Abstract
Manganese peroxidase (MnP) is an extracellular heme enzyme produced by the lignin-degrading white-rot fungus Phanerochaete chrysosporium. MnP catalyzes the peroxide-dependent oxidation of MnII to MnIII. The MnIII is released from the enzyme in complex with oxalate, enabling the oxalate-MnIII complex to serve as a diffusible redox mediator capable of oxidizing lignin, especially under the mediation of unsaturated fatty acids. One heme propionate and the side chains of Glu35, Glu39 and Asp179 have been identified as MnII ligands in our previous crystal structures of native MnP. In our current work, new 0.93 Å and 1.05 Å crystal structures of MnP with and without bound MnII, respectively, have been solved. This represents only the fourth structure of a protein of this size at 0.93 Å resolution. In addition, this is the first structure of a heme peroxidase from a eukaryotic organism at sub-Ångstrom resolution. These new structures reveal an ordering/disordering of the C-terminal loop, which is likely required for Mn binding and release. In addition, the catalytic Arg42 residue at the active site, normally thought to function only in the peroxide activation process, also undergoes ordering/disordering that is coupled to a transient H-bond with the Mn ligand, Glu39. Finally, these high-resolution structures also reveal the exact H atoms in several parts of the structure that are relevant to the catalytic mechanism.
Keywords: Manganese, peroxidase, crystallography, atomic resolution, refinement
INTRODUCTION
White-rot basidiomycetous fungi are the only organisms capable of degrading the phenylpropanoid, plant cell wall polymer, lignin [1-4]. The lignin-degrading system of these fungi also can oxidize a variety of economically and environmentally important aromatic pollutants [5-9] and will probably play an important role in processes for the conversion of lignocellulosics materials to ethanol. Under ligninolytic conditions, the best-studied lignin-degrading fungus, Phanerochaete chrysosporium, secretes two families of extracellular heme peroxidases, lignin peroxidase (LiP) and manganese peroxidase (MnP) [2-4, 10] and a hydrogen peroxide generating system [4, 6, 11, 12].
MnP from P. chrysosporium has been studied by a variety of biochemical and biophysical methods [2, 13-16]. The crystal structure of MnP illustrates that the heme environment of this enzyme is similar to that of other plant and fungal peroxidases [17, 18]. However, MnP is the only heme peroxidase capable of the one-electron oxidation of MnII in a typical peroxidase reaction cycle:
| (1) |
| (2) |
| (3) |
where MnPI and MnPII are the oxidized intermediates MnP compounds I and II, respectively.
Our earlier crystal structures of MnP show that the substrate, MnII, binds to one heme propionate and the side chains of three amino acids, Glu35, Glu39, and Asp179, as well as two solvent ligands [17, 18]. This site was proven by kinetic and biophysical studies of wild-type MnP and of proteins containing point mutations in the putative binding site. Alteration of the proposed amino acid ligands in the Mn-binding site significantly affects Mn-binding and oxidation [19-24] and crystals of both the single variant, D179N, and the double variant, E35Q-D179N, lack electron density at the proposed Mn-binding site [25] suggesting that MnII is not bound. Furthermore, competitive inhibitors such as CdII, bind at the identical site, although with alternative geometry [18, 24].
MnP is unique among enzymes, using manganese as a redox cofactor. Rather than permanently sequestering MnII in an interior binding site, MnP selectively binds MnII near the surface of the protein, oxidizes it and then releases the MnIII product in complex with organic acids such as oxalate. The relatively stable MnIII-oxalate complex acts as a diffusible mediator to oxidize the terminal substrate lignin, usually in the presence of a radical mediator such as an unsaturated fatty acid [26, 27]. Whether a MnII-chelator complex binds to the enzyme to form a ternary complex or the chelator simply facilitates release of MnIII via ligand displacement has now been resolved. Both NMR and crystal structure studies of the MnP-MnII complex in the absence of chelators indicate that the later alternative occurs [16, 18, 28-30].
Crystals of MnP are very stable and are good candidates for high resolution structure determination [18]. Although a good deal is known about the catalytic mechanism of MnP and of other plant and fungal peroxidases, atomic level structures could help reveal details of heme geometry and H-bonding critical to the catalytic cycle of this enzyme. Atomic-detail structures can also show the exact stereochemical properties of various prosthetic groups bound to proteins. Protein crystallographers normally rely on stereochemical parameters from crystal structures of small molecules to constrain prosthetic groups during refinement and hence, the final results are prejudiced toward the small molecule model. This is an important problem since it is a common axiom in structural biology that the protein modulates the reactivity of the prosthetic group by altering both electronic and geometric properties. Heme proteins provide a classic example of how the same heme, iron protoporphyrin IX, is used in a vast majority of heme proteins including the globins, cytochromes, and many heme enzymes, yet these heme proteins exhibit widely different functions. A detailed comparison of known heme protein structures illustrates that the heme group deviates significantly from planarity and that heme proteins, which are functionally related, exhibit similar types of non-planarity [31]. Thus, the ability to detail the geometry of such prosthetic groups is another benefit of ultrahigh resolution protein structures.
The use of a new generation of synchrotron radiation sources coupled with cryogenic techniques has enabled the solution of an increasing number of protein structures at true atomic resolution. This has provided an unprecedented level of detail in analyzing functionally important features of protein structures, including the identification of individual H atoms. The precise location of H atoms is important in understanding enzyme reactions, where H-bonds and acid-base catalysis are often utilized. Recent examples include the serine protease structures from Bacillus lentus [32] and Titiachium album limber [33] solved at 0.78Å and 0.98Å, respectively. For Bacillus lentus subtilisin an H atom was found between the essential catalytic His and Asp which forms part of the well-documented serine protease Ser-His-Asp catalytic triad. In addition, the His-Asp H-bond distance was found to be relatively short, 2.62Å [32]. Such results have important implications for the details of enzyme catalyzed reactions and, in the subtilisin example, suggest the role that “low-barrier” H-bonds [34] play in the reaction.
