Abstract
Mass spectrometric identification of cross-linked peptides can provide valuable information about the structure of protein complexes. We describe a straightforward database search scheme that identifies and assigns statistical confidence estimates to spectra from cross-linked peptides. The method is well suited to targeted analysis of a single protein complex, without requiring an isotope labeling strategy. Our approach uses a SEQUEST-style search procedure in which the database is comprised of a mixture: single peptides with and without linkers attached, and cross-linked products. In contrast to several previous approaches, we generate theoretical spectra that account for all of the expected peaks from a cross-linked product, and we employ an empirical curve-fitting procedure to estimate statistical confidence measures. We show that our fully automated procedure successfully re-identifies spectra from a previous study, and we provide evidence that our statistical confidence estimates are accurate.
Keywords: protein-protein interaction, peptide identification, calibration, cross-linked peptides
1 Introduction
Proteins are the primary functional molecules in the cell, and most protein functions are carried out by multiprotein complexes. However, understanding how a protein complex works often requires knowing the 3D structure of the complex, and discovering this structure is notoriously difficult. Therefore, mass spectrometry protocols that are capable of providing even partial information about the structure of a protein complex are in high demand [Young et al., 2000].
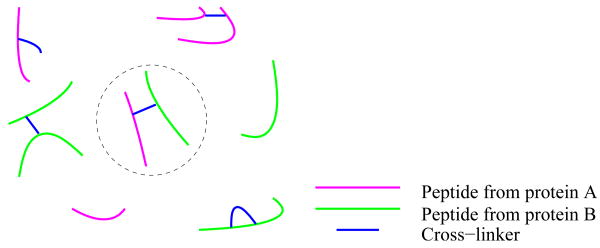
Perhaps the most straightforward protocol involves three steps: (1) cross-linking protein-protein interactions using a linker of known molecular weight, (2) enzymatically digesting the cross-linked proteins into peptides, and (3) subjecting the peptides to micro-liquid chromatography coupled with tandem mass spectrometry analysis. The resulting collection of fragmentation spectra correspond to various types of ions, illustrated in Figure 1: linear peptides, peptides with one cross-linker attached (either dead-end products or self-loops), intra-protein cross-links and inter-protein cross-links. The last class of molecules provides information about the 3D structure of the protein complex, because these molecules give information about the proximity of amino acids in two interacting proteins. Although more complex cross-linked peptide products can exist, they are usually not considered [Schilling et al., 2003].
Figure 1. Various types of molecules produced by a cross-linking protocol.
The molecule inside the dotted circle is an inter-protein cross-link.
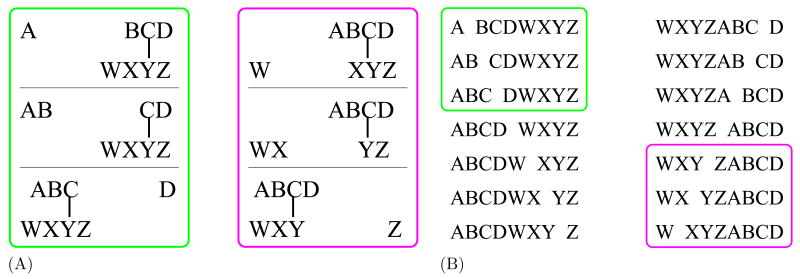
Identifying spectra produced by cross-linked products is challenging, because each spectrum contains a mixture of fragment ions from single peptides and from the cross-linked peptides. Two research groups have proposed protocols for mapping cross-linked peptides to observed spectra using existing database search tools. Maiolica et al. [2007] use the Mascot search tool [Perkins et al., 1999], coupled with a database of concatenated peptide pairs. The pairwise database is created by extracting, from a given peptide database, all possible peptide pairs and concatenating each pair in both possible orders. Thus, a cross-linked pair of peptides A and B will share ions with both of the corresponding peptide pairs A:B and B:A. Indeed, A:B and B:A jointly account for all of the single-bond cleavages of the corresponding cross-linked product; however, as illustrated in Figure 2(B), neither concatenated peptide pair alone matches all of the expected ions, and both concatenated peptide pairs contain additional ions that do not occur in the cross-linked product. Because of these deficiencies, Maiolica et al. [2007] only use Mascot to identify candidate cross-linked products. The authors then apply a second, probabilistic function to re-score high-scoring matches identified by Mascot. Fundamentally, this approach is hampered by its reliance on an existing database search engine, because any cross-linked pair that does not score well, according to Mascot, against one of its two corresponding concatenated peptides will never be considered by the second-tier score function.
Figure 2. Comparison of product ions.
(A) The figure shows the ions that result from fragmenting each b/y bond in a fictitious cross-linked product. The two cross-linked peptides are each of length four, resulting in six possible cleavage locations and 12 product ions. In the protocol of Singh et al. [2008], the cross-linked product is represented as two distinct peptides, each with a single modification. One modified peptide gives rise to the six product ions in the green box, and the other gives rise to the six ions in the magenta box. (B) In the protocol of Maiolica et al. [2007], the cross-linked product is represented by two concatenated peptide pairs. Each pair gives rise to 14 product ions. In the figure, the left and right columns list the fragment ions produced from the two relative orientations. In each case, only six of the resulting fragment ions (boxed) would be produced by the original cross-linked product.
