Abstract
Prostaglandin biosynthesis is catalyzed by two spatially and functionally distinct active sites in cyclooxygenase (COX) enzymes. Despite the crucial role of COXs in biology, molecular details regarding the function and regulation of these enzymes are incompletely defined. Reactive nitrogen species (RNS), formed during oxidative stress, produce modifications that alter COX functionalities and prostaglandin biosynthesis. We previously established that COX-1 undergoes selective nitration on Tyr385 via a mechanism that requires the presence of bound heme cofactor. As this is a critical residue for COX-1 catalysis, nitration at this site results in enzyme inactivation. We now show that occupancy of the COX-1 active site with substrate protects against Tyr385 nitration and redirects nitration to alternative Tyr residues on COX-1, preserving catalytic activity. This study reveals a novel role for substrate in protecting COX-1 from inactivation by nitration in pathophysiological settings.
Keywords: Cyclooxygenase, Arachidonic acid, Eicosapentanoic acid, Prostaglandins, Reactive nitrogen species, Nitric oxide, NSAIDS, Nitrotyrosine
Introduction
Cyclooxygenases (COXs) -1 and -2 are dimeric integral membrane proteins that catalyze the stereospecific conversion of arachidonic acid (AA) into biologically active prostaglandins (PGs). Importantly, PGs play a critical role in mammalian physiology and are intimately linked to the pathophysiology of numerous inflammatory conditions, such as cancer, cardiovascular, neurodegenerative and rheumatoid diseases. The diverse and often opposing biological effects of PGs have propelled structure-based pharmacological studies for the design of safe and effective COX inhibitors, namely nonsteroidal anti-inflammatory drugs (NSAIDs) and selective COX-2 inhibitors that obstruct the binding of substrate and consequently suppress PG biosynthesis1-4.
Mechanisms of COX catalysis remain incompletely understood. At present, it is widely accepted that COXs catalyze two spatially separate and independent reactions that couple through Tyr385, a residue that is strategically positioned to synchronize catalysis between the two active sites5-7. The cyclooxygenase reaction catalyzes the conversion of AA into prostaglandin G2 (PGG2) while the hemeperoxidase reaction reduces PGG2 to the unstable endoperoxide PGH2, the substrate for downstream enzymes in the PG biosynthetic pathway8-10. In addition to substrate bioavailability11, PG biosynthesis appears to be dependent on the cellular oxidant content; a component that is required for oxidizing Fe+3 into a catalytically competent Fe+4 in COX12,13. This catalytic profile of COX is consistent with a coordinated movement of substrate between the cyclooxygenase and the peroxidase site. As a result, structural methods that seek to reveal subtle dynamic changes in protein environment associated with catalysis or ligand binding have been difficult as catalysis is dependent on peroxidase turnover, and is set in motion by very small levels of peroxide and/or hydroperoxide formed in either aerobic or anaerobic settings14. In fact, high-resolution crystallographic depictions of COX as stable substrate-bound or inhibitor-bound COX complexes have shed little light on the sequence of molecular events that culminate in catalysis3,15,16. Although solution state nuclear magnetic resonance (NMR) is an ideal method for obtaining dynamic measurements of proteins in solution, the applicability of this method to COXs has been precluded by the fact that these membrane proteins are stable homodimers of 70 KD subunits and exceed the maximal size limit for effective determination of intact protein structures. Therefore, there is a continuing effort to elucidate structural features in COX that can explain fundamental, yet unresolved aspects of COX catalysis, including the basis for sensitivity, selectivity and affinity of isoform-specific inhibitors17-19, requirements for substrate recognition7,20, interactions between monomeric subunits19,21,22 and the coupling of cyclooxygenase and peroxidase activities via Tyr38523,24.
Another unique characteristic of COXs is its capacity to undergo irreversible self-inactivation during catalysis25, likely via a process following the formation of the catalytically competent ferryl-oxo porphyrin intermediate at the heme peroxidase site and a tyrosyl radical at nearby Tyr3858,26. Radical formation at Tyr385 and alternate sites can also form during oxidative stress27,28. In fact, an oxidative milieu is known to generate reactive nitrogen species (RNS), e.g. peroxynitrite (ONOO-), which engender NO-dependent modifications of Cys and Tyr residues that impact COX activities and overall PG production29-32. Numerous proteins undergo NO-dependent modifications of Cys and Tyr residues in a highly selective manner33-36. While the basis of selectivity remains largely hypothetical and poorly defined, chemical reactivity and the concentration of NO and NO-derived species, as well as structural features of the target protein likely play a dominant role in dictating the functional impact of these modifications. To this end, we have demonstrated that ONOO--induced COX-1 nitration is selectively targeted to Tyr385 by a mechanism that requires bound heme-cofactor37 and results in loss of catalytic function5,37,38. Interestingly, ONOO--induced nitration occurs in the absence of Fe+3 whereby Tyr385 is left unscathed and thus catalytic activity preserved37. We have also shown that Tyr nitration in COX-1 occurs in vivo and is elevated in human and murine atherosclerotic blood vessels29,30,32. To this end, reactions between RNS and COX in its dynamic state may constitute a previously unrecognized pathway to alter PG production in inflammatory disease.
We have employed a multifaceted approach in an attempt to detect subtle dynamic movements in COX-1 that may not be readily accessed using standard X-ray and NMR structure-based approaches. Herein, we used: i) ONOO-, a reactive nitrogen species and source for Tyr nitration of COX, ii) a pan-3-nitrotyrosine antibody to assess global Tyr nitration in COX, of which there are 27 Tyr residues per ovine COX-1 monomer (555 amino acid residues), iii) a site-specific antibody that uniquely recognizes 3-nitro-Tyr385 in COX, iv) HPLC with electrochemical detection (HPLC-EC) for absolute quantification of Tyr nitration in ONOO--treated COX complexes, v) liquid chromatography-tandem mass spectrometry (nLC-MS/MS) to identify specific sites of Tyr nitration in ONOO--treated COX-1, and vi) circular dichroism (CD) spectroscopy to assess conformational changes in ONOO--treated COX-1 complexes. Our findings indicate that although the peroxidase heme plays an obligate role in targeting ONOO--induced Tyr385 nitration in COX-1, active-site channel occupancy allows for allosteric modifications that redirect nitration to alternative Tyr residues while protecting against heme-catalyzed Tyr385 nitration and enzyme inactivation. This paradigm shift suggests that working COX-1 is shielded from inactivation by nitrative modifications and provides new insight into enzyme function.
Methods
Sample preparation
Ram seminal vesicles were obtained from Pel Freeze. Microsomal COX (~ 70 % endogenous FePPCOX-139) and purified COX-1 were prepared as previously described37,40. Tween-20, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), diethyldithiocarbamate (DEDTC), hemin (FePP), 3-nitrotyrosine (3-NT), sodium octanesulfonate, acetonitrile, proteinase K, arachidonic acid (AA; 99% purity), eicosapentanoic acid (EPA; 99% purity), N,N,N’,N’-tetramethylphenylenediamine (TMPD) and all other reagents were obtained from Sigma-Aldrich (minimum 95% purity; HPLC grade). AA and EPA were dissolved in ethanol and stored in aliquots (168 mM and 330 mM respectively) at −80°C. TMPD was prepared fresh at 1 mg/ml for each experiment. N-octyl- β-D-glucopyranoside (βOG) was from Anatrace. Cobalt (III) protoporphyrin IX chloride (CoPP) was purchased from Alexis Biochemicals. Aspirin, indomethacin, and monoclonal COX-1 antibody were purchased from Cayman Chemicals. Aspirin solutions (10 mM) were prepared fresh, dissolved in 70% ethanol then diluted with 0.9% NaCl. Indomethacin solutions (70 μM) were prepared by solubilization in a minimum volume (~ 10 μL) of DMSO followed by dilution with 50 mM Tris, 150 mM NaCl, 0.01 mM EDTA, 20 mM CHAPs, pH 7.4. Sodium peroxynitrite (100 - 200 mM in 4.7% sodium hydroxide) was obtained from Calbiochem, aliquoted, stored at -80°C and used freshly for each reaction. Monoclonal 3-nitrotyrosine antibody was obtained from Upstate Biotechnology. Anti-mouse horseradish peroxidase conjugated IgG and ECL Plus reagents were obtained from GE Healthcare. Materials for SDS-PAGE, Western blotting and anti-rabbit horseradish peroxidase conjugated IgG were obtained from Bio-Rad Laboratories. A custom-made antibody (New England Peptide, Inc.) was raised in rabbits that recognizes nitrated Tyr385 in the sequence (NRIAMEFNHLYNO2HWHPLMPNSF) with >90% sequence homology in COX-1 across species (humans, mice, rat and sheep). This polyclonal anti-3-nitro-Tyr385 COX-1 antibody was found to be selective for 3-nitro-Tyr385 in COX-1 when used for Western blotting and not recognize other proteins (nitrated or non-nitrated) under the conditions employed herein. Furthermore, the antibody preferentially binds the nitrated COX-1 peptide antigen relative to the non-nitrated peptide homolog. The selectivity of this reagent is documented in Supporting Information (Figure S1b).
