Abstract
All living systems contain naturally occurring nanoparticles with unique structural, biochemical and mechanical characteristics. Specifically, human saliva exosomes secreted by normal cells into saliva via exocytosis, are novel biomarkers showing tumor-antigen enrichment during oral cancer. Here we show the substructure of single human saliva exosomes, using a new ultra sensitive low force Atomic Force Microscopy (AFM) exhibiting sub-structural organization unresolvable in Electron Microscopy. We correlate the data with Field Emission Scanning Electron Microscopy (FESEM) and AFM images to interpret the nanoscale structures of exosomes under varying forces. Single exosomes reveal reversible mechanical deformation displaying distinct elastic, 70-100nm tri-lobed membrane with sub-structures carrying specific trans-membrane receptors. Further, we imaged and investigated, using force spectroscopy with antiCD63 IgG functionalized AFM tips, highly specific and sensitive detection of antigenCD63, potentially useful cancer markers on individual exosomes. The quantitative nanoscale morphological, biomechanical and surface biomolecular properties of single saliva exosomes, are critical for the applications of exosomes for cancer diagnosis and as a model for developing new cell delivery systems.
Keywords: Exosomes, saliva nanoparticles, nanocharacterization, Force spectroscopy, CD63 membrane receptors
Characterization of naturally occurring sub-100nm cellular nanostructures such as – vaults,1 viruses2 or lipid vesicles3 has recently gained interest due to their emerging role in drug delivery or immunotherapy. In particular, exosomes are 50-100nm particles secreted by a range of normal mammalian cells into body fluids such as saliva,5 blood,6 and urine7 via exocytosis8. Although their physiological functions are unclear, recently salivary exosomes have gained significance as biomarkers for oral cancer.4 Exosomes possess a cell-type specific lipid-bilayer membrane containing cytosolic proteins, formed by exocytosis through an inward budding of the cell membrane.9 Their role in specialized functions such as antigen presentation, inter-cellular communication, shuttling m-RNA/ mi-RNA6, 10or infectious agents reflects a well developed structure and functional organization of these nano-vesicles.11 Investigating individual exosomes also has important implications in understanding their fundamental physiological role, biomarker functions and for therapeutic encapsulation.12
Exosome characterization typically includes morphological analysis13 which given their nanoscale size, has been exclusively visualized with Transmission Electron Microscopy (TEM) using stained samples precluding the possibility to obtain additional biochemical and mechanical information. Exosomes vary widely in cell-type specific proteins and physiological conditions of the originating cell.14 Semi-quantitative proteomic14 and transcriptional analysis based on exosome populations15 rather than single vesicles are inadequate for analysis of proteins/receptors on single exosomes, thus structural and surface molecular characterization on exosomes are generally lacking.
In this article, we report the substructure of human saliva exosomes based on new ultra sensitive low force AFM and correlate their nanoscale structure obtained via AFM with high resolution (~1 nm) low voltage, chromatic aberration corrected FESEM images. We find that the mechanical deformation of single exosomes under varying applied forces reveals a unique insight into their substructure. We detect cell-type specific markers such as CD63 on individual exosomes via antiCD63 IgG functionalized AFM tips using force spectroscopy as well as antibody-gold beads.