The protocol proposed by Singh et al. [2008] relies upon a combination of two different types of search. First, the spectra are searched against a standard sequence database, and spectra that match with high confidence are eliminated from consideration. Second, the remaining unmatched spectra are analyzed using an open modification search tool called Popitam [Hernandez et al., 2003], which is designed to identify chemically modified peptides when the modification mass is not known in advance. The key idea behind the Singh et al. protocol is to find a spectrum that matches well against two different peptides with complementary modifications. Say that the first peptide has a mass of p1 and a modification of m1, and the second peptide has corresponding masses of p2 and m2. Then, because both peptides match the same spectrum, we know that
Furthermore, because we know the mass c of the cross-linker, we know that if these two peptides are cross-linked to one another, then the mass of the modification on the first peptide should equal the mass of the second peptide plus the mass of the cross-linker, and vice versa; i.e.,
If we identify two strongly matching peptides for which the modification masses obey these arithmetic rules, then we have a good candidate for a true cross-linked pair.
This idea has intuitive appeal, but the protocol suffers from one significant drawback: by treating one peptide as a single modification on the other peptide, each individual search effectively ignores all of the fragmentation peaks associated with one of the two peptides. The situation is illustrated in Figure 2(A). Popitam must be capable of recognizing matches to theoretical spectra that contain only half of the expected ions. Therefore, a spectrum may not match any single modified peptide very well, even though the spectrum matches the pair of peptides quite well indeed. Additionally, since Popitam scores the two peptides with their respective modifications separately rather than jointly as a cross-linked candidate, the quality of the match and the observed spectrum are manually verified.
In this work, we propose an alternative and more direct approach to the problem of identifying cross-linked product spectra: we modify an existing search algorithm to use a database that contains a mixture of peptides, dead-end products, self-loops and cross-linked products. For the cross-linked peptides, we use theoretical spectra similar to the one shown in Figure 3. We then perform a SEQUEST-style search against this database. To make this search procedure useful in practice, we must be able to assign statistical confidence estimates to the assigned spectra. We compute these confidence values using a previously described empirical curve-fitting protocol [Klammer et al., 2009].
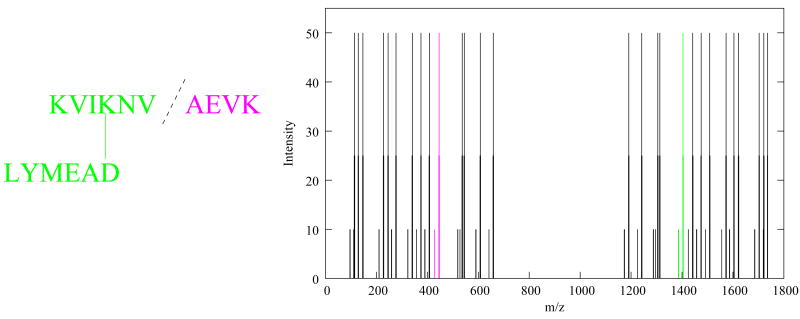
Figure 3. Theoretical spectrum for a cross-linked product.
The figure shows the theoretical spectrum for a cross-linked pair of +1 peptides, kviknvaevk and lymead, linked at positions 4 and 6, respectively. As in SEQUEST, b- and y-ions are arbitrarily assigned a height of 50, with flanking peaks of height 25 and neutral losses (ammonia, water and carbon monoxide) of height 10. The peaks corresponding to the fragmentation illustrated on the left are colored as shown.
Below, we demonstrate that the search protocol is capable of automatically re-discovering the correct cross-linked peptides from previously described spectra. We also demonstrate that the p-values estimated by our method are accurate, in the sense that they follow a uniform distribution when computed with respect to null data, and we show empirically that the method does not introduce false positive matches against spectra that correspond to unlinked peptides.
2 Materials and Methods
2.1 Data
The cross-linking data was collected as previously described in Singh et al. [2008]. Briefly, cross-linking reagent 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) was added to a 3:1 molar ratio of E. coli expressed recombinant human cytochrome b5 (b5) and cytochrome P450 2E1 (CYP 2E1). After quenching the cross-linking reaction, the sample was denatured with urea, reduced with DDT, alkylated with iodoacetamide, and finally digested with trypsin. The final sample was then analyzed using an LTQ-Orbitrap (Thermo-Fisher, San Jose, CA) equipped with a nanoflow HPLC system (NanoAcquity; Waters Corporation, Milford, MA). The raw data was extracted into peak lists (.dta files) using the instrument's software (extract msn.exe; Thermo Fisher, San Jose, CA). The peak lists were then converted into a single .ms2 file containing 3314 spectra using an in-house script. The precursor mass-to-charge and charge state is determined on-the-fly by the acquisition software.