FePPCOX-1 and CoPPCOX-1 were generated by reconstituting apoCOX-1 with metalloporphyrin (from a 500 μM solution) to a 1:1 molar ratio and measuring absorbance at 412 nm (ε = 1.42 × 105 M-1cm-1) using a Perkin-Elmer Lambda 20 spectrophotometer as described previously37.
Desired concentrations for COX inhibitors (described above) were reacted with COX samples for 40 min prior to performing further reactions. COX reactions with ONOO- were conducted at pH 8 in buffer (100 mM Tris or 20 mM NaHCO3, 0.3 mM DeDTC, 5 mM EDTA, 0.1% Tween or 0.3 % βOG) using concentrated ONOO- (110 mM). The concentration of ONOO- used for each experiment was spectroscopically determined (ε302nm = 1,670 M-1 cm-1).
Activity assays
A peroxidase assay was used to measure COX-1 activity as previously described37,41,42. A detailed description of this assay is provided in Supporting Information.
Quantification of 3-NT using HPLC with electrochemical (HPLC-EC) detection
The protocol for 3-NT quantification by HPLC-EC was previously described43 with modifications. A sample (30 μg of protein) is digested with proteinase K (150 U/mg; 8 hr at 55°C). The cooled digest is precipitated with 3 volumes of ice-cold buffer (0.1 M phosphoric acid and 0.23 M TCA), vortexed, incubated on ice for 5 min, and centrifuged at 12,000 × g for 15 min at 4°C. The resulting supernatant is concentrated at RT (SpeedVac) followed by extraction with 2 volumes of chloroform. The aqueous fraction containing 3-NT is dried at RT (SpeedVac) then reconstituted with vacuum-filtered (0.2-μm nylon membrane) and degassed HPLC mobile-phase buffer containing 90 mM sodium acetate, 35 mM citric acid, 130 μM EDTA, and 460 μM sodium octane sulfonate (pH 4.35) prepared in 18-MΩ resistance water. An isocratic HPLC system with a multichannel electrochemical (EC) CoulArray detector and EC cell (ESA, Inc.) is used to resolve 3-NT (+700 mV, RT = 15 min) from other species using a 100-mm C18 column (Microsorb-MV, Varian) and flow rate at 0.75 ml/min. For pure COX-1 complexes, results are represented as pmol 3NT/mg of COX, i.e. pmol 3-NT/14 × 103 pmol COX.
Western blot analysis
2 - 3 μg of protein/sample was treated with SDS/mercaptoethanol, separated on a 10% acrylamide gel and transferred onto a nitrocellulose membrane (Biorad). Conditions for probing with antibodies and band visualization are described in Supporting Information.
LC-MS/MS analysis and database search
Nanoflow liquid chromatography-tandem mass spectrometry (nLC-MS/MS) was used to identify sites of Tyr nitration in peptides from FePPCOX-1 and AA-bound FePPCOX-1 treated with ONOO- (0.5 mM). Analyses were performed using a 6520 accurate-mass quadrupole-time of flight (Q-TOF) mass spectrometer with a chip cube and C18 column on-chip (Agilent). The mobile phases were 0.1% formic acid in water (solvent A) and 0.1% formic acid in 90% acetonitrile (solvent B). 8 μl of the protein digest (preparation described in Supporting Information) was injected onto a 4 mm 40 nl Zorbax 300SB-C18 enrichment column at a flow rate of 5 μl/min and peptides were resolved on a 0.075 × 43 mm Zorbax 300SB-C18 analytical column (3.5 μm particle size) at a flow rate of 0.3 μl/min with a gradient of 3-50% solvent B for 20 min and 50-90% solvent B for 2 min. Mass spectra were acquired in the automated MS/MS mode, in which MS/MS scans were performed on the four most intense ions from each MS scan. The MS/MS spectra were used to identify the sites of Tyr nitration in COX-1 by a database search using SpectrumMill software (Agilent). Searching parameters were: minimum matched peak intensity of 50%, precursor mass tolerance of 20 ppm and product mass tolerance of 50 ppm. The program was instructed to account for Tyr nitration when matching peptide fragments to COX-1.
Circular dichroism (CD) spectroscopy
Structural perturbations in COX-1 mixtures (0.2 mg/ml) were recorded in the UV region by CD spectroscopy using a Jasco J-720 spectropolarimeter optically reconditioned and upgraded electronically to the J-715 model. Spectral differences in the far UV reflect contributions from the protein peptide backbone (i.e. α-helices, β-sheets, β-turns or random coils), therefore, this region is a gauge of protein structure and conformation. Thus, the CD spectrum of COX-1 which has a very high helical content and almost no β-pleated sheets 15 is characterized by two minima of almost similar intensity at ~222 nm and ~208 nm. Each spectrum represents data accumulated from 190 to 270 nm at 26°C over a 10 min scanning period in a 0.01 cm cuvette (Helma). Data were recorded with a 1 nm bandwidth, 0.1 nm pitch, 1 s response time and 100 nm/min scan speed. All results are reported as molar ellipticities and baseline corrected from buffer background and dilution effects using Jasco Spectra Manager Software.
Statistical analysis
All results were reproduced at least three times. Where appropriate, data are reported as averages ± SEM with significant differences determined by student’s t-Test. A p<0.05 is statistically significant. Image J (version 1.34s; NIH, USA) was used to quantify Western blot band densities in Figures S1b and S3a.
Results
ONOO--induced Structural Perturbations in COX-1 are Driven by Heme
We previously demonstrated that Tyr385 in COX-1 undergoes heme-dependent nitration37. Accordingly, incubation of FePP-containing COX-1 with ONOO- abolishes enzyme activity whereas incubation of apoCOX-1 (FePP-free) with ONOO-, prior to reconstitution with FePP, does not alter AA catalysis. To investigate whether this phenomenon is facilitated by steric perturbations in COX-1, the FePP moiety of COX-1 was substituted with cobalt-protoporphyrin (CoPP) and metalloporphyrin binding characteristics and the structural susceptibility of these complexes to ONOO- was examined. Notably, CoPPCOX-1 is structurally intact but catalytically incompetent16. As shown in Figure 1a, the absorbance spectrum of stoichiometrically reconstituted FePPCOX-1 (~1:1 molar ratio) includes a characteristic Soret band at ~412 nm. Likewise, reconstituted CoPPCOX-1 (Figure 1b) gives rise to a dominant Soret absorbance band (shifted to ~425 nm) that closely resembles native-like reconstituted FePPCOX-1 in its spectral properties. However, the resulting change in the heme electronic environment when Fe+3 is substituted with Co+3 prevents CoPPCOX-1 from initiating catalysis.
Figure 1.