RESULTS AND DISCUSSION
Structure of human saliva exosomes at the nanoscale level
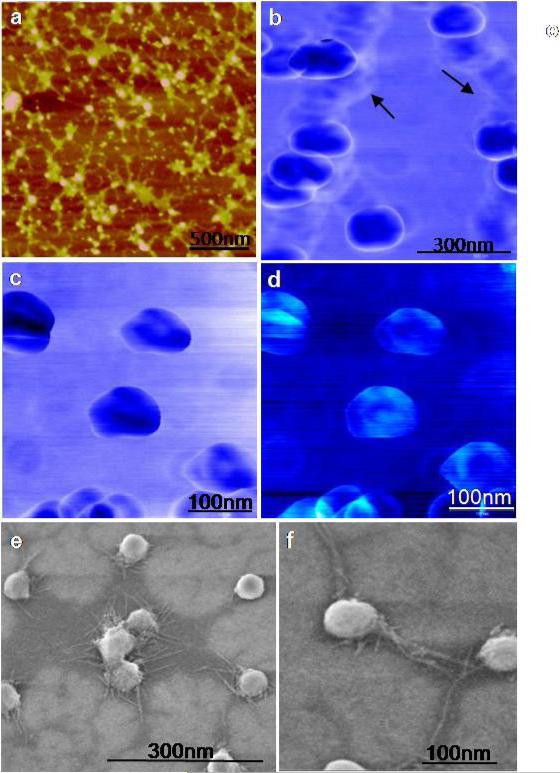
Previously, TEM imaging of numerous exosomes have revealed round vesicles with no substructure details.15 Here AFM has enabled critical structural data comparison with parallel imaging using high resolution FESEM while allowing additional details on physio-chemical properties of exosomes. We measured the three-dimensional structure and sub-vesicular organization of isolated exosomes using Tapping mode, AM-AFM (Amplitude Modulated) and PM-AFM (Phase Modulated) imaging, differing in the imaging force range and the feedback parameter used to generate images. Tapping mode (<1 nN force) height images of exosomes show fairly homogeneous round 50-70 nm vesicles surrounded by a network of extravesicular channels (Fig. 1a). Interestingly, at larger forces (~2nN), AM-AFM phase images reveal similar vesicle morphology (Fig. 1b) with diameters of 100±10 nm and indent of the vesicles in the centre indicative of mechanical deformation. Channel like elongations between exosomes appear without a prominent phase contrast while exosomes show some aggregation without inter-vesicular fusion. At ~2 nN forces, single exosomes (Fig. 1c) display a characteristic ring-like tri-lobed structure with one centre appearing as a depression with characteristic phase contrast suggesting role of heterogeneous density and/or viscoelastic image contrast mechanisms. Corresponding height images (Fig. 1d) reveal overall consistent flat ~100 nm vesicles. The observed AFM phase contrast of exosomes indicates non-homogenous surface which is tentatively attributable to the presence of proteins and/or mRNA enclosed inside the highly dense lipid membrane, consistent with previous proteomic and RNA analysis of saliva and other exosome populations.16 These images of single isolated sub-100 nm saliva exosome vesicles elucidate the sub-structural organization of exosomes unreported to date.
Figure 1. Ultrastructure of individual saliva exosomes observed under Tapping mode, AM-AFM and FESEM.
a, Tapping mode topographic AFM image showing round morphology of isolated exosomes. b, AM-AFM phase image of aggregated exosomes. Interconnections (arrows) lacking characteristic phase shift, probably indicate some extra-vesicular protein content. c, At higher forces under AM-AFM(~2nN) representative single exosome phase images reveal trilobed sub-structure within the centre of the vesicles. The contrast in images may be presumably attributed to variable constitutive elements (lipid, protein, RNA ratio) making up these structures. d, Corresponding height images show a central depression of the vesicles. e, FESEM exosome image showing multiple exosomes and f, single isolated vesicles as round bulging structures without a central depression and well resolved intervesicular connections.
High resolution FESEM exosome images show some aggregation (Fig. 1e) and single isolated vesicles show round bulging structures without a central depression (Fig. 1f). The images correlate well with the exosome structures obtained from AFM imaging where low imaging forces result in round, spherical shaped exosomes (Fig. 1a). The extra vesicular channels are well resolved and connect the exosomes as extensions (Fig. 1f). The higher magnification and lower voltage images, show a very spherical surface (Fig. 1f) suggesting these particles to have a round morphology unless an outside force is exerted on them, i.e. AFM probe. The different exosomes morphology from tri-lobed structures with a depression to round bulging vesicles with intervesicular connections reflect how topographical and biomechanical properties of exosomes can be explored by varying imaging feedback parameters such as force and phase setpoint.
