Abstract
Background
Leukotrienes play an important role in allergic and inflammatory diseases, but reports on the involvement of ALOX5AP and LTA4H in asthma have been inconclusive.
Objective
To determine whether polymorphisms in ALOX5AP and LTA4H genes are risk factors for asthma in two different Latino groups: Mexicans and Puerto Ricans.
Methods
The LTA4H gene was sequenced in individuals from both groups to identify novel polymorphisms. Single-nucleotide polymorphisms (SNPs) in the ALOX5AP and LTA4H genes were analyzed for associations with asthma and asthma-related phenotypes in 687 parent-child trios of Mexican and Puerto Rican origin.
Results
In LTA4H, five previously unknown polymorphisms were identified. Two SNPs within LTA4H (rs17525488 and rs2540493) were protective for asthma in Latinos (P = 0.007 and 0.05, respectively). Among Mexican patients, LTA4H polymorphisms were associated with baseline lung function and IgE levels. For ALOX5AP, the minor allele at SNP rs10507391 was associated with protection from asthma (OR = 0.78, P = 0.02) and baseline lung function (P = 0.018) in Puerto Ricans. A gene-gene interaction was identified between LTA4H (rs17525488) and ALOX5AP (rs10507391), (P = 0.003, in the combined sample).
Conclusion
Our results support the role of LTA4H and ALOX5AP variants as risk factors for asthma in Latino populations.
Keywords: Asthma, Leukotriene, Latino populations, Association study
INTRODUCTION
Asthma is a common disease resulting from a complex interaction of genetic, environmental and social factors. There are significant disparities in asthma prevalence, mortality and drug response between different ethnic groups.[1] In the US, asthma prevalence is highest in Puerto Ricans, African Americans, Filipinos and Native Hawaiians and lowest in Mexicans and Koreans. Furthermore, response to albuterol, the most commonly prescribed treatment for asthma, is lower in Puerto Ricans than in Mexicans.[2] Latinos are an admixed population, descendents of Native Americans, Africans, and Europeans in varying proportions. The analysis of admixed populations, such as Latino Americans, could provide the intrinsic variability needed to untangle complex gene-environment interactions in disease susceptibility, severity and drug responsiveness.[3]
The complexity in asthma diagnosis and treatment comes in part from the range of potential molecular pathways involved in asthma pathogenesis. Among these different mechanisms, leukotrienes (LTs) comprise a family of arachidonic acid metabolites which play an important role in the pathogenesis of allergic and inflammatory diseases.[4] The synthesis of leukotrienes is enhanced by arachidonate 5-lipoxygenase-activating protein (ALOX5AP) to produce leukotriene A4 (LTA4) from arachidonic acid. The resultant LTA4 can then be converted to leukotriene B4 (LTB4) by leukotriene A4 hydrolase (LTA4H) or it can be conjugated with reduced glutathione by the leukotriene C4 (LTC4) synthase. LTB4 and LTC4 are moved out of the cell and can exert their biologic influence through specific receptors in inflammatory cells. The synthesis of leukotrienes is increased in asthmatic patients by a transcriptional up-regulation of several genes.[5] Several drugs which block LT synthesis (zileuton) or LT receptors (montelukast, zafirlukast, and pranlukast) have been approved for the prophylaxis of exercise induced asthma and the treatment of asthma and allergic rhinitis. Although previous genetic association studies have evaluated the role of LT-related genes such as ALOX5 and LTC4S in asthma, the few reports on the involvement of the ALOX5AP or LTA4H genes in asthma have been inconclusive.[6–9] Because the two genes are coupled in the biologic pathway leading to the production of LTB4, we sought to clarify the involvement of LTA4H and ALOX5AP genetic polymorphisms and their interaction as risk factors for the development of asthma and related phenotypes.
METHODS
Samples
687 Latino trios consisting of asthmatic subjects and their biological parents from the Genetics of Asthma in Latino Americans (GALA) Study were analyzed. This sample includes 296 families of Mexican origin, recruited both in Mexico City and in the San Francisco Bay Area, and 391 families of Puerto Rican origin, collected in Puerto Rico and New York City. Probands were recruited if they had a diagnosis of asthma and were either taking a medication for asthma and had experienced two or more asthma-related symptoms (wheezing, coughing, and/or dyspnea) over the two years prior to enrollment. Spirometry, bronchodilator drug response (BDR) to albuterol, and total plasma IgE levels were collected for all patients. IgE levels were log10 transformed to normalize their distribution. Main clinical and demographic characteristics of these samples are shown in Table 1. Further details on these samples have been previously published.[2] All subjects provided written informed consent and local Institutional Review Boards (IRBs) approved all the studies.