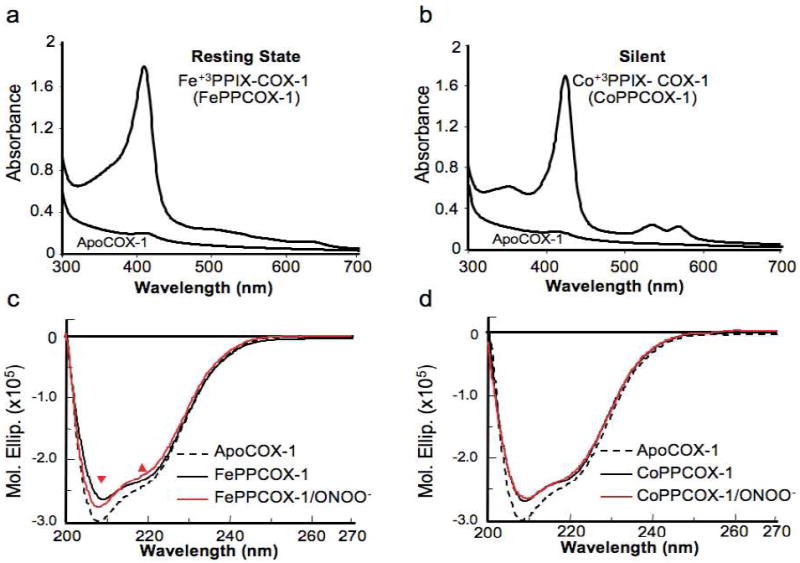
ONOO--induced structural perturbations in COX-1 are driven by heme. (a) Absorbance spectra were acquired for the reconstitution of apoCOX-1 (0.21 mg/ml) at 25°C with Fe+3PPIX (heme; FePP) and (b) Co+3PPIX (CoPP) from stock solutions (500 μM each). (c) Circular dichroism (CD) spectra acquired in the far UV region at 25°C compare the structure of apoCOX-1 (dashed) to FePPCOX-1 (solid black). The red CD spectrum was acquired following incubation of FePPCOX-1 with ONOO- (250 μM). (d) CD spectra were acquired as in (c), but for COX-1 reconstituted with CoPP.
COX-1 complexes in the absence of substrate were further analyzed by CD spectroscopy in the far UV region to gauge secondary structural movements associated with ligand binding. As shown in Figure 1c, CD spectral characteristics of FePPCOX-1 are markedly different from those of apoCOX-1 at equimolar concentrations. While the shape of both spectra, with two minima of almost identical intensities at 208 nm and 222 nm, suggest a dominant α-helical motif in both apoCOX-1 and FePPCOX-1, the increased total signal intensity in apoCOX-1 (a more negative signal) and the sharper minimum at 208 nm relative to 222 nm suggest helical disorder in the absence of bound FePP. ONOO--treatment of FePPCOX-1 induced a CD spectral change more closely resembling that of pure apoCOX-1 with predominant spectral band differences marked in Figure 1c by red arrows. Of note, spectral characteristics of apoCOX-1 were unaltered following exposure to ONOO-. In contrast to FePPCOX-1, the CD spectrum for ONOO--treated CoPPCOX-1 was indistinguishable from untreated CoPPCOX-1 (Figure 1d), clearly indicating that global ONOO--induced secondary structural perturbations in COX-1 are heme-dependent (i.e. require Fe+3 rather than Co+3 occupancy of protoporphyrin IX).
Substrate Identity Dictates ONOO--induced Structural Changes in FePPCOX-1
AA (ω-6 eicosatetraenoic acid) and EPA (ω-3 eicosapentanoic acid) are 20-carbon fatty acids that differ only in their double bond content (four vs. five, respectively). Of note, mediators derived from ω-3 fatty acids (such as EPA) are endogenous attenuators of inflammation and elicit cardioprotective actions in humans44. Whereas AA is the principal substrate of COX-1, EPA is a relatively poor substrate, owing to the double bond between C17 and C18 which creates a poor fit within the COX-1 active-site channel45 (Figure 2a). Furthermore, EPA oxygenation by COX-1 requires more hydroperoxide for the initiation of catalysis than does AA46. To this end, COX-1 peroxidase activity assays indicated that at the same concentration, EPA is 3-fold less active as a substrate compared with AA, using either purified FePPCOX-1 or native microsomal COX-1 (Figure 2a).
Figure 2.
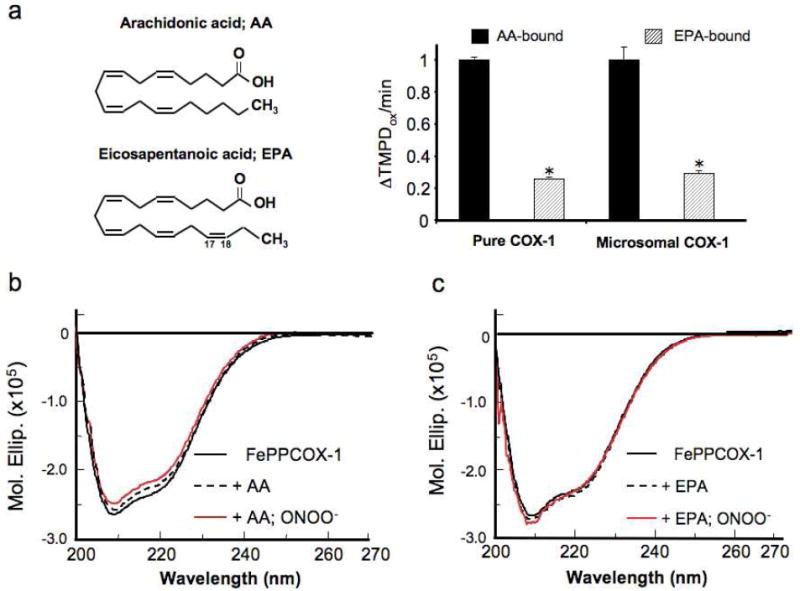
Substrate identity dictates ONOO--induced structural changes in FePPCOX-1. (a) Cyclooxygenase-coupled peroxidase activity measured in purified FePPCOX-1 (2 μg) and microsomal COX-1 (10.5 μg) following the immediate addition of arachidonic acid (AA; 100 μM) and eicosapentanoic acid (EPA; 100 μM) at pH 8. Results are monitored as ΔA611 during a 60 s interval. Data are averages ± SEM. *p < 0.0002 for EPA-activation relative to AA-activation. (b) CD spectra acquired in the far UV region at 25 °C compare the structure of FePPCOX-1 (0.21 mg/ml; solid black) to AA-bound FePPCOX-1 (dashed). The red CD spectrum was acquired following incubation of AA-bound FePPCOX-1 with ONOO- (250 μM). (c) CD spectra were acquired similarly to (b) but for EPA-bound FePPCOX-1.
Far UV CD spectral characteristics for substrate-bound FePPCOX-1 complexes were compared and analyzed for secondary structural changes following ONOO--incubation. Despite its initiation of catalysis, binding of AA to FePPCOX-1 caused very slight spectral changes relative to unbound FePPCOX-1 over a 20 m scanning period (Figure 2b). While incubation of AA-bound FePPCOX-1 with ONOO- caused an additional attenuation in signal intensity (Figure 2b), the observed change in AA-bound FePPCOX-1 was minimal compared to AA-free FePPCOX-1 (Figure 1c). Interestingly, neither EPA binding nor ONOO--treatment of EPA-bound FePPCOX-1 generated a marked spectral perturbation relative to substrate-free FePPCOX-1 (Figure 2c). We confirmed that the observed spectral changes were not an artifact of negligible dilution arising from substrate addition; and, that substrate occupancy of the COX-1 active site indeed determines the extent of ONOO--induced structural alterations.