In this study, the structure of MnP containing 357 amino acid residues was refined at 0.93 Å resolution. Of the total of about 37,000 protein structures determined by x-ray crystallography and deposited in the PDB, only five unique structures (a bacterial catalase, 1GWE [35]; PfluDING, a DING protein from Pseudomonas fluorescens, 3G63 [36]; cholesterol oxidase from Streptomyces sp., 1N4W [37]; xylose isomerase, 1MUW [38]; and pentaerythritol tetranitrate reducatase, 1VYR [39] are comparable to MnP in both chain length (> 350 residues) and resolution (≥ 0.93 Å). Thus MnP is the first eukaryotic heme peroxidase to be analyzed at sub-Angstrom resolution.
MATERIAL AND METHODS
Protein Purification and Crystallization
Wild-type MnP was purified from shaking cultures of Phanerochaete chrysosporium grown on high carbon, low nitrogen medium, as previously described [13, 14, 24]. MnII free MnP was prepared using a metal Chelax-100 column as described [24]. Crystals of MnII bound MnP (Mn-MnP) were grown at room temperature using the hanging drop vapor diffusion method with an excess of 5 mM MnCl2 as described [18, 40]. The reservoir contained 30% (w/v) of polyethylene glycol 8,000, 0.2 M ammonium sulfate, and 0.1 M sodium cacodylate buffer, pH 6.5. The crystallization drops were composed of 5 μl of the protein solution (10-15 mg/ml) mixed with an equal volume of reservoir solution. The crystallization was initiated by a seeding procedure, using serially diluted seed stocks prepared from old native MnP crystals. Crystals of MnII free MnP were grown, using similar reservoir conditions except that the drops contained 4 mM EDTA instead of MnCl2 and the crystallization temperature was 4 °C.
Data Collection and Processing
Crystals were harvested in synthetic mother liquor identical to the reservoir solution and transferred to the cryo-solution containing 10% (v/v) glycerol in the mother liquor. The cryo-soaked crystals were flash-frozen in an N2 cryostream and all data sets were collected at -160 °C.
The MnII free MnP data set was collected at the Stanford Synchrotron Radiation Laboratory (SSRL) beam line 7-1 at 1.08 Å wave length. The diffraction images were recorded in two steps using a single crystal on a MAR imaging plate at different detector settings and exposure times. The detector distance was 80 mm for the MAR300 plate to record high resolution reflections in dose mode with 3600 unit (~45 s) per frame and moved to 120 mm for the MAR180 plate to collect low resolution data with a 20 s exposure per frame. A total of 180 frames of 1° oscillation per frame were collected for each set. The images of each set were processed separately to extract raw intensities using MOSFLM [41] and were scaled together using SCALA to obtain a single data set at 1.05 Å (Table 1).
Table 1.
Data collection statistics
| Dataset | MnP (-Mn) | Mn-MnP |
|---|---|---|
| Resolution range (Å) | ∞ - 1.05 | ∞ - 0.93 |
| No. of observations | 653,391 | 1,246,031 |
| Unique reflections | 150,666 | 264,958 |
| Completeness (%) | 91.2 (79.3) | 97.5 (93.0) |
| Redundancy | 4.1 (2.7) | 4.9 (2.0) |
| <I>/<σ(I)> | 9.0 (3.4) | 26.3 (1.9) |
| Rsym | 0.05 (0.21) | 0.066 (0.512) |
The data set for the MnII bound MnP (Mn-MnP) was collected at SSRL Beam Line 9-1 at 0.78 Å wave length, using a single crystal. High resolution frames were collected using the MAR345 imaging plate at a 110 mm detector distance in dose mode with 6,000 units per frame. A total of 180 frames of 0.75° oscillation angle were collected to record high resolution data. Low resolution frames were collected using the MAR240 at 120 mm distance with 10 s exposure per frame. A total of 180 frames of 1° per frame were collected for each set. The intensities of each set were integrated separately using DENZO [42] and merged using SCALEPACK to obtain a single data set 0.93 Å (Table 1).
Refinement
Coordinates of the native MnP structure previously refined at 1.45 Å resolution (PDB Accession Code: 1YYD) were used as the starting model for the refinement with the MnII free MnP data set which was collected first. The reflections used for Rfree calculation in the 1.45 Å data set were flagged in the present data set and extended to full resolution at 1.05 Å using XPLOR [43]. However, in order to keep the unused reflections to a minimum, only 2% of the reflections were flagged for the test set (Table 2). The first round of refinement was carried out using data in the 8.0-1.45 Å resolution range by conjugate gradient least squares (CGLS) protocol in SHELXL [44]. The model was examined from amino- to carboxy-terminus guided by σA-weighted 2Fo-Fc and Fo-Fc maps. The electron density for the previously disordered C-terminal loop was more ordered for this data set and residues 342-352 were rebuilt. The resolution was extended gradually in a few rounds and the automatic water picking option (PLAN 50 2.4) was used to add new waters to the model. The refined model and new water positions were examined and manually corrected at each stage. The iterative cycles of manual model adjustment and refinement coupled with resolution extension was continued until the highest resolution of 1.05 Å was reached. Water molecules refined with a B-factor > 50 Å2 (with occupancy set to unity) were removed from the refinement throughout this exercise. At the end of 10 rounds of refinement the Rcryst (Fo > 4 σ) was 0.159 and the corresponding Rfree was 0.177. At this stage, anisotropic B-factor or atomic displacement parameter (ADP) refinement was turned on using ANIS keyword for all atoms and the model was refined in 20 CGLS cycles. The Rcryst and Rfree dropped to 0.122 and 0.154, respectively. Following this step, a few disordered sides chains were rebuilt in multiple conformations and the model was further refined. When the refinement converged, 20 cycles of CGLS refinement was run with riding hydrogen atoms for the polypeptide, sugar residues, and heme group. Both Rcryst and Rfree dropped to 0.110 and 0.123, respectively. Finally, 10 cycles of CGLS refinement was carried out with all reflections (i.e. working set + test set) to the final R-factor of 0.111.
Table 2.