Biomechanical properties of exosomes under variable forces
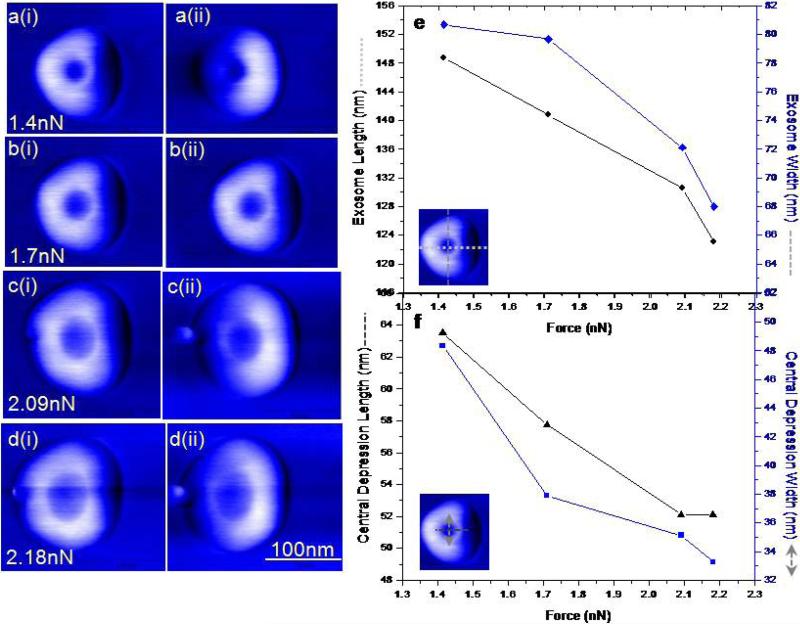
Biomechanical properties of vesicles17,18, 19 may play important role in exocytosis and inter-cellular transport. We applied PM-AFM as a force nano-manipulation technique to study exosome mechanics, local deformation and rupture characteristics. AFM image contrast exhibit combined local forces and micromechanical properties of the sample. The apparent exosome morphology changes under increasing loading force, where the force was gradually increased and then reduced, to test for elastic vesicle shape recovery. Under the highest applied load of 2.18 nN, the vesicle appears to have the largest lateral dimensions with the central depression occupying the greatest area of the exosomes (Fig. 2). The change in dimensions of the exosomes, central depression and changes in apparent height are shown (Fig. 2e, f). During high force imaging, we also observed blebbing of exosome (Fig. 2) suggesting a force induced structural perturbation. As the force was lowered during imaging their overall lateral dimensions decrease and the central depression becomes less apparent (see Supplementary Fig. S1). Although the decrease in apparent lateral dimension can be attributed to changes in average tip-sample contact area, change in shape and disproportionate growth of the central depression is consistent with their mechanical deformation. Above 5 nN forces, exosomes reproducibly rupture into three major fragments through disintegration of the vesicular structure as well as small fragments~10nm wide and 30nm in length, providing an insight into dynamic modifications under applied stress (see Supplementary Fig. S1).
Figure 2. Mechanical deformation of single exosomes during increased imaging force under PM-AFM.
a-d, Consecutive phase images of same exosome showing vesicle size as a function of applied force imaged under forward (i) and backward(ii) scan direction for each force setpoint. Increasing imaging forces (a to d) cause the overall lateral dimensions to increase and the central depression occupies more of the apparent structure. Blebbing of exosomes at high forces(c and d) a bleb or pinch observed on exosome surfaces suggest structural perturbation. Forward tip movement possibly results in backward folding of the bleb on the exosome structure whereas during backward scan the bleb is extended farther away from the exosome displayed as a protrusion. Bleb may result from a combination of mechanical stress well as inherent structural configuration of the exosomes. e, Cross sectional analysis of exosome structure under varying forces. f, Corresponding changes in the size of the central depression. The depth of the depression, changes from 1.1nm (at1.41 nN) to 1.7nm (at 2.18 nN).