Arachidonic Acid Binding Redirects ONOO--induced COX-1 Nitration
Conversion of AA to eicosanoid products by COXs require both cyclooxygenase and peroxidase functionalities. Previously, we demonstrated that ONOO- promotes heme-catalyzed Tyr385 nitration and subsequent inactivation of FePPCOX-137. While global COX-1 nitration was also observed in the absence of heme (apoCOX-1), the catalytically essential Tyr385 residue did not undergo nitration and thus activity was preserved. To determine whether substrate binding sterically hinders ONOO- access to Tyr385 for heme-catalyzed nitration, we compared the susceptibility of AA-incubated (active) and AA-free (resting) COX-1 to global and Tyr385-specific nitration by ONOO-. HPLC-EC and Western blot analysis with an anti-nitrotyrosine (anti-3-NT) antibody were used to measure global levels of ONOO--induced protein Tyr-nitration in the presence and absence of substrate, in two types of COX-1 complexes: 1) FePPCOX-1 which possesses both cyclooxygenase and peroxidase functionalities and is capable of metabolizing substrate, and 2) CoPPCOX-1, which possesses only peroxidase activity and is incapable of metabolizing substrate. Of note, the use of HPLC-EC for measurement of global protein 3-NT levels circumvents the detection of false signals that can arise from non-specific interactions involving the anti 3-NT antibody or diminished signals that can arise from COX-1 multimer formation under certain reaction conditions (described in Figure 6). In the absence of substrate, global Tyr nitration was reduced by about 3-fold in CoPPCOX-1 relative to FePPCOX-1, as measured by HPLC-EC (Figure 3a). Consistent with this HPLC-EC finding, Tyr nitration was also observed by Western blotting of both ONOO--treated FePPCOX-1 and CoPPCOX-1 and levels were markedly reduced when Fe+3 was substituted with Co+3 (Figure 3b). Of note, Western blotting results indicate that AA-binding enhanced total levels of nitration in ONOO--treated COX-1 (Figure 3b). Additional aliquots of these samples were probed by Western blotting with a selective anti-3-nitro-Try385 antibody. While AA-binding to FePPCOX-1 was associated with a global increase in ONOO--induced FePPCOX-1 nitration, this was accompanied by a remarkable decrease in Tyr385-specific nitration (Figure 3c). Substitution of Fe+3 with Co+3 had little impact on levels of Tyr385 nitration in AA-bound COX-1 (Figure 3c) emphasizing the fundamental role heme plays in directing this modification. Notably, HPLC-EC results showed an apparent slight decrease in global Tyr nitration of AA-bound CoPPCOX-1 relative to CoPPCOX-1 (Figure 3a) while Western blotting suggested the opposite result (Figure 3b). These fluctuations were not statistically significant.
Figure 6.
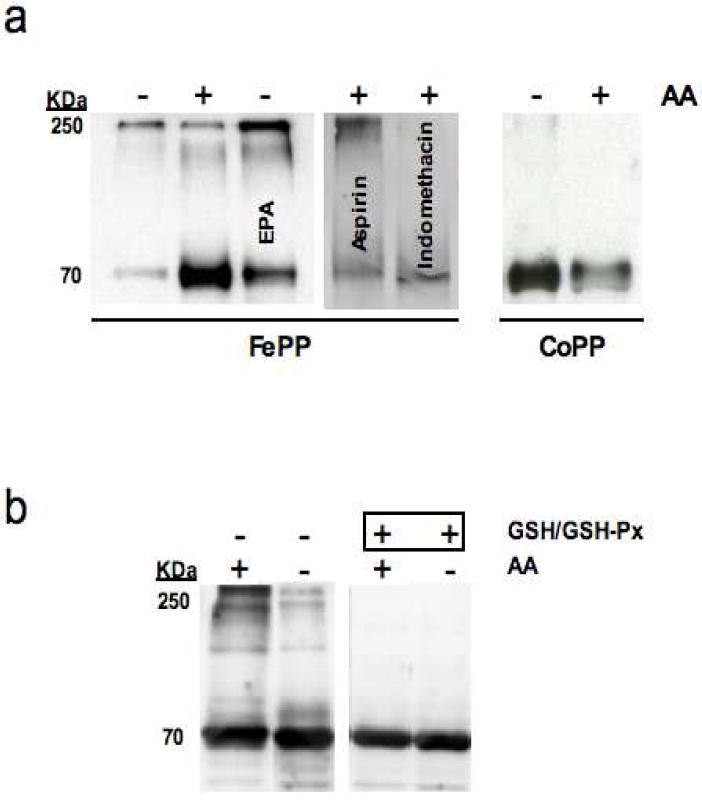
ONOO- is a stimulus for heme-dependent COX-1 multimer formation. Western blots for the reaction of reconstituted COX-1 (0.12 mg/ml) with ONOO- (250 μM) in the presence (+) and absence (-) of (a) arachidonic acid (AA; 100 μM), eicosapentanoic acid (EPA; 100 μM), aspirin (70 μM), and indomethacin (14 μM) and (b) AA (100 μM) and glutathione (GSH; 0.25 mM) in combination with glutathione peroxidase (GSH-Px: 25 U/ml). Blots were probed with monoclonal anti-nitrotyrosine (anti-3-NT).
Figure 3.
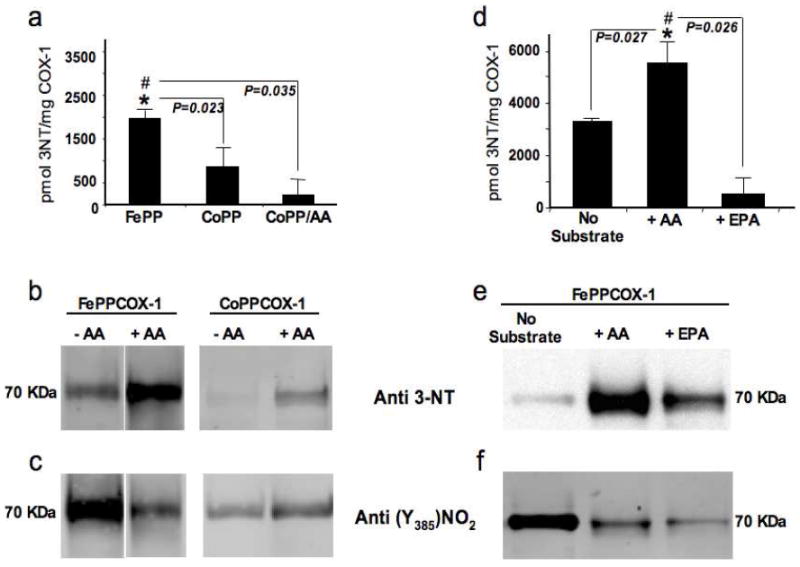
Arachidonic acid (AA) binding redirects ONOO--induced COX-1 nitration. In all reactions, COX-1 complexes were at 0.21 mg/ml, AA was at 100 μM and ONOO- was at 250 μM. (a) Quantification of total 3-NT by HPLC-EC for the reaction of FePPCOX-1 and CoPPCOX-1 with ONOO-. Total 3-NT for the reaction of AA-bound CoPPCOX-1 with ONOO- was also quantified. Data are averages ± SEM. *p ≤ 0.023 for FePPCOX-1 relative to CoPPCOX-1. #p ≤ 0.035 for AA-bound CoPPCOX-1 relative to FePPCOX-1. (b) Western blots compare the reactions of FePPCOX-1 and CoPPCOX-1 with ONOO- ± AA. Blots were probed with monoclonal anti-3-NT and with (c) polyclonal anti-3-nitro-Tyr385-peptide in COX-1. (d) Quantification of total 3-NT by HPLC-EC for the reaction of FePPCOX-1 with ONOO- ± AA and eicosapentanoic acid (EPA; 100 μM). Data are averages ± SEM. *p ≤ 0.027 for AA-bound FePPCOX-1 relative to substrate-free FePPCOX-1. #p ≤ 0.026 for EPA-bound FePPCOX-1 relative to AA-bound FePPCOX-1. (e) Western blots for the reaction of FePPCOX-1 with ONOO- ± AA and EPA. Blots were probed similarly to (b) and (c).
Interestingly, substitution of AA with EPA in these complexes was associated with a reduced susceptibility to nitration (Figure 3d-3f). Whereas AA-binding elicited a ~2-fold increase in ONOO--induced FePPCOX-1 nitration, EPA-binding significantly diminished ONOO--induced nitration of this complex (Figure 3d). This HPLC-EC finding of decreased total nitration of COX-1 by ONOO- when occupied by EPA, compared with AA was further corroborated by Western blotting (Figure 3e). Importantly, both AA- and EPA-binding within the active-site channel of COX-1 were associated with specific protection of the catalytically-essential Tyr385 from ONOO--induced nitration (Figure 3f). Thus, strong levels of global Tyr nitration are observed with AA as substrate (active state) while robust Tyr385 nitration is observed in the resting state of COX-1 when substrate does not occupy the active site channel. These findings indicate that both AA and EPA binding to COX-1 may prevent access of the active site Tyr385 to nitration, while concomitantly redirecting nitration to other Tyr residues in COX-1, presumably via a substrate-induced allosteric interaction. The ability of substrate occupancy of the active site to prevent Tyr385 from nitration (i.e., prevention of an inactivating process) and redirect nitration to other Tyr residues was similarly found in studies of crude native COX-1 (i.e. sheep microsomes), where the heme moiety is endogenously bound in the COX-1 peroxidase site (Supporting Information; Figure S1).