Refinement statistics
| MnP (-Mn) | Mn-MnP | |
|---|---|---|
| Resolution range (Å) | 8.0 – 1.05 | 8.0 – 0.93 |
| No of atoms | ||
| Amino acid residues | 2,622 | 2,622 |
| Sugar residues | 39 | 39 |
| Heme | 43 | 43 |
| CaII | 2 | 2 |
| MnII | - | 1 |
| Glycerol | 24 | 12 |
| Water | 549 | 477 |
| Rwork [working set, Fo > 4σ(Fo)] | 11.1 (144,730) | 10.7 (191,300) |
| Rwork (working set, all reflections) | 11.7 (156,881) | 12.4 (243,170) |
| Rfree [test set, Fo > 4σ(Fo)] | 13.6 (3,181) | 12.4 (3,967) |
| Rfree (test set, all reflections) | 13. 9 (3,352) | 13.4 (4,736) |
| Rcryst [working + test set, Fo > 4σ(Fo)] | 11.1 (147,911) | 10.8 (195,267) |
| Rcryst (working + test set, all reflections) | 11.6 (160,232) | 12.4 (247,906) |
| No. of Parameters | 30,135 | 29,226 |
| No. of observations/No. of parameters | 5.3 | 8.5 |
| No. of restraints | 36,426 | 35,548 |
| Mean isotropic B-factor (Å2) | ||
| All atoms | 12.973 | 13.670 |
| Main chain atoms | 9.279 | 10.098 |
| Side chain atoms | 11.776 | 13.276 |
| Sugar residues | 17.171 | 16.416 |
| Heme and metal ions | 8.394 | 7.150 |
| Solvent | 24.909 | 25.687 |
| RMS deviation from ideal geometry | ||
| Bond length (1-2) (Å) | 0.017 | 0.018 |
| Angle distance (1-3) (Å) | 0.038 | 0.033 |
| Chiral volume (Å3) | 0.129 | 0.096 |
| Non-zero chiral volume (Å3) | 0.219 | 0.105 |
| Deviation from planes (Å) | 0.029 | 0.028 |
For the 0.93 Å Mn-MnP data set, test reflections were flagged using the 1.05 Å data set as the reference and extended to full resolution. The refined model of the MnII free structure was used as the starting model for the refinement. However, the refinement was started at 1.5 Å with isotropic B-factors. The substrate MnII ion and its amino acid ligands were modeled using σA-weighted 2Fo-Fc and Fo-Fc maps. The resolution was increased to 0.93 Å in five rounds (1.2 Å, 1.05 Å, 1.00 Å, 0.95 Å and 0.93 Å) of CGLS refinement and the STIR command was used to extend the resolution in finer steps in several cycles (20-35) in each round. The maps were examined at the end of each refinement and minor changes were made to the model. New waters were added in each round but those refined with high B-factors (> 50 Å2) were removed in the subsequent refinement. The refinement converged with an Rcryst of 0.167 and an Rfree of 0.179 with isotropic B-factors. Subsequently anisotropic B-factor refinement was turned on and after extensive refinement the Rcryst and Rfree values converged to 0.12 and 0.143, respectively. At this stage, a few disordered side chains were identified and modeled in multiple conformations and refined further. When the refinement converged, 20 cycles of CGLS refinement was run with riding H atoms for the polypeptide, sugar residues and heme group (Rcryst = 0.109 and Rfree = 0.128). However, riding H atoms were not included for the proximal heme ligand, His173, the residue H-bonded to this histidine, Asp242, the distal base catalyst His46, and the residue H-bonded to this histidine, Asn80. Finally, 10 cycles of CGLS refinement was carried out with all reflections (i.e. working set + test set) to the final R-factor of 0.109.
The refinement statistics are shown in Table 2. The metal-ligand distances for the heme FeIII, the substrate MnII, and two structural CaII ions, were unrestrained throughout the refinement. After the anisotropic B-factor refinement began, side chain disorders were examined and several side chains were modeled in multiple conformations with appropriate partial occupancies and total occupancy was restrained to unity using the FVAR instruction. A few other residues were found to be highly disordered but their side chains were not truncated. For the calculation of estimated standard deviations (ESDs) for the atomic positions, one cycle of full-matrix least-squares refinement was performed (BLOC 1).
RESULTS AND DISCUSSION
Quality of the Maps and Models
Crystals of MnP from P. chrysosporium are very robust with low mosaicity ( < 0.2°) and can tolerate prolonged exposure as well as freeze-thaw cycles as observed in our previous study [18]. These well ordered crystals can diffract to 1.0 Å or better at a synchrotron source. The maps calculated using ultrahigh resolution data show high clarity that is typical of such data sets (Figure 1). The previously reported 1.45 Å structure showed an O-glycosylation site at Ser336 with a β-mannose residue and extra density at the N-glycosylation site at Asn131 that could be due to a potential β-mannose residue [18]. These features are observed in the ultrahigh resolution maps as well, but the maps do not show additional sugar residues at these sites nor do they reveal new glycosylation sites. The electron density for most of the bound waters is very clear and all of them were refined with unit occupancy. There remain small pieces of uninterpretable density in several places in the solvent regions.
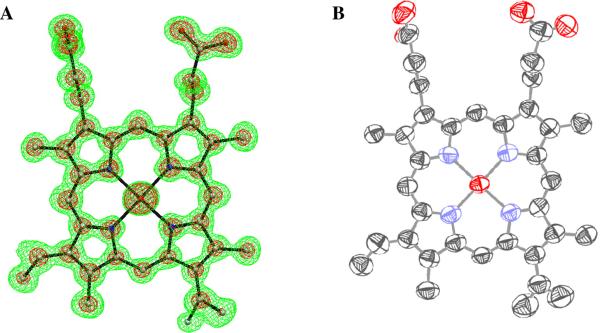
Figure 1.
(A) 2Fo-Fc map density for the refined heme group, calculated using the 0.93 Å native Mn-MnP data set. The contours are drawn at 1.0 σ (green) and 4.0 σ (magenta). (B) Thermal ellipsoid diagram for the heme group drawn using ORTEP. The structure, including anisotropic B-factors, was refined using the SHELXL program [44].
The final model of the substrate-bound MnP (Mn-MnP) consists of 357 amino acid residues, three sugar residues (GlcNac,GlcNac at Asn131 and a single mannose at Ser336) , a heme prosthetic group, two structural calcium ions, a substrate MnII ion, and 478 solvent molecules, including two glycerol molecules. The substrate-free MnP model differs only in lacking the MnII ion in the Mn binding site and in the number of solvent molecules, 549, which includes two glycerol molecules. The two models superimpose with an overall RMS deviation of 0.253 Å for all 357 Cα atoms and of 0.146 Å for all residues minus the C-terminal loop (residues 344-350). There is no significant difference between these structures and the previously reported 1.45 Å cryo structure or the 2.06 Å room temperature structure in overall topology (Figure 2). About 91% of the non-glycine residues are in the most favored regions of the Ramachandran plot and the remaining are in either allowed or generously allowed regions.