2.2 Generating theoretical spectra
For a given cross-linked product, we generate a SEQUEST-style theoretical spectrum. The spectrum includes peaks corresponding to b- and y-ions for both peptides. Depending upon the location of the cleavage site relative to the cross-linker, half of the ions include a “modification” whose mass is equal to the mass of the cross-linker plus the mass of the second peptide, as illustrated in Figure 3. For multiply charged products, cleavage ions from all possible lower charge states are generated. All b- and y-ions are arbitrarily assigned a theoretical height of 50. In addition, the spectrum includes two flanking peaks per b- or y-ion. These are assigned to 1 Da bins on either side of the corresponding primary peak, and are assigned a height of 25. Finally, the spectrum includes three types of neutral loss peaks—water, ammonia and carbon monoxide (a-ions)—with a fixed height of 10. The theoretical spectrum is created with 1 Da resolution. If a single 1 Da bin contains more than one peak, then the peak height is assigned as the maximum value of the overlapping peaks. Since we are only considering single b-y fragmentation events and we are assuming that the cross-link itself does not fragment, self-loops will have many ions of the same mass for the peptide's amino acids that lie in between the linker sites.
The database of candidate molecules is built in-memory from the supplied fasta file of proteins. Our method generates all possible product molecules—linear peptides, inter-/intra- cross-links, dead-ends, and self-loops—from the tryptic peptides with up to one missed cleavage site. In general, our model considers all of the fragment ions that result from a single fragmentation of a molecule. Self-linked peptides are included in the database. However, fragmentation events that occur anywhere along the peptide backbone between the two amino acids that are cross-linked linked, fragment ions with the same mass, so these are not considered. Our database does not include fully cyclic peptides, since cleaving them into two fragments would require two fragmentation events, which is not very common during typical CID experiments. Furthermore, fully cylic peptides are rarely generated from amine-reactive cross-linkers such as EDC, because trypsin fails to cleave at the Lysine residues that are cross-linked. Finally, dead-end molecules are included in our database, with the linker treated like a modification on the amino acid. Again, if the linked amino acid is a lysine for cross-linked or dead-end products, then the cleavage by trypsin is prohibited.
2.3 Search and calibration
For a given spectrum S, the search procedure consists of three steps. First, the spectrum itself is normalized according to the SEQUEST protocol [Eng et al., 2008]. Second, we extract from the database the set of peptides and cross-linked products whose total mass lies within a specified range of the precursor mass inferred from the possible charge states provided by the acquisition software. In the experiments reported here, we use a 2.1 Da precursor mass range. Third, these candidate peptides and cross-linked products are ranked according to the SEQUEST score function XCorr [Eng et al., 1994, 2008]:
where x and y are the observed and theoretical spectra, respectively. The result, for each spectrum, is a ranked list of peptides and cross-linked products.
In general, the XCorr assigned to a theoretical spectrum—either from a peptide or from a cross-linked product—depends upon properties of the spectrum as well as properties of the theoretical spectrum. Thus, an XCorr of 2.0 may be more surprising or less surprising, in a statistical sense, depending upon the properties of the spectrum. To account for the spectrum-specific distribution of XCorr scores, we use an empirical calibration scheme, as described previously [Klammer et al., 2009]. That procedure works by fitting, for each spectrum, a three-parameter Weibull distribution to the observed distribution of scores. The estimated Weibull parameters are then used to convert the maximal score to a p-value.
In the current study, we use a modified version of this calibration protocol. Because the database of cross-linked peptides is relatively small, we augment the observed score distribution with additional decoy scores. These decoys are generated by extracting candidate peptides using a larger precursor mass range (20 Da, rather than 2.1 Da). We then shuffle the non-terminal amino acids in each of these candidate peptides. To achieve an accurate fit, we require a minimum of 4000 scores (targets plus decoys), so we continue re-shuffling the decoys and re-scoring them until this minimum is achieved.
Each resulting p-value must be subjected to multiple testing correction, to account for the number of candidate peptides that were considered during the search. For multiply charged spectra, the p-values for all possible charge states are merged into a single list. We then select the top-ranked p-value, and adjust it for multiple tests via the following transformation:
where p is the initial p-value, p̂ is the adjusted p-value and c is the total number of candidates (not including decoys).
For a collection of n spectra, our search procedure produces a ranked list of n peptides and cross-linked products, each with an associated p-value. To account for multiple testing with respect to this collection of spectra, we use established methods [Storey, 2002] to convert the p-values into q-values, where the q-value is defined as the minimal false discovery rate (FDR) at which a given p-value is deemed significant.
3 Results
3.1 Search successfully identifies known cross-linked peptides
As an initial validation, we used our search tool to assign cross-linked peptides to 10 spectra that had been identified in the context of a previous study [Singh et al., 2008]1. Our input consisted of the two target proteins, human cytochrome P450 2E1 (CYP2E1) and cytochrome b5 (b5), which contain 34 and 9 tryptic peptides, respectively. The complete database contained 92 linear peptides, 11,709 intra- and inter- protein cross-links, 252 dead-end products, and 153 self-loop products. Enforcing a 2.1 Da mass window identified an average of 12 intra- and inter- cross-linked candidates per spectrum. Among these candidates, many cross-linked products differ only in the location of the cross-linker. On average, each of these ten spectra has four distinct peptide pairs as potential candidates within the mass range.