Surface- biomolecular characteristics of exosomes observed under force spectroscopy
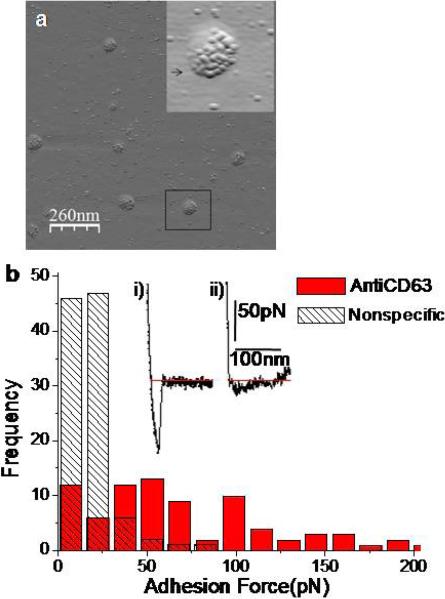
Structural EM probes can identify receptors on vesicles20 but limited in structure -function studies. Two quantitative approaches to biochemical characterization of exosomes are AFM imaging of bound biofunctionalized gold-beads and force spectroscopy with antiCD63 IgG functionalized tips for highly specific and sensitive detection of CD63 receptor cancer markers. Several EM techniques used functionalized gold-beads to identify specific target molecules on the surface structures.21 We used anti-CD63 IgG functionalized gold-beads for identification of exosome receptors by imaging under AFM. Visualization of labeled exosomes via anti-CD63 and secondary antibody coated gold beads (Fig. 3a) clearly indicates specific recognition of CD63 molecules. Multiple beads bound to exosomes indicate the presence of multiple CD63 molecules over a single membrane. This was verified by using non-specific primary antibody as control showing no preference for exosome binding.
Figure 3. Biochemical characterization of exosomes via AFM immunogold imaging and force spectroscopy showing presence of CD63 receptors on exosome surface.
a, Multiple CD63 receptor sites identified with antiCD63 monoclonal antibodies and secondary antibody-gold beads. AFM topographic image showing 5-8nm functionalized beads bound specifically to exosomes. The inset shows zoomed out 3D image of individual beads bound to the surface of an isolated single exosome. b, Distribution of rupture events. Non-specific interactions occur mostly at forces <50pN while specific CD63 antibody induced forces were distributed in the range of 30−200 pN. Sampled forces (n = 80 each) had each data point representing a single force measurement at any position on the exosomes surface (bin size 15pN). Typical curves showing force (pN) as a function of separation (nm) for a single pull with (i) strong adhesive event between AntiCD63 (ii) no-event for non-specific antibody functionalized tip and exosome.
Figure 3b shows interaction forces between antibody coated AFM tips and the exosome surface. The highest rupture force was used as a measure for direct determination of the strength of the bond formed between one or multiple individual CD63 and antiCD63 pairs. Much weaker adhesive forces (<50pN) only were observed by non-specific antibody tips, confirming the specificity of force measurements (Fig. 3b). Specific antibody tip forces were in 30−200 pN range, with majority of the data falling between 40 and 115 pN. The mean interaction force from a normal distribution of the histogram was calculated from bins 40 to 115pN as F = 73.01± 23.4 pN (mean± SD) and was used to estimate unbinding force between a single CD63 tetraspanin molecule and antibody. Though the exact orientation and state of antibody-antigen interaction is likely to vary, the mean interaction forces are comparable to the energy spectra of the force distribution histogram. The unbinding force is given by F= ΔH/d where ΔH is free enthalpy and d is the effective range of the potential. Typical antibody/antigen complexes with Kass from 102 to 1010 M-1 having free enthalpies of about 30 to 100 × 10-24 KJ/pair22 and binding pocket approximated as 0.93 nm from biotin/streptavidin data gives a force of about 100pN23 which corresponds well with our mean force F = 73±23 pN(mean± SD). Consequently, we estimate that specific adhesion peaks result from single antigen–antibody complex bond ruptures. Even higher forces (~150-200pN) indicate the presence of multiple tetraspanin molecules constituting the vesicle surface. Presence of CD63 molecules on exosomes also suggests, their endosomal instead of plasma membrane origin.24
CONCLUSIONS