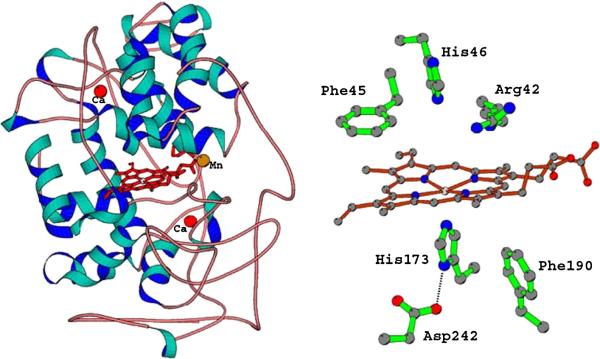
Figure 2.
(A) The overall structure of MnP. The red spheres are structural CaII ions conserved in extracellular heme peroxidases. The location of the substrate, MnII, near the heme, is indicated. (B) The active site structure of MnP. This architecture is highly conserved in heme peroxidase. The main variations are the Phe residues which are Trp in the intercellular peroxidases, cytochrome c and ascorbate peroxidase. The Asp242-His173 pair is conserved.
Disorder
The N-terminal disorder persists for the first 10 residues in the maps of both the 1.05 Å and the 0.93 Å data sets. However, there is an improvement in the quality of density near the C-terminal loop encompassing Gly344-Gly350 in the 1.05 Å map of the substrate-free MnP data set. Interestingly, this region is still disordered in the 0.93 Å map of the Mn-MnP data set (Figure 3). This difference in the quality of electron density between the substrate-free and substrate-bound forms is probably not due to the difference in their resolution. Instead, this dynamic disorder is likely due to the binding of the substrate with implications for the catalytic mechanism of the enzyme. This C-terminal loop traverses near the Mn binding pocket and must be sufficiently flexible to allow binding of the substrate MnII, the entry of dicarboxylic acid chelators such as oxalate to bind the oxidized MnIII product [16, 29], and the release of the MnIII-chelator complex. When we revisited the maps of the previously reported data sets of native MnP (1.45 Å), CdII-MnP complex (1.6 Å), SmIII-MnP complex (1.6 Å) and oxalate soaked SmIII-MnP complex (1.4 Å) similar differences are observed [18]. Whereas all three metal bound structures show disorder, the map of oxalate soaked SmIII-MnP complex, which lacks a metal ion, shows a more ordered C-terminal loop (data not shown).
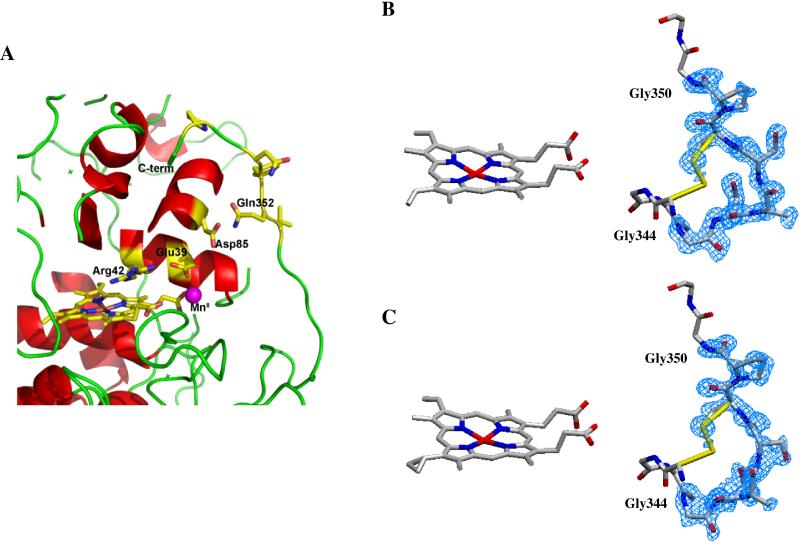
Figure 3.
(A). C-terminal region of the MnP structure. The loop encompassing residues 342-352 traverses close to the MnII binding site. This loop is disordered when MnII is bound in the 0.93 Å structure of native Mn-MnP but is ordered in the substrate free MnP structure at 1.05 Å. 2Fo-Fc electron density for the ordered (B) and disordered (C) C-terminal loop is shown contoured at 1.0 σ.
Besides the disorder at the N- and C-termini, there are small sections of surface loops and the side chains of a few surface residues that are disordered. Some of the disordered side chains could be modeled in two conformations with partial occupancies in each. Notable among them are a few cysteine residues, (Cys3, Cys14 and Cys253) which are found to be partially reduced due to radiation, and the active site residue Arg42. In addition, two of the MnII ligands, Glu35 and Glu39, are disordered and exhibit multiple conformations in the MnII free structure. Other disordered residues show a complete lack of density which could not be modeled with confidence. Nevertheless, the side chains were not truncated and were modeled according to the known sequence information [45, 46]. These residues are characterized by high isotropic B-factors and exhibit unrealistic atomic displacement parameters (ADPs) as analyzed by PARVATI [47].
Mn Binding Site
The manganese binding site in both the substrate free MnP and Mn-MnP structures is shown in Figure 4 and Table 3 provides ligand coordination distances. As described in our earlier study, treating MnP with EDTA did not completely remove the metal from the substrate binding site but did lead to a substantial decrease in electron density [25]. In our current work, however, we were able to crystallize metal free MnP protein and the structure confirms the dynamic nature of the active site described in our previous studies [18, 25]. Two metal ligands, Glu35 and Glu39, move from their original MnII binding conformations and this provides insights into the mechanism of MnP. Once MnP oxidizes MnII to MnIII, the MnIII acts as a diffusible oxidant of lignin and other oxidizable substrates only when complexed with a suitable chelator [16, 29]. The latter is required to stabilize the highly reactive MnIII. The question is how such a complex forms.