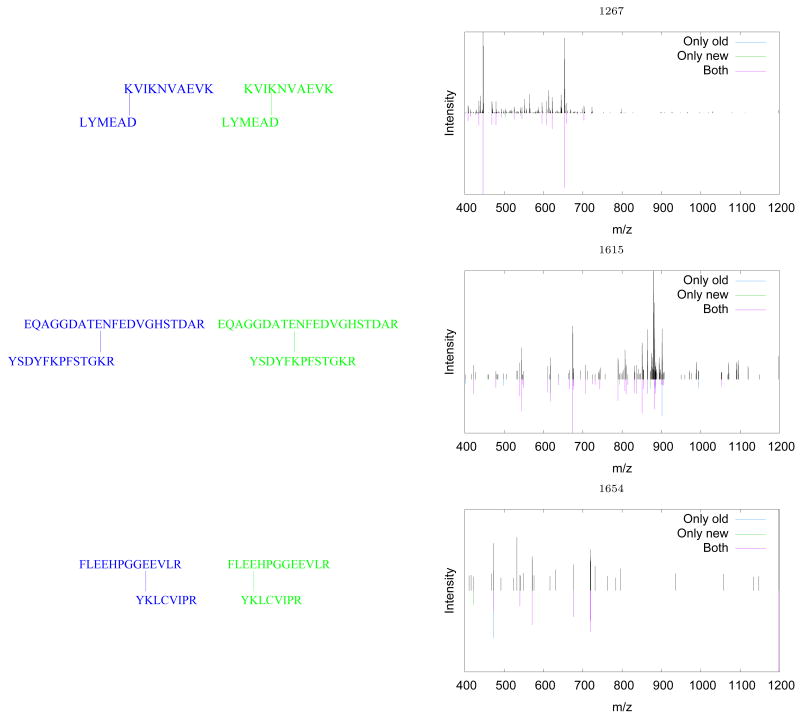
For each spectrum, we ranked the candidates by XCorr and examined the top-scoring candidate. In seven out of 10 cases, this procedure successfully identified the same pair of matched peptides, with the same cross-linker location. For the remaining three spectra, the two methods identify the same pair of peptides but disagree on the location of the cross-linker relative to one of the two linked peptides. These three spectra each correspond to distinct pairs of peptides. To better understand the differences in predicted cross-linker location, we compared each observed spectrum to the two theoretical spectra produced by the two cross-linker locations. Figure 4 shows the results of this analysis. In the figure, peaks are colored according to whether they are matched by one or both of the theoretical spectra. Scan 1267 is the only one of the 10 spectra that maps to this particular peptide pair (KVIKNVAEVK and LYMAED). As indicated by the preponderance of magenta peaks in Figure 4A, the shift in the location of the cross-linker by three amino acids has a very small effect on the two theoretical spectra. Scan 1615 (Figure 4B), on the other hand, contains a mixture of blue and green peaks, indicating that the two theoretical spectra provide different but almost equally good matches to the observed spectrum. Accordingly, the corresponding XCorrs are similar—1.89 and 2.22. Furthermore, scan 1605 (not shown) maps to the same pair of peptides, but assigns a different location to the cross-linker. Thus, for both of these scans, the true location of the cross-linker is difficult to ascertain. For scan 1615, the cross-linker location differs by one amino acid from the assignment given in the four other scans (1653, 1657, 1658, and 1662). Because scan 1615 contains so few matching ions, the precise cross-linker location is more difficult to ascertain.
Figure 4. Disagreements in the location of the cross-linker.
Each panel shows one spectrum for which the two methods disagreed on the location of the cross-linker. The complete spectrum is shown as positive peaks; negative peaks correspond to peaks that are matched by one or both theoretical spectra. Each such peak is colored according to whether it is matched by both theoretical spectra, only the spectrum corresponding to the previously identified cross-linker location, or only the spectrum corresponding to the new location. Listed above the spectrum is the scan number, and below is the pair of linked peptides.
3.2 Decoy p-values follow a uniform distribution
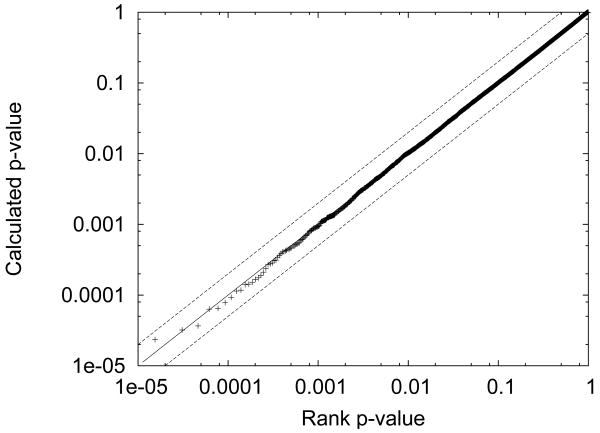
Having established that the method can successfully rank candidate peptides with respect to individual spectra, we next investigated whether the empirical curve-fitting procedure could successfully convert the XCorr scores into p-values. To do so, we searched a previously described set of 3314 spectra [Singh et al., 2008] against a database derived from shuffled peptides from the cytochrome P450 2E1 and cytochrome b5 proteins. Among these spectra, 2797 had at least one candidate peptide within 2.1 Da of the inferred precursor mass. Because the peptides have been shuffled, we do not expect any true matches to occur; therefore, the observed p-values should be uniformly distributed. We demonstrate this uniformity in Figure 5, which plots the calculated p-value as a function of the rank p-value, where the rank p-value of a score x is defined as the fraction of scores that are greater than or equal to x. The linear relationship in Figure 5 shows that the distribution of observed decoy p-values are uniform and that we have successfully calibrated the XCorr values.