Table 1.
Clinical characteristics of the Mexican and the Puerto Rican subjects with asthma from the GALA study. Values are expressed as median (25th: 75th percentile).
| Mexicans | Puerto Ricans | |
|---|---|---|
| Number of Subjects | 296 | 391 |
| Characteristics: | ||
| Age (years) | 16.5 (11:22) | 14.0 (9:19) |
| Gender (% male) | 53.8% | 55.9% |
| BMI (kg/m2) | 24.5 (20:29) | 22.3 (18:27) |
| Plasma IgE (IU/ml) | 470.4 (50:891) | 487.9 (58:918) |
| Baseline Spirometry: | ||
| Pre-FEV1 (% pred) | 88.9 (76:101) | 83.7 (72:95) |
| Pre-FEV1 (% pred) < 80% | 30.7% | 39.9% |
| Bronchodilator Responsiveness: | ||
| ΔFEV1 (relative % change) | 10.1 (2:19) | 6.2 (0:15) |
BMI = body mass index, Pre-FEV1 = baseline FEV1 expressed as percentage of predicted, ΔFEV1 = relative percent change in Pre-FEV1 after albuterol administration.
SNP Screening and Selection
Latinos display different patterns of genetic variation than populations included in public databases such as NCBI or HapMap. Therefore, before performing large scale genotyping, we sought to determine the allele frequencies in a subset of individuals with asthma. Sixteen LTA4H gene amplicons and 8 SNPs in the ALOX5AP gene were analyzed in a panel of 24 unrelated asthmatic subjects of each ethnic group (Mexican and Puerto Rican). The inclusion of 48 chromosomes for each group provides greater than 90% power to detect any polymorphic variant with a minor allele frequency (MAF) of greater than 5%. Only SNPs that had a MAF greater than 5%, and were not in tight linkage disequilibrium with other selected SNPs (assessed as r2 > 0.80) were genotyped. To avoid an excessive number of analyses, which would require strict multiple testing corrections and a consequent loss of statistical power, only SNPs with a minor allele involved in haplotypes with a frequency higher than 10% were included in the final analyses.
Genetic analyses
A total of 10,294 bp in the LTA4H gene were PCR-amplified and sequenced through 16 amplicons designed to cover all exons, exon-intron boundaries and the promoter region. Primers were located in neighboring flanking intronic sequences (see Supplementary Table 1) and PCR products were analyzed by direct sequencing, using the BigDye Terminator v3.1 Kit on an ABI 3700 Genetic Analyzer (Applied Biosystems, Foster City, CA). Exact locations of single-nucleotide polymorphisms (SNPs) were established in accordance to NCBI sequence NC_000012 and new variants were named after their relative position to the start codon (used as basepair +1).
In the ALOX5AP gene, eight SNPs covering most of the gene region were genotyped (SNP information is summarized in Supplementary Table 2). These SNPs were selected because they form two haplotypes, which have been previously shown to have an effect on LTB4 production in neutrophils.[10]
All selected SNPs were genotyped using the AcycloPrime-FP method.[11] All plates included blank and duplicate wells as controls. Enzymatic cleanup and single base extension genotyping reactions were performed with AcycloPrime-FP kits (Perkin Elmer, Waltham, MA) and plates were read on an EnVision fluorescence polarization plate reader (Perkin Elmer).
Statistical analyses
Hardy-Weinberg equilibrium (HWE) was assessed in each different ethnic group (with parents and probands analyzed separately) by means of a Pearson’s chi-square test. Families showing Mendelian inconsistencies were detected using the program PedCheck [12] and were excluded from further analyses.
FBAT software was used to perform transmission disequilibrium tests (TDT) to detect associations between individual SNPs and haplotypes with asthma and quantitative asthmarelated phenotypes.[13] Given the number of comparisons performed, a Monte Carlo procedure implemented in FBAT was used to assess the robustness of the significant results. This test computes p-values in the univariate case using 100,000 Monte Carlo samples from the null distribution of no linkage and no association and obtains the maximally significant statistic out of the set of individual statistics. Odds ratios (OR) and their 95% confidence intervals for associations between individual SNPs and asthma were estimated using the program UNPHASED.[14]
The presence of gene-gene interactions was determined using the likelihood algorithms implemented in the program UNPHASED. Specifically, individual SNPs that were significantly associated with asthma were chosen for further analysis of gene-gene interactions. SNPs of LTA4H were paired with SNPs of ALOX5AP to determine if the presence of both variants modified the risk for asthma differently than each gene alone.