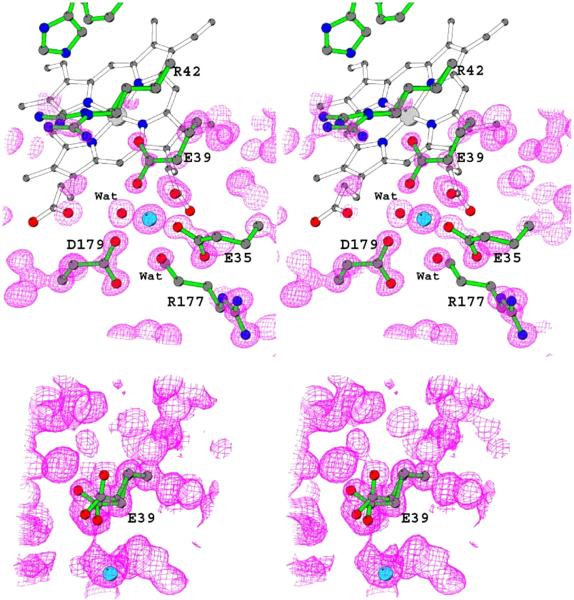
Figure 4.
Stereo views of the electron density maps obtained from the 0.93 Å Mn-MnP structure contoured at 1.6σ (top) and 1.05Å Mn free MnP structure obtained from EDTA-treated crystal contoured at 1.0σbottom). MnII (cyan) and all its ligands including two water molecules are labeled for the MnII-bound structure. Glu39 is in a single conformation in this structure where it is in two conformations in the MnII-free structure, as shown in the figure.
Table 3.
Metal-ligand distances
| Metal – ligand | MnP (-Mn) (Å) | Mn-MnP (Å) |
|---|---|---|
| H2O* | MnII | |
| Heme propionate | 2.25 | 2.12 |
| Glu35 OE1 | 2.36 | 2.14 |
| Glu39 OE1 | -- | 2.27 |
| Asp179 OD1 | -- | 2.28 |
| Water 1 (1040) | 2.56 | 2.25 |
| Water 2 (1108) | 2.64 | 2.26 |
| Proximal CaII | ||
| Carbonyl O-Ser174 | 2.36 | 2.37 |
| Side chain OG1-Ser174 | 2.47 | 2.47 |
| Side chain carboxyl Asp191 | 2.42 | 2.44 |
| Carbonyl O-Thr193 | 2.37 | 2.38 |
| Side chain OG1-Thr193 | 2.50 | 2.49 |
| Carbonyl O-Thr196 | 2.49 | 2.51 |
| Side chain carboxyl Asp198 | 2.46 | 2.48 |
| Distal CaII | ||
| Carbonyl O-Asp47 | 2.44 | 2.44 |
| Side chain carboxyl Asp47 | 2.31 | 2.31 |
| Carbonyl O-Gly62 | 2.44 | 2.46 |
| Side chain carboxyl Asp64 | 2.39 | 2.42 |
| Side chain O-Ser66 | 2.49 | 2.49 |
| Water 1 (1085) | 2.39 | 2.41 |
| Water 2 (1027) | 2.35 | 2.33 |
A water or unidentified monovalent cation occupies the Mn-binding site in the substrate free structure.
In the structures of the single mutant (D179N) and the double mutant (E35Q, D179N) MnPs, which are essentially inactive and, which do not bind MnII, the side chain of Glu39 moves away (“open” conformation) from the metal binding site [25]. Similarly, the metal free structure of oxalate soaked SmIII-MnP complex structure showed Glu39 in the “open” conformation [18]. Another Mn ligand, Glu35, undergoes a less dramatic conformational change in these structures as its movement is restricted by a salt bridge with the side chain of Arg177 [21]. Whereas the mobility of Glu39 is effected by a rotation of the Cα–Cβ bond resulting in a major movement, Glu35 side chain is rotated about the Cβ–Cγ bond resulting in variable, but minor changes in the wild-type and mutant structures depending upon the electronic environment of the site. Other ligands—the heme propionate and Asp179—do not move from their original positions whether a metal ion is bound or not, strongly suggesting that precise geometry is required for efficient MnII-binding and oxidation. This is confirmed by steady-state and transient-state kinetic analyses of a MnP E39D single mutant and an E35D-E39D-D179E triple mutant [23]. The single and triple mutant variants exhibit 20- and 40-fold increase in Km, and 103 and 104 decrease in catalytic efficiency, respectively. Although the overall charge is retained, a decrease in the chain length of one ligand could not be compensated for by an increase in the chain length of another ligand for either MnII binding or oxidation. Our previous study using the non-reducible/oxidizable trivalent cation SmIII showed that it could bind at the Mn binding site of MnP [18]. In addition, the release of SmIII by oxalate from the pregrown SmIII-MnP crystals provides insights into how the Mn binding site can bind both divalent and trivalent cations and how the trivalent cation can be released from the resting enzyme by organic acids [18]. Taken together, our past studies and the present high resolution structures imply that MnII binding is precise and the site is relatively rigid, except for the ability of Glu35 and Glu39 to adopt two conformations—“closed” conformations in the metal bound state and “open” conformations in the metal free state, possibly acting as a “gate”, enabling a small carboxylic acid like oxalate or malonate to remove MnIII from the binding site. Without some flexibility in the Mn ligands, it is difficult to envisage how oxalate could remove MnIII from its coordination shell unless MnIII is first freely released.
Another residue, Arg42, is also disordered, and this has significance for peroxidase function. This arginine residue is conserved in all peroxidases and is implicated in stabilizing the compound I and II intermediates by forming a hydrogen bond with the oxyferryl oxygen [48, 49]. This active site arginine residue appears disordered or in multiple conformations in this high resolution structures of MnP and in cytochrome c peroxidase (CcP) [48] but is in a well defined single conformation in the compound I structure of CcP [48]. Arg42 is disordered in both substrate-bound and -free structures of MnP and is modeled in two conformations—“in” and “out”, in which Arg42 moves closer to and away from the oxyferryl center. However, the main difference occurs in its interaction with Glu39. Besides providing a ligand to MnII, the carboxylate group of Glu39 also forms a salt bridge with the guanidium group of Arg42 in the “out” conformation in the substrate-bound structure. This is similar to the salt bridge between Glu35 and Arg177 described earlier [17, 18, 21, 50]. However, in the substrate-free structure, movement of Glu39 to the “open” conformation breaks this salt bridge and a water molecule occupies the position of the carboxylate oxygen of Glu39. Thus, Arg42 may also stabilize MnII binding in the resting state through its interaction with Glu39, but its movement to the “in” conformation to stabilize the oxyferryl group in compounds I and II may destabilize Glu39 enabling it to move out for the oxidized MnIII to be chelated by oxalate. In contrast, the other two amino acid ligands for MnII, Glu35 and Asp179, are held in place by the rigid guanidium group of the Arg177 side chain and backbone amide group of residue Ala187, respectively. In support of this idea, disruption of the salt bridge between Glu35 and Arg177, through mutation of the arginine residue, has been shown to significantly lower the efficiency of MnII binding and oxidation in our previous work [21, 50]. This new role for Arg42 is revealed for the first time in this high resolution structure of MnP.