Figure 5. Decoy p-values follow a uniform distribution.
The figure plots the calculated p-value as a function of the rank p-value for all candidates and all spectra in the data set. The rank p-value of a score x is defined as the fraction of scores that are greater than or equal to x. The distribution of p-values is compared against y = x (solid line), y = 2x and y = x/2 (dotted lines).
3.3 Analysis of a larger data set
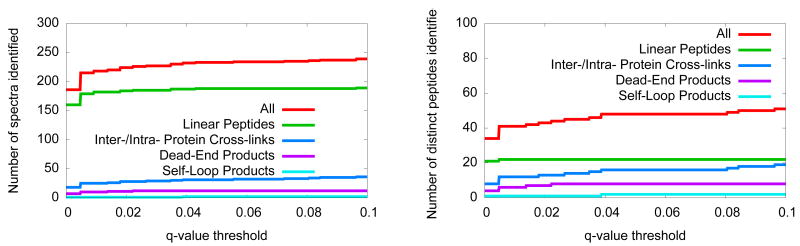
Next, we applied our search and calibration procedure to the larger data set of 3314 spectra, this time using the unshuffled protein sequences. To correct for multiple testing with respect to these spectra, we use established methods [Storey, 2002] to convert the p-values into q-values, where the q-value is defined as the minimal false discovery rate (FDR) at which a given score is deemed significant. Figure 6 plots the number of spectra that are successfully identified as a function of q-value threshold. At a threshold of q < 0.01, we identify 218 spectra. Of these 218 spectra 182 are from linear peptides, 25 are from inter- or intra-protein cross-links, six are from dead-end products, and one is from a self-loop product.
Figure 6. Results of a large-scale search.
(A) The figure plots the number of spectra identified, as a function of q-value threshold. The number of linear peptides, inter-/intra- protein cross-links, dead-end products, and self-loop products are on this plot. (B) This panel is similar to (A), except that the lines correspond to the number of unique species found versus q-value threshold.
Reassuringly, many of the same pairs of cross-linked peptides are identified multiple times. Table 2 lists the distinct products identified in the search. In many cases, the same pair of peptides is identified with multiple linker locations for different spectra. As previously discussed in section 3.1, finding the exact location of the cross-linker is difficult, since many of the ions between the two products are similar. This problem is especially true in the cases of two particular cross-linked pairs of peptides: (fkpehflnengk, gtvvvptldsvlydnqefpdpek) and (fleehpggeevlr, hnhskstwlilhhk). In these cases, the predicted cross-link sites differ by only one or two amino acids—respectively, (2,22) versus (2,20) and (3,5) versus (4,5).
Table 2. Distinct products identified in the large-scale search.
The table lists each peptide pair identified by the search at q < 0.01, along with the cross-linker locations and the number of spectra that mapped to that pair.
| Peptide1 | Peptide2 | Loc1 | Loc2 | Num | % by | % insensity | mass error(ppm) |
|---|---|---|---|---|---|---|---|
| fkpehflnengk | gtvvvptldsvlydnqefpdpek | 2 | 22 | 6 | 16.2 | 36.2 | 250.2 |
| fkpehflnengk | gtvvvptldsvlydnaefpdpek | 2 | 20 | 1 | 11.0 | 49.1 | 246.0 |
| fkpehflnengk | gtvvvptldsvlydnaefpdpek | 2 | 9 | 1 | 7.2 | 51.9 | 252.2 |
| fleehpggeevlr | hnhskstwlilhhk | 3 | 5 | 3 | 18.0 | 37.1 | 7.8 |
| fleehpggeevlr | hnhskstwlilhhk | 4 | 5 | 1 | 16.8 | 48.4 | 6.7 |
| fleehpggeevlr | yklcvipr | 3 | 2 | 5 | 19.5 | 37.6 | 4.7 |
| fleehpggeevlr | yklcvipr | 9 | 2 | 3 | 18.5 | 36.1 | 8.4 |
| kviknvaevk | lymaed | 4 | 6 | 1 | 34.8 | 44.5 | 7.2 |
| eqaggdatenfedvghstdar | ysdyfkpfstgk | 6 | 6 | 1 | 11.0 | 34.2 | 1.8 |
| eqaggdatenfedvghstdar | ysdyfkpfstgkr | 1 | 6 | 1 | 20.3 | 37.9 | 9.0 |
| fleehpggeevlr | viknvaevk | 4 | 3 | 1 | 13.1 | 36.8 | 6.2 |
| lytmdgitvtvadlffagtettsttlr | ygllilmkypeieek | 20 | 8 | 1 | 6.3 | 30.6 | 6.8 |
To produce the results in Table 2, we only considered tryptic peptides that result from at most one missed cleavage. We investigated the behavior of the algorithm when we relax this requirement, allowing multiple missed cleavages. In this case, the last entry in Table 2, intra-protein cross-linked product (lytmdfitvtvadlffagtettsttlr, ygllilmkypeieek), is assigned to a linear peptide with two missed cleavages. In addition, allowing multiple missed cleavages produces a new identification: the intra-protein cross-linked product a new identification is produced: the intra-protein cross-linked product (dtifrgylipkgtvvvptldsvlydnqefpdpek, fkysdyfkpfstgkr) is assigned to a scan that previously was not identified. These results suggest that allowing more than one missed cleavage may be beneficial.