RESULTS
Polymorphism screening and SNP selection
Supplementary Table 3 summarizes the information on all polymorphisms identified in this study. Twenty-one polymorphisms were identified in the LTA4H gene. Five SNPs were new to the NCBI database and most of these were population-specific. Puerto Ricans had a higher number of polymorphisms than Mexicans.
Most of the polymorphisms found in the LTA4H gene (66.6%) were located in exon-intron boundaries, five SNPs were located in the promoter region, and two SNPs were found in exons 1 and 2, but both of them were synonymous SNPs that did not result in an amino acid substitution.
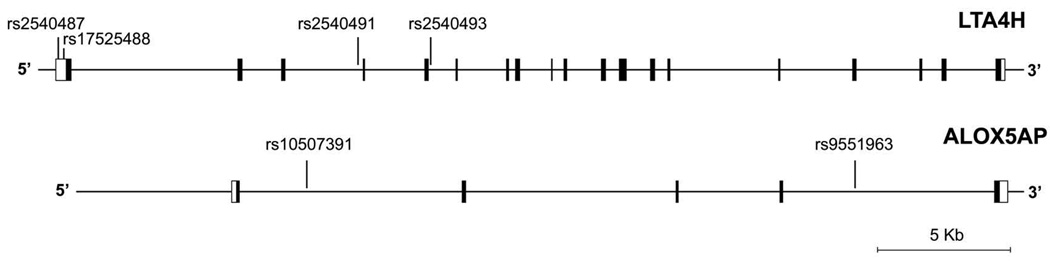
According to the aforementioned criteria, a total of 4 SNPs in the LTA4H gene were selected for analysis in the Latino families: rs2540487, rs17525488, rs2540491, and rs2540493. Two SNPs in the ALOX5AP gene, SNPs rs10507391 and rs9551963, were selected for analysis. Figure 1 depicts their location in the gene region and Supplementary Figure 1 plots the linkage disequilibrium (LD) between the selected SNPs. All markers were in Hardy-Weinberg equilibrium.
Figure 1.
Schematic diagrams of LTA4H and ALOX5AP genes and the genotyped single nucleotide polymorphisms in relation to the gene. Full boxes depict exons and empty boxes represent untranslated regulatory regions, according to public databases.
LTA4H association results
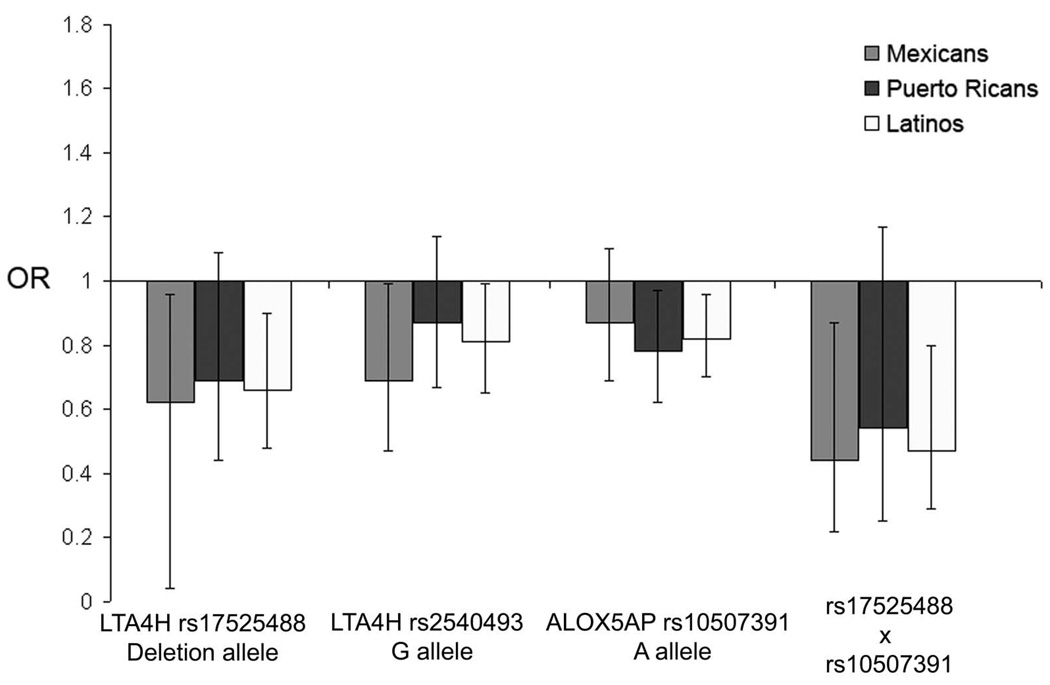
In the LTA4H gene, SNPs rs17525488, in the promoter region, and rs2540493, in an exonintron boundary, were significantly associated with a decreased risk for asthma in Mexicans and a similar trend was observed in Puerto Ricans (Table II and Figure 2). The minor allele of marker rs17525488 (deletion of an adenine) conferred an OR of 0.62 among Mexicans (95% CI = 0.04-0.96, p=0.035) and 0.69 among Puerto Ricans (95% CI = 0.44-1.09, p=0.088). In addition, the G allele at rs2540493 conferred an OR of 0.69 (95% CI = 0.47-0.99, p=0.049) in Mexicans. Analysis of these two markers in both Latino populations combined were significantly associated with a decreased risk for asthma (p=0.007 and p=0.049, respectively). After 100,000 Monte Carlo cycles, these results in Mexicans and in the combined sample remained significant.