Heme Stereochemistry
The overall structure of Mn-MnP (Figure 2) at 0.93Å appears essentially the same as that in our earlier lower resolution work [17, 18]. Given the favorable ratio of data to parameters to be refined, the least squares matrix could be inverted which provides an accurate estimate of bond distances and angles. The electron density for the heme and proximal heme ligand together, shown in Figure 5 and Table 4, provides specific heme parameters. As expected from spectroscopic studies [51], the heme Fe is pentacoordinate and high-spin. A functionally important parameter of heme geometry is the displacement of the iron from the porphyrin core. In MnP the Fe is 0.26Å out of plane compared to 0.48Å for typical model heme complexes [52]. The ability of the Fe atom to move in and out of the porphyrin core as a function of ligation, redox state, and spin state is a central feature of the hemoglobin allosteric mechanism proposed by Perutz [53]. Compared to the globins, the Fe atom in MnP and CcP is closer to the porphyrin core. We have attributed this difference [54] to the surrounding protein environment which in the peroxidases places the proximal helix containing the heme His ligand much closer to the protein. This prevents the spring-like up/down motion of the Fe-His-helix as seen in hemoglobin. The functional relevance of this difference is that in peroxidases, the iron is oxidized from FeIII to FeIV=O. The higher oxidation state, lower-spin state, and strong FeIV=O bond all favor the FeIV closer to the heme plane. Hence, the resting FeIII state with only partial displacement of the Fe from the porphyrin plane is poised for oxidation to FeIV, which is reminiscent of the entatic state [55], where the kinetic and/or thermodynamic barriers required for changing redox and spin-state are lowered owing to protein-metal interactions.
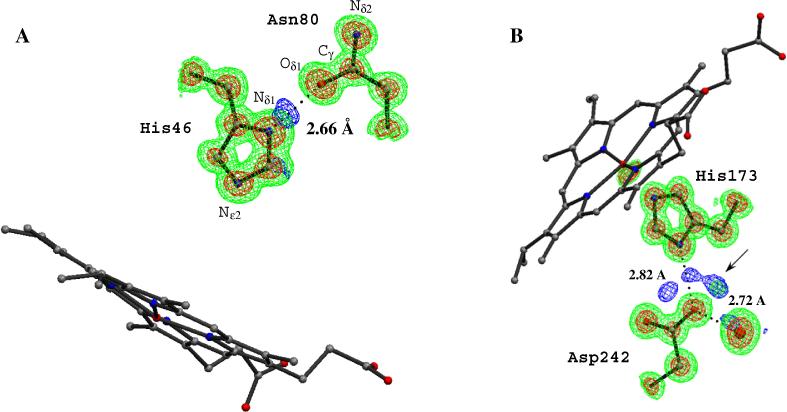
Figure 5.
2Fo – Fc (green, 1 σ and red, 3 σ and Fo – Fc (blue, 3 σ) electron density maps in the distal (A) and proximal (B) regions. The peaks (blue) in Fo – Fc map indicate potential hydrogen peaks near the proximal Asp242 and distal His46.
Table 4.
Heme parameters
| Distance (Å) | MnP (-Mn) (Å) | Mn-MnP (Å) | CcP (Å) |
|---|---|---|---|
| Fe—His N | 2.10 | 2.07 | 2.07 |
| Fe—pyrrole N | 2.02 | 2.04 | 2.05 |
| Fe—pyrrole N plane | 0.28 |
Mechanism and H atoms
As with many enzyme systems, H-bonds and proton transfer play a critical role in peroxidase catalysis. The original stereochemical mechanism proposed for heme peroxidases [56] indicates that the distal histidine residue (His 46 in MnP, Figure 2) acts as an acid-base catalyst by removing a proton from the iron-linked peroxide O atom and delivering it to the leaving OH moiety to produce water. As shown in Figure 5A, which of the N atoms of the imidazole moiety in His46 carries the proton is clearly visible in Fo-Fc electron density maps. The Nδ1atom is protonated and donates an H-bond to Asn80. This His-Asn H-bonding arrangement is conserved in all peroxidases and is thought to play an important role in orientating the distal His for catalysis as well as ensuring that the Nε2 is free to accept a proton from hydrogen peroxide. Another level of detail at 0.93 Å is the unambiguous orientation of amide side chains such as that for Asn80. At lower resolution, it is not possible to differentiate between the Nδ2 and Oδ1 side chain atoms. However, as shown in Figure 5A the amide side chain Oδ1 atom of Asn80 exhibits smeared electron density with the Cγ side chain atom while the Cδ-Nε2 density is much shaper. This indicates a Cγ-Oδ1 double bond and an H bond between the Nε1 of His 46 and the Oδ1 of Asn80.
The role of H-bonds involving the proximal His ligand is not well documented. In all known heme peroxidase structures, the His ligand is within H-bonding distance of a buried Asp, Asp242 in MnP (Figure 2). A number of important functional properties have been attributed to this H-bond, one of which involves redox potential. A strong His-Asp H-bond favors a lower redox potential by stabilizing the additional positive charge on FeIV compared to FeIII. In the globins the histidine ligand forms an H-bond with a peptide carbonyl O atom, which presumably is weaker than the His-Asp H-bond in peroxidases. This difference helps to explain why the peroxidases exhibit lower redox potentials than globins. This view is supported by both model heme systems [57-60], protein NMR [61], and site directed mutagenesis [62, 63].