Evidence for Specificity in ONOO- Induced Nitration of COX-1
To confirm that AA binding is associated with a significant increase in ONOO--induced global nitration of FePPCOX-1, despite protection against Tyr385 nitration, we sequence-mapped Tyr-nitration in reaction mixtures by nLC-MS/MS. We had previously demonstrated by mass spectral studies that ONOO--treatment of FePPCOX-1 caused selective heme-driven nitration of Tyr385 that was absent in apoCOX-137. Herein, we have extended these studies to investigate the impact of AA-binding in COX-1. Figure 4a depicts overlays of deconvoluted extracted ion chromatograms (EICs) with a mass-to-charge ratio (m/z) of 649.06 for ONOO--treated FePPCOX-1 in the presence and absence of AA. The identified peptide ion, which elutes at ~15 min, represents the 649.06 m/z peptide from substrate-free FePPCOX-1 which is nitrated at Tyr385 (377-IAMEFNQLYNO2HWHPLMPDSFR-396), establishing that Tyr385 is indeed susceptible to nitration when AA is absent (Fig. 4a; top EIC). However, this peptide ion was not detected in ONOO--treated AA-bound FePPCOX-1 (Figure 4a; lower EIC), indicating that AA selectively prevents Tyr385 nitration. Notably, identification of the m/z 649.06 peptide ion as containing nitrated Tyr385 was confirmed by collision-induced-dissociation and analysis of y- and b- production fragments (Figure 4b). As expected, untreated FePPCOX-1 in the presence or absence of AA did not result in a m/z 649.06 peptide ion, in accord with a lack of Tyr385 nitration. Additionally, the peptide ion containing unmodified Tyr385 (i.e., Ile377-Arg396) could be detected at m/z 634.30 in all samples. Together, these results demonstrate that COX-1 occupancy with AA selectively protects against heme-catalyzed Tyr385 nitration, while promoting collateral nitration.
Figure 4.
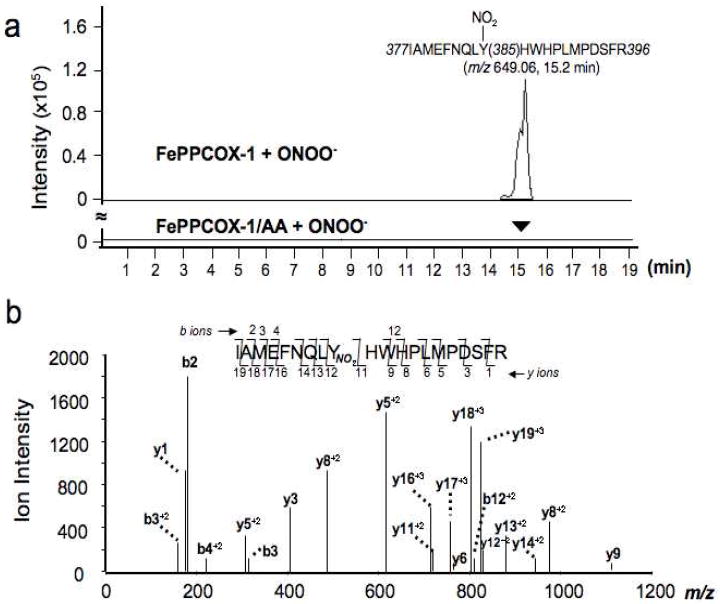
Evidence for specificity in ONOO- induced nitration of COX-1. FePPCOX-1 and AA-bound FePPCOX-1 (0.21 mg/ml each) were incubated with ONOO- (500 μM) for 1 h at RT. Trypsinized reaction mixtures were analyzed by nLC-MS/MS as described in Methods. (a) Overlays of deconvoluted extracted ion chromatograms that monitor the elution of the tryptic peptide fragment (m/z 649.06) representing the quadruply-charged peptide ion containing nitrated Tyr385. The arrow at ~15 min in the lower elution profile denotes the missing peptide of m/z 649.06 for AA-bound FePPCOX-1 treated with ONOO- that is present in the top elution profile of ONOO--treated FePPCOX-1. (b) MS/MS analysis of the peptide ion eluting at ~15 min from ONOO--treated FePPCOX-1 for confirmation of peptide identity. Daughter ions that arise from the parent ion m/z 649.06 are depicted.
An additional COX-1-derived peptide, containing nitrated Tyr254 (254-YNO2QMLNGEVYPPSVEEAPVLMHYPRG-278), was present at similar levels in the absence and presence of AA in ONOO--treated FePPCOX-1, eluting at ~14 min with m/z 955.79 (Supporting Information; Figure S2). The non-selectivity of ONOO--induced Tyr254 nitration is not surprising given the fact that it is surface exposed as compared to Tyr385, which is buried in the enzyme core. Of note, treatment of AA-bound FePPCOX-1 with increasing concentrations of ONOO- (0.5 mM - 2 mM) enhanced levels of methionine oxidation (detected residues include Met256, Met273, Met522, and Met525) and Tyr254 nitration relative to substrate-free FePPCOX-1.
A body of work exists on the chemical reactivity of nitrosocarbonate, the reaction product of ONOO- with carbon dioxide and a powerful nitrating agent that can form under physiological bicarbonate levels47,48. Thus, we repeated experiments in the presence of millimolar bicarbonate levels to determine whether nitration patterns could be modified under these conditions. Western blotting results demonstrate a remarkable increase in nitration levels of apoCOX-1 as well as FePPCOX-1 in bicarbonate buffer relative to Tris buffer. Importantly, the increase in apoCOX-1 nitration in the presence of bicarbonate did not interfere with the enzyme’s ability to reconstitute heme and regain full catalytic function (Supporting Information; Figure S3). These results support our findings that despite extensive increases in COX-1 nitration and oxidation under certain conditions, only nitration at Tyr385 results in loss of catalytic function. Additionally, substrate-occupancy and ensuing allosteric consequences determine the sites and extent of COX-1 nitration by ONOO-.
COX Inhibitors Attenuate ONOO--induced Nitration and Prevent Conformational Changes in COX-1
We then investigated whether COX-1 active site occupancy by pharmacological inhibitors alter global COX-1 nitration and protect against heme-catalyzed Tyr385 nitration. Levels of ONOO--induced FePPCOX-1 nitration were compared following AA addition, in the presence and absence of aspirin or indomethacin. HPLC-EC quantification of global COX-1 nitration levels showed a ~6-fold reduction in aspirin-pretreated FePPCOX-1 and a ~10-fold reduction in indomethacin pre-treated FePPCOX-1, relative to FePPCOX-1 in the presence of AA alone (Figure 5a). Consistent with HPLC-EC measurements, global nitration levels were markedly reduced in aspirin- and indomethacin-pretreated COX-1 in both the presence and absence of AA, as shown by Western blotting (Figure 5b). Notably, these reagents suppressed tyrosine nitration in a non-specific manner as demonstrated by their ability to alter ONOO--induced nitration levels in bovine serum albumin, an un-related protein (Supporting Information; Figure S4a). Interestingly, while aspirin pretreatment of FePPCOX-1 protected Tyr385 from ONOO--induced nitration, indomethacin pre-treatment did not attenuate nitration of Tyr385 (Figure 5b).
Figure 5.