To further validate our search method, we also report in Table 1 the estimated q-values for the previously identified spectra. Using our reported threshold of q < 0.01 we successfully identify 8 of the 10 spectra. One other spectrum receives a low q-value of 0.03. The remaining, extremely high q-value for scan 1654 is indicative of a problematic spectrum (see Figure 4C). The spectrum has few peaks, with a low proportion of peaks matched by either theoretical spectrum, indicating that this is likely an incorrect identification.
Table 1. Searching with 10 previously identified spectra.
The table lists, for each of the 10 spectra, the scan number, charge state (“+”), number of candidate products (all peptides pairs with every possible link), number of candidate peptide pairs, the two peptides, the inferred location of the cross-linker according to the previous method (“Loc (old)”) and the proposed method (“Loc (new)”), and the q-value assigned to the match. Spectra for which the two methods disagree on the location of the linker are marked with an asterisk in the “Diff” column.
| Scan | + | #Prod | #Pairs | Peptide 1 | Peptide 2 | Loc (old) | Loc (new) | q-val | Diff |
|---|---|---|---|---|---|---|---|---|---|
| 1267 | 4 | 7 | 2 | kviknvaevk | lymaed | (1, 6) | (4, 6) | 0.005 | * |
| 1370 | 4 | 6 | 2 | fleehpggeevlr | viknvaevk | (4, 3) | (4, 3) | 0.005 | |
| 1605 | 5 | 14 | 2 | eqaggdatenfedvghstdar | ysdyfkpfstgkr | (1, 6) | (1, 6) | 0.000 | |
| 1615 | 5 | 14 | 2 | eqaggdatenfedvghstdar | ysdyfkpfstgkr | (9, 12) | (9, 6) | 0.030 | * |
| 1758 | 5 | 18 | 5 | eqaggdatenfedvghstdar | ysdyfkpfstgk | (6, 6) | (6, 6) | 0.000 | |
| 1653 | 4 | 12 | 6 | fleehpggeevlr | yklcvipr | (3, 2) | (3, 2) | 0.000 | |
| 1654 | 5 | 12 | 6 | fleehpggeevlr | yklcvipr | (3, 2) | (4, 2) | 0.838 | * |
| 1657 | 5 | 12 | 6 | fleehpggeevlr | yklcvipr | (3, 2) | (3, 2) | 0.000 | |
| 1658 | 4 | 12 | 6 | fleehpggeevlr | yklcvipr | (3, 2) | (3, 2) | 0.000 | |
| 1662 | 5 | 12 | 6 | fleehpggeevlr | yklcvipr | (3, 2) | (3, 2) | 0.005 |
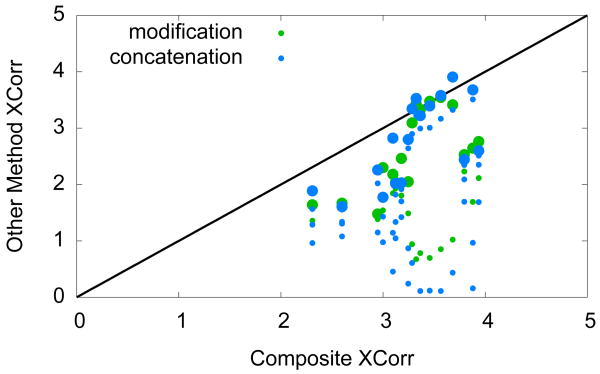
Finally, we investigated the extent to which the cross-linked peptides identified by our method could have been identified using theoretical spectra like the ones employed by Singh et al. [2008] and Maiolica et al. [2007]. For this analysis, we started with the 25 spectra identified with inter- or intra-protein cross-links at q < 0.01, and we eliminated any spectrum for which the identified cross-linked peptide was only observed once. This procedure yielded 17 high-confidence identifications. We then scored each spectrum against our composite theoretical spectrum, as well as the four degenerate theoretical spectra shown in Figure 2; i.e., we considered each peptide with the other peptide represented as a single large modification, and we considered the pair of peptides concatenated in both orientations. Figure 7 compares the XCorr scores computed using a composite theoretical spectrum versus the two types of degenerate theoretical spectra. In order to detect a viable cross-linked peptide, the modification method of Singh et al. [2008] involves scoring two degenerate theoretical spectra for each cross-linked product. The concatenation method, on the other hand, searches using a variable modification for the cross-link mass with the peptides linearized in the two different orientations. With this protocol, each candidate cross-linked peptide candidate results in four separate match scores, two for the order of the two concatenated peptides times two for the cross-link modification existing on either peptide. As seen in Figure 7, most of the XCorr scores for either method are lower than the corresponding scores from our composite method. From this observation, we conclude that our composite score is less likely to introduce false negative identifications than the two other methods we compared against.
