi: 10.1073/pnas.91.9.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Li Y, Chen E, Hall DL, Darke PL, Culberson C, Shafer JA, Kuo LC. J. Biol. Chem. 1994;269:26344–26348. [PubMed] [Google Scholar]
- 30.Wallace AC, Laskowski RA, Thornton JM. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 31.Vargas R, Garza J, Dixon DA, Hay BP. J. Am. Chem. Soc. 2000;122:4750–4755. [Google Scholar]
- 32.Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS. J. Med. Chem. 2008;51:769–779. doi: 10.1021/jm070549+. [DOI] [PubMed] [Google Scholar]
- 33.Guthrie JP. J Phys Chem B. 2009;113:4501–4507. doi: 10.1021/jp806724u. [DOI] [PubMed] [Google Scholar]
- 34.Marenich AV, Cramer CJ, Truhlar DG. J. Phys. Chem. B. 2009;113:4538–4543. doi: 10.1021/jp809094y. [DOI] [PubMed] [Google Scholar]
- 35.Congreve M, Chessari G, Tisi D, Woodhead AJ. J Med Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 36.Moghaddam S, Inoue Y, Gilson MK. J. Am. Chem. Soc. 2009;131:4012–4021. doi: 10.1021/ja808175m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W, Chang CE, Gilson MK. Biophys. J. 2004;87:3035–3049. doi: 10.1529/biophysj.104.049494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyce SE, Mobley DL, Rocklin GJ, Graves AP, Dill KA, Shoichet BK. J. Mol. Biol. 2009;394:747–763. doi: 10.1016/j.jmb.2009.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA. J. Mol. Biol. 2007;371:1118–1134. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Deng Y, Roux B. Biophys. J. 2006;91:2798–2814. doi: 10.1529/biophysj.106.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jayachandran G, Shirts MR, Park S, Pande VS. J. Chem. Phys. 2006;125 doi: 10.1063/1.2221680. [DOI] [PubMed] [Google Scholar]
- 42.Fujitani H, Tanida Y, Ito M, Jayachandran G, Snow CD, Shirts MR, Sorin EJ, Pande VS. J. Chem. Phys. 2005;123:084108. doi: 10.1063/1.1999637. [DOI] [PubMed] [Google Scholar]
- 43.Kollman P. Chem. Rev. 1993;93:2395–2417. [Google Scholar]
- 44.Knight JL, Brooks CL. J. Comput. Chem. 2009;30:1692–1700. doi: 10.1002/jcc.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Phys. Chem. Chem. Phys. 2006;8:5166–5177. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- 46.Foloppe N, Hubbard R. Curr. Med. Chem. 2006;13:3583–3608. doi: 10.2174/092986706779026165. [DOI] [PubMed] [Google Scholar]
- 47.Jorgensen WL. Acc. Chem. Res. 2009;42:724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R, Lu Y, Wang S. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 49.Charifson PS, Corkery JJ, Murcko MA, Walters WP. J. Med. Chem. 1999;42:5100–5109. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- 50.Oda A, Tsuchida K, Takakura T, Yamaotsu N, Hirono S. J. Chem. Inf. Model. 2006;46:380–391. doi: 10.1021/ci050283k. [DOI] [PubMed] [Google Scholar]
- 51.Wang R, Wang S. J Chem Inf Comput Sci. 2001;41:1422–1426. doi: 10.1021/ci010025x. [DOI] [PubMed] [Google Scholar]