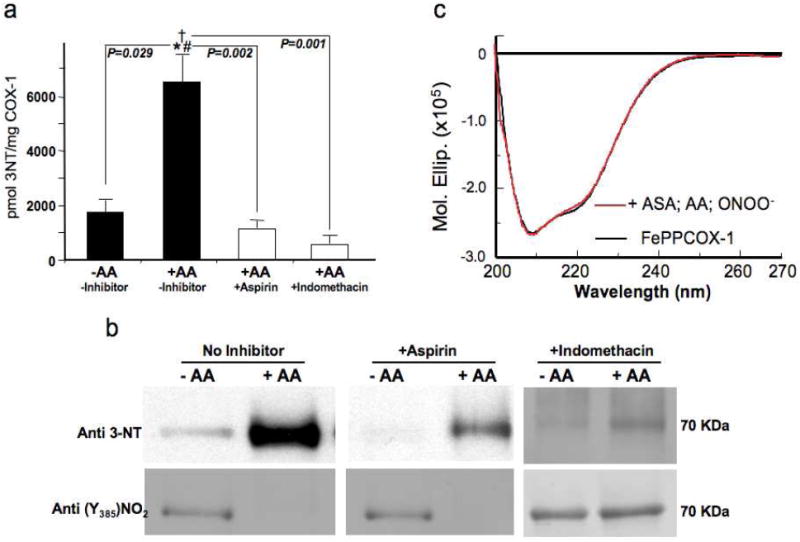
COX inhibitors attenuate ONOO--induced nitration and prevent conformational changes in COX-1. In all reactions, COX-1 complexes were at 0.21 mg/ml and ONOO- was at 250 μM (a) Quantification of total 3-NT by HPLC-EC for the reaction of FePPCOX-1 with ONOO- ± arachidonic acid (AA; 100 μM) and the COX inhibitors aspirin (70 μM) and indomethacin (14 μM). Data are averages ± SEM. *p ≤ 0.029 for AA-bound FePPCOX-1 relative to substrate-free FePPCOX-1. #p ≤ 0.002 for aspirin-treated FePPCOX-1 relative to AA-bound FePPCOX-1. †p ≤ 0.001 for indomethacin-treated FePPCOX-1 relative to AA-bound FePPCOX-1. (b) Western blots for the reaction of FePPCOX-1 with ONOO- ± AA (100 μM) and the COX inhibitors aspirin (70 μM) and indomethacin (14 μM). Blots were probed with monoclonal anti-3-NT (top) and with polyclonal anti-3-nitro-Tyr385-peptide in COX-1 (bottom). (c) CD spectra recorded in the far UV region at 25°C compare the structure of FePPCOX-1 (black) to aspirin-treated FePPCOX-1 following reaction with ONOO-.
Structural evaluation by CD spectroscopy showed that aspirin-pretreated FePPCOX-1 is resistant to ONOO--induced structural perturbations, relative to untreated FePPCCOX-1 (Figure 5c). Binding of aspirin to FePPCOX-1 in the absence of ONOO--treatment did not result in detectable spectral changes relative to untreated FePPCOX-1 (Supporting Information; Figure S4b). Solutions containing indomethacin contributed interference in the far UV region, which prevented the acquisition of CD spectra. Nevertheless, COX-1 occupancy by aspirin appears to limit the access of ONOO- into the active-site channel, thereby protecting against heme-catalyzed Tyr385 nitration.
ONOO- is a Stimulus for Heme-dependent COX-1 Multimer Formation
We previously demonstrated that treatment of purified and reconstituted FePPCOX-1 with ONOO- results in the appearance of high molecular weight bands at ~200-250 KDa (when probed with an anti-3-NT Ab), suggesting the presence of nitrated COX-1 multimers which were absent in ONOO--treated apoCOX-137. Furthermore, substrate-binding to FePPCOX-1 amplified nitration levels at an apparent mass of 70 KDa as well as in higher molecular weight COX-1 species37. Although EPA binding to FePPCOX-1 attenuated total nitration by ONOO- relative to AA binding (at 70 KDa), the presence of nitrated COX-1 multimers were similarly observed with EPA and AA (Figure 6a). On the other hand, aspirin, and to a greater extent indomethacin, reduced the levels of AA-engendered nitrated COX-1 multimers (Figure 6a). Interestingly, treatment of reaction mixtures with a hydroperoxide scavenging system (glutathione and glutathione peroxidase), abolished the formation of COX-1 multimers (Figure 6b). Collectively, these findings suggest that exposure of FePPCOX-1 to peroxide, generated from catalysis or by the direct addition of ONOO-, promotes COX-1 multimer formation. Notably, nitrated multimers were absent in ONOO--treated FePPCOX-1 when using bicarbonate buffer (Supporting Information Figure S3a). Furthermore, substitution of Co+3 for Fe+3 in protoporphyrin IX of ONOO--treated COX-1 abrogated the formation of nitrated COX-1 multimers in the presence and absence of AA (Figure 6a), consistent with the absence of multimers in apoCOX-137, thereby implicating heme as a mediator of COX-1 multimer formation.
Discussion
Reactive nitrogen species (RNS)-dependent posttranslational modifications of proteins are increasingly appreciated for their potential to alter protein and cellular functions. The results presented herein demonstrate that COX nitration occurs in a manner in which substrate binding determines the modification site and extent, as well as degree to which catalytic function is consequently lost. Furthermore, we demonstrate the use of nitration as a probe for allosteric changes in COX-1 protein structure.
The heme moiety in the solvent exposed peroxidase site of COXs can be easily replaced with other metalloporphyrins e.g. CoPP, to yield structurally native-like complexes but with altered functionality16. Additionally, the cyclooxygenase active site channel can accommodate an array of chemically distinct fatty acid substrates and NSAIDs that produce COX complexes with or without productive catalysis2,3. It is likely that some degree of molecular motion is associated with ligand binding regardless of whether the end result is activation or inhibition. Although crystallographic images do not reveal tangible structural variations among the various COX complexes, it is inferred that the entry site to the hydrophobic channel may undergo conformational changes that allow for substrate binding and product exit, or may exhibit structural rigidity as a result of inhibitor binding3,49.
By using a transient pulse of ONOO- as a reactive probe that accesses unhindered sites on COX-1 and can be detected as stable covalent modifications, we could distinguish very subtle changes in secondary structure with CD spectroscopy. Notably, in the absence of ligand binding, ONOO- resulted in heme-dependent structural perturbations that were undetectable when COX-1 heme was substituted with CoPP (Figure 1). The observed spectral distortions in resting state heme-bound COX may indicate helical flexibility at the peroxidase site that could arise from heme alterations by the oxidant ONOO-37. Interestingly, COX-1 adopted a more rigid helical structure with ligand binding in the active site channel (Figures 2 and 5). Whereas aspirin inhibition and binding of a poorly metabolized substrate (EPA) resulted in no apparent ONOO--induced structural perturbations, binding of AA, the preferred COX substrate, resulted in a very modest but reproducible spectral change that may suggest protein aggregation following catalysis. Thus, the identity of the COX-1 ligand, either substrate or inhibitor, confers rigidity to the molecule that is detectable by CD spectroscopy following exposure to ONOO-.
The susceptibility of Tyr residues to nitration due to peroxidase metal and ligand identity was also determined (Figure 3). Substitution of Fe+3 with Co+3 significantly diminished global levels of Tyr nitration that were further reduced by substrate binding. The static nature of substrate-bound CoPPCOX-1 likely limits ONOO- access to Tyr sites. In contrast, occupancy of the COX-1 channel with AA, a catalytically productive complex, generates the highest levels of ONOO--induced Tyr nitration. A lower amount of Tyr nitration was observed with EPA-bound FePPCOX-1 relative to AA-bound FePPCOX-1, where the substrate is misaligned in the COX channel and the complex is catalytically incompetent46. Surprisingly, our results demonstrate that while Tyr385, residing in the conduit between peroxidase and cyclooxygenase functionalities, is a target for nitration in the absence of substrate binding, AA protects Tyr385 from nitration. We further found that AA binding protects COX-1 from inactivation by ONOO- by redirecting the nitrative insult away from Tyr385 and towards alternate Tyr residues on COX-1. This is evident from the increase in global Tyr nitration coupled with a specific decrease in Tyr385 nitration during AA-initiated catalysis (Figures 3 and 4). Additionally, while the powerful and physiological nitrating agent nitrosocarbonate remarkably increased nitration levels in COX-1, it did not prevent heme reconstitution and regeneration of full catalytic function (Figure S3). As expected, inhibition of COX function with aspirin and indomethacin attenuated global ONOO--induced nitration (Figure 5). While aspirin protected Tyr385 from nitration, this effect was not observed for indomethacin for reasons that are unclear. Of note, aspirin and NSAIDs have been previously shown to display antioxidant properties and possess the ability to scavenge reactive oxygen species50-52. Thus, a reduction in global COX-1 nitration in the presence of NSAIDs may be due to ONOO- scavenging, a phenomenon that was underlined by the ability of aspirin to reduce global nitration in bovine serum albumin, a non-relevant protein (Figure S4). Interestingly aspirin, but not other NSAIDs, has been shown to trigger cell-cell interactions that can shunt arachidonic acid metabolism and form novel anti-inflammatory mediators53.