Figure 7. Comparison of scoring methods.
In the figure, each point corresponds to a single spectrum that has been identified as a cross-linked product with high confidence. The y-axis is the XCorr computed with respect to the composite theoretical spectrum, and the x-axis is the XCorr computed with respect to a degenerate theoretical spectrum. Green dots correspond to degenerate spectra created by considering one peptide as a modification on the second. Blue dots correspond to degenerate spectra created by concatenating the two peptides in the two different orders as well as the cross-link modification existing on either peptide of the linearized pair. Larger dots signify the maximum score assigned to a given cross-linked product.
3.4 Negative control: non-cross-linked spectra
To further test the robustness of our search procedure, we performed one additional negative control experiment. In this test, we used a previously described collection of 35,236 spectra derived from a yeast whole-cell lysate. Based on previous analyses using Percolator [Käll et al., 2007], we collected a set of 756 high-confidence proteins, each containing at least five peptide identifications with confidence q < 0.01. We then randomly selected five of these high-confidence proteins and used them to construct a database. The resulting database contains 1244 linear peptides (allowing at most one missed cleavage), 2,835,438 inter- and intra- cross-link products, 3839 dead-end products, and 2481 self loop products. Searching the 35,236 spectra against this database and applying a q-value threshold < 0.01, our procedure identifies 91 spectra. This set includes 83 linear peptides, 6 inter- and intra-protein cross-links, 2 dead-end products, and no self-loop products. The 83 linear peptides are highly redundant, corresponding to only 29 distinct peptides. In contrast, the 8 identifications for the cross-linked products are unique for each spectrum. The small number of cross-link products shows that our method is robust, i.e., the method prefers linear peptides rather than cross-link products from spectra that contain only linear peptides.
4 Discussion
We have described a straightforward method for identifying cross-linked peptides by comparing observed spectra to theoretical spectra derived from the cross-linked products. In contrast to previous, multi-step methods, our approach automatically produces a single, ranked list of matched spectra. We use an empirical calibration procedure, coupled with two types of multiple testing correction, to compute false discovery rate estimates. Thus, each matched spectrum is reported along with a q-value, allowing the researcher to choose a confidence threshold appropriate for their study.
While we demonstrated our method's utility over two previously described protocols [Maiolica et al., 2007, Singh et al., 2008], many other algorithms exist for finding cross-linked peptides from tandem mass spectra [Chu et al., 2010, Gao et al., 2006, Hojrup, 1990, Koning et al., 2006, Rinner et al., 2008, Schilling et al., 2003, Seebacher et al., 2006, Singh et al., 2008, Tang et al., 2005, Lee et al., 2007]. However, most of these methods are not automatic or are designed to work with cross-links or peptides that have been isotopically labeled. Although not addressed in this work, a similiar method could be used to find cross-linked peptides that have been isotopically labeled.
Our results suggest that our method correctly identifies matched peptides but is less precise about the location of the cross-linker. This observation is not surprising, because the effect on the theoretical spectrum when the cross-linker moves can be relatively small. Additionally, double fragmentation is shown to occur on either side of the cross-linked peptide [Lee et al., 2007, Schilling et al., 2003] and would produce ions which were not included in our current method. In the future, we will determine if the addition of these ions to the theoretical spectrum assists in precisely locating the position of the cross-linker. Another direction we are actively investigating is methods to make use of high resolution MS/MS spectra.
In the future, we plan on testing out method on datasets with different cross-linkers and with more proteins. Unlike some other methods [Singh et al., 2008, Chu et al., 2010], the approach we have described here will not scale directly to very large databases. If we consider a database of n peptides, then we must consider approximately n2 pairs of peptides. Multiplying by the number of distinct cross-linker locations can quickly lead to a very large database. With this increase in the number of candidates, the search time and discrimination power will be affected.
In this proof-of-concept investigation, we used unoptimized code to carry out the database searches. Accordingly, the search times are quite large—for Section 3.3 approximately one CPU day, and for Section 3.4 approximately seven CPU days. In the former case, much of the running time was devoted to achieving accurate calibration. The requirement of 4000 scores to fit a three-parameter Weibull distribution is quite conservative. If speed is an issue, this requirement could be relaxed, at the expense of higher variance in the resulting q-values. For the negative control experiment, the selection of candidate peptides dominates the search time. This time could be decreased by at least an order of magnitude simply by making use of Crux's existing database indexing scheme [Park et al., 2008]. Thus, through the use of straightforward optimizations of our existing code, scaling up the computations to relatively large complexes should be straightforward.
Of course, as we increase the search space, the discrimination task will also become more difficult. To address these issues, we can employ machine learning methods [Käll et al., 2007, Spivak et al., 2009] to achieve better separation of correct from incorrect identifications.