In summary, we have demonstrated unique structural, biochemical and mechanical characteristics of natural, biologically important, human saliva exosomes at the single vesicle level. We report distinct substructure of single isolated sub-100 nm human saliva exosomes in the form of tri-lobed structures and demonstrate their reversible elastic nanomechanical properties, which are constitutively secreted by normal mammalian salivary gland cells and useful as novel biomarkers for detection of pathologies such as cancer. This is demonstrated by quantitative single receptor level detection of the specific markers, CD63 receptors, on individual exosomes from human saliva via targeted antibody tip coated force spectroscopy and antibody labeled gold-beads. The recognition of single receptor molecules on biological fluid derived exosomes such as saliva can potentially detect patho-physiology, and thereby should enable early cancer diagnosis, where conventional methods may prove ineffective due to sensitivity limitations. Our study shows the reversible mechanical stability of exosomes under low stress while high applied forces cause structural deformation and disintegration. These data are coherent with high resolution FESEM images. As biological nanostructures, exosomes may also provide a means to encapsulate, mimic and enhance drug delivery systems. The characterization of naturally occurring nano-structures such as exosomes individually through size, substructure and mechanics also provides critical data for developing engineered exosome -drug delivery carriers with inherent biocompatibility and nano-architecture.
METHODS
Exosome isolation and purification
Saliva samples were obtained from healthy volunteers from the Division of Otolaryngology, Head and Neck Surgery, at the Medical Center, University of California, Los Angeles (UCLA), CA, in accordance with a protocol approved by the UCLA Institutional Review Board. All participants gave written informed consent, and the ethics committee of UCLA approved the study. The mean age of the volunteers was 31 years (range, 26 – 43 years). The volunteers had no history of malignancy, immune-deficiencies, autoimmune disorders, hepatitis, or HIV infection. Exosomes were prepared as described15 with slight modifications. Briefly, 50 ml of saliva was equally mixed with Phosphate Buffer Saline and spun at 2600 × g for 15 min to remove cells. The supernatants were then sequentially centrifuged at 12000 × g for 20 min and 120,000 × g for 3 hours. The 120,000 × g pellet was re-suspended in PBS and used for AFM analysis.
AFM imaging
Purified exosomes were diluted 1:100 in de-ionized water and adsorbed to freshly cleaved mica sheets, rinsed with de-ionized water and dried under a gentle stream of Nitrogen. The exosomes were stable without any lysis or degradation. A home built AFM under tapping mode, AM-AFM and PM-AFM and silicon probes (MPP-13220-W, Spring Constant, k~200 Nm-1; Veeco) were used. Tapping-mode AFM is based on AM-AFM, which detects the change in the vibration amplitude of the oscillating cantilever and uses it as a feedback signal to generate high resolution images25. Phase images in which the phase change of the cantilever relative to the excitation signal is recorded while the feedback maintains constant vibration amplitude of the cantilever, mapping variations in material properties such as sample density and viscoelasticity. Additionally, PM-AFM enabled high resolution images of exosomes. PM-AFM detects phase change of the oscillating silicon cantilever relative to the excitation signal and uses it as the feedback signal to obtain topographic images. Although previous studies demonstrated PM-AFM for high resolution imaging26, it has not been extensively used for imaging soft biological samples. Topographic height and phase images were recorded simultaneously at 512×512 pixels at a scan rate of 1Hz. Image processing was performed using Gwyddion or SPIP™ software.