Table 2.
Association tests between asthma and LTA4H and ALOX5AP SNPs in Mexicans and Puerto Ricans.
| Mexicans | Puerto Ricans | |||
|---|---|---|---|---|
| OR (95%CI) | P | OR (95%CI) | P | |
| LTA4H | ||||
| rs2540487 | 0.72 (0.52-1.01) | 0.059 | 1.14 (0.89-1.47) | 0.298 |
| rs17525488 | 0.62 (0.04-0.96) | 0.038 | 0.69 (0.44-1.09) | 0.088 |
| rs2540491 | 0.93 (0.71-1.21) | 0.586 | 0.92 (0.75-1.13) | 0.443 |
| rs2540493 | 0.69 (0.47-0.99) | 0.050 | 0.87 (0.67-1.14) | 0.318 |
| ALOX5AP | ||||
| rs10507391 | 0.87 (0.69-1.10) | 0.277 | 0.78 (0.62-0.97) | 0.027 |
| rs9551963 | 1.02 (0.80-1.31) | 0.849 | 1.09 (0.88-1.35) | 0.453 |
Odds ratios are indicated, with 95% confidence intervals in parentheses. P values after 100,000 Monte Carlo steps. P values <0.05 are bolded.
Figure 2.
Plot of odds ratios of association analysis of LTA4H and ALOX5AP single-nucleotide polymorphisms (SNPs) and LTA4H-ALOX5AP gene-gene interaction with asthma in Mexicans, Puerto Ricans, and combined Mexicans and Puerto Ricans. Error bars indicate 95% confidence intervals.
Significant results were also observed between SNP rs17525488 and baseline lung function (pre-bronchodilator FEV1) and IgE levels in Mexican patients (Table III). Lung function (expressed as a percentage of the predicted normal value) in Mexican patients carrying at least one deletion allele at rs17525488 was 5.3% lower than patients without this allele (p = 0.011 after Monte Carlo test). This allele was also associated with lower IgE levels (p=0.015 after Monte Carlo test) among Mexican patients. Combined analysis of Latino asthmatic patients revealed the same significant associations (p = 0.002 and p=0.015 for Pre-FEV1 and IgE, respectively, after Monte Carlo tests). Similar results were observed for marker rs2540493 and Pre-FEV1 and IgE levels in Mexicans, although at a lower level of statistical significance (p = 0.08 and p=0.04, respectively).
Table 3.
Association analysis between quantitative asthma traits with LTA4H and ALOX5AP SNPs.
| Trait | LTA4H SNPs | ALOX5AP SNPs | |||||
|---|---|---|---|---|---|---|---|
| rs2540487 | rs17525488 | rs2540491 | rs2540493 | rs10507391 | rs9551963 | ||
| Pre-FEV1 | MEX | (−) 0.07 | (−) 0.01* | (−) 0.45 | (−) 0.08 | (−) 0.46 | (+) 0.81 |
| PR | (+) 0.82 | (−) 0.08 | (−) 0.21 | (−) 0.21 | (−) 0.02* | (+) 0.70 | |
| ΔFEV1 | MEX | (−) 0.92 | (−) 0.49 | (+) 0.36 | (−) 0.10 | (−) 0.53 | (+) 0.49 |
| PR | (−) 0.95 | (−) 0.43 | (−) 0.86 | (−) 0.61 | (−) 0.09 | (−) 0.22 | |
| Log10IgE | MEX | (−) 0.10 | (−) 0.01* | (−) 0.38 | (−) 0.04* | (−) 0.41 | (+) 0.78 |
| PR | (+) 0.84 | (−) 0.32 | (−) 0.12 | (−) 0.20 | (−) 0.06 | (+) 0.99 | |
P-values are indicated. (+) and (−) indicate direction of trait association with minor allele, that is higher or lower values of the quantitative traits. MEX and PR state for Mexicans and Puerto Ricans. P values <0.05 are denoted with an asterisk.