The specificity and differential nitration observed for ONOO--induced Tyr385 nitration, a residue that is buried in the enzyme core, was not observed for any of the other Tyr residues in COX-1. By mass spectrometry, we identified and determined that the surface exposed Tyr254 is almost equally nitrated in substrate-bound and substrate-free COX-1 complexes (Figure S2). Thus, COX-1 conformation, Tyr residue environment, steric hindrance to nitrating agent and degree of solvent exposure, all emerge as physical parameters that determine the specificity of nitration in COX-1, and ultimately characteristics of enzymatic function.
Previously, we demonstrated that heme catalyzed the formation of ONOO--induced nitrated COX-1 multimers but their levels were increased with AA binding by a mechanism that remains unclear37. In this study, nitrated multimers were also observed with EPA binding and were abrogated when the heme moiety was replaced by CoPP (Figure 6). Aspirin, and to a greater extent indomethacin, also reduced the levels of nitrated multimers. Of note, heme catalyzed COX-1 multimer formation was observed with bound substrate and in the absence of ONOO--treatment. We can rule out disulfide formation as the mechanism for this observed cross-linking in substrate-bound samples since all samples were treated with a thiol reductant prior to Western blotting. As treatment of reaction mixtures with a hydroperoxide scavenging system abolished the formation of COX-1 multimers, we can conclude that exposure of COX-1 to peroxide generated from either catalysis or by the direct addition of ONOO-, may be a stimulus for COX-1 multimer formation in the presence of heme (Figure 6). An alternative process leading to aggregate formation could involve dityrosine cross-linking, which has been observed in proteins identified in neurodegenerative disorders54. Nevertheless, as the identity of substrate did not modify COX-1 aggregates and bicarbonate attenuated the capacity of these multimers to form, it remains unclear whether high molecular weight COX forms contribute to enzymatic deactivation or other pathophysiological events in vivo.
Conclusion
We and others have previously demonstrated that ONOO- can provide the peroxide tone for heme activation of COX12,13. Subsequent results suggested that ONOO- can inactivate COX-1 catalysis by nitration37. We now know that substrate can regulate the access of inactivating nitrating species to COX-1. While heme-driven nitration of the centrally located Tyr385 by ONOO- in substrate-free COX inhibits all catalytic function, AA-bound COX is structurally and functionally preserved with nitration directed away from Tyr385 and to alternate Tyr sites. EPA similarly protects Tyr385 nitration, albeit with attenuated catalytic activity and distinct product generation versus that observed with AA. Alternatively, NSAIDs inhibit catalysis and prevent Tyr385 nitration, yet these resultant COX-inhibitor complexes retain their peroxidase activity. During inflammatory conditions, ONOO- formation is enhanced, increasing the biological relevance of interactions between reactive nitrogen species (RNS) and COX enzymes that appear to be directed by the bioavailability of substrate and/or the presence of inhibitors. A relevant paradigm has been shown in cardiovascular disease models whereby the bioavailability of functional NO is decreased and may be compensated for by an elevation in COX function and a concomitant increase in vasorelaxant prostanoid synthesis (i.e. prostacyclin)55,56. It is known that blood vessels are prone to alterations in NO chemistry in the setting of chronic inflammation whereby reactive oxygen species (ROS; i.e. superoxide) production is elevated and may consume NO to generate RNS (e.g. N2O3, NO2 and ONOO-). This study reveals a novel role for substrate in protecting COX-1 from inactivation by nitration in pathophysiological settings.
Supplementary Material
Acknowledgments
We are grateful to Ms. Barbara Summers, Mr. Albert Morrishow and Dr. Quiying Chen for expert technical assistance, and to Dr. Scott Blanchard for his critical review of the manuscript. This work was supported by National Institutes of Health Grants to D.P.H. (P01 HL046403, R01 HL091101 and 5T-32 HL07423), National Institutes of Health Grants to S.S.G. (P01 HL46403 and R01 HL80702), a Grant-in-Aid from the American Heart Association to R.S.D. (AHA655783T), a Pfizer award to R.S.D. (Pfizer Inc.), a predoctoral fellowship from the National Institute of Health to T.N. (F31 AG32195), a Philip Morris USA Inc. and Philip Morris International award to R.K.U, an Alice Bohmfalk Charitable Trust award to R.K.U, and grants to D.P.H. from the Julia and Seymour Gross Foundation, and by the Abercrombie Foundation.
Footnotes
Supporting Information Available. Supporting information includes an expansion of methods essential to experimental reproducibility as well as additional results that will allow the presentation of all findings that are essential to conclusions of the study. This information is available free of charge via the internet at http://pubs.acs.org/.
References
- 1.Moncada S. Arterioscler Thromb Vasc Biol. 1982;2:193–207. [Google Scholar]
- 2.Kurumbail RG, Kiefer JR, Marnett LJ. Curr Opin Struct Biol. 2001;11:752–60. doi: 10.1016/s0959-440x(01)00277-9. [DOI] [PubMed] [Google Scholar]
- 3.Garavito RM, Malkowski MG, DeWitt DL. Prostaglandins Other Lipid Mediat. 2002;68-69:129–152. doi: 10.1016/s0090-6980(02)00026-6. [DOI] [PubMed] [Google Scholar]
- 4.McGiff JC. Pharmacol Rep. 2006;58(Suppl):47–51. [PubMed] [Google Scholar]
- 5.Shimokawa T, Kulmacz RJ, De Witt DL, Smith WL. J Biol Chem. 1990;265:20073–20076. [PubMed] [Google Scholar]
- 6.Smith WL, Song I. Prostaglandins Other Lipid Mediat. 2002;68-69:115–128. doi: 10.1016/s0090-6980(02)00025-4. [DOI] [PubMed] [Google Scholar]
- 7.Kozak KR, Prusakiewicz JJ, Rowlinson SW, Prudhomme DR, Marnett LJ. Biochemistry. 2003;42:9041–9. doi: 10.1021/bi034471k. [DOI] [PubMed] [Google Scholar]
- 8.Dietz R, Nastainczyk W, Ruf HH. Eur J Biochem. 1988;171:321–328. doi: 10.1111/j.1432-1033.1988.tb13793.x. [DOI] [PubMed] [Google Scholar]
- 9.Smith WL, Marnett LJ. Biochim Biophys Acta. 1991;1083:1–17. doi: 10.1016/0005-2760(91)90119-3. [DOI] [PubMed] [Google Scholar]
- 10.Tsai AL, Palmer G, Kulmacz RJ. J Biol Chem. 1992;267:17753–17759. [PubMed] [Google Scholar]
- 11.Marshall PJ, Kulmacz RJ, Lands WE. J Biol Chem. 1987;262:3510–3517. [PubMed] [Google Scholar]
- 12.Landino LM, Crews BC, Timmons MD, Morrow JD, Marnett LJ. Proc Natl Acad Sci U S A. 1996;93:15069–15074. doi: 10.1073/pnas.93.26.15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Upmacis RK, Deeb RS, Hajjar DP. Biochemistry. 1999;38:12505–12513. doi: 10.1021/bi983049e. [DOI] [PubMed] [Google Scholar]