Force spectroscopy
Exosome immobilization- Exosomes were diluted 1:50 times in PBS buffer containing 10mM MgCl2/5mM CaCl2 (pH 7.4) and adsorbed to mica surface overnight at 4°C for stable immobilization. Samples were rinsed and imaged (Bioscope II; Veeco) under PBS in tapping mode using soft cantilevers (MSCT; Veeco) at 0.25 Hz. No significant changes were observed in the shape and size of exosomes under these conditions (see Supplementary Fig. S2). Tip functionalization: Anti-CD63 antibodies were attached to gold coated AFM probes (MSCT; Veeco) via thiol ester linkage as described previously27 with experimentally determined k= 0.02 Nm-1. The probes were washed in PBS, blocked with 1% BSA- PBS for 1 hour followed by rinsing with PBS. All measurements were recorded in physiological buffer conditions. Exosomes were electrostatically immobilized over mica and AFM tip was positioned on top of the exosomes. Force-separation curves were recorded at a ramp size of 1 μm, tip velocity 1 μm/sec under low forces (<500 pN). Nanoscope software was used for data analysis. No antibody or a non-specific mouse antibody (Anti HUR, Santa Cruz Biotechnology) coated AFM tips were used as control.
Immuno-bead AFM imaging
Exosomes prepared as above were incubated with 1:100 dilution of antiCD63 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), followed by 20 minute incubation with 1:1000 dilution secondary antibody (Molecular Probes, Invitrogen, CA) coated 10nm gold particles (Sigma Aldrich) with consecutive PBS washes after each binding step. Samples were rinsed with deionized water, dried and imaged under tapping mode (Bioscope II; Veeco) with silicon probes (f=305 KHz, OTESPA, Veeco). Absence or non-specific primary antibody served as controls.
Electron Microscopy
Isolated exosomes were immobilized over UV cleaned silicon wafers. Samples were coated with iridium for fifteen seconds at a current of twenty mA and the wafer edges were grounded using silver paint. Vesicles were examined under low beam energies (1.5kV @ 3.1 pA) with a Magellan™ 400L Extreme High-Resolution FESEM (FEI Co., Hillsboro, OR).
Supplementary Material
Acknowledgements
This work was supported by WPI Center for Materials NanoArchitectonics (MANA), NIMS, Japan ( JKG) and National Institutes of Health grants RO1 DE017170, UO1- DE016275 (DTW) and R00DE018165(VP). We thank W Klug and J Rim for helpful discussions.
Footnotes
Supporting Information available: S1Reversible mechanical deformation and disintegration of single exosome under varying forces during PM-AFM imaging. S2 AFM 3D topographic image of exosomes immobilized over mica surface imaged in buffer. This material is available free of charge via the Internet at http://pubs.acs.org
Author contributions: SS designed and performed AFM experiments, analyzed data and wrote the manuscript. HR performed AFM experiments, analyzed data; VP extracted purified exosomes and analyzed the data; CM, MS performed SEM experiments; DTW helped design experiments, JKG conceived and designed experiments, analyzed results, co-wrote the manuscript and provided all AFM characterization resources used. All authors discussed the results and commented on the manuscript.