Haplotype analyses identified associations between the two main haplotypes and asthma, Pre-FEV1 and IgE (see Supplementary Table 5). These results reflect the associations detected for the individual SNPs, but they did not remain statistically significant after Monte Carlo tests.
ALOX5AP association results
In the ALOX5AP gene, SNP rs10507391 revealed significant associations with asthma (Table II and Figure 2). The A allele was significantly associated with a decreased risk for asthma in Puerto Rican patients (OR=0.78, 95% CI = 0.62-0.97, p=0.027 after Monte Carlo test), but not Mexicans (p=0.28). Overall, combining both Latino groups, the A allele was associated with a decreased risk for asthma (p=0.017 after Monte Carlo test).
The same allele was also associated with lower baseline lung function in Puerto Ricans (p = 0.018, after Monte Carlo tests) but not in Mexicans (Table III). Puerto Rican patients carrying at least one A allele at rs10507391 had a baseline lung function 2.8% lower than patients without this allele. Bronchodilator responsiveness (BDR) and IgE levels showed borderline statistical association in Puerto Ricans, but did not reach significance after Monte Carlo tests (p=0.09 and p=0.06, respectively).
Association results between haplotypes in the ALOX5AP gene and asthma and asthmarelated phenotypes did not reveal any significant association after Monte Carlo tests were applied (Supplementary Table 5).
Gene-gene interaction
Given that polymorphisms in ALOX5AP and LTAH4 were individually associated with asthma, and are both in the leukotriene pathway, we tested for the presence of gene-gene interactions on asthma. Gene-gene interaction was analyses were limited to SNPs that were individually determined to be significantly associated with asthma, (i.e. SNPs rs17525488 and rs2540493 in the LTA4H gene, and rs10507391 of the ALOX5AP gene). The likelihood algorithm implemented in the program UNPHASED detected a gene-gene interaction between LTA4H and ALOX5AP, which was significantly associated with a decreased risk for asthma in Latino populations (Figure 2).
The deletion allele of SNP rs17525488 in the LTA4H gene significantly interacted with the A allele of ALOX5AP SNP rs10507391 to decrease the risk of asthma (OR = 0.44, 95% CI=0.22-0.87, p=0.01) in Mexicans and the same trend was observed in Puerto Ricans (OR = 0.54, 95% CI=0.25-1.17, p=0.10). The odds ratio for asthma in the combined analysis of these two groups for the LTA4H-ALOX5AP (rs17525488-rs10507391) interaction is 0.47 (95% CI=0.29-0.80, p=0.003). There was also a trend towards lower asthma risk for the interaction between LTA4H SNP rs2540493 and ALOX5AP SNP rs10507391, but it did not reach statistical significance in any group (p=0.17, p=0.26, and p=0.09 for Mexicans, Puerto Ricans, and both populations combined, respectively).
DISCUSSION
In the present study we find strong evidence for the involvement of the LTA4H and ALOX5AP genes in the pathophysiology of asthma. The inclusion of two ethnically diverse populations (Mexicans and Puerto Ricans) have allowed us to identify alleles in these two genes that are associated with a differential risk for asthma and asthma-related phenotypes, such as baseline lung function and IgE levels. Furthermore, we have also identified an interaction between polymorphisms at both loci that confers a decreased risk for asthma beyond what would have been expected from main effects alone. These results confirm the importance of the leukotriene pathway in asthma and allergic processes and identify a specific role in these processes for LTB4 production pathways.
These two genes were selected as potential candidate genes for asthma because of the involvement of leukotriene (LT) pathways in the inflammatory response of asthma. Some LT pathway genes are up-regulated in asthmatic patients and LTB4 production is significantly higher in asthmatic patients compared to healthy or atopic nonasthmatic patients.[5,15] Drugs blocking ALOX5 or LT receptors, such as zileuton or montelukast, have been approved for the treatment of chronic asthma. However, it is estimated that as many as 58–78% of all patients receiving montelukast do not respond to the medication.[16–17] This interpatient variability in drug response may be partially attributable to genetic variation.
Relative to other genes, LTA4H and ALOX5AP have not been thoroughly analyzed in genetic association studies. A limited number of investigators have analyzed the relationship between polymorphisms in these two genes and asthma and asthma-related phenotypes.[6–9,18–20] To our knowledge, this investigation is the first to analyze these genes (including sequencing of the LTA4H gene) and compare the results between admixed populations from different ethnic backgrounds. The comparison of association test results in different populations can provide insight not only on shared genetic risk factors for disease and drug response, but also on population-specific risk factors.