The present structure provides a somewhat complex picture of the His-Asp interaction. As shown in Figure 5B there is a strong lobe of electron density in both Fo-Fc and 2Fo-Fc maps at about 1.34Å from Oδ2 of Asp242. Initially we considered that this peak might be an H atom which would mean that Asp242 carries the proton and not His173. However, the O-H distance, 1.34Å, is too long, compared to other H atoms in the structure, and the electron density peak too large for an H atom. Two additional data sets to 1.15Å and 1.05Å have been collected and maps generated from these data sets also exhibit additional electron density between Asp242 and His173 (data not shown). The prospects of a low-barrier H-bond seems unlikely since the O-N hydrogen bonding distance is quite normal at 2.83Å while low-barrier H-bonds are much shorter.. The exact explanation for this extra electron density remains unclear.
ACKNOWLEDGEMENT
This research was supported by a grant GM42614 (to T.L.P.) from the National Institutes of Health and grants MCB-9808420 from the National Science Foundation and DE-03-96ER20235 from the Division of Energy Biosciences, U.S. Department of Energy (to M.H.G). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
ABBREVIATIONS
- MnP
manganese peroxidase
- Mn-MnP
manganese-enzyme complex
- L.S.
least squares
- CGLS
conjugate gradient least squares
- ESD
estimated standard deviation
- ADP
atomic displacement parameter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Atomic coordinates have been submitted to the Protein Data Bank as entries XXX and YYY.
REFERENCES
- 1.Buswell JA, Odier E. CRC Crit. Rev. Biotechnol. 1987;6:1–60. [Google Scholar]
- 2.Gold MH, Alic M. Microbiol. Rev. 1993;57:605–622. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold MH, Wariishi H, Valli K. ACS Symp. Ser. 1989;389:127–140. [Google Scholar]
- 4.Kirk TK, Farrell RL. Annu. Rev. Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 5.Bumpus JA, Aust SD. BioEssays. 1989;6:166–170. [Google Scholar]
- 6.Hammel KE. Enzyme Mirob. Technol. 1989;11:776–777. [Google Scholar]
- 7.Reddy GV, Gelpke MD, Gold MH. J Bacteriol. 1998;180:5159–5164. doi: 10.1128/jb.180.19.5159-5164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valli K, Brock BJ, Joshi DK, Gold MH. Appl Environ Microbiol. 1992;58:221–228. doi: 10.1128/aem.58.1.221-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valli K, Wariishi H, Gold MH. J Bacteriol. 1992;174:2131–2137. doi: 10.1128/jb.174.7.2131-2137.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuwahara M, Glenn JK, Morgan MA, Gold MH. FEBS Lett. 1984;169:247–250. [Google Scholar]
- 11.Kersten PJ, Kirk TK. J Bacteriol. 1987;169:2195–2201. doi: 10.1128/jb.169.5.2195-2201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perie FH, Sheng D, Gold MH. Biochim. Biophys. Acta. 1996;1297:139–148. doi: 10.1016/s0167-4838(96)00096-9. [DOI] [PubMed] [Google Scholar]
- 13.Glenn JK, Akileswaran L, Gold MH. Arch. Biochem. Biophys. 1986;251:688–696. doi: 10.1016/0003-9861(86)90378-4. [DOI] [PubMed] [Google Scholar]
- 14.Glenn JK, Gold MH. Arch. Biochem. Biophys. 1985;242:329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- 15.Gold MH, Youngs HL, Sollewijn Gelpke MD. In: Manganese and Its Role in Biological Processes. Sigel A, Sigel H, editors. Macel Dekker; New York: 2000. pp. 559–586. [Google Scholar]
- 16.Wariishi H, Valli K, Gold MH. J. Biol. CHem. 1992;267:23688–23695. [PubMed] [Google Scholar]
- 17.Sundaramoorthy M, Kishi K, Gold MH, Poulos TL. J Biol Chem. 1994;269:32759–32767. [PubMed] [Google Scholar]
- 18.Sundaramoorthy M, Youngs HL, Gold MH, Poulos TL. Biochemistry. 2005;44:6463–6470. doi: 10.1021/bi047318e. [DOI] [PubMed] [Google Scholar]
- 19.Kishi K, Kusters-van Someren M, Mayfield MB, Sun J, Loehr TM, Gold MH. Biochemistry. 1996;35:8986–8994. doi: 10.1021/bi960679c. [DOI] [PubMed] [Google Scholar]
- 20.Kusters-van Someren M, Kishi K, Lundell T, Gold MH. Biochemistry. 1995;34:10620–10627. doi: 10.1021/bi00033a037. [DOI] [PubMed] [Google Scholar]
- 21.Sollewijn Gelpke MD, Moenne-Loccoz P, Gold MH. Biochemistry. 1999;38:11482–11489. doi: 10.1021/bi990943c. [DOI] [PubMed] [Google Scholar]
- 22.Whitwam RE, Brown KR, Musick M, Natan MJ, Tien M. Biochemistry. 1997;27:5365–5370. doi: 10.1021/bi9708794. [DOI] [PubMed] [Google Scholar]
- 23.Youngs HL, Sollewijn Gelpke MD, Li D, Sundaramoorthy M, Gold MH. Biochemistry. 2001;40:2243–2250. doi: 10.1021/bi002104s. [DOI] [PubMed] [Google Scholar]
- 24.Youngs HL, Sundaramoorthy M, Gold MH. Eur J Biochem. 2000;267:1761–1769. doi: 10.1046/j.1432-1327.2000.01173.x. [DOI] [PubMed] [Google Scholar]
- 25.Sundaramoorthy M, Kishi K, Gold MH, Poulos TL. J Biol Chem. 1997;272:17574–17580. doi: 10.1074/jbc.272.28.17574. [DOI] [PubMed] [Google Scholar]
- 26.Bao W, Fukushima Y, Jensen KA, Jr., Moen MA, Hammel KE. FEBS Lett. 1994;354:297–300. doi: 10.1016/0014-5793(94)01146-x. [DOI] [PubMed] [Google Scholar]