REFERENCES
- 1.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, Rome LH. Engineering of Vault Nanocapsules With Enzymatic and Fluorescent Properties. Proc Natl Acad Sci U S A. 2005;102:4348–4352. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamada T, Ueda M, Seno M, Kondo A, Tanizawa K, Kuroda S. Novel Tissue and Cell Type-Specific Gene/Drug Delivery System Using Surface Engineered Hepatitis B Virus Nano-Particles. Curr Drug Targets Infect Disord. 2004;4:163–167. doi: 10.2174/1568005043341037. [DOI] [PubMed] [Google Scholar]
- 3.Cevc G. Lipid Vesicles and Other Colloids As Drug Carriers on the Skin. Adv Drug Deliv Rev. 2004;56:675–711. doi: 10.1016/j.addr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 4.Simpson RJ, Lim JWE, Moritz RL, Mathivanan S. Exosomes: Proteomic Insights and Diagnostic Potential. Expert Review of Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-Like Vesicles With Dipeptidyl Peptidase IV In Human Saliva. Biol Pharm Bull. 2008;31:1059–1062. doi: 10.1248/bpb.31.1059. [DOI] [PubMed] [Google Scholar]
- 6.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr., Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisitkun T,SR, Knepper MA. Identification and Proteomic Profiling of Exosomes in Human Urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. Epub 2004 Aug 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson RJ, Jensen SS, Lim JW. Proteomic Profiling of Exosomes: Current Perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 9.Thery C, Ostrowski M, Segura E. Membrane Vesicles as Conveyors of Immune Responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-Mediated Transfer of mRNAs And microRNAs is a Novel Mechanism of Genetic Exchange Between Cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Willem Stoorvogel MJKHJGGR. The Biogenesis and Functions of Exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 12.Scheerlinck JP, Greenwood DL. Virus-sized Vaccine Delivery Systems. Drug Discov Today. 2008;13:882–887. doi: 10.1016/j.drudis.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Hegmans JP, Gerber PJ, Lambrecht BN. Exosomes. Methods Mol Biol. 2008;484:97–109. doi: 10.1007/978-1-59745-398-1_7. [DOI] [PubMed] [Google Scholar]
- 14.Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, Biogenesis and Function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 15.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 16.Février B, Raposo G. Exosomes: Endosomal-derived Vesicles shipping Extracellular Messages. Current Opinion in Cell Biology. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nanomechanical Analysis of Cells from Cancer Patients. Nat Nano. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 18.Laney DE, Garcia RA, Parsons SM, Hansma HG. Changes in the Elastic Properties of Cholinergic Synaptic Vesicles As Measured by Atomic Force Microscopy. Biophys J. 1997;72:806–813. doi: 10.1016/s0006-3495(97)78714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin AJ, Prasad K, Smith PD, Lafer EM, Nossal R. Measuring the Elasticity of Clathrin-coated Vesicles via Atomic Force Microscopy. Biophys J. 2006;90:3333–3344. doi: 10.1529/biophysj.105.068742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melo RC, Spencer LA, Perez SA, Neves JS, Bafford SP, Morgan ES, Dvorak AM, Weller PF. Vesicle-mediated Secretion of Human Eosinophil Granule-derived Major Basic Protein. Lab Invest. 2009;89:769–781. doi: 10.1038/labinvest.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panyi G, Bagdany M, Bodnar A, Vamosi G, Szentesi G, Jenei A, Matyus L, Varga S, Waldmann TA, Gaspar R, Damjanovich S. Colocalization and Nonrandom Distribution of Kv1.3 Kotassium Channels and CD3 molecules in the Plasma Membrane of Human T Lymphocytes. Proc Natl Acad Sci U S A. 2003;100:2592–2597. doi: 10.1073/pnas.0438057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Oss C. Nature of Specific Ligand-receptor Bonds, in particular the Antigen-Antibody Bond. Marcel Dekker; New York: 1994. pp. 581–614. [DOI] [PubMed] [Google Scholar]
- 23.Moy VT, Florin EL, Gaub HE. Intermolecular Forces and Energies between Ligands and Receptors. Science. 1994;266:257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 24.Cocucci E, Racchetti G, Meldolesi J. Shedding Microvesicles: Artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Paulo AS, Garcia R. Tip-surface Forces, Amplitude, and Energy Dissipation In Amplitude-Modulation (Tapping Mode) Force Microscopy. Phys. Rev. B. 2001;64:193411–193414. [Google Scholar]
- 26.Naritaka Kobayashi YJL. Yoshitaka Naitoh, Masami Kageshima and Yasuhiro Sugawara High-Sensitivity Force Detection by Phase-Modulation Atomic Force Microscopy. Jpn. J. Appl. Phys. 2006;45:L793–795. [Google Scholar]
- 27.Aizawa H, Gokita Y, Jong-Won P, Yoshimi Y, Kurosawa S. Antibody Immobilization on Functional Monolayers Using a Quartz Crystal Microbalance. Sensors Journal, IEEE. 2006;6:1052–1056. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.