Sequencing of the LTA4H gene has revealed the presence of five previously unknown polymorphisms, many of which are population-specific. LTA4H association results identified minor alleles at SNPs rs17525488 and rs2540493 as protective factors for asthma in Mexicans and Puerto Ricans. This protective effect may be exerted through a lower allergic profile, as the same alleles are also associated with lower IgE levels. Among Mexicans, all identified associations are robust to Monte Carlo procedures, which account for multiple testing. However, the associations were not unequivocally significant in Puerto Ricans. Similarly, we found that the rs10507391 polymorphism in intron 1 of the ALOX5AP gene was associated with a differential risk for asthma in these two populations. Thus, the A allele at rs10507391 locus confers a protection from asthma to its carriers in Puerto Ricans, but not Mexicans. This study suggests that the effect appears to be mediated through allergic response as the A allele at the rs10507391 locus is associated with lower IgE levels, but the magnitude of this effect did not reach statistical significance after adjustment for multiple testing.
SNP rs17525488 is located in the promoter region of the LTA4H gene and therefore may alter transcriptional regulation. In addition, SNPs in the LTA4H (rs2540493) and ALOX5AP (rs10507391) genes map to exon-intron boundaries, which may affect the efficiency of gene splicing.[21] Furthermore, these SNPs may be in linkage disequilibrium with yet undiscovered genetic variants that alter gene expression or enzymatic activity. Genetic variation which alters the structure of the catalytic site of LTA4H may result in a decrease or selective loss of the enzyme’s ability to produce LTB4.[22–24] Moreover, it has been determined that specific mutations in the ALOX5AP gene prevent enzyme dimerization thereby decreasing leukotriene biosynthesis.[25]
The alleles in LTA4H and ALOX5AP genes which confer a lower risk for asthma are also associated with lower baseline lung function. Although this last finding may seem contradictory the same association has been previously described for both genes in a recent study of British participants.[20]
Our results are not entirely consistent with the previous report by Holloway and colleagues.[20] Even though the authors found a SNP in intron 11 of the LTA4H gene that conferred a protective role for asthma and lower baseline lung function, variants in the promoter region did not demonstrate an association with the disease or related traits. Their results also showed that the A allele at rs10507391 in the ALOX5AP gene was a risk factor for asthma and was associated with related phenotypes, such as atopy and increased lung function, while in the populations we studied, the allele was protective. However, the contradictory findings should not be interpreted as a spurious finding. There are several reasons that can account for this observation. LD patterns within these genes in Puerto Ricans and Mexicans differed from the British sample. If the observed associations are non-causal, differences in LD may explain the observation that a given SNP shows significant results and opposite effects in different populations.
Furthermore, the heterogeneous effects of rs10507391 may be due to differences in genetic and/or environmental background between Latino and European populations. It has been demonstrated that opposite directions of a genetic association can occur when the analyzed polymorphism is correlated with additional causal variants at other loci or when it contributes to multilocus effects.[26] For these reasons, it has been previously suggested that gene-based approaches should capture all of the potential risk-conferring variations. [27] Consequently, replication of genetic association studies may be more informative on a gene-based level than on a polymorphism-based level.
ALOX5AP and LTA4H are key proteins in the sequential enzymatic steps leading to LTB4 production. ALOX5AP enhances LTA4 production from arachidonic acid, and LTA4 can then be converted to LTB4 by LTA4H. Consequently, there is biological plausibility for hypothesizing an interaction between these two genes in their effect on the risk of developing asthma. Taken together, when the minor alleles at polymorphisms LTA4H rs17525488 and ALOX5AP rs10507391 occurred together, the risk for asthma is much lower than when either SNP occurred alone. This suggests that synergistic polymorphisms of LTA4H and ALOX5AP are protective factors for asthma in Latino populations. This effect is likely to be mediated through an overall decrease in LTB4 production. This gene-gene interaction together with other known genetic interactions in asthma, such as IL-4RA / IL-13 and TNC / NPSR1, [28–29] gives additional support to the joint analysis of different genes in the same biological pathway instead of single-gene analysis in association studies.