- 27.Reddy GV, Sridhar M, Gold MH. Eur J Biochem. 2003;270:284–292. doi: 10.1046/j.1432-1033.2003.03386.x. [DOI] [PubMed] [Google Scholar]
- 28.Banci L, Bertini I, Dat Pozzo L, Del Conte R, Tien M. Biochemistry. 1998;37:9009–9015. doi: 10.1021/bi972879+. [DOI] [PubMed] [Google Scholar]
- 29.Kishi K, Wariishi H, Marquez L, Dunford HB, Gold MH. Biochemistry. 1994;33:8694–8701. doi: 10.1021/bi00195a010. [DOI] [PubMed] [Google Scholar]
- 30.Kuan IC, Tien M. Proc. Natl. Acad. Sci. USA. 1993;90:1242–1246. doi: 10.1073/pnas.90.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jentzen W, Ma JG, Shelnutt JA. Biophys J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn P, Knapp M, Soltis SM, Ganshaw G, Thoene M, Bott R. Biochemistry. 1998;37:13446–13452. doi: 10.1021/bi9813983. [DOI] [PubMed] [Google Scholar]
- 33.Betzel C, Gourinath S, Kumar P, Kaur P, Perbandt M, Eschenburg S, Singh TP. Biochemistry. 2001;40:3080–3088. doi: 10.1021/bi002538n. [DOI] [PubMed] [Google Scholar]
- 34.Frey PA, Whitt SA, Tobin JB. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Grebenko AA, Brannigan JA, Anston AA, Barynin VV, Dodson GG, Dauter Z, Wilson KS, Melik-Adamyan WR. Acta Crystallogr D Biol Crystallogr. 2002;58:1972–1982. doi: 10.1107/s0907444902016566. [DOI] [PubMed] [Google Scholar]
- 36.Liebschner D, Elias M, Moniot S, Fournier B, Scott K, Jelsch C, Guillot B, Lecomte C, Chabriere E. Journal of American Chemical Society. 2009;131:7879–7886. doi: 10.1021/ja901900y. [DOI] [PubMed] [Google Scholar]
- 37.Lyubimov AY, Lario PI, Moustafa I, Vrielink A. Nature Chemical Biology. 2006;2:259–264. doi: 10.1038/nchembio784. [DOI] [PubMed] [Google Scholar]
- 38.Fenn TD, Ringe D, Petsko GA. Biochemistry. 2004;43:6464–6474. doi: 10.1021/bi049812o. [DOI] [PubMed] [Google Scholar]
- 39.Khan H, Barna T, Harris RJ, Bruce NC, Barsukov I, Munro AW, Moody PC, Scrutton NS. J Biol Chem. 2004;279:30563–30572. doi: 10.1074/jbc.M403541200. [DOI] [PubMed] [Google Scholar]
- 40.Sundaramoorthy M, Kishi K, Gold MH, Poulos TL. J Mol Biol. 1994;238:845–848. doi: 10.1006/jmbi.1994.1338. [DOI] [PubMed] [Google Scholar]
- 41.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 42.Otwinowski Z, Minor W. Methods in Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT. X-PLOR Manual Version 3.1: A system for X-ray Crystallography and NMR. Yale University; New Haven, CT: 1992. [Google Scholar]
- 44.Sheldrick GM, Scheider TR. Methods in Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 45.Godfrey BJ, Mayfield MB, Brown JA, Gold MH. Gene. 1990;93:119–124. doi: 10.1016/0378-1119(90)90144-g. [DOI] [PubMed] [Google Scholar]
- 46.Pribnow D, Mayfield MB, Nipper VJ, Brown JA, Gold MH. J Biol Chem. 1989;264:5036–5040. [PubMed] [Google Scholar]
- 47.Merritt EA. Acta Crystallogr D Biol Crystallogr. 1999;55:1109–1117. doi: 10.1107/s0907444999003789. [DOI] [PubMed] [Google Scholar]
- 48.Bonagura CA, Bhaskar B, Shimizu H, Li H, Sundaramoorthy M, McRee DE, Goodin DB, Poulos TL. Biochemistry. 2003;42:5600–5608. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 49.Edwards SL, Nguyen HX, Hamlin RC, Kraut J. Biochemistry. 1987;26:1503–1511. doi: 10.1021/bi00380a002. [DOI] [PubMed] [Google Scholar]
- 50.Gelpke MD, Youngs HL, Gold MH. Eur J Biochem. 2000;267:7038–7045. doi: 10.1046/j.1432-1327.2000.01798.x. [DOI] [PubMed] [Google Scholar]
- 51.Mino Y, Wariishi H, Blackburn NJ, Loehr TM, Gold MH. J Biol Chem. 1988;263:7029–7036. [PubMed] [Google Scholar]
- 52.Hoard JL. Science. 1971;174:1295–1302. doi: 10.1126/science.174.4016.1295. [DOI] [PubMed] [Google Scholar]
- 53.Perutz MF. Q Rev Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 54.Poulos TL. Nat Struct Biol. 1996;3:401–403. doi: 10.1038/nsb0596-401. [DOI] [PubMed] [Google Scholar]
- 55.Williams RJ. Eur J Biochem. 1995;234:363–381. doi: 10.1111/j.1432-1033.1995.363_b.x. [DOI] [PubMed] [Google Scholar]
- 56.Poulos TL, Kraut J. J Biol Chem. 1980;255:8199–8205. [PubMed] [Google Scholar]
- 57.Chang YT, Stiffelman OB, Loew GH. Biochimie. 1996;78:771–779. doi: 10.1016/s0300-9084(97)82535-7. [DOI] [PubMed] [Google Scholar]
- 58.Doef MM, Sweigart DA, O'Brien P. Inorg. Chem. 1983;22:851–852. [Google Scholar]
- 59.Nappa M, Valentine JS, Snyder PA. Journal of American Chemical Society. 1977;99:5799–5800. doi: 10.1021/ja00459a045. [DOI] [PubMed] [Google Scholar]
- 60.Valentine JS, Sheridan RP, Allen LC, Kahn PC. Proceedings of National Academy of Sciences U S A. 1979;76:1009–1013. doi: 10.1073/pnas.76.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banci L, Bertini I, Pease EA, Tien M, Turano P. Biochemistry. 1992;31:10009–10017. doi: 10.1021/bi00156a021. [DOI] [PubMed] [Google Scholar]
- 62.Choudhury K, Sundaramoorthy M, Hickman A, Yonetani T, Woehl E, Dunn MF, Poulos TL. J Biol Chem. 1994;269:20239–20249. [PubMed] [Google Scholar]
- 63.Goodin DB, McRee DE. Biochemistry. 1993;32:3313–3324. [PubMed] [Google Scholar]