We did consider several alternative methods for performing the calibration procedure. For example, one could imagine omitting the peptide shuffling procedure and instead extracting a large number of candidate peptides (or peptide pairs) from a large, auxiliary database. This alternative method has the advantage of using a set of decoy sequences that should be more diverse in their amino acid content while having mass closer to the target candidates. This approach will be explored in the future.
Acknowledgments
This work was supported by NIH/NCRR awards R01 EB007057 and P41 RR0011823 as well as NIH/NIAID award 5 U54 AI057141-03.
Footnotes
Based on independent, prior analyses, the location of the cross-linker for the pair (FLEEHPGGEEVLR, YKLCVIPR) was found to be in error in the original manuscript; the correct assignment is (3, 2).
Supporting Information Available: The following supporting information is available at http://noble.gs.washington.edu/proj/xhhc:
- the 10 spectra referred to in Table 1,
- the collection of 3314 spectra from [Singh et al., 2008],
- the sequences of proteins CYP2E1 and b5,
- the results of the large-scale search described in Section 3.3,
- the collection of 35,236 yeast spectra from [Käll et al., 2007], and
- the sequences of the 5 randomly selected yeast proteins used in Section 3.4.
Software for performing the cross-linked search procedure will be made available as part of the Crux software toolkit [Park et al., 2008], available at http://noble.gs.washington.edu/proj/crux.
Contributor Information
Sean McIlwain, Department of Genome Sciences, University of Washington.
Pragya Singh, Department of Medicinal Chemistry, University of Washington.
Paul Draghicescu, Department of Computer Science, University of Washington.
David R. Goodlett, Department of Medicinal Chemistry, University of Washington
William Stafford Noble, Department of Genome Sciences, Department of Computer Science, University of Washington.
References
- Chu F, Baker PR, Burlingame AL, Chakley RJ. Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Molecular and Cellular Proteomics. 2010;9(1):25–31. doi: 10.1074/mcp.M800555-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Eng JK, Fischer B, Grossman J, MacCoss MJ. A fast SEQUEST cross correlation algorithm. Journal of Proteome Research. 2008;7(10):4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- Gao Q, Xue S, Doneanu C, Shaffer S, Goodlett D, Nelson S. Pro-crosslink. software tool for protein cross-linking and mass spectrometry. Analytical Chemistry. 2006;78:2145–2149. doi: 10.1021/ac051339c. [DOI] [PubMed] [Google Scholar]
- Hernandez P, Gras R, Frey J, Appel RD. Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proteomics. 2003;3(6):870–878. doi: 10.1002/pmic.200300402. [DOI] [PubMed] [Google Scholar]
- Hojrup P. Ion Formation from Organic Solvents. Vol. 6. John Wiley & Sons; New York: 1990. pp. 1–66. [Google Scholar]
- Käll L, Canterbury J, Weston J, Noble WS, MacCoss MJ. A semi-supervised machine learning technique for peptide identification from shotgun proteomics datasets. Nature Methods. 2007;4:923–25. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Klammer AA, Park CY, Noble WS. Statistical calibration of the sequest XCorr function. Journal of Proteome Research. 2009;8(4):2106–2113. doi: 10.1021/pr8011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning L, Kasper P, Back J, Nessen M, Vanrobaeys F, Beeumen J, Gherardi E, Koster C, Jong L. Computer assisted mass spectrometric analysis of naturally occurring and artifically introduced cross-links in proteins and protein complexes. FEBS Journal. 2006;273:281–291. doi: 10.1111/j.1742-4658.2005.05053.x. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lackner L, Nunnari J, Phinney B. Shotgun cross-linking analysis for studying quaternary and tertiary protein structure. Journal of Proteome Research. 2007;6:3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Molecular and Cellular Proteomics. 2007;6(12):2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- Park CY, Klammer AA, Käll L, MacCoss MP, Noble WS. Rapid and accurate peptide identification from tandem mass spectra. Journal of Proteome Research. 2008;7(7):3022–3027. doi: 10.1021/pr800127y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rinner O, Seebacher J, Waltzhoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nature Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Row R, Gibson B, Guo X, Young M. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. jasms. 2003;14(8):834–850. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- Seebacher J, Mallick P, Zhang N, Eddes J, Aebsersold R, Gelb M. Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and intergrated computational data processing. Journal of Proteome Research. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Analytical Chemistry. 2008;80(22):8799–8806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak M, Weston J, MacCoss MJ, Noble WS. Direct maximization of protein identifications from tandem mass spectra. 2009 doi: 10.1074/mcp.M111.012161. Manuscript under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society. 2002;64:479–498. [Google Scholar]
- Tang Y, Chen Y, Lichti C, Hall R, Raney K, Jennings S. CLPM: A cross-linked peptide mapping algorithm for mass spectrometric analysis. BMC Bioinformatics. 2005:S9. doi: 10.1186/1471-2105-6-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Tang N, Hempel J, Oshiro C, Taylor E, Kuntz I, Gibson B, Dollinger G. High throughput protein fold identification by using experimental constraint derived from intramolecular cross-links and mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2000 May;97(11) doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]