We conclude from our results that LTA4H and ALOX5AP constitute genetic risk factors for the development of asthma in Mexican and Puerto Rican populations, possibly through allergy-related mechanisms. These results together with the recent survey in a British cohort support the role of LTB4 production in the pathophysiology of asthma. Further approaches should focus on the potential clinical implications of these findings. Thus, pharmacogenetic studies analyzing the joint effect of molecular variation in these genes on the response to antileukotriene drugs should be performed in large-scale and ethnically diverse cohorts. Interpatient variability in response to antileukotriene drugs is considerably wide among asthmatic patients. Pharmacogenetic studies including LTA4H and ALOX5AP genetic variation may serve to identify the subset of asthmatic patients most likely to benefit from these therapies.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by the National Institutes of Health (HL078885, AI077439, HL088133, U01 GM61390, ES015794), the Flight Attendant Medical Research Institute (FAMRI) and RWJF Amos Medical Faculty Development Award to Esteban Gonzalez Burchard, Tobacco-Related Disease Research Program New Investigator Award (15KT-0008) to Shweta Choudhry, the Ernest S. Bazley Trust to Pedro C. Avila, Beatriu de Pinos Postdoctoral Grant (2006 BP-A 10144) to Marc Via, and the Sandler Center for Basic Research in Asthma and the Sandler Family Supporting Foundation. The authors would like to acknowledge the families and the patients for their participation. The authors would also like to thank the numerous health care providers and community clinics for their support and participation in the GALA Study. The authors would like to especially thank Jeffrey M. Drazen, M.D., Scott Weiss, M.D., Ed Silverman, M.D., Ph.D., Homer A. Boushey, M.D., and Jean G. Ford, M.D., for all of their effort toward the creation of the GALA study. The authors also would like to thank two anonymous reviewers for their useful comments that helped improve the manuscript.
REFERENCES
- 1.Drake KA, Galanter JM, Burchard EG. Race, ethnicity and social class and the complex etiologies of asthma. Pharmacogenomics. 2008;9:453–462. doi: 10.2217/14622416.9.4.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, Toscano M, Sylvia JS, Alioto M, Salazar M, Gomez I, Fagan JK, Salas J, Lilly C, Matallana H, Ziv E, Castro R, Selman M, Chapela R, Sheppard D, Weiss ST, Ford JG, Boushey HA, Rodriguez-Cintron W, Drazen JM, Silverman EK. Genetics of Asthma in Latino Americans (GALA) Study. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169:386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 3.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok PY, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG. Genetics of Asthma in Latino Americans GALA Study. Population stratification confounds genetic association studies among Latinos. Hum Genet. 2006;118:652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 4.Peters-Golden M, Henderson WR., Jr. Leukotrienes. N Engl J Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 5.Zaitsu M, Hamasaki Y, Matsuo M, Ichimaru T, Fujita I, Ishii E. Leukotriene synthesis is increased by transcriptional up-regulation of 5-lipoxygenase, leukotriene A4 hydrolase, and leukotriene C4 synthase in asthmatic children. J Asthma. 2003;40:147–154. doi: 10.1081/jas-120017985. [DOI] [PubMed] [Google Scholar]
- 6.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, Wang J, Sylvester J, Holbrook J, Wise R, Weiss ST, Barnes K. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173:379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koshino T, Takano S, Kitani S, Ohshima N, Sano Y, Takaishi T, Hirai K, Yamamoto K, Morita Y. Novel polymorphism of the 5-lipoxygenase activating protein (FLAP) promoter gene associated with asthma. Mol Cell Biol Res Commun. 1999;2:32–35. doi: 10.1006/mcbr.1999.0147. [DOI] [PubMed] [Google Scholar]
- 8.Heinzmann A, Grotherr P, Jerkic SP, Lichtenberg A, Braun S, Kruse S, Forster J, Kuehr J, Deichmann KA. Studies on linkage and association of atopy with the chromosomal region 12q13-24. Clin Exp Allergy. 2000;30:1555–1561. doi: 10.1046/j.1365-2222.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- 9.Klotsman M, York TP, Pillai SG, Vargas-Irwin C, Sharma SS, van den Oord EJ, Anderson WH. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics. 2007;17:189–196. doi: 10.1097/FPC.0b013e3280120043. [DOI] [PubMed] [Google Scholar]