- 14.Smith WL, Eling TE, Kulmacz RJ, Marnett LJ, Tsai A. Biochemistry. 1992;31:3–7. doi: 10.1021/bi00116a001. [DOI] [PubMed] [Google Scholar]
- 15.Picot D, Loll PJ, Garavito RM. Nature. 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 16.Malkowski MG, Theisen MJ, Scharmen A, Garavito MR. Arch Biochem Biophys. 2000;380:39–45. doi: 10.1006/abbi.2000.1906. [DOI] [PubMed] [Google Scholar]
- 17.Harman CA, Turman MV, Kozak KR, Marnett LJ, Smith WL, Garavito RM. J Biol Chem. 2007;282:28096–105. doi: 10.1074/jbc.M701335200. [DOI] [PubMed] [Google Scholar]
- 18.Tosco P, Lazzarato L. Chem Med Chem. 2009;4:939–45. doi: 10.1002/cmdc.200800438. [DOI] [PubMed] [Google Scholar]
- 19.Prusakiewicz JJ, Duggan KC, Rouzer CA, Marnett LJ. Biochemistry. 2009;48:7353–5. doi: 10.1021/bi900999z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chubb AJ, Fitzgerald DJ, Nolan KB, Moman E. Biochemistry. 2006;45:811–20. doi: 10.1021/bi051973k. [DOI] [PubMed] [Google Scholar]
- 21.Yuan C, Rieke CJ, Rimon G, Wingerd BA, Smith WL. Proc Natl Sci U S A. 2006;103:6142–6147. doi: 10.1073/pnas.0601805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Seibold SA, Rieke CJ, Song I, Cukier RI, Smith WL. J Biol Chem. 2007;282:18233–44. doi: 10.1074/jbc.M701235200. [DOI] [PubMed] [Google Scholar]
- 23.Lassmann G, Odenwaller R, Curtis JF, DeGray JA, Mason RP, Marnett LJ, Eling TE. J Biol Chem. 1991;266:20045–55. [PubMed] [Google Scholar]
- 24.Tsai A, Hsi LC, Kulmacz RJ, Palmer G, Smith WL. J Biol Chem. 1994;269:5085–91. [PubMed] [Google Scholar]
- 25.Smith WL, Lands WEM. Biochemistry. 1972;11:3276–3285. doi: 10.1021/bi00767a024. [DOI] [PubMed] [Google Scholar]
- 26.Teruhiko S, Kulmacz RJ, DeWitt DL, Smith WL. J Biol Chem. 1990;265:20073–20076. [PubMed] [Google Scholar]
- 27.Hsi LC, Hoganson CW, Babcock GT, Garavito RM, Smith WL. Biochem Biophys Res Commun. 1995;207:652–660. doi: 10.1006/bbrc.1995.1237. [DOI] [PubMed] [Google Scholar]
- 28.Shi W, Hoganson CW, Espe M, Bender CJ, Babcock GT, Palmer G, Kulmacz RJ, Tsai A-L. Biochemistry. 2000;39:4112–4121. doi: 10.1021/bi992561c. [DOI] [PubMed] [Google Scholar]
- 29.Boulos C, Jiang H, Balazy M. J Pharm Exp Ther. 2000;293:222–229. [PubMed] [Google Scholar]
- 30.Deeb RS, Resnick MJ, Mittar D, McCaffrey T, Hajjar DP, Upmacis RK. J Lipid Res. 2002;43:1718–1726. doi: 10.1194/jlr.m200199-jlr200. [DOI] [PubMed] [Google Scholar]
- 31.Kim SF, Huri DA, Snyder SH. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 32.Deeb RS, Shen H, Gamss C, Gavrilova T, Summers BD, Kraemer R, Hao G, Gross SS, Laine M, Maeda N, Hajjar DP, Upmacis RK. Am J Pathol. 2006;168:349–362. doi: 10.2353/ajpath.2006.050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groszmann RJ, Shah VH, Sessa WC. Proc Natl Acad Sci U S A. 2006;103:19777–82. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szabo C, Ischiropoulos H, Radi R. Nat Rev Drug Discov. 2007;6:662–80. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 35.Derakhshan B, Hao G, Gross SS. Cardiovascular Research. 2007;75:210–219. doi: 10.1016/j.cardiores.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deeb RS, Upmacis RK, Lamon BD, Gross SS, Hajjar DP. Hypertension. 2008;51:1–7. doi: 10.1161/HYPERTENSIONAHA.107.092866. [DOI] [PubMed] [Google Scholar]
- 37.Deeb RS, Hao G, Gross SS, Laine M, Qiu JH, Resnick B, Barbar EJ, Hajjar DP, Upmacis RK. J Lipid Res. 2006;47:898–911. doi: 10.1194/jlr.M500384-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Goodwin DC, Gunther MR, Hsi LC, Crews BC, Eling TE, Mason RP, Marnett LJ. J Biol Chem. 1998;273:8903–8909. doi: 10.1074/jbc.273.15.8903. [DOI] [PubMed] [Google Scholar]
- 39.Takeguchi C, Kono E, Sih CJ. Biochemistry. 1971;10:2372–6. doi: 10.1021/bi00788a030. [DOI] [PubMed] [Google Scholar]
- 40.Van der Ouderaa FJ, Buytenhek M, Nugteren DH, Van Dorp DA. Biochim Biophys Acta. 1977;487:315–331. doi: 10.1016/0005-2760(77)90008-x. [DOI] [PubMed] [Google Scholar]
- 41.Kulmacz RJ. Prostaglandins. 1987;34:225–240. doi: 10.1016/0090-6980(87)90246-2. [DOI] [PubMed] [Google Scholar]
- 42.Loll PJ, Sharkey CT, O’Connor SJ, Dooley CM, O’Brien E, Devocelle M, Nolan KB, Selinsky BS, Fitzgerald DJ. Mol Pharmacol. 2001;60:1407–1413. doi: 10.1124/mol.60.6.1407. [DOI] [PubMed] [Google Scholar]
- 43.Nuriel T, Deeb RS, Hajjar DP, Gross SS. Methods Enzymol. 2008;441:1–17. doi: 10.1016/S0076-6879(08)01201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dona M, Fredman G, Schwab JM, Chiang N, Arita M, Goodarzi A, Cheng G, von Andrian UH, Serhan CN. Blood. 2008;112:848–55. doi: 10.1182/blood-2007-11-122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malkowski MG, Thuresson ED, Lakkides KM, Rieke CJ, Micielli R, Smith WL, Garavito RM. J Biol Chem. 2001;276:37547–37555. doi: 10.1074/jbc.M105982200. [DOI] [PubMed] [Google Scholar]
- 46.Smith WL. Curr Opin Cell Biol. 2005;17:174–182. doi: 10.1016/j.ceb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Lymar SV, Jiang Q, Hurst JK. Biochemistry. 1996;35:7855–7861. doi: 10.1021/bi960331h. [DOI] [PubMed] [Google Scholar]
- 48.Radi R, Denicola A, Freeman BA. Methods Enzymol. 1999;301:353–367. doi: 10.1016/s0076-6879(99)01099-x. [DOI] [PubMed] [Google Scholar]
- 49.Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Nature. 1996;384:644–8. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 50.Wolin MS. Circ Res. 1998;82:1021–2. doi: 10.1161/01.res.82.9.1021. [DOI] [PubMed] [Google Scholar]
- 51.Shi X, Ding M, Dong Z, Chen F, Ye J, Wang S, Leonard SS, Castranova V, Vallyathan V. Mol Cell Biochem. 1999;199:93–102. doi: 10.1023/a:1006934612368. [DOI] [PubMed] [Google Scholar]
- 52.Petersen A, Carlsson T, Karlsson JO, Zetterberg M. Ophthalmic Res. 2008;40:77–85. doi: 10.1159/000113885. [DOI] [PubMed] [Google Scholar]
- 53.Claria J, Serhan CN. Proc Natl Acad Sci U S A. 1995;92:9475–9. doi: 10.1073/pnas.92.21.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souza JM, Giasson BI, Chen Q, Lee VM-Y, Ischiropoulos H. J Biol Chem. 2000;275:18344–18349. doi: 10.1074/jbc.M000206200. [DOI] [PubMed] [Google Scholar]
- 55.Osanai T, Fujita N, Fujiwara N, Nakano T, Takahashi K, Guan WP, Okumura K. Am J Physiol-Heart Circ Physiol. 2000;278:H233–H238. doi: 10.1152/ajpheart.2000.278.1.H233. [DOI] [PubMed] [Google Scholar]
- 56.Drelicharz L, Kozlovski V, Skorka T, Heinze-Paluchowska S, Jasinski A, Gebska A, Guzik T, Olszanecki R, Wojnar L, Mende U, Csanyi G, Chlopicki S. Basic Res Cardiol. 2008;103:417–30. doi: 10.1007/s00395-008-0723-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