- 10.Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SF, Thorgeirsson G, Sveinbjornsdottir S, Valdimarsson EM, Matthiasson SE, Johannsson H, Gudmundsdottir O, Gurney ME, Sainz J, Thorhallsdottir M, Andresdottir M, Frigge ML, Topol EJ, Kong A, Gudnason V, Hakonarson H, Gulcher JR, Stefansson K. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Kwok PY. Homogeneous genotyping assays for single nucleotide polymorphisms with fluorescence resonance energy transfer detection. Genet Anal. 1999;14:157–163. doi: 10.1016/s1050-3862(98)00016-3. [DOI] [PubMed] [Google Scholar]
- 12.O’Connell JR, Weeks DE. Pedcheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montuschi P, Martello S, Felli M, Mondino C, Barnes PJ, Chiarotti M. Liquid chromatography/mass spectrometry analysis of exhaled leukotriene B4 in asthmatic children. Respir Res. 2005;6:119. doi: 10.1186/1465-9921-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malmstrom K, Rodriguez-Gomez G, Guerra J, Villaran C, Piñeiro A, Wei LX, Seidenberg BC, Reiss TF. Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. A randomized, controlled trial. Montelukast/Beclomethasone Study Group. Ann Intern Med. 1999;130:487–495. doi: 10.7326/0003-4819-130-6-199903160-00005. [DOI] [PubMed] [Google Scholar]
- 17.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, Zeiger RS, Larsen G, Spahn JD, Bacharier LB, Bloomberg GR, Guilbert TW, Heldt G, Morgan WJ, Moss MH, Sorkness CA, Taussig LM. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115:233–242. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Choi JH, Park HS, Oh HB, Lee JH, Suh YJ, Park CS, Shin HD. Leukotriene-related gene polymorphisms in ASA-intolerant asthma: an association with a haplotype of 5-lipoxygenase. Hum Genet. 2004;114:337–344. doi: 10.1007/s00439-004-1082-1. [DOI] [PubMed] [Google Scholar]
- 19.Sayers I, Barton S, Rorke S, Sawyer J, Peng Q, Beghé B, Ye S, Keith T, Clough JB, Holloway JW, Sampson AP, Holgate ST. Promoter polymorphism in the 5-lipoxygenase (ALOX5) and 5-lipoxygenase-activating protein (ALOX5AP) genes and asthma susceptibility in a Caucasian population. Clin Exp Allergy. 2003;33:1103–1110. doi: 10.1046/j.1365-2222.2003.01733.x. [DOI] [PubMed] [Google Scholar]
- 20.Holloway JW, Barton SJ, Holgate ST, Rose-Zerilli MJ, Sayers I. The role of LTA4H and ALOX5AP polymorphism in asthma and allergy susceptibility. Allergy. 2008;63:1046–1053. doi: 10.1111/j.1398-9995.2008.01667.x. [DOI] [PubMed] [Google Scholar]
- 21.Hull J, Campino S, Rowlands K, Chan MS, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, Kwiatkowski D. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudberg PC, Tholander F, Thunnissen MM, Haeggström JZ. Leukotriene A4 hydrolase/aminopeptidase. Glutamate 271 is a catalytic residue with specific roles in two distinct enzyme mechanisms. J Biol Chem. 2002;277:1398–1404. doi: 10.1074/jbc.M106577200. [DOI] [PubMed] [Google Scholar]
- 23.Rudberg PC, Tholander F, Thunnissen MM, Samuelsson B, Haeggstrom JZ. Leukotriene A4 hydrolase: selective abrogation of leukotriene B4 formation by mutation of aspartic acid 375. Proc Natl Acad Sci U S A. 2002;99:4215–4220. doi: 10.1073/pnas.072090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeggström JZ, Tholander F, Wetterholm A. Structure and catalytic mechanisms of leukotriene A4 hydrolase. Prostaglandins Other Lipid Mediat. 2007;83:198–202. doi: 10.1016/j.prostaglandins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Plante H, Picard S, Mancini J, Borgeat P. 5-Lipoxygenase-activating protein homodimer in human neutrophils: evidence for a role in leukotriene biosynthesis. Biochem J. 2006;393:211–218. doi: 10.1042/BJ20060669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battle NC, Choudhry S, Tsai HJ, Eng C, Kumar G, Beckman KB, Naqvi M, Meade K, Watson HG, Lenoir M, Burchard EG. SAGE Investigators. Ethnicity-specific gene-gene interaction between IL-13 and IL-4Ralpha among African Americans with asthma. Am J Respir Crit Care Med. 2007;175:881–887. doi: 10.1164/rccm.200607-992OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orsmark-Pietras C, Melén E, Vendelin J, Bruce S, Laitinen A, Laitinen LA, Lauener R, Riedler J, von Mutius E, Doekes G, Wickman M, van Hage M, Pershagen G, Scheynius A, Nyberg F, Kere J. PARSIFAL Genetics Study Group. Biological and genetic interaction between tenascin C and neuropeptide S receptor 1 in allergic diseases. Hum Mol Genet. 2008;17:1673–1682. doi: 10.1093/hmg/ddn058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.